Abstract
Since methods for analysing chromosome conformation in mammalian cells are either low resolution or low throughput and are technically challenging they are not widely used outside of specialised laboratories. We have re-designed the Capture-C method producing a new approach, called next generation (NG) Capture-C. This produces unprecedented levels of sensitivity and reproducibility and can be used to analyse many genetic loci and samples simultaneously. Importantly, high-resolution data can be produced on as few as 100,000 cells and SNPs can be used to generate allele specific tracks. The method is straightforward to perform and should therefore greatly facilitate the task of linking SNPs identified by genome wide association studies with the genes they influence. The complete and detailed protocol presented here, with new publicly available tools for library design and data analysis, will allow most laboratories to analyse chromatin conformation at levels of sensitivity and throughput that were previously impossible.
Introduction
Our ability to annotate gene regulatory elements and investigate their function has been driven by technologies such as RNA-seq1, ChIP-seq2,3, DNase-seq4 and ATAC-seq5. An outstanding challenge is to understand the mechanisms by which regulatory elements control specific gene promoters at a distance (10s to 1,000s kb). Conventional chromosome conformation capture (3C), allows for the detailed analysis of the interactions between regulatory elements and promoters at individual loci6-11. Recently, we have shown, using a high-throughput approach (Capture-C), the interrogation of cis-interactions, at hundreds of loci at high-resolution in a single experiment12. Such approaches are of immediate value in defining the regulatory landscapes of many loci, and identifying the genes and the functional effects of SNPs that are associated with complex diseases, the majority of which lie in intergenic cis-acting regulatory elements13-15.
The original Capture-C protocol12 uses oligos synthesized on a microarray with a design minimum of 40,000, irrespective of the number of desired viewpoints, so the cost per sample is very high for small designs. Experimental designs often require much smaller subsets of regions but from multiple samples. Furthermore, its sensitivity does not readily allow for the analysis of very weak cis or trans-interactions.
To address these limitations we redesigned the Capture-C protocol to use biotinylated DNA oligos so that each set of capture oligos can be designed specifically to capture from one to many hundreds of regions, in a single experiment and designs can be easily expanded by addition of new oligos to existing pools. Importantly, multiple independent 3C libraries from different samples can now be processed in a single reaction greatly increasing throughput, minimising experimental variation and allows for meaningful subtractive analysis of chromosome conformation in different cell types.
Using Next Generation (NG) Capture-C, we have defined the smallest number of cells required to identify robust interactions and shown how SNP specific interaction profiles can be generated.
RESULTS
Overview and experimental workflow
3C libraries were made using standard methods similar to in situ Hi-C16 (Fig. 1a, Supplementary methods). Prior to oligonucleotide capture, the 3C libraries were sonicated to 200 bp followed by the addition of Illumina paired-end sequencing adaptors. Sonication randomly generates unique fragments which is an important advantage of Capture-C compared to 4C and 5C as over-amplified PCR duplicates can be removed bioinformatically allowing the number of unique ligation junctions present in the 3C library to be quantified accurately (Fig. 1b).
Figure 1. Overview of the method.
a. Experimental workflow. High-resolution 3C libraries generation:, crosslinking live cells (1); digestion of chromatin, optimized for four cutter restriction enzymes (eg Dpn II) (2); ligation (3); de-crosslinking and DNA extraction (4). This 3C library is sonicated to produce random ~200 bp fragments (5) followed by; sequencing adaptor ligation and indexing (6); pooling of indexed samples (7) hybridization with biotinylated oligonucleotides to the pool of indexed samples (8); pull down using streptavidin beads (9) and PCR from beads using adapter P5&7 sequences (10). Steps 8-10 are repeated, resulting in enrichments up to 3,000,000-fold over the uncaptured 3C library, and sequenced (11).
b. Analysis. 1. Raw data (FASTQ). 2. Reconstruction of paired reads into original fragments. 3. in silico digestion into component restriction fragments to allow for mapping. 4. Reads not containing a restriction site or a captured viewpoint are discarded as background. 5. Reads that are not unique are collapsed into a single representative read. 6. Interactions are only reported if a read pair maps within a captured fragment and maps outside all of the capture fragments and proximity exclusion regions in the experiment (usually 1 kb on either side of the captured viewpoint fragments). This is done to prevent undigested material being reported as interacting and to prevent the reporting of fragments captured by two different oligonucleotides. The data are then filtered to remove regions with problematic mappability due to copy number differences33 and mis-mapped reads from the proximity exclusion region.
Three factors influence the number of unique interactions that can be determined from each viewpoint in a 3C library. First, a maximum of only four interactions can be detected from each region per cell (one from each end of the captured viewpoint fragment on each allele); so available cell numbers determines the maximum number of interactions that can be detected. Second, the hybridisation efficiency of the capture probe is important, and this is largely dictated by the underlying sequence. Third, the efficiency of the assay and depth of sequencing required, is determined by the proportion of background fragments from non-captured DNA contaminating the library.
To maximise the number of unique interactions defined, NG Capture-C was optimized to analyse 3C material containing eight times more ligation junctions than the previous protocol. This was achieved by minimising losses during the addition of sequencing adaptors and by mixing material from two parallel library preparations; allowing a total input of 10 μg 3C library to be used. This at least doubled the complexity of the material used for the hybridisation reaction. Additionally the amount of this material used in the hybridisation reaction was increased four-fold (from 500 ng to 2 μg).
To reduce the amount of background fragments we implemented two changes. First, the library design was simplified so that single 120 bp biotinylated DNA oligonucleotides, which include the restriction sites, were used to capture each end of the target restriction fragment (rather than multiple overlapping oligos, Supplementary Figure 1a) maximizing the capture of informative junction fragments. This made additional steps, such as biotin fill in, unnecessary and avoided losses in library complexity which is the critical component of sensitivity, particularly at low cell numbers17,18. Crucially, a second, sequential round of capture was introduced, which markedly reduced the background of uncaptured material and reduced the need for prohibitively deep sequencing.
We tested a minimal design containing probes to only the Hba-a1 and Hba-a2 promoters, equivalent to a 4C-seq analysis and found that a single oligonucleotide capture step enriched the targets ~ 5-20,000 fold. Despite this, the captured DNA from this single region only made up less than 1% of the sequenced reads; the remainder being uncaptured background (Fig. 2a i). In NG Capture-C the use of two sequential oligonucleotide capture steps resulted in up to 1,000,000 fold enrichment so that captured material now made up approximately 50% of the sequenced material (Fig. 2a i and ii). This second step increased the number of PCR cycles (20 to 34) and the number of PCR duplicates sequenced (Fig. 2a iii and iv) because the library complexity (i.e. the number of interactions available to capture) limited the number of unique interactions that could be sequenced. The greatly improved enrichment meant that sequencing depth was no longer limiting and PCR duplicates could be easily excluded bioinformatically. This is demonstrated by the fact that we saw no differences in the local interaction profiles (Fig. 2b) and there was little change in GC content or read length (Supplementary Figure 1b) when comparing single captured and double captured libraries.
Figure 2. Comparison of single and double oligonucleotide capture.
3C material generated from erythroid cells was captured using a single set of oligonucleotides designed to the α globin promoters (Supplementary Data). The two copies of the gene are virtually identical, therefore interaction profiles were generated from both genes simultaneously. After the first oligonucleotide capture step some of the material was sequenced using the Illumina MiSeq. The remaining library was used as input for a second round of oligonucleotide capture and then sequenced.
a. Comparison of the enrichment (to scale) resulting from the single (i) and double capture (iii) and the subsequent sequence read categorization following alignment (iii and iv). (i) Single capture resulted in 5-20,000 fold enrichment but only 0.3% of the reads contained a sequence that mapped to the captured fragment. (ii) Double capture increased the enrichment markedly; producing up to 3,000,000 fold enrichment. This dramatically increased the percentage of reads containing a restriction fragment that map to the capture region from 0.3% to 48.6%. The number of unique interactions was increased around 30-fold following double capture (from 10,832 to 327,787) (iii & iv) as library complexity now becomes the limiting factor.
b. Comparison of the raw informative interactions count per restriction enzyme fragment for single and double capture. The red vertical lines denote the location of captured viewpoints. The light blue lines highlight the five well described regulatory elements in the mouse (R1, R2, R3, R4 and HS-12). This showed that double capture did not notably alter the local interaction profile yet has 30-fold increased sensitivity.
To demonstrate the scalability of the approach, we combined the Hba-a1 and Hba-a2 capture probes with capture probes for Hbb-b1 and Hbb-b2 (the adult β globin genes) and Slc25A37 (Mitoferrin 1). The α and β globin genes are amongst the most extensively characterized genes and their regulatory interactions have been interrogated by almost every 3C based method to date7,9,10,12,19-22 so they provide important controls for the validation of any new methodology. The interaction profiles of all three control genes were almost identical in the biological replicates (Supplementary Figure 2) and matched their previously determined patterns of interactions (Fig. 2b, Supplementary Figures. 3-5). Importantly, for the same depth of sequencing the double capture increased the sensitivity of the profile 30 fold (Fig. 2b, Supplementary Figures. 6 and 7, Supplementary Data file). We next scaled up to a 35 gene design and increased the number of samples analysed in a single experiment, capturing seven pooled indexed libraries in a single assay. The efficiency that resulted from the double capture step, allowed us to sequence these 245 interaction profiles (35 genes, seven samples) using a single Illumina HiSeq run (177 million reads).
After normalization of individual profiles for the total number of unique interactions across the genome from each viewpoint in each sample, the genome-wide correlation of the two replicates for all genes exceeded an R2 value of 0.97, showing exceptional levels of correlation across biological replicates (Supplementary Figure 1c). The coefficient of variation fell substantially (CV < 50%) when more than ten normalised interactions mapped to any individual restriction fragment (Supplementary Figure 1d). Thus ligation junctions present at 1 part in 10,000 in the 3C library can be detected reproducibly, since the data are normalised to a total of 100,000 unique interactions across the genome. Furthermore, the pattern of both short-range interactions (Supplementary Figures. 2 and 3) and long-range cis-interactions (Supplementary Figures. 6 and 7) were highly reproducible. In addition, NG Capture-C produced a more comprehensive profile than existing 4C-seq10 (Supplementary Figures. 6 and 8) and did so regardless of restriction enzyme fragment size (Supplementary Figure 1e).
We developed a set of tools for design and analysis of NG Capture-C experiments (Fig. 1b). An online tool to generate oligonucleotide design for multiple targets can be found at http://apps.molbiol.ox.ac.uk/CaptureC/cgi-bin/CapSequm.cgi and analysis scripts are available via github (https://github.com/telenius/captureC/releases). The depth of NG Capture-C data allowed unique interactions to be reported per individual restriction fragment or half fragment, the highest possible resolution for such experiments (Supplementary Figure 8b); there was no requirement to integrate data by using a moving window.
In summary, the substantial increase in signal allows multiple 3C libraries (e.g. from different cell types or replicates) to be indexed and pooled prior to capture. This greatly increases the throughput of the assay, and importantly allows biological replicates and different experimental conditions to be processed and analysed together, removing sources of experimental variation.
Identification of regulatory elements using comparative analysis
Currently there is no ideal way to consistently call all likely important interactions from chromosome conformation data. Sequences from any capture point will interact with the surrounding genome, in a distance dependant manner, whether it is active or inactive. Therefore, current analysis of 3C data typically includes approaches to normalise interaction data taking into account the distance from the viewpoint. In practice, the outputs from such approaches are highly dependent on the normalisation model and input parameters used, with a tendency to under call cis-interactions with genuine regulatory sequences lying close to the capture point, where normalisation is most stringent (see Supplementary Note).
The reproducibility of NG Capture-C profiles enabled us to test a complementary approach to identify regulatory interactions by comparing different cell types. Subtractive analysis of normalised data from erythroid and non-erythroid (mES) cells successfully identified all known regulatory elements in well characterised test loci (Figs. 3 and 4, Supplementary Figures. 3-5.) and, in the same data, identified similar interactions in the other less well characterised loci in the capture design which included clinically important genes (CD47 Supplementary Figure 9) and complete regulatory networks (Myc, Sox2, Oct4/Pou5f1, Klf4 and Nanog) (Supplementary Figures. 10-14). Interestingly we identified interactions with regulatory elements over 1 Mb from the capture point, consistent with previously reported high-resolution Hi-C data (Supplementary Figure 10 and 12).
Figure 3. Identification of regulatory elements using comparative analysis.
Top panels show the overlaid normalized mean Capture-C profiles from eythroid (genes active in red) and ES cells (genes inactive in blue) at three erythroid specific loci α globin, β globin and Slc25A37 (Mitoferrin 1) in (erythroid n=4 and ES cells n=3). These data were generated along with the profiles for another 32 gene promoters simultaneously from seven samples in a single capture reaction (making a total of 245 interaction profiles from one oligonucleotide capture reaction). The Y-axis denotes the mean number of unique interactions per restriction fragment, scaled to a total of 100,000 interactions genome-wide.
The captured viewpoint fragments are highlighted in red and the interactions with the well-known enhancers (as annotated by DNAseI hypersensitivity) are highlighted as black hatched lines. The differential track (Δ Capture-C) shows that interactions with the local erythroid enhancers are clearly and specifically increased in erythroid cells when the genes are active. Below this DESeq2 analysis of the differential enrichment (minus log10 adjusted p-values) mapped across the three loci. The DESeq2 analysis shows the highly significant (< 10−30) enrichment of the known regulatory interactions.
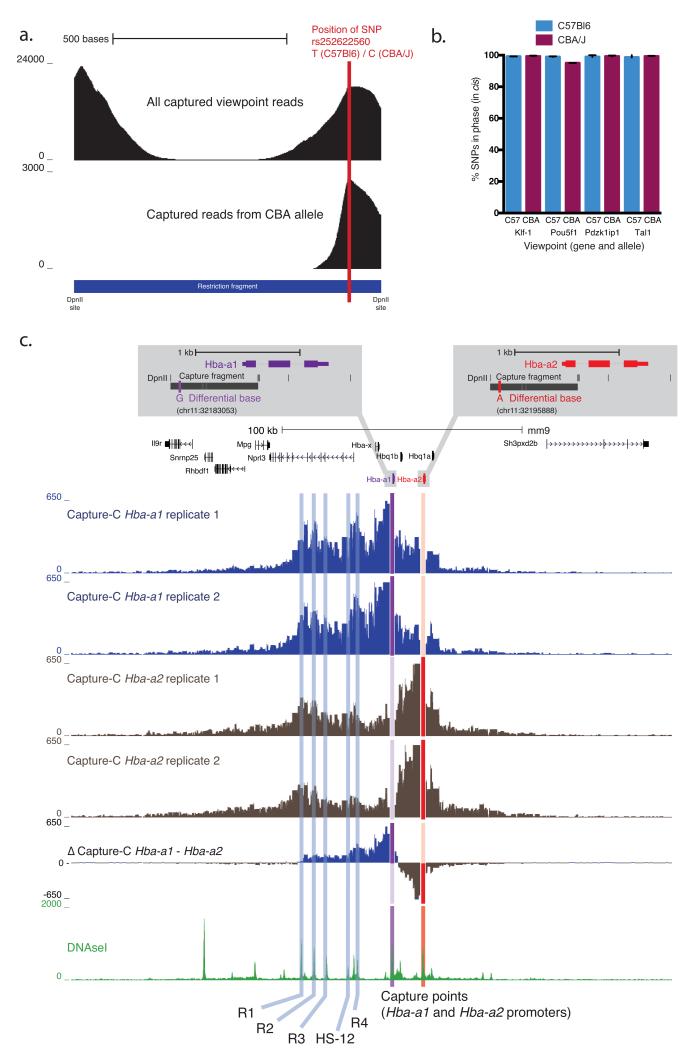
Figure 4. SNP specific interaction profiles.
a. Required positioning of SNP. Density plot of the total reads and CBA SNP allele reads from mapping to the Tal-1 captured restriction fragment (the Tal-1 promoter fragment is shown). SNPs under the captured regions allowed for the generation of allele specific interaction profiles in F1 crosses between C57BL/6 and CBA/J mice (see also Supplementary Figure 22). In the example locus the SNP rs252622560 has been used to separate interactions from the two different alleles.
b. Interactions occur in cis. Graphical representation of the percentage of SNPs in phase in the interacting reads compared with the strain of the captured allele in cis. This demonstrates that the chromosome predominately interacts with itself in cis rather than its sister chromatid.
c. SNP specific NG Capture-C. Using this approach we generated specific interaction profiles for Hba-a1 and Hba-a2 paralogous genes. A single nucleotide difference between the two genes allowed the generation of specific tracks (see inset). Hba-a1 is the more active of the two genes, producing around 70% of the total mRNA. Comparison of the two biological replicates showed that the SNP specific profiles are highly reproducible. The Δ Capture-C track showed the difference of the mean Hba-a1 and Hba-a2 profiles. This revealed that that the Hba-a1 gene preferentially interacts with the enhancers, particularly proximal HS-12 and R4 elements. The Hba-a2 gene interacts much more strongly with the chromatin between the two genes. Interestingly Hba-a2 interacts with the most distal enhancer (R1) to a very similar degree as the Hba-a1 gene.
Subtractive analysis also uncovered fine details of tissue-specific regulation of genes that are active in multiple cell types. For example, the Pnpo gene encodes Pyridoaxime 5′-phosphate Oxidase which is a rate-limiting enzyme in vitamin B6 metabolism producing an essential cofactor in the heme synthetic pathway23. Pnpo is specifically up-regulated in mouse erythroid cells by an erythroid-specific enhancer (HS-26)12. Comparison of ES and erythroid data precisely and specifically identified HS-26 at a resolution sufficient to distinguish it from the promoter of a neighbouring gene (Cdk5rap3) ~1 kb away (Supplementary Figure 15).
Subtractive analyses not only showed new interactions in the specific cell type under investigation but also identified new patterns of interaction in the cell type used for comparison. For example analysis of the Tal1 locus revealed one pattern of interaction in ES cells and another in erythroid cells; these cells acting as reciprocal controls for each other (Supplementary Figure 16). It is important to note that as this approach relies on changes between active states its goal is to find regulatory elements rather than constitutive structural interactions.
The subtractive profiles can be additionally statistically interrogated using common approaches for the differential analysis of sequence count based data, such as the Bioconductor package DEseq224. We compared the effectiveness of this approach at identifying known regulatory elements with two tools commonly used for 3C analysis (Fig. 3, Supplementary Figures. 3-5 and Supplementary Results), FourCseq25 and r3C-seq26 which also use replicates and comparative analysis but additionally normalise for genomic distance using different models. We tested all approaches on the well-characterized test loci; α globin, β globin and Slc25A37, using default parameters to simulate the output at uncharacterized loci. Of these three loci α and β globin are used as gold standards in the 3C field due to the depth of the functional knowledge of their regulation. These tools called the known elements in the β globin and Slc25A37 loci, but each variably missed the most proximal elements in the α globin locus, unlike the comparative approach which called all of the known elements in each locus (Supplementary Note).
Reproducible megabase scale cis and trans-interactions
The original Capture-C method does not readily detect weak long-range interactions. NG Capture-C enabled us to investigate such interactions and, importantly, evaluate their strength relative to local interactions. Analysis of interaction frequencies across a whole chromosome containing a captured region showed that interactions with the entire chromosome were not easily seen when viewed on the same scale as interactions with the more local regulatory elements. However, reproducible, low level (<100 fold) cis-interactions were detected with other active regions of the chromosome (Supplementary Figure 7). Similar patterns of general interactions could also been seen in trans but these were a further ten fold weaker than the long-range cis interactions (Supplementary Figures. 17-19). The patterns of trans interactions became visible when the threshold for any interaction was reduced to fewer than 250 interactions per 100 kb, these interactions had similar distributions independent of the gene promoter used as the view point (Supplementary Figures. 18 and 19) and were correlated with gene density, the number of active promoters, enhancers and CTCF sites (Supplementary Figure 20). This is of particular interest in the case of the α and β globin genes as they have been reported to interact with each other in erythroid cells. Some have suggested that these interactions are frequent27 whereas others have shown them to be rare28. Now, with the sensitivity and robust quantitation provided by the NG Capture-C approach, the trans-interaction between these two genes are shown to be rare (~1,000 fold less than local cis-interactions) and on the same scale as those with most other active regions of the genome. These weak interactions are unlikely to be functional but this allows us to be confident that all functional interactions quantifiable by 3C approaches can be detected using NG Capture-C.
In summary we have shown that real, reproducible albeit weak very long-range interactions exist in cis and trans. Furthermore, all analytical approaches tested discriminate appropriately between these weak interactions and the much stronger interactions with known regulatory elements (See Supplementary note).
Robust interaction profiles from low cell numbers
In human primary tissues cell numbers are often limited and so NG Capture-C was further adapted to analyse small numbers of cells (see Supplementary Methods). Low cell number did not alter digestion efficiency and the amount of DNA extracted per cell was constant, hence preparation of material for the hybridisation reaction was optimised for the reduced DNA content of the 3C libraries. Using 100,000 cells we generated ~19,000 interactions compared to an average of 137,000 when cell number was not limiting, however, the interaction profiles at the α and β globin loci remained virtually unchanged (Supplementary Figure 21) although weak, long-range interactions became difficult to determine reproducibly (Supplementary Figure 6) when using only 100,000 cells.
Generation of SNP specific interaction profiles
SNPs underlying GWAS traits are frequently heterozygous and may affect the regulatory interactions of the affected allele. Therefore, it would be of great value to generate allele-specific interaction tracks. Due to the greatly improved depth of signal provided by the NG Capture-C protocol we could distinguish between separate alleles when a SNP is included within a capture point and hence sequenced (Fig. 4a). These allele-specific interaction profiles showed that over 95% of the strain-specific SNPs in cis were in phase with the captured strain specific SNP, showing that interactions with the sister chromatid were relatively rare (Fig. 4b). For the examined genes, the interaction profiles were very similar probably because none of the SNPs were of functional importance (Supplementary Figure 22).
This type of analysis also applies to non-allelic SNPs. The parologous Hba-a1 and Hba-a2 genes are almost exact copies differing at only a few positions, one of which lies near the 5′ promoter capture point (Fig. 4c). This allowed separate interaction profiles for the Hba-a1 and Hba-a2 genes to be generated (Fig. 4c). The 5′ Hba-a1 interacted with the proximal regulatory elements (HS-12 and R4) more frequently than Hba-a2. Interestingly, Hba-a2 and Hba-a1 had very similar interactions with the MCSR1 and R2 regulatory elements, which are thought to have stronger enhancer function than the other elements29,30.
Discussion
NG Capture-C was developed to generate a completely flexible assay allowing researchers to analyse interactions involving single or many genes and multiple samples, simply and cheaply. NG Capture-C is able to detect interactions present 1 in 5-10,000 cells, which far exceeds the current reasonable limit of detection by fluorescence in-situ hybridisation31.
The investigation of gene regulation is not only limited by the number of genes or elements that can be interrogated, but also by the number of replicates, conditions, cell types and genetic variants that can be easily analysed. The huge increase in signal of NG Capture-C allows for the simultaneous capture of multiple samples in a single reaction, greatly increasing the throughput and economy of the assay. In practice this allows complete networks of important genes, such as those encoding the Yamanaka pluripotency factors32 (Myc, Sox2, Oct4, Klf4) to be analysed simultaneously in multiple cell types. These data are compatible with standard analytical tools and their reproducibility and comparability between active and inactive states of NG Capture-C provides a complementary approach to the statistical identification of regulatory elements. This approach identifies all known regulatory elements at characterised test loci at levels of resolution previously not possible. Importantly, mindful of the current challenges in the analysis of GWAS and regulatory variants, the NG Capture-C method has been optimized for smaller cell numbers (~100,000) and to generate SNP-specific interaction profiles.
It is important to note that unlike most other high-resolution chromosome conformation methods, NG Capture-C provides sufficient depth of data that the output is expressed as “raw counts” per fragment with no need to integrate interactions via a moving window10,12,16. Furthermore, the sensitivity provided by double capture together with the ability to remove PCR duplicates means that the interaction data faithfully represent all interactions within the library allowing researchers to make estimates of relative quantitation between weak and strong interactions. The complete and detailed protocol presented here, with new publically available tools for library design and data analysis are intended to allow any laboratory to perform chromatin conformation capture analysis of the highest quality and at levels of throughput that were previously impossible.
Supplementary Material
Acknowledgements
James Davies would like to thank the Wellcome Trust for funding his work (Wellcome Trust Clinical Research Training Fellowship ref 098931/Z/12/Z). The work was also supported by a Wellcome Trust Strategic Award (reference 106130/Z/14/Z) and the Medical Research Council (MRC Core Funding and Centenary Award reference 4050189188).
We would like to thank E. Repapi for statistical advice and L. Hanssen, M. Oudelaar, D. Jeziorska, B. Graham, M. Kassouf, M. Suciu, H. Long, S. Pasricha, V. Buckle, T. Milne, T. Fulga, T. Sauka-Spengler, D. Downes and A. Drakesmith for their critique of the manuscript. We would like to thank S. Thongjuea for discussions on analysis.
Footnotes
Accession codes
All of the Capture-C data sets are available at the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) accession code GSE67959.
References
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 4.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–9. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 7.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 8.Noordermeer D, et al. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–5. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 9.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Werken HJ, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9:969–72. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- 11.de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 12.Hughes JR, et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet. 2014 doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 13.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46:136–43. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker SC, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–6. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jager R, et al. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat Commun. 2015;6:6178. doi: 10.1038/ncomms7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoenfelder S, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015;25:582–97. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–51. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes JR, et al. High-resolution analysis of cis-acting regulatory networks at the alpha-globin locus. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120361. doi: 10.1098/rstb.2012.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bau D, et al. The three-dimensional folding of the alpha-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol. 2011;18:107–14. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 23.Kang JH, et al. Genomic organization, tissue distribution and deletion mutation of human pyridoxine 5′-phosphate oxidase. Eur J Biochem. 2004;271:2452–61. doi: 10.1111/j.1432-1033.2004.04175.x. [DOI] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein FA, et al. FourCSeq: analysis of 4C sequencing data. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thongjuea S, Stadhouders R, Grosveld FG, Soler E, Lenhard B. r3Cseq: an R/Bioconductor package for the discovery of long-range genomic interactions from chromosome conformation capture and next-generation sequencing data. Nucleic Acids Res. 2013;41:e132. doi: 10.1093/nar/gkt373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 28.Noordermeer D, et al. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol. 2011;13:944–51. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernet A, et al. Targeted inactivation of the major positive regulatory element (HS-40) of the human alpha-globin gene locus. Blood. 1995;86:1202–11. [PubMed] [Google Scholar]
- 30.Anguita E, et al. Deletion of the mouse alpha-globin regulatory element (HS - 26) has an unexpectedly mild phenotype. Blood. 2002;100:3450–6. doi: 10.1182/blood-2002-05-1409. [DOI] [PubMed] [Google Scholar]
- 31.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.