Abstract
Nanog, a core pluripotency factor in the inner cell mass of blastocysts, is also expressed in unipotent primordial germ cells (PGC) in mice1, where its precise role is yet unclear2-4. We investigated this in an in vitro model, where naïve pluripotent embryonic stem cells (ESCs) cultured in bFGF/ActivinA develop as epiblast-like cells (EpiLCs), and gain competence for PGC-like fate5. Consequently, bone morphogenetic protein (BMP4), or ectopic expression of key germline transcription factors Prdm1/ Prdm14/ Tfap2c, directly induce PGC-like cells (PGCLCs) in EpiLCs, but not in ESCs6-8. Here we report an unexpected discovery that Nanog alone can induce PGCLCs in EpiLCs, independently of BMP4. We propose that following the dissolution of the naïve ESC pluripotency network during establishment of EpiLCs9,10, the epigenome is reset for cell fate determination. Indeed, we found genome-wide changes in NANOG binding pattern between ESCs and EpiLCs, indicating epigenetic resetting of regulatory elements. Accordingly, we show that NANOG can bind and activate enhancers of Prdm1 and Prdm14 in EpiLCs in vitro; BLIMP1 (encoded by Prdm1) then directly induces Tfap2c. Furthermore, while SOX2 and NANOG promote the pluripotent state in ESCs, they show contrasting roles in EpiLCs since Sox2 specifically represses PGCLC induction by Nanog. This study demonstrates a broadly applicable mechanistic principle for how cells acquire competence for cell fate determination, resulting in the context-dependent roles of key transcription factors during development.
Transcription factors and epigenetic changes confer competence for somatic and PGC fates when naïve pluripotent inner cell mass (ICM) from embryonic day (E) 3.5-4.5 blastocysts develop to primed epiblast at ~E 6.011. Similarly, naïve pluripotent embryonic stem cells (ESCs) in ‘2i’ acquire competency within ~48h following culture in bFGF/ActivinA in vitro, when day 2 epiblast-like cells (D2 EpiLCs) differentiate into PGC-like cells (PGCLCs) in response to BMP45. These putative PGCLCs show expression of ΔPE-Oct4-GFP (hereafter called GOF-GFP) and Blimp1-GFP reporters (Fig. 1a and Extended Data Fig. 1a-c), following upregulation of the three key regulators of PGCLCs; Prdm1 (encoding BLIMP1), Prdm14 and Tfap2c (encoding AP2Ɣ)5,7,8.
Figure 1. Nanog induces PGCLCs in EpiLCs.
a, Brightfield/GFP representing D4 male PGCLCs induced by Nanog (+Dox); % GFP+ve cells after FACS; scale bar, 200 μm.
b, FACS plots for GOF-GFP+ve D4 PGCLCs induced by BMP4 or Nanog (+Dox) and +/−Noggin.
c, Analysis of male GOF-GFP cells (qPCR) as indicated. GFP+ve cells were FACS sorted. ΔΔCt +/− s.d. (n=3 biological replicates).
d, Microarray analyses of GOF-GFP ESCs and PGCLCs; unsupervised hierarchical clustering, and principal component (PC)1 scores.
e, IF of Nanog-induced BLIMP1-GFP+ve PGCLCs. Arrowheads highlight single GFP+ve cells; two biological replicates; scale bar, 10 μm; two-sided/unpaired t-test; n.s.=not significant; s=significant; n=number of cells.
NANOG and PRDM14 share similar binding profiles in ESCs and contribute to pluripotency12. While Prdm14 is also a key regulator of PGC fate13,14, the role of Nanog is unclear, although Nanog is detected in E6.5 posterior proximal epiblast15,16, the site of PGC induction, and thereafter in the early germline1,7. However, we unexpectedly found that Doxycycline (Dox) induced expression of Nanog alone, stimulated GOF-GFP and Blimp1-GFP expression in D2 EpiLCs, indicating specification of putative PGCLCs (Fig. 1a, Extended Data Fig. 1a, d-f, 2a-e). Furthermore, Nanog apparently acts synergistically with BMP4 to increase the number of GFP+ve cells, which we did not see with Oct4 (Extended Data Fig. 2f-h). Nanog induced PGCLCs in the presence of Noggin, a BMP signalling inhibitor, demonstrating that it acts independently of BMP-SMAD signalling (Fig. 1b). Physiological (equivalent to ESCs) or higher levels of NANOG induced PGCLCs with similar efficiency (Extended Data Fig. 3a-c).
We analysed FACS-sorted Nanog-induced GFP+ve cells, which showed upregulation of the key PGC regulators; Prdm1/Prdm14/Tfap2c as well as Nanos3 and Dppa3, but ESC-specific Klf4 was downregulated (Fig. 1c, Extended Data Fig. 3d-f). This mirrors the response seen with BMP4-mediated PGCLC induction5. Notably, PCA analysis of global gene expression confirmed that Nanog- and BMP4-induced D4 PGCLCs are highly similar, and closely match with the previously reported D6 PGCLCs5 (Fig. 1d, Extended Data Fig. 3g-j). Further, BLIMP1, PRDM14 and AP2Ɣ (but not KLF4) were detected in PGCLCs by immunofluorescence (IF) (Fig. 1e, Extended Data Fig. 4). Thus, Nanog clearly induces PGC-like fate in EpiLCs and not their reversion to ESCs.
The Nanog-induced PGCLCs also showed unique early germline-specific epigenetic modifications; global enrichment of H3K27me3 and erasure of H3K9me217,18 (Fig. 1e, Extended Data Fig. 4), together with the initiation of DNA demethylation through the repression of Uhrf1, Dnmt3a and Dnmt3b (Fig. 1c, Extended Data Fig. 3e, i), and upregulation of 5-hydroxymethylcytosine (5hmC) and TET119 (Extended Data Fig. 4). Expression of Dazl also indicated progression of DNA demethylation in PGCLCs (Extended Data Fig. 4a, b), which is reminiscent of BMP4-induced PGCLCs5.
Next, we asked if Nanog- and cytokine-induced PGCLCs could dedifferentiate into pluripotent embryonic germ cells (EGCs) as seen with E8.5 PGCs11. We first subjected PGCLCs to a selection with retinoic acid and bFGF for 5 days, which promotes PGCLC proliferation, but not of ESC-like cells20,21 (Extended Data Fig. 5a, b). The resulting PGCLCs were transferred to 2i/LIF to promote their dedifferentiation into EGC-like cells (EGCLCs), which after several passages produced self-renewing GFP+ve EGCLCs (Extended Data Fig. 5b). These EGCLCs when introduced into blastocysts contributed extensively (in 27/29 embryos) to chimeric fetuses at E9.5 (Extended Data Fig. 5c), unlike ‘unipotent’ PGCLCs/PGCs, which neither integrated nor contributed to the fetus (Extended Data Fig. 5d-g).
We then sought genetic evidence that Nanog induces bona fide PGCLCs using ESCs with a mutation in Prdm1, which is obligatory for PGC specification, but not for the pluripotent state22,23. Consistently, no PGCLCs were induced from Prdm1−/− D2 EpiLCs, nor did they revert to ESCs (Fig. 2a, Extended Data Fig. 6a, b). Instead, the aggregates showed somatic gene expression, including Hoxa1 and Hoxb1, which is reminiscent of the aborted PGC fate in Prdm1−/− embryos in vivo22. Furthermore, H3S10ph and ƔH2A.X analysis by IF of D6 aggregates indicated that while cell proliferation was unaffected, the rate of apoptosis increased, presumably as the differentiated cells could not survive in the culture conditions (Extended Data Fig. 6c, d).
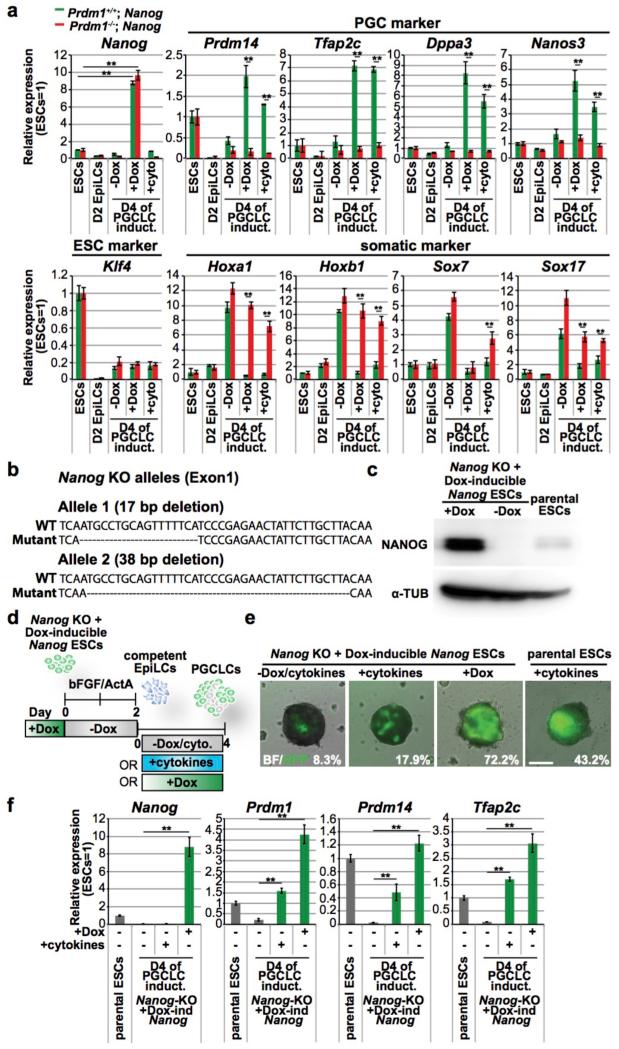
Figure 2. Loss of Prdm1 and Nanog affects PGCLC specification.
a, Analysis (qPCR) of mutant (Prdm1−/−) and control (Prdm1+/+) unsorted cells following Nanog expression (+Dox). ΔΔCt +/− s.d (n=2 technical replicates each from 2 biological replicates); two-sided/unpaired t-test: **p<0.01; *p<0.05.
b, Nanog frameshift mutant alleles.
c, Western blot for NANOG and α-TUBULIN (α-TUB) as depicted. +/−Dox for 2 days; gel source data in Supplementary Fig.1.
d, Experimental design for e-f.
e, PGCLC induction in Nanog-KO cells, and rescue by Nanog (+Dox). Merged brightfield/GFP at D4; GFP+ve cells (%) after FACS; scale bar, 200μm.
f, Analysis (qPCR) of ESCs and D4 PGCLC aggregates shown in (e). ΔΔCt +/− s.d. (n=2 technical replicates each from 2 biological replicates); two-sided/unpaired t-test: **p<0.01.
To further investigate PGCLC induction by Nanog, we generated CRISPR/Cas9-mediated Nanog knockout alleles in GOF-GFP ESCs with Dox-inducible Nanog (Fig. 2b, c). We found a significant reduction in the induction of PGCLCs from Nanog mutant cells in response to BMP4 (Fig. 2d-f), but ectopic Nanog expression rescued this deficit, suggesting complementary roles for BMP4 and Nanog in PGCLC induction.
Next, we investigated if the Wnt-BRACHYURY pathway is important for PGCLC induction by Nanog as is the case with BMP424. We induced PGCLCs in the presence of XAV939 tankyrase inhibitor, which promotes degradation of β-catenin25 resulting in the repression of Brachyury (Extended Data Fig. 6e-g). PGCLC induction with BMP4 was repressed by XAV939 but not when induced with Nanog (Extended Data Fig. 6h, i). Furthermore, Wnt had no detectable effect on Nanog expression (Extended Data Fig. 6g, i), indicating that Nanog acts independently of Wnt-BRACHYURY.
We then asked when during the transition of ESCs to EpiLCs, cells become responsive to Nanog for PGCLC induction. We found a large majority of D1 EpiLCs (63.8%) reverted to ESCs when transferred to 2i/LIF medium, and Nanog enhanced this response (to 84.7%), as confirmed by expression of Klf4 and repression of PGC genes (Fig. 3a-c). This reversion to ESCs diminished significantly in D2 EpiLCs (28.4%), and Nanog repressed it further (to 9.8%); instead these cells exhibited a distinct phenotype with expression of Brachyury and Wnt3 mesodermal genes (Fig. 3a-c). Thus, D2 EpiLCs do not revert to ESCs but acquire competence for PGCLC fate in response to Nanog.
Figure 3. Competence for PGCLCs versus reversion to ESCs.
a, Experimental design for b, c.
b, Brightfield/GFP depicting D1/D2 EpiLCs in 2i/LIF+Nanog (+Dox); note D1 EpiLCs revert to ESCs, and attempted PGCLC induction in D1 EpiLCs also results in ESC reversion (see c). Scale bar, 200 μm.
c, qPCR analysis; ΔΔCt +/− s.d. (n=2 technical replicates each from 2 biological replicates); ESCs as reference for p-values (two-sided/unpaired t-test): **p<0.01; *p<0.05.
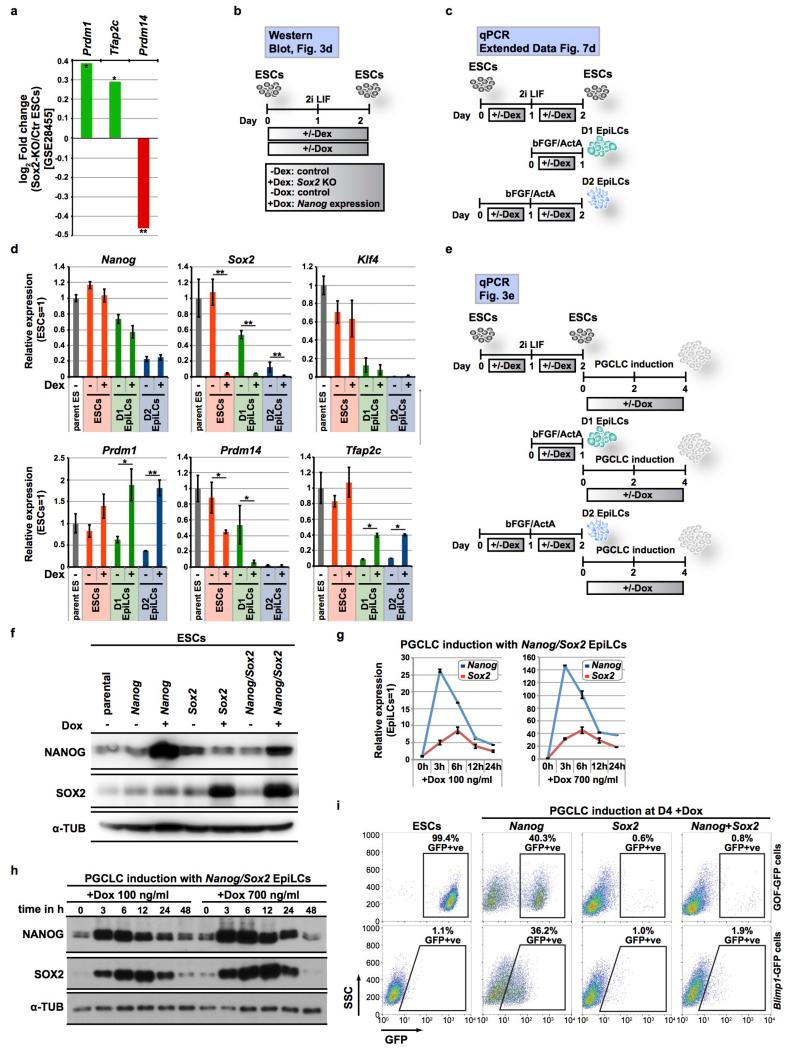
d, Western blot for NANOG, SOX2 and α-TUBULIN (α-TUB) with ESCs as depicted; +/−Dex/Dox for 2 days. Experimental design in Extended Data Fig. 7b; gel source data in Supplementary Fig. 1.
e, Analysis (qPCR) after Sox2-KO (+Dex) and Nanog induction (+Dox); ΔΔCt +/− s.d. (n=2 technical replicates each from 2 biological replicates); parental ESCs as reference for p-values (two-sided/unpaired t-test): **p<0.01; *p<0.05. Experimental design in Extended Data Fig. 7e.
f, PGCLC induction with Dox-inducible transgenes (Nanog, Sox2 or Nanog/Sox2); D4 brightfield/GOF-GFP+ve cells (%) after FACS; scale bar, 200 μm.
Nanog and Sox2 promote pluripotency in ICM, but thereafter Nanog is detected in the E6.25 posterior epiblast where PGCs arise15,16, and Sox2 in the anterior epiblast where it promotes neuronal fate and inhibits mesodermal specification16. Sox2 also represses germline genes in ESCs26 (Extended Data Fig. 7a). We tested their roles in our experimental model using ESCs with Dexamethasone (Dex)-inducible knockout of Sox227, in conjunction with Dox-inducible Nanog (Fig. 3d, Extended Data Fig. 7b). Loss of Sox2 caused a moderate upregulation of Prdm1/Tfap2c in ESCs without affecting Nanog expression (Extended Data Fig. 7c, d). Notably, Nanog induced Prdm1/Prdm14/Tfap2c in Sox2 knockout D1 EpiLCs but not in wildtype cells (Fig. 3e, Extended Data Fig. 7e). Since there is a gradual decline in Sox2 during development of EpiLCs5 (Extended Data Fig. 7d), residual Sox2 in D1 EpiLCs might repress competency for PGCLCs, and accounts for their reversion to ESCs in response to Nanog (Fig. 3a-c). By contrast, Dox-induced expression of Sox2 strongly repressed PGCLC specification in D2 EpiLCs in response to Nanog but not BMP4 (Fig. 3f, Extended Data Fig. 7f-i). Sox2 however caused rapid proliferation of PGCLCs after their induction by BMP4 (Extended Data Fig. 8), consistent with its role in PGCs in vivo28. Thus, progressive downregulation of Sox2 in EpiLCs contributes to competency for PGCLCs, but thereafter Sox2 supports proliferation of early germ cells. This further confirms that Nanog and BMP-SMAD act independently during PGCLC induction.
While both Nanog and BMP4 induce PGCLCs, the temporal sequence of Prdm1/Prdm14/Tfap2c induction differs slightly. Nanog induces Prdm14 first at 3h, that increases rapidly over ~18h (Fig. 4a). This is followed by Prdm1 at ~12h that increases over the following 12h, and finally Tfap2c expression at ~18h. Thus, all the three regulators of PGCLCs are upregulated within ~24h. While the response of D2 EpiLCs to BMP4 is similar, Prdm1 expression is detected first and slightly ahead of Tfap2c, followed by Prdm14 (Extended Data Fig. 9a). Tfap2c, a direct target of BLIMP17,8, is rapidly induced by Prdm1 alone within 6h (Extended Data Fig. 9b).
Figure 4. Context-dependent NANOG binding in ESCs/EpiLCs.
a, qPCR analysis; induction of Prdm1/Prdm14/Tfap2c by Nanog (+Dox) in EpiLCs; ΔΔCt +/− s.d. (n=3 technical replicates each from 2 biological replicates).
b, Genome-wide NANOG binding in D2 EpiLCs 3h after Nanog (+Dox). ‘Distal’ intergenic’ peaks: +/−50kb of coding gene, and those further away designated as ‘intergenic’.
c, NANOG ChIP-seq in ESCs and D1/D2 EpiLCs, with specific or shared high-confidence peaks; n=2 biological replicates.
d, ChIP-seq tracks9,30 at Prdm1 and Prdm14 loci, with putative enhancers (boxed) analysed in (e-g). RPM = Reads per Million.
e, Analysis of Prdm1 enhancer-luciferase reporter in ESCs, EpiLCs, and after PGCLC induction (+Dox, EpiLC aggregations, unsorted). Mean luciferase activity normalised to protein quantity (luc/pro) +/− s.d. (n=3 technical replicates). Reference for p-value (two-sided/unpaired t-test): EpiLC aggregations −Dox; **p<0.01; *p<0.05. Controls and replicates in Extended Data Fig. 10c, e.
f, SOX2 ChIP-qPCR in ESCs and 6h after PGCLC induction +/−Dox (+/−Sox2) (unsorted EpiLC aggregations). Mean of fold enrichment over negative region +/− s.e. (n=2 technical replicates each from 2 biological replicates); reference for p-values (two-sided/unpaired t-test): IgG; *p<0.05.
g, Analysis of Prdm14 enhancer-luciferase reporter in ESCs, EpiLCs and after PGCLC induction (+Dox, EpiLC aggregations, unsorted). Mean luciferase activity normalised to protein quantity (luc/pro) +/− s.d. (n=3 technical replicates); reference for p-values (two-sided/unpaired t-test): EpiLC aggregations −Dox 24h; **p<0.01. Colour code as in (e); controls and replicates in Extended Data Fig. 10c, e.
To explore how NANOG promotes both pluripotency and the induction of PGCLCs, we performed NANOG ChIP-seq in ESCs and 3h after induction of physiological levels (equivalent to ESCs) of NANOG in EpiLCs (Extended Data Fig. 9c). We found NANOG binding primarily in the intergenic regions and introns (>90%), where enhancer elements reside (Fig. 4b, Extended Data Fig. 9d), with marked differences in binding patterns and enriched motifs in ESCs compared to EpiLCs (Fig. 4c, Extended Data Fig. 9e); this provides a basis for the context-dependent functions of NANOG. Overall, many D2 EpiLC enhancers bound by NANOG show enrichment of H3K27ac in D2 PGCLCs, indicative of active enhancers29 (Extended Data Fig. 9f-h). This shows that during PGCLC induction, NANOG might contribute to the activation of these elements together with BLIMP1/PRDM14/AP2Ɣ. Importantly, we also found and confirmed intergenic NANOG binding sites proximate to the Prdm14 and Prdm1 loci (Fig. 4d, Extended Data Fig. 10a, b). These sites were devoid of the promoter and gene-body associated H3K4me3 and H3K36me3 modifications, respectively. Instead, they were enriched for the enhancer associated H3K4me1 modification in EpiLCs, suggesting their priming before activation via NANOG and gain of H3K27ac in PGCLCs (Fig. 4d, Extended Data Fig. 10a). Since Prdm14 is critical for both ESCs and PGCLCs, its enhancer showed a similar H3K4me1/H3K27ac/NANOG enrichment profile in both cell types.
Next, we tested the putative Prdm1 enhancer in luciferase reporter assay, and found that following its low activity in ESCs and EpiLCs, Nanog activated the enhancer within 24h after PGCLC induction (Fig. 4e, Extended Data Fig. 10c, d). Notably, Sox2 strongly repressed this activity consistent with SOX2 binding to this enhancer (Fig. 4e, f, Extended Data Fig. 10c, d). By contrast, the putative Prdm14 enhancer, which did not bind SOX2 (Fig. 4f), was active in ESCs; this declined in EpiLCs but increased again within 12h after the induction of PGCLCs by Nanog (Fig. 4g, Extended Data Fig. 10c, e). This reflects the importance of Prdm14 for both pluripotency and PGCLC fate. Notably, while both BRACHYURY and NANOG bind to and activate the Prdm1 enhancer24, the latter acts independently of Wnt during PGCLC induction (Fig. 4e, Extended Data Fig. 6e-i and 10c, d). Thus, NANOG activates key regulators of PGCLCs independently of BMP4 and Wnt signalling. Additional regulatory elements associated with Prdm1/Prdm14 may respond similarly.
In conclusion, the resetting of the epigenome during the gain of competency for PGC-like fate is reflected in the differential NANOG binding pattern in ESCs and EpiLCs, consistent with its role in pluripotency and PGCLC specification (Extended Data Fig. 10f). Nanog is detected in the proximal epiblast and the early germline15,16. Transcription factors also affect competency, since SOX2 inhibits the induction of PGCLCs by NANOG, while NANOG and SOX2 cooperatively promote pluripotency in the ICM/ESCs. NANOG acts independently of BMP4 during PGCLC induction, but they might act cooperatively in vivo, since loss of Nanog significantly impairs the efficiency of PGCLC specification via BMP4. Notably, epigenome resetting during differentiation of competent EpiLCs establishes a mechanistic paradigm for context-dependent roles of transcription factors such as NANOG that could apply generally during development.
Methods
Animals
Timed natural matings were used for all experiments, where noon of the next day after the vaginal plugs of mated females were identified was scored as E0.5. Animal studies were authorized by a UK Home Office Project License and carried out in a Home Office-designated facility.
Mouse embryonic stem cells
ΔPE-Oct4-GFP (GOF-GFP)31,32, Blimp1-GFP and Prdm1−/− ESC lines were established previously14,19,22,23. Inducible Sox2 knockout (2CG2) ESC line was a gift by Dr. Hitoshi Niwa27. All ESC lines were maintained in naïve ‘ground state’33 condition; i.e. in N2B27 medium (R&D) with 2i (PD0325901, 1 μM; CHIR99021, 3 μM; Stemgent), LIF and 1% KnockOut Serum Replacement (KSR; Life Technologies) on Fibronectin-coated dishes (16.7 μg/ml; Millipore). Medium was changed daily. ESC colonies were passaged by dissociating with TrypLE (Life Technologies).
Establishment of PiggyBac based Tet-on expression system during PGCLCs induction
Oct4, Sox2, Nanog, Prdm1, Prdm14 and Brachyury cDNAs were cloned from mouse cDNA pool. cDNAs were inserted into PiggyBac based doxycycline (Dox) inducible vectors (a gift of Dr. Hitoshi Niwa). These vectors were transfected using Lipofectamine 2000 (Life Technologies) into ESCs together with a pPyCAG-PBase vector and a pPBCAG-rtTAIRESNeor vector, which harbours a neomycin resistance gene. After 1 week of neomycin (80 μg/ml; Life Technologies) selection, pooled or single clones were used for experiments. To induce transgene expression, various concentrations of Dox (Sigma-Aldrich) were added to the media.
Induction of EpiLCs and PGCLCs
EpiLCs and PGCLCs were induced as described previously5. Transgenes were induced by addition of Dox at day0 of PGCLC induction. 100 ng/ml Dox was used in experiments shown in Fig. 3f, 4e, g, Extended Data Fig. 3b, c, 7g-i, 8a-c. 200 ng/ml Dox was used in Fig. 4b-d, f. 700 ng/ml of Dox was used in all other experiments. PGCLCs were induced as described in the manuscript. For inhibition of the BMP-SMAD pathway, Noggin (200 ng/ml; R&D) was added in the media at day0 of PGCLC induction. For inhibition of Wnt signalling, XAV939 (1 μM; Sigma-Aldrich) was added to the media.
Reversion of epiblast-like cells into ES-like cells
D1 or D2 EpiLCs were transferred into GMEM 15%KSR 2i LIF with or without Dox in monolayer culture. In addition, D1 or D2 EpiLCs were aggregated in low-cell-binding U-bottom shaped 96-well plates (Thermo Scientific) (1000-2000 cells per well) in PGCLC induction media (GMEM with L-glutamine (Life Technologies), 15% KSR (Life Technologies), 1× MEM NEAA (Life Technologies), 1× Sodium Pyruvate (Life Technologies), 1× 2-mercaptoethanol (Life Technologies), 1× Penicillin/Streptomycin (Life Technologies)) and Dox, The medium was replaced daily. After 3 days, the GFP reporter signal was analysed with a fluorescence microscope and via Fluorescence-Activated Cell Sorting (FACS) analysis. RNA was collected from pooled cells for qRT-PCR.
Embryonic germ cell-like cell (EGCLCs) derivation
D4 aggregates were dissociated with TrypLE and plated on mouse embryonic feeder cells (MEFs) with PGC selection medium (DMEM with L-glutamine (Life Technologies), 15% Fetal Bovine Serum (FBS, Sigma-Aldrich)), LIF, 15 ng/ml bFGF, 30 ng/ml SCF (R&D) and 2 μM All trans-retinoic acid (Sigma-Aldrich). Retinoic acid promotes germ cell self-renewal while promoting differentiation of ESCs20,21. The media was replaced daily. After 5 days, proliferating GFP+ve cells were dissociated with TrypLE and plated on Fibronectin-coated dishes with ESC medium (N2B27 with 2i LIF).
Fluorescence-Activated Cell Sorting (FACS)
PGCLCs were dissociated with TrypLE, washed with DMEM containing 10% FBS and re-suspended with 1×PBS containing 0.1% BSA. Large clumps of cells were removed using a cell strainer (BD Biosciences). The cells were analysed and sorted on flow cytometers (FACS Calibur, BD Biosciences; MoFlo high speed cell sorter, Beckman Coulter; S3 cell sorter, Biorad). FACS plots show FL1; green on x-axis and SSC; side scatter on y-axis.
RT-qPCR
Total RNAs from ESCs, EpiLCs and FACS-sorted cells were extracted using the RNeasy Mini Kit (Qiagen) or Picopure RNA Isolation Kit (Life Technologies). The total RNAs were reverse transcribed by the Quantitect Reverse Transcription Kit (QIAGEN). The first-strand cDNAs were used for RT-qPCR analysis with SYBR Green PCR reagent (Sigma-Aldrich). The primer sequences used for the qRT-PCR are listed in Supplementary Table 1. Student’s t-test was used to test for significance.
Microarray
ESCs and D4 PGCLC were dissociated and sorted with a MoFlo high-speed cell sorter (Beckman Coulter). Total RNAs were extracted using the RNeasy Mini Kit (QIAGEN). Complementary RNA (cRNA) generation, quality control, hybridization and data analysis were performed by Cambridge Genomic Services at the University of Cambridge. Raw intensity values from Illumina MouseWG-6 v2.0 expression beadchip microarrays were pre-processed with the Bioconductor lumi and preprocessCore packages (www.bioconductor.org): Probes that were not detected in at least one sample were removed, Variance Stabilization Transformation (VST) was applied, and samples were quantile-normalized. Differential expression was evaluated with the Bioconductor limma package.
Comparison with published microarray data (Extended Data Fig. 3j): Our data set has been assayed on an Illumina MouseWG-6 v2.0 expression beadchip, the data set from Hayashi et al. (2011)5 on the Affymetrix Mouse Genome 430 2.0 Array platform. We therefore quantile-normalized the data sets to ensure that the data sets span comparable ranges of expression values. Principal component analysis (PCA) was performed on the center-scaled expression values, where systematic differences between platforms are mainly captured by the first principal component.
Immunofluorescence stainings
D3, D4 and D6 aggregates were fixed with 2% or 4% paraformaldehyde for 20 minutes at RT or for 2h at 4°C. Fixed aggregates, were washed several times in PBS and transferred into 10%sucrose/PBS (2h), 20% sucrose/PBS (2h) and finally into OCT embedding matrix (over night; CellPath). Next day, cell aggregates were embedded in OCT in tissue molds and stored at −80C. A Leica Cryostat CM3050S was used to cut the OCT blocks in 6-8μm thick sections, which were collected on SuperFrost Plus slides (VWR).
For immunofluorescence staining, the slides were washed with PBS, permeabilised with PBS/0.1-1% Triton X-100 and then incubated with primary antibodies in permeabilisation buffer including 5% donkey serum (Sigma-Aldrich) over night at 4°C. Next day, slides were washed with PBS and incubated with secondary antibodies in permeabilisation buffer for 2h at RT, washed with PBS, incubated with DAPI in PBS for 15-30 minutes, and mounted using Vectashield Mounting Medium (VECTOR Labs). Images were acquired using a Leica SP5 or SP8 confocal microscope. For 5-hmC stainings, it was required to perform an additional antigen retrieval step before incubation with primary antibodies: slides with sections were transferred into TE buffer, pH8, at ~95°C and microwaved at very low power for 45 minutes.
The following primary antibodies were used: mouse anti-OCT4 (1:100, BD Biosciences, O50808), rat anti-BLIMP1 (1:50, eBioscience, clone 6D3, 14-5963), rabbit anti-AP2γ (1:250, SantaCruz, sc-8977), rabbit anti-PRDM14 (1:250, a kind gift of Prof. Danny Reinberg), rabbit anti-DAZL (1:500, Abcam, ab34139), mouse anti-H3K9me2 (1:250, Abcam, ab1220 and 1:500, Millipore, 07-441), rabbit anti-H3K27me3 (1:500, Millipore, 07-449), rabbit anti-TET1 (1:500, Millipore, 09-872), rabbit anti-5hmC (1:500, Active Motif, 39791), goat anti-KLF4 (1:100, R&D, AF3158), rabbit anti-H3S10ph (1:500, Millipore, 06-5770), mouse anti-ƔH2A.X (1:250, Millipore, 05-636), rat anti-GFP (1:500, Nakalai Tesque, GF090R) and Alexa Fluor488 and 568 secondary antibodies (1:500, Life Technologies) were used.
Quantification of immunofluorescence data
All quantifications were preformed using Fiji34. The DAPI, H3S10ph and ƔH2A.X channels were processed by applying a Gaussian Blur (H3S10ph staining: DAPI/H3S10ph - σ 0.5/1.1; DAPI/ƔH2A.X: σ 1.0/1.5) to reduce noise. The images were then binarized using the Otsu thresholding algorithm and holes were filled before the total signal area was measured. In D6 Prdm1−/− +Dox aggregates, many cells underwent cell death. Therefore, nuclei with bright discrete spots of DAPI signal, which indicates chromatin condensation, were excluded from the analysis. The diameter of ~10 cells was measured and used to calculate the average area of one cell in order to estimate the number of cells in the field of view (DAPI+ve area/area of one cell).
For all other quantifications on a single cell level, we developed ‘Object Scan’, which is an object mapping and analysis plugin for Fiji that combines advanced functions with a user-friendly interface. Images are processed with a choice of feature enhancement algorithms, objects are identified by patch sampling to detect intensity edges based on the local energy gradient, and the generated two-dimensional masks are clustered in three dimensions to define the final object map for analysis. We used Object Scan to carry out DoG processing and contained signal analysis using the DAPI channel for object mapping, watershed segmentation, a scan radius of one and the following channel specific settings: edge gradient = 10, estimated object radius = 9 μm. The results were scale normalised (X-Xmin/Xmax-Xmin) to the range 0 to 1 for comparison. Student’s t-test was used to test for significance. The Object Scan plugin is available from this link: http://www.gurdon.cam.ac.uk/stafflinks/downloadspublic/imaging-plugins.
Chromatin immunoprecipitation (ChIP)
Low cell number ChIP-qPCR was performed as previously described35. 3×105 cells per ChIP were fixed in 1% formaldehyde (room temperature, 10 min), quenched with 1 vol. of 250 mM glycine (room temperature, 5 min), and rinsed with chilled TBSE buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA) twice before storage at −80°C. After thawing the cells on ice, fixed cells were lysed with 100 μl 1% SDS Lysis Buffer (50mM Tris-HCl pH8, 10mM EDTA, 1% SDS, Roche protease inhibitor cocktail; on ice, 5 min) and then centrifuged (2,000 rpm, 10 min). Pellet was resuspended in 100 μl of Dilution buffer (16.7mM Tris-HCl, pH8, 167mM NaCl, 1.2mM EDTA, 1.1% Triton X-100, 0.01% SDS, Roche protease inhibitor cocktail). Samples were sonicated 9 times (30 s pulses with 30 s break interval) using the Bioruptor water bath sonicator (Diagenode). Chromatin extracts were then precleared with Dynal Magnetic Beads (Invitrogen) (4°C, 1 hr) followed by centrifugation (2,000 rpm, 30 min). Supernatant (precleared chromatin) was immunoprecipitated overnight with Dynal Magnetic Beads coupled with anti-Nanog antibody (1 μg per ChIP, Cosmo Bio Co., RCAB0001P) or normal rabbit serum (1μg per ChIP). On the next day, beads were washed (nutate in wash buffer for 5 min at 4°C) in low salt buffer (0.1% SDS, 1% TritonX-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl), high salt buffer (0.1% SDS, 1% TritonX-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 300 mM NaCl) and LiCl buffer (0.25M LiCl, 1% NP400, 1% Na deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0), for a total of three washes. Following an additional wash in TE, elution was performed in a PCR machine (68°C, 10 min). After digesting and reverse crosslinking (with Proteinase K at 42°C for 2 hr and 68°C for 6 hr) DNA was purified (phenol-chloroform extraction) and used for qPCR analysis. For the negative control region, we used the Snai3 locus as described previously36. Student’s t-test was used to test for significance.
The same protocol was used for the SOX2 ChIP with some deviations. D2 EpiLCs were aggregated in low binding plates for 6h in the presence of 200 ng/ml of Doxycycline before collection. 5×106 ESCs and EpiLCs, respectively, were fixed and processed as described above. Samples were sonicated 20 times (30 s pulses with 30 s break interval) using a Bioruptor water bath sonicator (Diagenode). Samples were divided for immunoprecipitations with SOX2 antibody (10μg per ChIP, Santa Cruz, sc-17320 X) or normal rabbit IgG (10μg per ChIP, Santa Cruz, sc-2027 X) as a negative control. Beads were washed with low salt buffer, and 2× with high salt buffer for 10 minutes each. The beads were rinsed in TE, resuspended in Proteinase K digestion buffer (20mM HEPES, 1mM EDTA, 0.5% SDS) with 2μl of 10mg/ml Proteinase K and incubated for 15 min at 50C. In parallel, 2μl of 10mg/ml Proteinase K was added to the saved input samples. 3μl 5M NaCl was added to the supernatants and the input samples. To reverse the crosslinks, samples were incubated at 42C for 2h and 68C over night. Next day, the DNA was purified using Agencourt Ampure XP beads (Beckman Coulter) according to manufacturer’s instructions. The purified DNA was used for qPCR analysis. For the negative control region, we used the Snai3 locus. Student’s t-test was used to test for significance. The primer sequences used for RT-qPCRs are listed in Supplementary Table 1.
NANOG ChIP-seq
The NANOG ChIP for subsequent sequencing was performed as described above with some deviations. D1 or D2 EpiLCs were aggregated in low binding plates for 3h in the presence of 200 ng/ml of doxycycline. ESCs and EpiLCs were fixed and processed as described above. 3×106 fixed cells were lysed with 1 ml 1% SDS Lysis Buffer and then centrifuged (2,000 rpm, 15 min). Nuclear fraction was resuspended in 0.9 ml of dilution buffer. Samples were sonicated 10 times (30 s pulses with 30 s break interval) using a Bioruptor water bath sonicator (Diagenode). Immunoprecipitations were performed with anti-NANOG antibody (2μg per ChIP, Cosmo Bio Co., RCAB0001P). After elution samples were digested with Proteinase K and reverse crosslinked for 6 h at 68°C. 12 ng of purified DNA was used for library preparation using Ovation Ultralow DR Multiplex System (Nugen). Once prepared, library was size selected and sequenced using HiSeq2000 with single-end 50 nt read length.
ChIP-Seq analysis
ChIP-seq reads were aligned with the bwa aligner (bio-bwa.sourceforge.net) to the mouse reference genome (GRCm38/mm10). Peaks were called with MACS (version 2.1.0 https://github.com/taoliu/MACS) and visualised using the Integrative Genomics Viewer (https://www.broadinstitute.org/igv/). Peak regions from two biological replicates were intersected using bedops (bedops.readthedocs.org). Overlapping peak regions with peak summits within < 50 nt distance in both replicates were retained. Peak regions from the three cell types were merged. Differences in ChIP-seq read intensities on peak regions were evaluated by using diffReps (code.google.com/p/diffreps) and MACS (macs2 bdgdiff). High-confidence sets of differentially bound regions that were detected by both methods were selected for further analysis by applying the following thresholds for diffReps: pValue < 0.001 and abs(log2FC) > 1. Previously published H3K27ac ChIP-seq data sets9,30 were aligned to the mouse reference genome in a similar manner as above, and H3K27ac enrichment (log(ChIP/input) values were determined on NANOG peak regions.
De novo motif analysis
High-confidence MACS peaks, for which the distance of the peak summits in both replicates was <50 nt, were selected. De novo motifs were determined with HOMER (http://homer.salk.edu/homer) in the 2,000 top-enriched peaks in ESCs, D1 and D2 EpiLC for both repeat-masked and repeat-unmasked regions within +/− 50 nt of the peak summit.
Luciferase assay
Genomic regions containing putative enhancers of Prdm1 and Prdm14, as well as a negative control region depleted of enhancer signatures, were amplified from mouse E14 ESC genomic DNA. These regions were cloned into a PiggyBAC-based firefly luciferase reporter plasmid upstream of a minimal TK promoter. Stable luciferase reporter GOF-GFP ESC lines, which can overexpress Nanog, Nanog/Sox2 or Brachyury upon Dox addition, were established. Cell pellets were collected from ESCs cultured in N2B27 2i LIF, D2 EpiLCs and EpiLCs after PGCLC induction +/− Dox at 12/24 hours. Luciferase assays were performed with the ONE-Glo™ Luciferase Assay System (Promega). Protein concentration in each lysate was quantified by Pierce 660 nm Protein Assay (Thermo Scientific). Relative luciferase activities were obtained by dividing luciferase activity by protein concentration in each sample.
Blastocyst injections
ESC clones carrying both the Nanog transgene and a CAG monomeric Kusabira-orange (mKO) fluorescence reporter were selected by neomycin (Sigma-Aldrich) and zeocine (Life Technologies). D4 PGCLCs were induced from D2 EpiLCs with Nanog and used for derivation of EGCLC. For ESC or D4 PGCLC injections, GOF-GFP ESCs were co-transfected with a vector, which enabled inducible expression of Nanog and constitutive expression of Venus, a variant of EGFP. For D4 PGCLC, after induction of PGCLCs with Nanog, cells were stained with PE conjugated-CD61 antibody (1:10, Biolegend, 104308) and Alexa660 conjugated-SSEA-1 antibody (2.5 μl/ 105 cells, eBioscience, clone eBioMC-480, 50-8813) according to the manufacturer’s instructions. Double positive PGCLC cells were collected by using a S3 cell sorter (Biorad). Embryos for chimera experiments were obtained from CBA/C57BL/6 F1 crossed with C57BL/6 mice. Blind tests or randomization methods were not used. The sex of embryos was not determined. Manipulations of embryos were performed as described previously37. Briefly, five cells were injected into a morula, which were subsequently cultured in KSOM (Millipore). At the following day, the embryos were transferred into the uteri of pseudopregnant mice. All embryos were analysed one week after embryo transfer, which corresponded to embryonic day 9.5.
Generation of Nanog knockout ESCs
The CRISPR/Cas9 system was used to generate Nanog knock-out ESCs. gRNAs targeting exon1 of the Nanog gene were cloned into pX33038 (Addgene). 1μg of this plasmid was transfected with a pPyCAG-monomeric Kusabira Orange-IRES-Pac plasmid. Transfected cells were selected by puromycin (1 μg/ml) for 2 days. Clonal Nanog KO ES lines were established and mutations of Nanog alleles were confirmed by QPCR, Western blotting and DNA sequencing. Subsequently, pPBhCMV*1-Nanog-pA plasmid was transfected into those lines with pPyCAG-PBase and pPBhCMV*1-rtTA-IRESNeor to generate Nanog KO ESC lines carrying a Dox-inducible Nanog transgene. Loss of Nanog affected the growth of ESCs. Thus, these cell lines were maintained in N2B27 2i LIF with a low dose of Dox (100 ng/ml). gRNA sequences are listed in Supplementary Table 1.
Western blots
5 × 104 cells were lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 1% SDS, 10 mM EDTA). Protein concentration was measured by Bicinchoninic Acid Kit (Sigma-Aldrich). The protein amount was adjusted among samples, then 4× Laemmli buffer was added. Samples were boiled at 95°C for 5 min. Proteins were separated on 10% acrylamide gels, blotted on Immobilon-P transfer membrane (Millipore). The membrane was blocked with 5% skimmed milk and incubated with primary antibodies: anti-NANOG (1:500, mouse IgG, eBioscience, clone eBioMLC-51, 14-5761), anti-SOX2 (1:500, rabbit igG, Cell Signaling, 2748), anti-α-TUBULIN (1:1000, mouse IgG, Sigma-Aldrich, clone DM1A, T9026). Primary antibodies were detected on X-ray film with anti-rabbit or -mouse IgG conjugated with Horseradish peroxidase (Dako) followed by detection using Western Detection System (GE Healthcare). For gel source data, see Supplementary Figure 1.
Generation of Sox2 conditional knockout ESCs with Dox-inducible Nanog transgene
pPBhCMV*1-Nanog-pA, pPBCAG-rtTA-IRESNeor and pPyCAG-PBase were transfected into the Sox2 conditional knockout ESC line (2CG2)27. After one week of neomycin selection (80 μg/ml), pooled cells were used for the subsequent experiments. Dexamethasone-inducible Sox2 KO and Dox-inducible Nanog expression were confirmed by qPCR and Western blotting.
Extended Data
Extended Data Figure 1: GOF-GFP as a reporter for PGCLCs; Dox-inducible transgene system.
a, Experimental design for the induction of PGCLCs in vitro5.
b, Representative brightfield/GFP images of male/female GOF-GFP ESCs, D2 EpiLCs, D2 and D4 cytokine-induced PGCLCs. Scale bar, 200 μm.
c, FACS analysis for GFP with samples shown in (b).
d, Simplified scheme of PiggyBac (PB 5’TR and PB 3’TR) based plasmids for transgene overexpression using the Tet-On system. The rtTA protein activates the minimal promoter (hCMV*-1) driving the expression of the cDNA of interest only in the presence of doxycycline (Dox).
e, Proof of principle experiment to test the Dox-inducible expression of a transgene during the sequential differentiation of PGCLCs from ESCs. GOF-GFP ESCs carrying PiggyBac based Dox-inducible mCherry expression plasmids were differentiated into D2 EpiLCs and then induced into PGCLCs with cytokines in +/−Dox conditions. Representative brightfield, GFP and mCherry images are shown 12h after aggregation. Scale bar, 200μm.
f, Representative FACS analysis for GFP and mCherry of D2 cytokine-induced PGCLCs from male/female GOF-GFP ESCs carrying a Dox-inducible mCherry transgene. Most cells express mCherry after Dox addition.
Extended Data Figure 2: Nanog but not Oct4 induces GFP+ve cells from competent EpiLCs.
a, qPCR analysis of transgenic Nanog expression 24h after Dox addition in male/female GOF-GFP ESCs. ΔΔCt mean values +/− s.d. (n=2 technical replicates each from 2 biological replicates). Two-sided/unpaired t-test: **p<0.01. Related to Fig. 1a.
b, Representative brightfield/GFP images of male D2 PGCLCs induced from GOF-GFP D2 EpiLCs; +Dox for Nanog expression. Scale bar, 200 μm. Related to Fig. 1a.
c, Representative FACS analysis of male D4 PGCLCs (shown in Fig. 1a) induced from GOF-GFP D2 EpiLCs; +Dox for Nanog expression.
d, Representative brightfield/GFP images of female D2 and D4 PGCLCs induced from GOF-GFP D2 EpiLCs; +Dox for Nanog expression. Scale bar, 200 μm. Related to Fig. 1a.
e, FACS analysis for GFP with samples shown in (d). Related to Fig. 1a.
f, qPCR analysis of transgenic Oct4 expression 24h after Dox addition in male ESCs. ΔΔCt mean values +/− s.d. (n=2 technical replicates each from 2 biological replicates). Two-sided/unpaired t-test: **p<0.01.
g, Expression of Oct4 (unlike Nanog) does not result in the induction of GFP+ve cells. PGCLC induction from female GOF-GFP EpiLCs; +Dox for Oct4 or Nanog expression. Representative brightfield/GFP images at D4. Scale bar, 200 μm.
h, FACS analysis for GFP with samples shown in (g).
Extended Data Figure 3: The transcriptomes of D4 Nanog- and cytokine-induced PGCLCs are highly similar.
a, 100-200 ng/ml of Dox in EpiLCs results in NANOG expression levels similar to ESCs as shown by Western blot analysis for NANOG and α-TUBULIN (α-TUB) with GOF-GFP ESCs and D2 EpiLCs 24h after PGCLC induction (EpiLC aggregations) with Nanog (+Dox). For gel source data, see Supplementary Fig. 1.
b, PGCLC induction with 100 or 700 ng/ml Dox (for Nanog expression) with Noggin from GOF-GFP EpiLCs. Representative brightfield/GFP images at D4. GFP+ve cells are induced in both conditions. Scale bar, 200 μm.
c, Physiological (equivalent to ESCs) or higher levels of Nanog induce PGCLCs with comparable efficiency. FACS for GFP at D4 of PGCLC induction with 100 or 700 ng/ml Dox (for Nanog expression) with Noggin from GOF-GFP or Blimp1-GFP EpiLCs.
d, Alternative representation of qPCR data for Nanog, Prdm1 and Tfap2c shown in Fig. 1c. The induction of these genes in +cytokine conditions appears less evident, when compared to +Dox conditions. The data was log2-scaled, which allows a better comparison.
e, qPCR analysis of female GOF-GFP cells. GFP+ve cells were FACS-sorted. Note, the upregulation of PGC markers but not of the ESC marker Klf4. ΔΔCt mean values +/− s.d. (n=3 biological replicates). Colour code is shown in (d). Related to Fig. 1c.
f, qPCR analysis of male Blimp1-GFP cells. GFP+ve cells were FACS-sorted. Note, the upregulation of PGC markers but not of the ESC marker Klf4. ΔΔCt mean values +/− s.d. (n=3 biological replicates). Colour code is shown in (d). Related to Fig. 1c.
g, The transcriptomes of Nanog- and cytokine-induced PGCLCs are highly similar. Scatter plot showing the correlation of microarray data of ESCs, FACS-sorted D4 PGCLCs induced by cytokines or Nanog with Noggin. R indicates the Pearson correlation coefficient. n=2 biological replicates; related to Fig. 1d.
h, Nanog- and cytokine-induced PGCLCs cluster together as shown in unsupervised hierarchical clustering of microarray data described in (g). Related to Fig. 1d.
i, Heatmap showing the expression levels of selected genes from microarray data described in (g). Related to Fig. 1d.
j, Nanog-induced D4 PGCLCs are closely related to cytokine-induced D6 PGCLCs. PCA analysis with published microarray datasets5 (cross-platform comparison; see Methods for details). Note, that the separation of ESC samples is probably due to differences in genomic background and culture conditions.
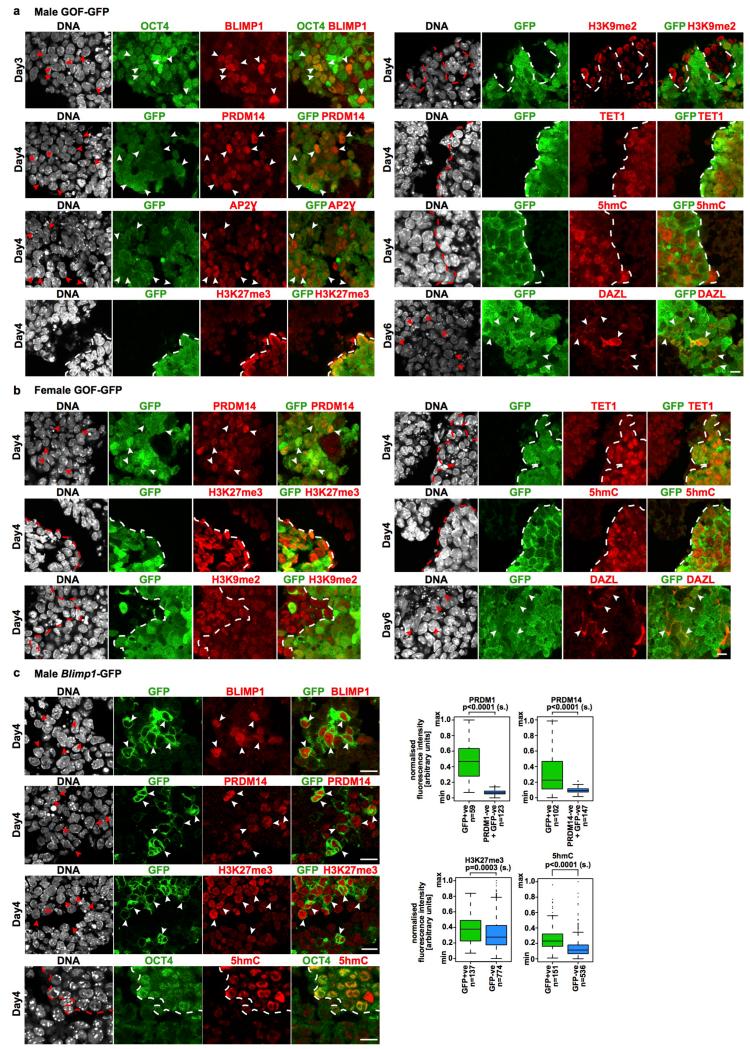
Extended Data Figure 4: Nanog-induced PGCLCs show hallmarks of PGC development.
a-c, IF analysis of PGC markers in GFP+ve cells induced by Nanog from male (a) and female (b) GOF-GFP and male Blimp1-GFP (c) EpiLCs shows expression of BLIMP1, PRDM14, and AP2Ɣ, TET1, enrichment of H3K27me3 and 5hmC and a decrease of H3K9me2 intensity; DAZL is detected in some cells on D6. Arrowheads and dashed lines highlight single or cluster of GFP+ve cells. n=2 biological replicates; scale bar, 10 μm. Quantification in (c) was scale normalised. Two-sided/unpaired t-test: n.s.=not significant; s=significant; n=number of cells analysed. Related to Fig. 1e.
Extended Data Figure 5: Functional analysis of Nanog-induced PGCLCs.
a, Experimental design (for b, c) for the derivation of EGC-like cells (EGCLCs). PGCLCs were induced with cytokines or by Nanog (+Dox) from male or female GOF-GFP EpiLCs carrying a constitutively active Kusabira-Orange reporter. On D4, aggregations were dissociated and cultured on MEF in PGC selection medium (LIF, SCF, bFGF, retinoic acid) for 5 days. After the selection, selected colonies were dissociated and transferred into ESC medium (2i LIF).
b, Experiment was performed as shown in (a). Left panel shows representative images of proliferating GFP+ve cells after 3 days of PGC selection. Right panel shows established EGCLCs after 3 passages in 2i LIF.
c, EGCLCs derived from D4 PGCLCs by Nanog expression were injected into blastocysts resulting in high contribution to chimeras at E9.5 as shown by Kusabira-Orange expression.
d, Experimental design (for e-g) for generating chimeras. PGCLCs were induced from a GOF-GFP ESC line expressing a fluorescent VENUS reporter constitutively and Nanog upon Dox addition (TVN2 cell line). On D4, aggregations were dissociated and SSEA1+ve and CD61+ve cells were sorted by FACS, injected into morulae and analysed on E9.5.
e, Representative brightfield, GFP/VENUS images of GOF-GFP or TVN2 cells during PGCLC induction by cytokines or Nanog (+Dox). Scale bars, 100μm.
f, FACS profile for SSEA1+ve and CD61+ve PGCLCs on D4 induced as described and shown in (d, e).
e, ESCs but not PGCLCs contribute efficiently to chimeras. ESCs or FACS-sorted Nanog-induced SSEA1+ve/CD61+ve PGCLCs were injected into morulae and representative brightfield/VENUS images from chimeras at E9.5 are shown.
Extended Data Figure 6: Prdm1−/− abrogates PGCLC induction by Nanog; Nanog and the Wnt pathway act independently.
a, PGCLC induction with cytokines or Nanog (+Dox) from Prdm1−/− ESCs (Prdm1−/−; Nanog). Representative brightfield images of D4 and D6 aggregations. Scale bar, 200 μm. Related to Fig. 2a.
b, Loss of Prdm1 abrogates PGCLCs induced by NANOG as shown by qPCR analysis of mutant (Prdm1−/−; Nanog) compared with control (Prdm1+/+; Nanog) cells with Nanog (+Dox) or cytokines (+cyto). Unsorted samples were used for analysis. Note that the data shown in Fig. 2a was combined with additional qPCR data on cells at D6 of PGCLC induction. ΔΔCt mean values +/− s.d. (n=2 technical replicates each from 2 biological replicates); two-sided/unpaired t-test: **p<0.01; *p<0.05. Related to Fig. 2a.
c, Nanog does not affect cell proliferation rate of Prdm1−/−; Nanog cells. IF staining for the mitotic marker H3S10ph in Prdm1−/−; Nanog cells at D6 of PGCLC induction; +Dox for Nanog expression. Scale bar, 10μm; two-sided/unpaired t-test; n.s.=not significant; n=estimated number of cells (see Methods for details). Related to Fig. 2a.
d, Induced expression of Nanog results in an increased number of cell death of Prdm1−/−; Nanog cells. IF stainings of the DNA double strand break marker ƔH2AX in Prdm1−/−; Nanog cells at D6 of PGCLC induction; +Dox for Nanog expression. Scale bar, 10μm; two-sided/unpaired t-test; s=significant; n=estimated number of cells (see Methods for details). Related to Fig. 2a.
e, Experimental design (for f-i) to test the interdependence of Nanog and the Wnt pathway for PGCLC induction. Blimp1-GFP ESCs were sequentially differentiated into PGCLCs +/− tankyrase inhibitor XAV939, which causes the degradation of β-catenin25; +Dox for Nanog expression.
f, XAV939 does not affect the morphology and proliferation of D2 EpiLCs. Representative brightfield/GFP images of D2 EpiLCs induced from GOF-GFP ESCs with 1μM XAV939. Scale bar, 200μm.
g, qPCR of D2 EpiLCs treated with XAV939 as shown in (a, b). The expression of Nanog and of the EpiLC markers Dnmt3a and Dnmt3b are not affected by XAV939. Brachyury, the downstream target of WNT, is most efficiently repressed with 1μm XAV939. ΔΔCt mean values +/− s.d. (n=2 technical replicates each from 2 biological replicates); two-sided/unpaired t-test: **p<0.01; *p<0.05.
h, The efficiency of PGCLC induction by cytokines but not by Nanog (+Dox) is markedly reduced upon XAV939 addition. PGCLCs were induced from 1μM XAV939-treated D2 EpiLCs. Representative FACS analysis for GFP with cells at D4 of PGCLC induction.
i, XAV939 does not affect the induction of PGC marker expression in Nanog-induced PGCLCs. Gene expression analysis by qPCR with FACS-sorted Nanog-induced D4 PGCLCs +/− 1μM XAV939. Mean ΔΔCt values +/− s.d. (n=2 technical replicates each from 2 biological replicates). Two-sided/unpaired t-test: **p<0.01; *p<0.05; n.s.=not significant.
Extended Data Figure 7: Sox2 inhibits PGCLC induction by Nanog.
a, Prdm1 and Tfap2c are upregulated and Prdm14 is downregulated in Sox2-KO ESCs from published microarray data26. **p<0.01; *p<0.05.
b, Experimental design for the Western blot shown in Fig. 3d. Conditional Sox2-KO ESCs carrying transgenes for Dox-inducible Nanog expression were treated with Dex to induce a Sox2-KO and/or Dox for Nanog expression for 2 days.
c, Experimental design for the qPCR analysis shown in (d). Sox2-KO ESCs: Conditional Sox2-KO ESCs carrying transgenes for Dox-inducible Nanog expression were treated +/− Dex for 2 days; Sox2-KO D1 EpiLCs: ESCs were cultured in 2i LIF medium with Dex for one day and in bFGF/ActivinA medium with Dex for one more day; Sox2-KO D2 EpiLCs: ESCs were transferred into bFGF/ActivinA medium containing Dex for two days.
d, Loss of Sox2 results in upregulation of Prdm1 and Tfap2c and downregulation of Prdm14 in ESCs, D1 and D2 EpiLCs; qPCR analysis following Sox2-KO (+Dex). ΔΔCt mean values +/− s.d. (n=2 technical replicates each from 2 biological replicates); two-sided/unpaired t-test: **p<0.01; *p<0.05. Experimental design is shown in (c). Related to Fig. 3e.
e, Experimental design for the qPCR analysis shown in Fig. 3e. Sox2-KO ESCs, D1 or D2 EpiLCs were generated as described in (c), and subsequently induced into PGCLCs +/− Nanog (+/− Dox).
f, Western blot for NANOG, SOX2 and α-TUBULIN (α-TUB) in GOF-GFP ESCs carrying Dox-inducible transgenes for Nanog, Sox2 or Nanog/Sox2 (+Dox for 24h). Related to Fig. 3f. For gel source data, see Supplementary Fig. 1.
g, Time-course qPCR analysis showing Nanog and Sox2 expression kinetics during PGCLC induction. PGCLCs were induced from GOF-GFP EpiLCs; +100 or 700 ng/ml Dox for Nanog/Sox2 expression. Related to Fig. 3f.
h, Time-course Western blot for NANOG, SOX2 and α-TUBULIN (α-TUB) showing Nanog and Sox2 protein kinetics during PGCLC induction. PGCLCs were induced from GOF-GFP EpiLCs; +100 or 700 ng/ml Dox for Nanog/Sox2 expression. Related to Fig. 3f.
i, FACS analysis for GFP at D4 of PGCLC induction from GOF-GFP or Blimp1-GFP EpiLCs; +Dox for Nanog, Sox2 or Nanog/Sox2 expression. Related to Fig. 3f. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 8: Sox2 positively affects cell proliferation rate of cytokine-induced PGCLCs.
a, Sox2 increases the number of GFP+ve cells induced by BMP4 alone. Representative FACS analysis for GFP at D4 of PGCLC induction from Blimp1-GFP EpiLCs; +Dox for Sox2 expression.
b, Sox2 does not affect the upregulation of PGC markers in cytokine-induced PGCLCs. qPCR analysis of FACS-sorted GFP+ve cells induced by BMP4 and/or Sox2 (+Dox). ΔΔCt mean values +/− s.d. (n=2 technical replicates each from 2 biological replicates). Reference sample for p-value calculations: ESCs; two-sided/unpaired t-test: **p<0.01; *p<0.05.
c, Time-course FACS analysis of GFP+ve cells after PGCLC induction with BMP4 and +/− Nanog or Sox2 (+/− Dox). The number of GFP+ve cells at D2 of PGCLC induction with or without Sox2 expression is comparable, but increased with Nanog. After D2, PGCLCs induced by BMP4 with Sox2 or Nanog increase their proliferation rate.
Extended Data Figure 9: Nanog shows a cell-type specific binding pattern and induces Prdm1/Prdm14/Tfap2c.
a, Time-course qPCR for Prdm1, Prdm14 and Tfap2c between 1-48h after PGCLC induction with cytokines from GOF-GFP EpiLCs. ΔΔCt mean values +/− s.d. (n=3 technical replicates). Related to Fig. 4a.
b, Prdm1 alone can induce the expression of Tfap2c. GOF-GFP EpiLCs with combinations of Dox-inducible transgenes encoding Prdm1, Prdm14 and/or Nanog +/− Dox for 6h were analysed by qPCR. The expression of Prdm1, Prdm14 and/or Nanog is upregulated in the corresponding EpiLCs upon Dox addition. ΔΔCt mean values +/− s.d. (n=2 technical replicates each from 2 biological replicates); two-sided/unpaired t-test: **p<0.01; *p<0.05.
c, To acquire sufficient numbers of cells for ChIP-seq studies, GOF-GFP D1 or D2 EpiLCs (~1×106 cells per 6 cm plate) with Dox-inducible Nanog transgenes were aggregated in low binding plates +Dox to induce PGCLCs. qPCR analysis of D1 and D2 EpiLCs after 3h with 100 or 200 ng/ml Dox is shown. The addition of 200 ng/ml of Dox results in Nanog expression levels comparable to ESCs after 3h. ΔΔCt mean values +/− s.d. (n=2 technical replicates each from 2 biological replicates); two-sided/unpaired t-test: **p<0.01; n.s.=not significant.
d, NANOG ChIP-seq analysis shows genomic distribution of NANOG in GOF-GFP ESCs and D1 EpiLCs +Nanog (+Dox) for 3h. ‘Distal’ refers to intergenic peaks, which are +/− 50kb of an annotated coding gene, while those further away are categorised as ‘intergenic’. Related to Fig. 4b.
e, De novo motif analysis with NANOG ChIP-seq data. Shown are the 5 top matches of the de novo motifs to known motifs. The analysed cell types show enrichment for the NANOG and SOX motifs. ESCs show additional enrichment for pluripotency motifs, while EpiLCs show a different set of motif enrichment.
f, D2 EpiLC-specific (D2 EpiLCs +Dox 3h) NANOG-bound enhancers become more enriched for H3K27ac than ESC-specific NANOG-bound enhancers in cytokine-induced D2 PGCLCs as compared to ESCs. Contour plots showing differential binding of NANOG in D2 EpiLCs vs ESCs (x-axis) compared to the differential enrichment of H3K27ac in D2 PGCLCs30 vs ESCs9 (y-axis).
g, NANOG binds enhancers that are enriched for H3K27ac in D1/D2 EpiLCs (D1/2 EpiLCs +Dox 3h). A subset of enhancers, however, becomes more enriched for H3K27ac in cytokine-induced D2 PGCLCs30 as compared to D2 EpiLCs30.
h, NANOG might contribute to the activation of enhancers associated with germline genes. Scatter plots show differential gene expression analysis between D6 PGCLCs5 and D2 EpiLCs5 (y-axis), and differential H3K27ac enrichment between D2 PGCLCs30 and D2 EpiLCs30 (x-axis) on NANOG binding sites. Top 40% of NANOG peaks were associated with the nearest gene in a 200kb window. Highlighted are candidate enhancers associated with germline genes. A selected set of genes associated with the germline and candidate enhancers, which become activated (H3K27ac-enriched) in PGCLCs, is indicated.
Extended Data Figure 10: Nanog induces PGC-like fate - a model.
a, ChIP-seq data tracks9,30,39 at the Prdm1 and Prdm14 loci for NANOG in ESCs, D1 and D2 EpiLCs (EpiLCs were collected after 3h with 200 ng/ml Dox for Nanog expression). Boxed are putative enhancer elements. The Prdm1 enhancer is enriched for H3K4me1 in D2 EpiLCs and gains H3K27ac in PGCLCs. The Prdm14 enhancer shows enrichment for H3K4me1 in ESCs and EpiLCs and becomes enriched for H3K27ac in ESCs and PGCLCs but not in EpiLCs. Note that these enhancer marks follow the expression pattern of Prdm1 or Prdm14, respectively. RPM = Reads per Million. Related to Fig. 4d.
b, ChIP-qPCR validation of NANOG ChIP-seq data with GOF-GFP D2 EpiLCs before and 3h after PGCLC induction by Nanog expression (+Dox). NANOG is enriched at putative enhancer regions, which are close to Prdm1 and Prdm14. Error bars indicate s.d. (n=2 technical replicates each from 2 biological replicates).
c, ESC lines with luciferase reporter plasmids with a genomic region, which does not show any enhancer marks and NANOG binding, and indicated Dox-inducible transgenes served as a negative control. Luciferase activity, measured in ESCs, D2 EpiLCs and 24h after PGCLC induction (EpiLC aggregations), was normalised to protein quantity (luc/pro). Mean values +/− s.d. (n=3 technical replicates each from 2 biological replicates).
d, Biological replicate experiment for the luciferase assay with the Prdm1 enhancer as shown and described in Fig. 4e. Luciferase activity, measured in ESCs, D2 EpiLCs and 24h after PGCLC induction, was normalised to protein quantity (luc/pro). Mean values +/− s.d. (n=3 technical replicates); colour code is shown in (c); reference for p-values (two-sided/unpaired t-test): EpiLC aggregations −Dox; **p<0.01; *p<0.05.
e, Biological replicate experiment for the luciferase assay with the Prdm14 enhancer as shown and described in Fig. 4g. Luciferase activity, measured in ESCs, D2 EpiLCs and 12/24h after PGCLC induction, was normalised to protein quantity (luc/pro). Mean values +/− s.d. (n=3 technical replicates); colour code is shown in (c); reference for p-values (two-sided/unpaired t-test): EpiLC aggregations −Dox 24h; **p<0.01; *p<0.05.
f, Model showing the role of NANOG during PGCLC induction in vitro. D1 EpiLCs are not competent to become PGCLCs, but retain the capability to revert to an ES-like state via 2i LIF and/or Nanog overexpression. D2 EpiLCs differentiate into PGCLCs upon Nanog expression. NANOG binds to putative enhancer elements of Prdm1 and Prdm14 to activate their transcription, which is sufficient to induce the PGCLC fate. This effect can be antagonized by SOX2, which co-binds the Prdm1 enhancer.
Supplementary Material
Acknowledgements
We thank Harry Leitch for ESC lines, Caroline Lee for help with animal husbandry, Hitoshi Niwa for vectors and conditional Sox2-KO ESCs, Nigel Miller, Rachel Walker and Andy Riddell for FACS sorting and Julien Bauer for analysis of microarray data. K.M. was supported by the JSPS Institutional Program for Young Researchers Overseas Visits. U.G. was supported by a Marie Sklodowska-Curie and a Newton Trust/Leverhulme Trust Early Career fellowship. J.J.Z. was a recipient of a Wellcome Trust PhD Studentship (RG44593). T.K. was supported by a JSPS Fellowship for research abroad. This research was supported by Gurdon Institute core grants from the Wellcome Trust (092096) and the Cancer Research UK (C6946/A14492) and a grant from the Wellcome Trust to M.A.S. (WT096738).
Author contribution
K.M. and U.G. designed and performed experiments, and wrote the paper; W.T. designed and carried out the luciferase assays; NANOG ChIP experiments were carried out by J.J.Z., while R.S. performed immunofluorescence analysis; T.K. and S.K. designed and carried out the chimera experiments; S.D. performed bioinformatic analysis; R.B. developed the ‘Object Scan’ plugin; M.A.S. supervised the project, designed experiments and wrote the paper. All authors discussed the results and contributed to the manuscript.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Supplementary Table 1: Oligonucleotide sequences
Supplementary Figure 1: Uncropped scans of Western blot gels
Author information
The accession number for the microarray data presented in this study is available from the Gene Expression Omnibus (GEO) database under accession GSE71933.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T. Nanog expression in mouse germ cell development. Gene Expr. Patterns. 2005;5:639–646. doi: 10.1016/j.modgep.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi S, et al. Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development. 2009;136:4011–4020. doi: 10.1242/dev.041160. [DOI] [PubMed] [Google Scholar]
- 4.Carter AC, Davis-Dusenbery BN, Koszka K, Ichida JK, Eggan K. Nanog-independent reprogramming to iPSCs with canonical factors. Stem Cell Reports. 2014;2:119–126. doi: 10.1016/j.stemcr.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 6.Lawson KA, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magnúsdóttir E, et al. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakaki F, et al. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- 9.Buecker C, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zylicz JJ, et al. Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. eLife. 2015 doi: 10.7554/eLife.09571. 10.7554/eLife.09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat. Struct. Mol. Biol. 2011;18:120–127. doi: 10.1038/nsmb.2000. [DOI] [PubMed] [Google Scholar]
- 13.Yamaji M, et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 14.Grabole N, et al. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 2013;14:629–637. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun LT, et al. Nanog co-regulated by Nodal/Smad2 and Oct4 is required for pluripotency in developing mouse epiblast. Dev. Biol. 2014;392:182–192. doi: 10.1016/j.ydbio.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman JA, Wu CI, Merrill BJ. Tcf7l1 prepares epiblast cells in the gastrulating mouse embryo for lineage specification. Development. 2013;140:1665–1675. doi: 10.1242/dev.087387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seki Y, et al. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 18.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackett JA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koshimizu U, Watanabe M, Nakatsuji N. Retinoic acid is a potent growth activator of mouse primordial germ cells in vitro. Dev. Biol. 1995;168:683–685. doi: 10.1006/dbio.1995.1113. [DOI] [PubMed] [Google Scholar]
- 21.West JA, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 23.Bao S, et al. The germ cell determinant Blimp1 is not required for derivation of pluripotent stem cells. Cell Stem Cell. 2012;11:110–117. doi: 10.1016/j.stem.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aramaki S, et al. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell. 2013;27:516–529. doi: 10.1016/j.devcel.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Huang S-MA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 26.Adachi K, et al. Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol. Cell. 2013;52:380–392. doi: 10.1016/j.molcel.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Masui S, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 28.Campolo F, et al. Essential role of Sox2 for the establishment and maintenance of the germ cell line. Stem Cells. 2013;31:1408–1421. doi: 10.1002/stem.1392. [DOI] [PubMed] [Google Scholar]
- 29.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurimoto K, et al. Quantitative Dynamics of Chromatin Remodeling during Germ Cell Specification from Mouse Embryonic Stem Cells. Cell Stem Cell. 2015;16:517–532. doi: 10.1016/j.stem.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Yeom YI, et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 32.Yoshimizu T, et al. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 1999;41:675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- 33.Ying Q-L, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng J-H, et al. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev. Cell. 2013;24:324–333. doi: 10.1016/j.devcel.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Gillich A, et al. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell. 2012;10:425–439. doi: 10.1016/j.stem.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behringer R, Gertsenstein M, Nagy KV, Nagy A. Manipulating the Mouse Embryo. A Laboratory Manual. 3rd edition 2003. pp. 453–506. [Google Scholar]
- 38.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marks H, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.