We report a dual functional system for bacterial nitrate (NO3−) assimilation and nitric oxide (NO) detoxification. The assimilatory NO3− reductase (NasC) can generate nitric oxide (NO). Co-expression of an NO-detoxification system acts to counteract accumulation of cytotoxic NO during anaerobic NO3−-dependent growth.
Keywords: bacterial denitrification, bacterial haemoglobin, nitrate reduction, nitric oxide reductase, nitrite reduction
Abstract
Rhizobia are recognized to establish N2-fixing symbiotic interactions with legume plants. Bradyrhizobium japonicum, the symbiont of soybeans, can denitrify and grow under free-living conditions with nitrate (NO3−) or nitrite (NO2−) as sole nitrogen source. Unlike related bacteria that assimilate NO3−, genes encoding the assimilatory NO3− reductase (nasC) and NO2− reductase (nirA) in B. japonicum are located at distinct chromosomal loci. The nasC gene is located with genes encoding an ABC-type NO3− transporter, a major facilitator family NO3−/NO2− transporter (NarK), flavoprotein (Flp) and single-domain haemoglobin (termed Bjgb). However, nirA clusters with genes for a NO3−/NO2−-responsive regulator (NasS-NasT). In the present study, we demonstrate NasC and NirA are both key for NO3− assimilation and that growth with NO3−, but not NO2− requires flp, implying Flp may function as electron donor to NasC. In addition, bjgb and flp encode a nitric oxide (NO) detoxification system that functions to mitigate cytotoxic NO formed as a by-product of NO3− assimilation. Additional experiments reveal NasT is required for NO3−-responsive expression of the narK-bjgb-flp-nasC transcriptional unit and the nirA gene and that NasS is also involved in the regulatory control of this novel bipartite assimilatory NO3−/NO2− reductase pathway.
INTRODUCTION
Fixation of atmospheric dinitrogen (N2) by plant-associated symbiotic soil bacteria, collectively termed rhizobia, is a significant agricultural process that reduces dependence on synthetic nitrogen (N) containing fertilizers in crop production. This protects water quality and human health as well as the wider environment. In addition to N2 fixation, the soybean endosymbiont Bradyrhizobium japonicum USDA110 is capable of growing anaerobically with the water-soluble nitrate (NO3−) anion, as an alternative terminal electron acceptor to oxygen (O2), which is reduced to N2 gas by respiratory denitrification. During this process, several free N-containing intermediates are produced, including: (i) the oxyanion nitrite (NO2−), (ii) the gaseous cytotoxic free-radical nitric oxide (NO) and (iii) the potent and long-lived greenhouse gas nitrous oxide (N2O). In B. japonicum, the denitrification apparatus is encoded by the napEDABC, nirK, norCBQD and nosRZDFYLX genes, which express the periplasmic NO3− reductase (NapABC), copper-containing NO2− reductase (NirK), cytochrome-c NO reductase (NorCB) and N2O reductase (NosZ) enzymes, respectively [1]. This bacterium is distinguished by the ability to denitrify under both free-living and symbiotic conditions [2–4].
Several reports suggest that rhizobial denitrification is the main driver for production and release of the environmentally damaging agents NO and N2O from alfalfa and soybean nodules [5–8]. NO is a highly reactive and well-studied ozone-depleting agent, whereas N2O is increasingly recognized as a powerful greenhouse gas with an estimated 300-fold higher radiative potential for global warming, molecule for molecule, compared with carbon dioxide [9–11]. Importantly, in active root nodules, NO also acts as a potent inhibitor of nitrogenase, the central enzyme of symbiotic N2-fixation [8,12]. Under free-living denitrifying conditions, the B. japonicum proteins NirK and NorCB are physiologically important for the synthesis and detoxification of NO, respectively [1]. However, several studies suggest the involvement of other sinks for NO that are distinct from the recognized denitrification pathway in nodules [8,13]. For example, in related bacteria, NO may be oxidized to NO3− or reduced to N2O by cytoplasmic detoxification enzymes. These systems include single-domain haemoglobins (sdHbs), truncated haemoglobins (trHbs), flavohaemoglobins (FHbs) and flavorubredoxin (FlRd) [14–18].
Following sequencing of the B. japonicum USDA110 genome [19], several studies have investigated the involvement of a putative bacterial sdHb, termed Bjgb, in NO-detoxification, under free-living conditions [3,20]. This bacterial haemoglobin is encoded by the ORF blr2807 and resides within a cluster of other uncharacterized ORFs (blr2803–09) predicted to encode components of a NO3− assimilation (Nas) pathway, including: an ABC-type NO3− transport system (blr2803–05), a major facilitator superfamily (MFS)-type NO3−/NO2− transporter (blr2806), an FAD-dependent NAD(P)H oxidoreductase (blr2808) and the catalytic subunit of the assimilatory NO3− reductase (blr2809), termed NasC (we note the gene for the assimilatory NO3− reductase in B. japonicum was previously termed nasA, but here we unify the gene nomenclature for α-proteobacteria). The genome also contains a putative ferredoxin-dependent assimilatory NO2− reductase (NirA) that is present at bll4571, a distinct locus on the chromosome. This putative nirA gene lies immediately downstream of genes recently reported to code for a NO3−/NO2− responsive regulatory system (NasS-NasT), similar to that characterized in the model NO3−-utilizing soil bacterium Paracoccus denitrificans PD1222 [21,22]. However, to date, a role for the proteins encoded at blr2803–09 and bll4571–73 loci in NO3−/NO2− assimilation and conceivably NO management in B. japonicum remains to be established.
Although the biochemical components for Nas systems may be highly modular, in related α-proteobacteria such as P. denitrificans and Rhodobacter capsulatus E1F1, genes encoding regulatory and structural elements for the NO3− assimilation pathway are typically found together [23,24]. For example, in P. denitrificans, the genes required for import and reduction of NO3− and/or NO2− are encoded by nasABGHC and the nasTS genes required for NO3−/NO2−-responsive regulatory control are found immediately upstream [25]. Here, the assimilatory NO3−/NO2− reductase apparatus includes a: NO3−/NO2− transporter (NasA), NO2− reductase (NasB), ferredoxin (NasG), NO2− transporter (NasH) and NO3− reductase (NasC). Notably, the nasG gene is highly conserved in NO3−/NO2− assimilation gene clusters, which is consistent with a key role for the NasG ferredoxin in mediating electron flux from the NADH-oxidizing site in NasB to the sites of NO3− and NO2− reduction in NasC and NasB respectively, in order to prevent intracellular accumulation of NO2− [25]. In P. denitrificans, the RNA-binding protein NasT has been recently shown to positively and directly regulate nas expression (i.e. nasABGHC) by interacting with the nasA-leader mRNA. The NO3−/NO2−-binding sensor NasS controls NasT activity and the NasS and NasT proteins co-purify as a stable heterotetrameric regulatory complex, NasS-NasT in the absence of inducer [21]. The NasS-NasT system from B. japonicum has now been characterized by Sánchez et al. [22] and shown to regulate expression of napE and nosZ genes for the dissimilatory denitrification pathway.
The processes and enzymes for rhizobial denitrification have been well studied [1–4]; however, the biochemical apparatus for NO3−/NO2− assimilation by plant-associated rhizobia has yet to be characterized. In the present work, we demonstrate a dual role for the blr2806–09 and bll4571–73 loci for NO3−/NO2− assimilation and NO management in B. japonicum.
EXPERIMENTAL
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in the present study are listed in Supplementary Table S1. Gene deletion and transcriptional reporter construction used previously established methods [26,27], with key modifications as outlined below. B. japonicum strains were grown routinely under aerobic conditions at 30°C in peptone-salts-yeast extract (PSY) medium supplemented with 0.1% (w/v) L-arabinose [28]. Growth curves for different N-sources were performed in Bergersen minimal medium [29] supplemented with 10 mM KNO3 (BN3), 1 mM NaNO2 (BN2) or 10 mM KNO3 plus 6.5 mM L-glutamate (BGN3) as sole N-sources and incubated aerobically or anaerobically. Anaerobic conditions were reached by incubating the cells in completely filled glass serum tubes. Growth was followed by measuring attenuance (D) of cell cultures at 600 nm.
To test growth inhibition by nitrosative stress, cells were grown in Bergersen minimal medium with 6.5 mM L-glutamate (BG) as sole N-source and incubated under microaerobic conditions in serum tubes sealed with rubber septa. The headspace of these tubes was filled with a gas mixture of 2% (v/v) O2 and 98% (v/v) N2 and was replaced with fresh gas mixture every 24 h. Nitrosative stress was induced by adding 1 mM sodium nitroprusside (SNP) or spermine NONOate to the cultures 24 h after inoculation. To test cell survival after nitrosative stress induction, cells were grown to early stationary growth phase under the same conditions as for growth inhibition experiments (final D value at 600 nm was ∼0.5). Then, 1 mM SNP or spermine NONOate was added to the cultures (replica cultures were not subjected to nitrosative stress as controls). Cell cultures were incubated at 30°C and 0.1 ml of samples were taken periodically, serially diluted in growth medium and plated into PSY-agar. Colonies were counted after incubation for 7 days under aerobic conditions. The capacity for colony formation of control cells was considered as 100% survival.
To induce the expression of NorCB as a cellular marker for NO, B. japonicum was grown in Bergersen minimal medium where glycerol was replaced with 10 mM succinate as carbon source (BS) [30,31]. The medium was supplemented with 10 mM KNO3 as sole N-source (BSN3). In these experiments, the headspace was filled with 2% (v/v) O2 and 98% N2 (v/v). In contrast with microaerobic growth conditions, the atmosphere for cultures was not replaced. As such, cells consumed the O2 present and reached anoxic conditions after 24 h incubation.
Antibiotics were added to B. japonicum cultures at the following concentrations (μg·ml−1): chloramphenicol 20, tetracycline 100, spectinomycin 200, streptomycin 200 and kanamycin 200. Escherichia coli strains were cultured in LB medium [32] at 37°C including tetracycline 10, spectinomycin 20, streptomycin 20, kanamycin 30 and ampicillin at 200 μg·ml−1. E. coli S17-1 served as the donor for conjugative plasmid transfer [33].
Construction of B. japonicum narK-lacZ and nirA-lacZ transcriptional fusions
For construction of transcriptional fusion reporter plasmids, DNA fragments for the narK (520 bp) and nirA (563 bp) promoter regions were amplified using primers narK-lacZ_For/narK-lacZ_Rev and nirA-lacZ_For/nirA-lacZ_Rev, respectively (see Supplementary Table S2 for oligonucleotide sequences). Fragments were digested with PstI or PstI-EcoRI and cloned into the lacZ fusion vector pSUP3535, which is derived from pSUP202 [33] to give plasmids pDB4009 and pDB4018 respectively (Supplementary Table S1). The correct orientation of cloned inserts was verified by sequencing.
Transcriptional fusion plasmids pDB4009 and pDB4018 were integrated by homologous recombination into the chromosome of wild-type (WT) B. japonicum USDA110 and nasS and nasT mutants to produce strains 4009, 4012-4009, 4013-4009, 4018, 4012-4018 and 4013-4018 detailed in Supplementary Table S1. Correct recombination was checked by PCR analysis of genomic DNA isolated from each strain.
Growth conditions for β-galactosidase activity assay of narK-lacZ, nirA-lacZ and norC-lacZ fusions
Strains 4009, 4012-4009, 4013-4009, 4018, 4012-4018 and 4013-4018 containing the narK-lacZ or nirA-lacZ reporter-fusion constructs (Supplementary Table S1) were grown aerobically in PSY medium. Cells were harvested by centrifugation at 8000 g for 10 min, washed twice with nitrogen-free Bergersen medium and cultured aerobically in the same medium or in BN3 medium, for 48 h (until a D value of ∼0.5 at 600 nm was obtained). To measure β-galactosidase activity from the norC-lacZ fusion, plasmid pRJ2499 (Supplementary Table S1) was integrated by homologous recombination into the chromosome of bjgb, nasC, napA and nasC, napA mutants to produce strains 4001-2499, 4003-2499, GRPA1-2499 and GRPA1-4003-2499 respectively (Supplementary Table S1). In order to induce expression of nor genes, cells were cultured in BSN3 medium with 2% (v/v) initial O2 concentration.
Construction and complementation of B. japonicum mutants
Genomic and plasmid DNA isolation was carried out using the REALPURE Genomic DNA purification Kit (Real) and Qiagen Plasmid Kit (Qiagen) respectively. Custom oligonucleotide primers were supplied by Sigma, PCR was performed using the High Fidelity DNA polymerase Phusion enzyme (Thermo) and DNA digestions were carried out using Fast digest enzymes (Fermentas). All mutants constructed in the present work were made by in frame deletion of the corresponding gene using the mobilizable pK18mobsacB suicide vector that conferred kanamycin resistance and sucrose sensitivity on the host (Supplementary Table S1). To generate mutant strains, upstream and downstream regions of relevant target genes were amplified by PCR using the gene-specific primer sets detailed in Supplementary Table S2.
For the narK deletion mutant, upstream (834 bp) and downstream (981 bp) DNA fragments flanking blr2806 were amplified by PCR using blr2806_up_For/ blr2806_up_Rev and blr2806_down_For/blr2806_down_Rev primer pairs (Supplementary Table S2). The 981-bp fragment was inserted into pK18mobsacB as an XbaI-HindIII fragment that contained a new unique XhoI restriction site immediately downstream of the XbaI site. Subsequently, the 834-bp fragment was inserted into this plasmid as a BamHI-XhoI fragment yielding plasmid pDB4000. Double recombination of pDB4000 with the B. japonicum genome led to the replacement of the WT blr2806 gene encoding a 459 amino acid (aa) protein for an in frame truncated version encoding 38 aa.
To generate bjgb and nasT deletion mutants, upstream and downstream regions flanking blr2807 (824 and 884 bp fragments) and bll4573 (696 and 736 bp fragments) were amplified by PCR using gene-specific primer pairs, i.e. blr2807_up_For/blr2807_up_Rev and blr2807_down_For/blr2807_down_Rev for blr2807 and bll4573_up_For/bll4573_up_Rev and bll4573_down_For/bll4573_down_Rev for bll4573 (Supplementary Table S2). The PCR products containing the upstream regions of blr2807 and bll4573 were inserted separately into pK18mobsacB as EcoRI-XbaI fragments and subsequently the downstream PCR products were inserted into the respective plasmids as XbaI-HindIII DNA fragments yielding plasmids pDB4001 and pDB4013 (Supplementary Table S1). Double recombination of pDB4001 and pDB4013 with the B. japonicum genome led to the replacement of the blr2807 gene encoding a 142 aa protein and the bll4573 gene encoding a 196 aa protein with in-frame truncated versions of 33- and 27-aa for blr2087 or bll4573, respectively.
To generate the flp and nasC mutants, upstream and down-stream regions of blr2808 (983 and 856 bp) and blr2809 (829 and 840 bp) were amplified by PCR using gene-specific primer pairs blr2808_up_For/blr2808_up_Rev and blr2808_down_For/blr2808_down_Rev for blr2808 and bll2809_up_For/bll2809_up_Rev and bll2809_down_For/bll2809_down_Rev for bll2809 (Supplementary Table S2). The PCR product containing the upstream regions of blr2808 and blr2809 were cloned into separate pK18mobsacB plasmids as EcoRI-BamHI fragments and subsequently the downstream DNA regions were inserted into the relevant plasmid as BamHI-HindIII fragments to give pDB4002 and pDB4003. Double recombination of either pDB4002 or pDB4003 with the B. japonicum genome led to the replacement of the blr2808 gene (encoding a 418-aa protein) or the blr2809 gene (encoding a 901-aa protein) for in-frame truncated versions encoding a 37- or 27-aa peptide respectively.
The ntrABC mutant was generated by double recombination of B. japonicum USDA110 genomic DNA with plasmid pDB4004 (Supplementary Table S1). To generate pDB4004, the upstream region of blr2803 (773 bp) and the downstream region of blr2805 (721 bp) were amplified by PCR using primer pairs blr2803_up_For/blr2803_up_Rev and blr2805_down_For/blr2805_down_Rev (Supplementary Table S2). Firstly, the PCR product containing the upstream region was digested with BamHI and XbaI restriction enzymes and cloned into pK18mobsacB. Then, the PCR product corresponding to the downstream region was inserted into the plasmid as an XbaI-PstI fragment to give plasmid pBD4004 (Supplementary Table S1). Double recombination of pBD4004 with the B. japonicum genome replaced blr2803–05 genes for an in-frame truncated version encoding a 31-aa peptide.
The nirA and nasS mutants were generated by double recombination of B. japonicum USDA110 genomic DNA with plasmids pDB4011 and pDB4012 respectively. To generate pDB4011 and pDB4012, the upstream and downstream regions of bll4571 (641 and 692 bp) and bll4572 (687 and 664 bp) were amplified by PCR using the primer pairs bll4571_up_For/bll4571_up_Rev and bll4571_down_For/bll4571_down_Rev for bll4571 and bll4572_up_For/bll4572_up_Rev and bll4572_down_For/bll4572_down_Rev for bll4572 (Supplementary Table S2). PCR products containing the upstream regions were cloned first into pK18mobsacB as EcoRI-KpnI DNA fragments. Then, amplified downstream regions were inserted into the plasmid as KpnI-XbaI fragments yielding plasmids pDB4011 and pDB4012 (Supplementary Table S1). Finally, double recombination replaced bll4571 and bll4572 genes (encoding 625- and 388-aa proteins respectively) for in-frame truncated versions encoding 17- and 23-aa peptides respectively.
All plasmids constructed for mutagenesis were sequenced and transferred via conjugation into B. japonicum USDA110 using E. coli S17-1 as donor strain. Double recombination events were favoured by first selecting single recombinants for kanamycin resistance and screening candidates by PCR. Then, selected clones containing the plasmid co-integrated in the genome were grown in PSY-agar medium supplemented with sucrose 10% (w/v) to select for double recombinants. Sucrose resistant colonies were checked for kanamycin sensitivity. Double recombinants were confirmed by PCR.
B. japonicum GRC131-4001 containing a double mutation in bjgb and norC was generated by transferring plasmid pJNOR43M2 [34] via conjugation into the B. japonicum bjgb mutant (Supplementary Table S1). Double recombinants were selected for kanamycin resistance and tetracycline sensitivity. The correct replacement of the WT norC gene by kanamycin resistance gene (aphII) insertion was checked by PCR.
B. japonicum GRPA1-4003 containing a double mutation in napA and nasC was generated by transferring plasmid pBG602Ω [26] via conjugation into the B. japonicum nasC mutant (Supplementary Table S1) using E. coli S17-1 as donor strain. Double recombination events were favoured by growth on agar plates containing sucrose. Mutant strains resistant to spectinomycin/streptomycin, but sensitive to kanamycin were checked by PCR for correct replacement of the WT fragment by the Ω interposon.
The bjgb, flp and nasC strains were complemented with pDB4014, pDB4015, pDB4017 expression constructs containing the corresponding intact genes (Supplementary Table S1). For this, bjgb, flp and nasC genes were amplified by PCR using primer sets blr2807_For/blr2707_Rev, blr2808_For/blr2808_Rev and blr2809_For/blr2809_Rev (Supplementary Table S2). DNA fragments containing the relevant ORF and Shine–Dalgarno sequence were cloned separately into pTE3 vector [35]. All complementation constructs were sequenced and transferred via conjugation into the relevant B. japonicum mutant using E. coli S17-1 as donor strain. Complemented strains 4001-pDB4014, 4002-pDB4015 and 4003-pDB4017 (Supplementary Table S1) were confirmed by plasmid extraction and checked by restriction analyses and PCR.
Analysis of gene expression by RT-PCR
Total RNA was isolated from B. japonicum cells grown anaerobically to a D of ∼0.4 (at 600 nm) in BN3 medium, as previously described [36]. First strand cDNA synthesis was performed with the SuperScript II reverse transcriptase (Invitrogen) according to the supplier's guidelines, using 1 μg of total RNA and primer e (Supplementary Table S2). cDNA generated was used for amplification of putative intergenic regions between blr2805 and blr2809, using primers pairs a1/a2 to d1/d2 (Supplementary Table S2), essentially as described by Sambrook and Russell [32]. In negative controls, reverse transcriptase was omitted and for positive controls PCR was performed with B. japonicum USDA110 genomic DNA as template.
β-Galactosidase assays
β-Galactosidase activity was determined using permeabilized cells from at least three independently grown cultures assayed in triplicate for each strain and condition, as previously described [37]. The absorbance data at 420 and 600 nm were determined for all samples and cultures in a plate reader (SUNRISE Absorbance Reader, TECAN), using the software XFluor4 (TECAN) and specific activities were calculated in Miller units.
Determination of NO3− reductase and NO2− reductase activity
B. japonicum was grown under aerobic conditions in PSY medium, harvested by centrifugation at 8000 g for 10 min at 4°C, washed twice with BN3 medium and inoculated to a D value of ∼0.4 (at 600 nm) in fresh minimal medium of the same composition. Following 72 h incubation under relevant conditions, cells were harvested, washed with 50 mM Tris/HCl buffer (pH 7.5) to remove excess NO2− and then resuspended in 1 ml of the same buffer prior to assay for enzymatic activity. Methyl viologen-dependent NO3− reductase (MV-NR) and NO2− reductase (MV-NIR) activity was measured essentially as described by Delgado et al. [26]. The reaction mixture contained 50 mM Tris/HCl buffer (pH 7.5), 200 μM MV, 100 μl of cell suspension (with 0.02–0.04 mg of protein) and 10 mM KNO3 or 100 μM NaNO2 for MV-NR or MV-NIR activity respectively. Methyl viologen was reduced by the addition of freshly prepared sodium dithionite (dissolved in 300 mM NaHCO3 solution) at a final concentration of 14.4 mM.
Haem-staining analysis
Aerobically grown B. japonicum cells were harvested by centrifugation, washed twice with BSN3 medium and resuspended in 500 ml of fresh medium of the same composition. Microaerobic conditions were then established with 2% (v/v) initial O2 concentration and cells were cultured for 48 h until a final D of ∼0.5 (at 600 nm) was reached. Cells were disrupted using a French pressure cell (SLM Aminco, Jessup) and membranes were isolated as described previously [26]. Membrane protein aliquots were diluted in sample buffer [124 mM Tris/HCl, pH 7.0, 20% (v/v) glycerol, 5% (v/v) SDS and 50 mM 2-mercaptoethanol] and incubated at room temperature for 10 min. Membrane proteins were separated at 4°C by SDS-PAGE [12% (w/v) acrylamide resolving gel with 20 μg of protein per lane], transferred to a nitrocellulose membrane and stained for haem-dependent peroxidase activity [38], using the chemiluminescence detection kit ‘SuperSignal’ (Pierce, Thermo Fisher Scientific). Protein concentration was estimated using the Bio-Rad assay (Bio-Rad Laboratories).
Intracellular NO2− determination
B. japonicum cells were harvested, washed and lysed by using a French pressure cell (SLM Aminco, Jessup). Soluble cell extracts were prepared by centrifugation at 10000 g for 30 min at 4°C and assayed for NO2− using the method of Nicholas and Nason [39].
NO consumption activity
NO consumption rates were determined using intact B. japonicum cells [obtained from BSN3 cultures with 2% (v/v) initial O2 and a D of ∼0.5 (at 600 nm)] with a 2 mm ISONOP NO electrode APOLLO 4000® (World Precision Institute). The reaction chamber (2 ml) was temperature-controlled, magnetically stirred and contained: 760 μl of 25 mM phosphate buffer (pH 7.4), 900 μl of cell suspension (4–5 mg protein), 100 μl of an enzyme mix containing Aspergillus niger glucose oxidase (40 units·ml−1) and bovine liver catalase (250 units·ml−1) (Sigma-Aldrich), 90 μl of 1 M sodium succinate and 100 μl of 320 mM glucose. Once a steady base line was obtained, 50 μl of a saturated NO solution (1.91 mM at 20°C) was added to the cuvette to start the reaction. Each assay was monitored until the NO detection had dropped to zero, i.e. when all NO was consumed.
N2O measurements
B. japonicum cells were cultured as indicated above for NO consumption experiments, except that in addition to 2% (v/v) initial O2, the headspace of the cultures also contained 10% (v/v) acetylene in order to inhibit N2O reductase activity. After 96 h growth, gaseous samples were taken from the headspace of cultures. N2O was measured using an HP 4890D gas chromatography instrument equipped with an electron capture detector (ECD). The column was packed with Porapak Q 80/100 MESH and the carrier gas was N2 at a flow rate of 23 ml·min−1. The injector, column and detector temperatures were 125, 60 and 375°C respectively. The samples were injected manually using a Hamilton® Gastight syringe. Peaks corresponding to N2O were integrated using GC ChemStation Software (Agilent Technologies©) and the concentrations of N2O in each sample were calculated using N2O standards (Air Liquid).
RESULTS
Genetic basis for NO3− and NO2− assimilation in endosymbiotic denitrifying rhizobia
B. japonicum USDA110 contains a putative assimilatory NO3− reductase encoded at blr2809 (Figure 1A) [19,20]. Experiments confirmed that B. japonicum is able to grow aerobically or anaerobically using NO3− as sole N-source with values for μmax (app) (apparent maximum growth rate) of approximately 0.06 and 0.04 h−1 respectively (Figures 2A and 2B; Supplementary Table S3).
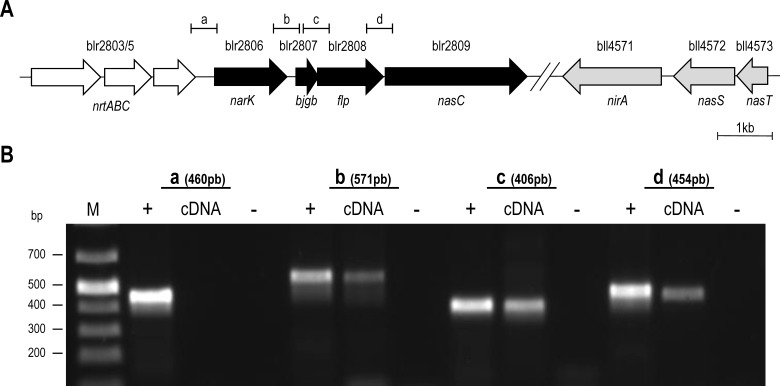
Figure 1. Organization of regulatory and structural genes for the assimilatory NO3−/NO2− reductase pathway in B. japonicum.
(A) Schematic representation of the blr2803-5, blr2806-09 and bll4571-73 ORFs investigated in the present study. Putative intergenic regions probed by RT-PCR to determine the transcriptional architecture of the blr2806-09 region (i.e. narK-bjgb-flp-nasC) are labelled a–d. (B) The results for RT-PCR analysis obtained by agarose gel electrophoresis for regions a–d. Total RNA isolated from cells grown anaerobically with NO3− served as the template for cDNA synthesis, whereas PCR amplifications using genomic DNA and without reverse transcriptase enzyme served as positive and negative controls respectively (as indicated above lanes).
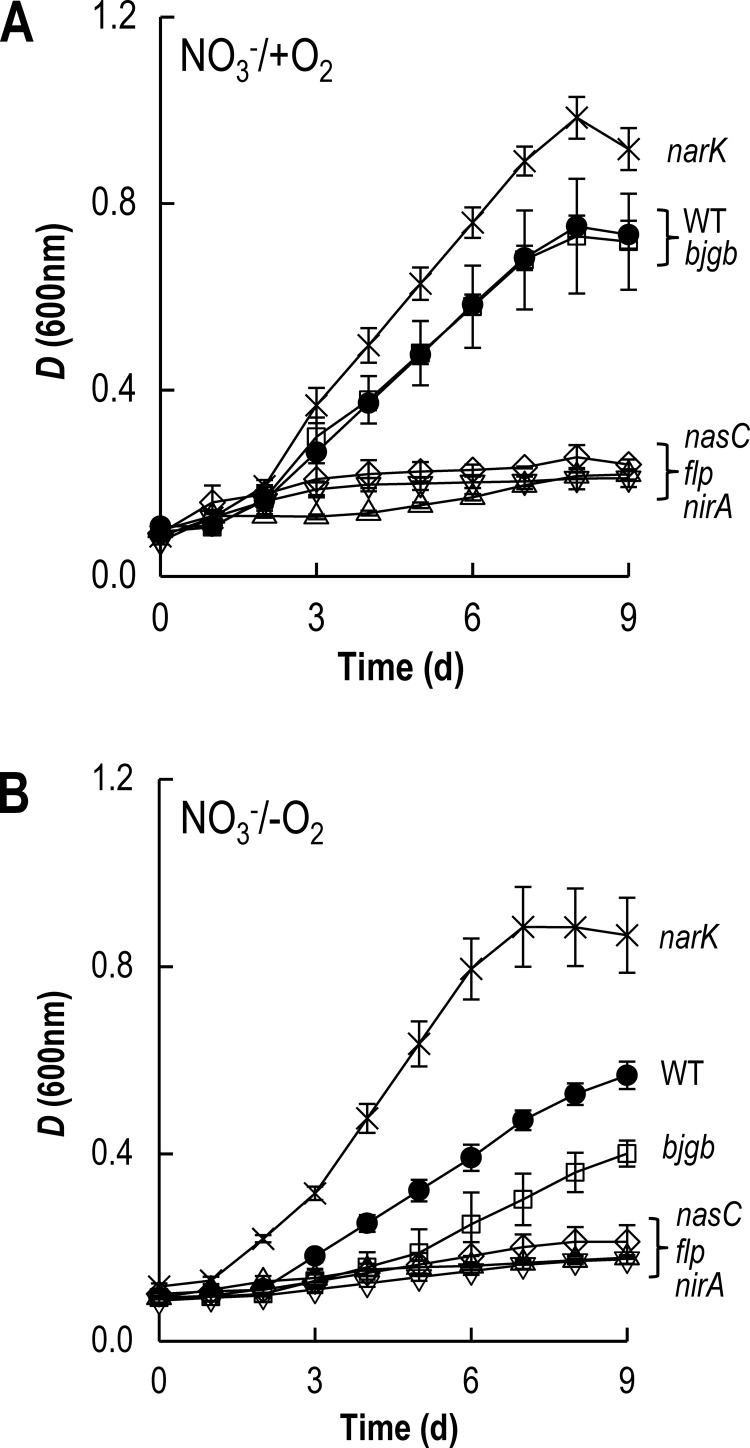
Figure 2. NO3−-dependent growth of B. japonicum.
Growth curves for WT (●), narK (×), bjgb (□), flp (▽), nasC (◇) and nirA (△) strains were measured under aerobic (A) and anaerobic (B) conditions in BN3 minimal medium with NO3− as sole N-source. The results presented are the mean of two biological replicates assayed in triplicate.
Notably, blr2809 lies downstream of several putative ORFs with predicted roles in N-metabolism (Figure 1A). To investigate the transcriptional architecture of this region, RT-PCR experiments were performed to detect intergenic regions (a–d). Here, specific cDNA was obtained for all regions except ‘a’ (Figure 1B). These findings reveal that blr2806–09 constitute a transcriptional unit. Thus, in-frame deletion strategies for subsequent molecular genetics experiments were adopted to prevent possible polar effects on co-transcribed genes (see ‘Experimental’ section for details).
Analysis of the amino acid sequence of blr2809 suggests the protein is a member of the molybdenum bis-molybdopterin dinucleotide cofactor binding superfamily and contains consensus motifs for co-ordination of an N-terminal [4Fe-4S] cluster and a C-terminal [2Fe-2S] cluster. This general organization is similar to other assimilatory NO3− reductases, including P. denitrificans NasC and Klebsiella oxytoca NasA from the α- and γ-proteobacterial clades, respectively [25]. Accordingly, we adopt the α-proteobacterial nomenclature, NasC, for the B. japonicum protein encoded at blr2809 hereafter. A B. japonicum strain that was mutated by in-frame deletion of nasC lost the capacity for aerobic or anaerobic growth with NO3− as sole N-source (Figures 2A and 2B; Supplementary Table S3). However, this strain retained the ability to grow using NO2− as sole N-source and displayed similar growth kinetics to WT [μmax (app) ∼ 0.03 h−1] (Figure 3A; Supplementary Table S3).
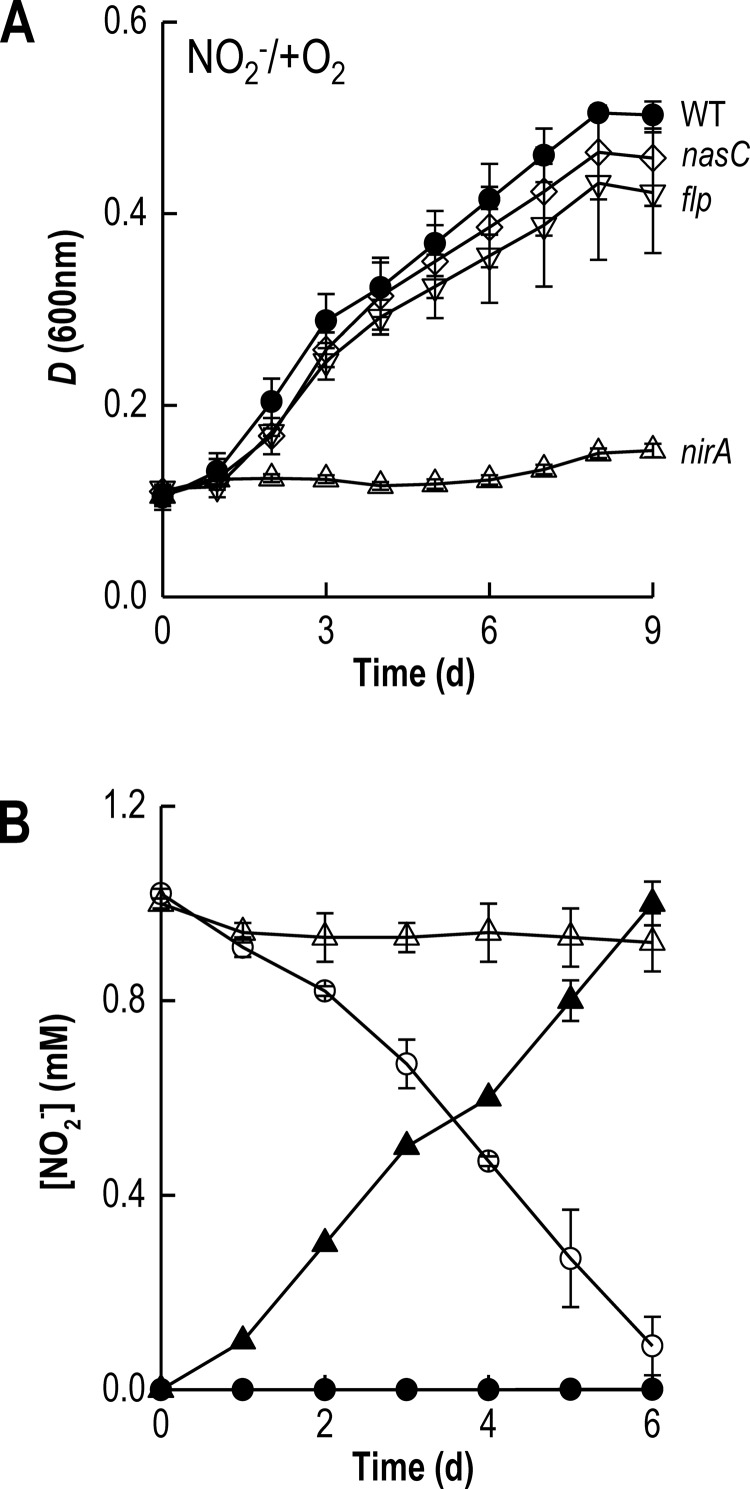
Figure 3. NO2−-dependent growth of B. japonicum.
(A) Growth curves for WT (●), flp (▽), nasC (◇) and nirA (△) strains were measured under aerobic conditions in BN2 minimal medium with NO2− as sole N-source. (B) Extracellular NO2− consumption (open symbols) and accumulation (solid symbols), using 1 mM NO2− or 10 mM NO3− as sole N-source respectively measured during growth of WT (circles) and nirA (triangles) strains. The results presented are the mean of two biological replicates assayed in triplicate.
The genome of B. japonicum also contains an ORF for a putative assimilatory NO2− reductase (nirA) at bll4571, a distinct locus situated ∼2 Mb from nasC on the chromosome (Figure 1A). NirA contains canonical cysteine-rich motifs in central and C-terminal sequence regions for iron–sulfur co-ordination and formation of the sirohaem NO2− reductase/sulfite reductase ferredoxin half-domain respectively. However, NirA lacks N-terminal FAD- and NAD(P)H-binding domains present in bacterial NirB-type NAD(P)H-dependent NO2− reductases [25]. Deletion of nirA resulted in B. japonicum being unable to grow aerobically or anaerobically with either NO3− or NO2− as sole N-source (Figures 2A, 2B and 3A; Supplementary Table S3). The ability of WT and nirA cells to consume 1 mM NO2− during incubation experiments was tested (Figure 3B, open symbols). Whereas all NO2− was removed from minimal medium after ∼6 days by WT cells, no significant decrease in extracellular NO2− was observed in nirA mutant cultures (Figure 3B). Conversely, NO2− production experiments using 10 mM NO3−, as sole N-source, revealed that WT cells did not accumulate NO2− in the extracellular medium (Figure 3B, closed symbols). However, accumulation of ∼1 mM NO2− was observed following incubation of the nirA mutant with NO3− (Figure 3B). Thus, pre-cultured cells of the nirA mutant retained the capacity to reduce NO3− to NO2−, but no further.
The putative flavoprotein (Flp), encoded at blr2808, contains canonical FAD- and NAD(P)H-binding domains typical of cytoplasmic NAD(P)H-dependent oxidoreductases present in several bacterial Nas operons [23] and is a strong candidate for mediating electron transfer to NasC and/or NirA. A B. japonicum flp mutant was unable to grow aerobically or anaerobically with NO3− as the sole N-source (Figures 2A and 2B; Supplementary Table S3). However, the flp mutant displayed similar growth kinetics and yields [μmax (app) ∼ 0.03 h−1, maximum D (at 600 nm)=0.43±0.08] to that observed for WT [μmax (app) ∼ 0.03 h−1, maximum D (at 600 nm)=0.51±0.01] when cultured aerobically with NO2− (Figure 3A; Supplementary Table S3). These findings suggest that Flp mediates electron transfer to NasC, but not to NirA. In order to confirm that deletion of flp did not influence expression of downstream genes, relevant strains were complemented with either pDB4017 (nasC) or pDB4015 (flp) constructs. The presence of pDB4017 and pDB4015 plasmids restored both aerobic and anaerobic growth of the nasC and flp mutants in the presence of NO3− to near WT levels, thereby verifying the phenotypes observed (Supplementary Table S3).
Deletion of the blr2803–05 ORFs, predicted to encode an NrtABC-type NO3− transporter, did not affect the capacity of the cells to grow with NO3− as sole N-source (Supplementary Table S3). Bioinformatics analysis of blr2806 revealed that it encodes a putative member of the MFS of membrane proteins, sharing 66% and 59% amino acid similarity with the NO3−/NO2− antiporters E. coli NarK [40] and P. denitrificans NarK2 [41] respectively (Supplementary Figure S1). Thus, we term this MFS-type transporter NarK rather than the generic ‘nitrite extrusion protein’ genome annotation currently assigned (http://genome.kazusa.or.jp/rhizobase/).
A B. japonicum narK mutant showed improved growth kinetics and yields when cultured aerobically [μmax (app) ∼ 0.09 h−1, maximum D (at 600 nm)=0.98±0.05] or anaerobically [μmax (app) ∼ 0.07 h−1, maximum D (at 600 nm) 0.89±0.09] with NO3− as sole N-source when compared with aerobic [μmax (app) ∼ 0.06 h−1, maximum D (at 600 nm)=0.73±0.12] or anaerobic [μmax (app) ∼ 0.04 h−1, maximum D (at 600 nm)=0.61±0.03] growth of WT under the same conditions (Figures 2A and 2B; Supplementary Table S3). Furthermore, following 24 h aerobic growth, the narK mutant accumulated ∼2-fold higher levels of intracellular NO2− than that accumulated by WT cells, i.e., 5.3±0.7 compared with 2.2±0.1 nmol NO2− mg·protein−1 for the narK and WT strains respectively (Supplementary Figure S2). The addition of L-glutamate to minimal growth medium restored the inability of the nasC, nirA and flp mutants to grow with NO3− under aerobic or anaerobic conditions (Supplementary Table S3). Under these conditions, growth yields obtained from the narK mutant were also similar to those obtained from WT cells (Supplementary Table S3). Collectively, these results confirm the importance of NarK, Flp, NasC and NirA for NO3− assimilation by B. japonicum.
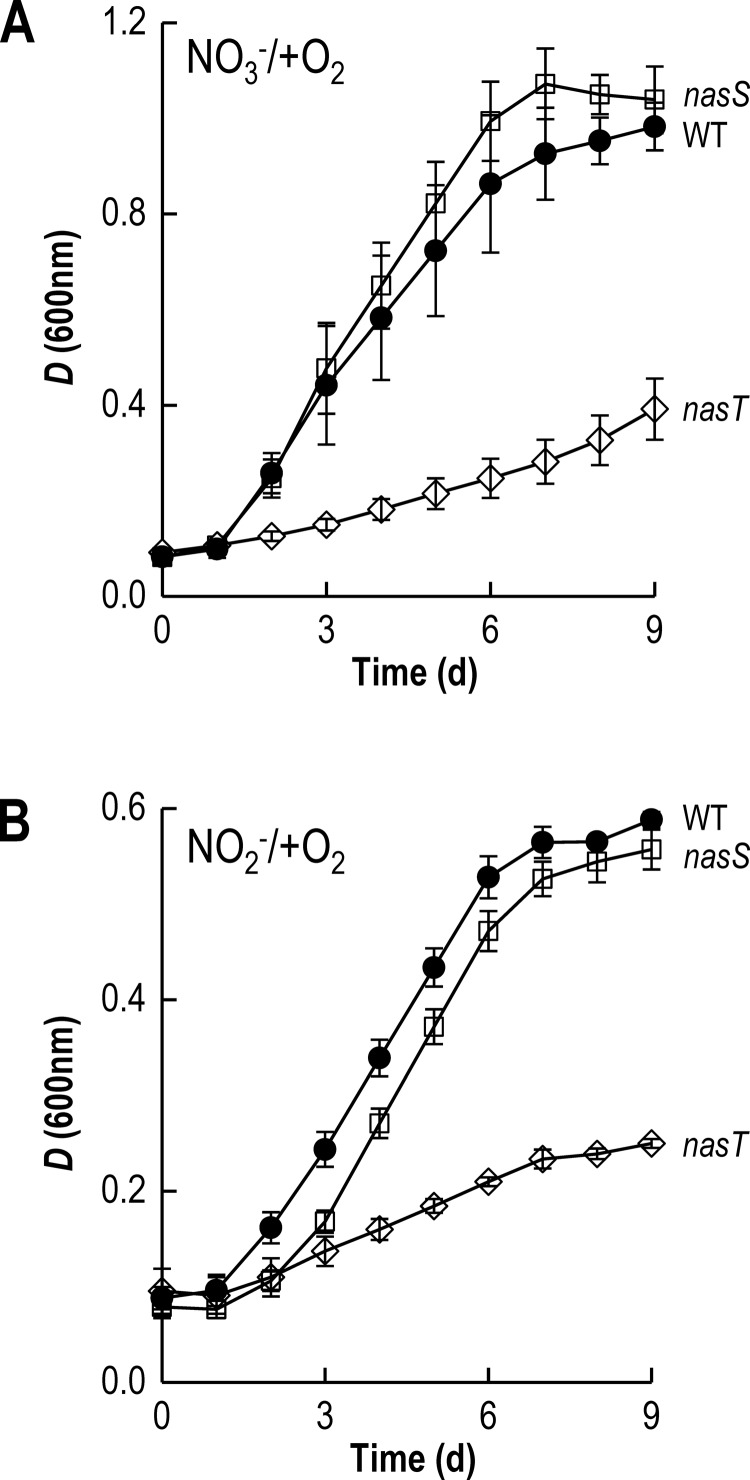
The regulatory proteins encoded by bll4573 (nasT) and bll4572 (nasS) constitute a NO3−/NO2− responsive two-component system, NasS-NasT, which has been recently reported in B. japonicum [22]. A B. japonicum nasT mutant strain showed significant growth attenuation compared with the WT cells when cultured aerobically with either NO3− (Figure 4A; Supplementary Table S3) or NO2− (Figure 4B; Supplementary Table S3) as sole N-source, but growth of this strain was unaffected when cells were grown in the presence of L-glutamate (Supplementary Table S3). By contrast, a strain in which the nasS gene was mutated did not show a clear growth defect with respect to WT (Figure 4; Supplementary Table S3).
Figure 4. Growth curves for the B. japonicum nasS and nasT mutants.
Growth of WT (●), nasS (□) and nasT (◇) strains was measured in minimal medium, under aerobic conditions, with either NO3− (A) or NO2− (B), as sole N-source. The results presented are the mean of two biological replicates assayed in triplicate.
A biochemical pathway for assimilation of NO3− and NO2−
The biochemical basis of growth phenotypes observed for the various deletion strains was examined by enzymatic activity assay of whole cells, using dithionite-reduced methyl viologen, as an artificial electron donor. Here, MV-NR and MV-NIR activities were measured in WT and nasC, nirA, flp, bjgb and narK mutants, following aerobic incubation with NO3− as sole N-source (Table 1). Since B. japonicum has periplasmic respiratory NO3− reductase (NapABC) and NO2− reductase (NirK) systems that might also use methyl viologen as an electron donor [26,42], control experiments using napA and nirK mutants were also performed in the present study. Importantly, and as expected, the respective MV-NR and MV-NIR activity levels observed in napA and nirK cells were similar to those observed in WT cells (Table 1), indicating that the contribution of the NapABC or NirK respiratory enzymes was not significant in cells cultured under aerobic conditions. This provided a solid platform for subsequent experiments.
Table 1. MV-NR and MV-NIR activities of B. japonicum strains incubated aerobically in minimal medium with NO3− as sole N-source.
| Activities | |||
|---|---|---|---|
| B. japonicum str. | Genotype | MV-NR* | MV-NIR† |
| USDA 110 | WT | 32.0±5.2 | 6.9±0.9 |
| GRPA1 | napA | 32.1±0.5 | – |
| GRK308 | nirK | – | 7.5±0.8 |
| 4003 | nasC | n.d. | 7.2±1.2 |
| 4003-pDB4017 | nasC (pDB4017) | 30.5±4.8 | – |
| 4011 | nirA | 49.7±1.8 | n.d. |
| 4002 | flp | 68.3±6.7 | 6.1±0.8 |
| 4001 | bjgb | 28.4±5.0 | 6.1±0.4 |
| 4000 | narK | 35.8±2.2 | 10.9±1.5 |
*MV-NR and †MV-NIR activities are expressed as nanomoles of NO2− produced or consumed min−1·mg·protein−1. Data are expressed as the mean value±S.D. from at least two different cultures assayed in triplicate;–, not determined; n.d., not detectable.
Significantly, MV-NR activity was not detectable in nasC cells, but a similar level of MV-NIR activity was observed compared with WT cells. This was consistent with the loss of assimilatory NO3− reductase expression, but not NO2− reductase expression, in nasC cells (Table 1). MV-NR activity could be restored to WT levels in the nasC mutant, when the deletion was complemented with a corresponding plasmid-borne gene copy. Also, MV-NIR activity was absent from the nirA mutant (Table 1), consistent with the loss of assimilatory NO2− reductase expression. However, the nirA mutant showed similar levels of MV-NR activity present in the parental strain following incubation with NO3−. Additional experiments revealed that MV-NR levels of flp cells showed an apparent ∼2-fold increase in activity compared with WT incubation with NO3−, but MV-NIR activity was relatively similar in both flp and WT cells (Table 1). That the absence of Flp (i.e. the proposed electron donor and partner to NasC) should increase MV-NR activity may result from modulation in catalytic activity of the isolated NasC protein. Alternatively, without Flp, the artificial chemical electron donor could have greater access to NasC and thus may enhance NO3− reductase activity. Finally, as shown in Table 1, MV-NR and MV-NIR activities of bjgb or narK mutants were similar to those observed in WT cells.
Regulation of the narK-bjgb-flp-nasC operon and nirA by NasS-NasT
In order to test the involvement of the NasT regulatory protein in NO3−-dependent induction of the narK-bjgb-flp-nasC operon and nirA gene, we examined expression of narK-lacZ and nirA-lacZ transcriptional fusion constructs in WT and nasT mutant cells following aerobic culture in the presence or absence of the inducer NO3− (Table 2). Whereas similar low levels of β-galactosidase activity were observed from both fusions in WT cells incubated without NO3−, the presence of this molecule induced expression of the narK-lacZ and nirA-lacZ transcriptional fusions by approximately 5- and 3-fold respectively. However, β-galactosidase activity from the narK-lacZ reporter was undetectable in the nasT strain regardless of whether NO3− was present or not (Table 2). Although similar basal levels of nirA-lacZ expression were observed in WT and nasT cells incubated without NO3−, a decrease of approximately 2-fold was found in nasT compared with WT when cells were incubated in the presence of NO3− (Table 2).
Table 2. β-Galactosidase activity for narK-lacZ and nirA-lacZ fusions in B. japonicum WT, nasS or nasT strains.
Cells were cultured under aerobic conditions, in minimal medium, with or without NO3− as sole N-source. Data are means±S.D. from at least three independent cultures, assayed in triplicate; n.d., not detectable.
| Miller units | |||
|---|---|---|---|
| B. japonicum str. | Relevant genotype | –NO3− | +NO3− |
| 4009 | WT::narK-lacZ | 153±40 | 759±54 |
| 4012-4009 | nasS::nark-lacZ | 972±132 | 897±66 |
| 4013-4009 | nasT::narK-lacZ | n.d. | n.d. |
| 4018 | WT::nirA-lacZ | 137±22 | 395±56 |
| 4012-4018 | nasS::nirA-lacZ | 412±37 | 372±31 |
| 4013-4018 | nasT::nirA-lacZ | 163±34 | 203±13 |
Additional studies to examine the role of NasS in NasT-dependent induction of the narK-bjgb-flp-nasC operon and nirA gene were also performed, using narK-lacZ or nirA-lacZ reporters. Here, β-galactosidase assays revealed that, in the absence of NO3−, the activity of each reporter fusion was significantly higher (approximately 6- and 3-fold for narK-lacZ and nirA-lacZ respectively) in nasS cells compared with WT cells (Table 2). These results imply that, in the absence of NO3−, NasS is a repressor of narK-bjgb-flp-nasC and nirA transcription. When equivalent experiments were performed in WT and nasS cells that had been pre-exposed to NO3−, expression levels for each reporter-fusion were very similar (Table 2).
Collectively, the reporter-fusion results suggest an inhibitory role for NasS in NasT-dependent induction of gene expression in B. japonicum and that NO3−-responsive control of both narK-bjgb-flp-nasC and nirA assimilatory gene expression is lost in vivo without NasS. This mode of regulation is analogous to NO3−/NO2−-responsive control of nas gene expression by NasS-NasT in the related α-proteobacterium P. denitrificans [21].
Involvement of Bjgb and Flp in nitrosative stress defence
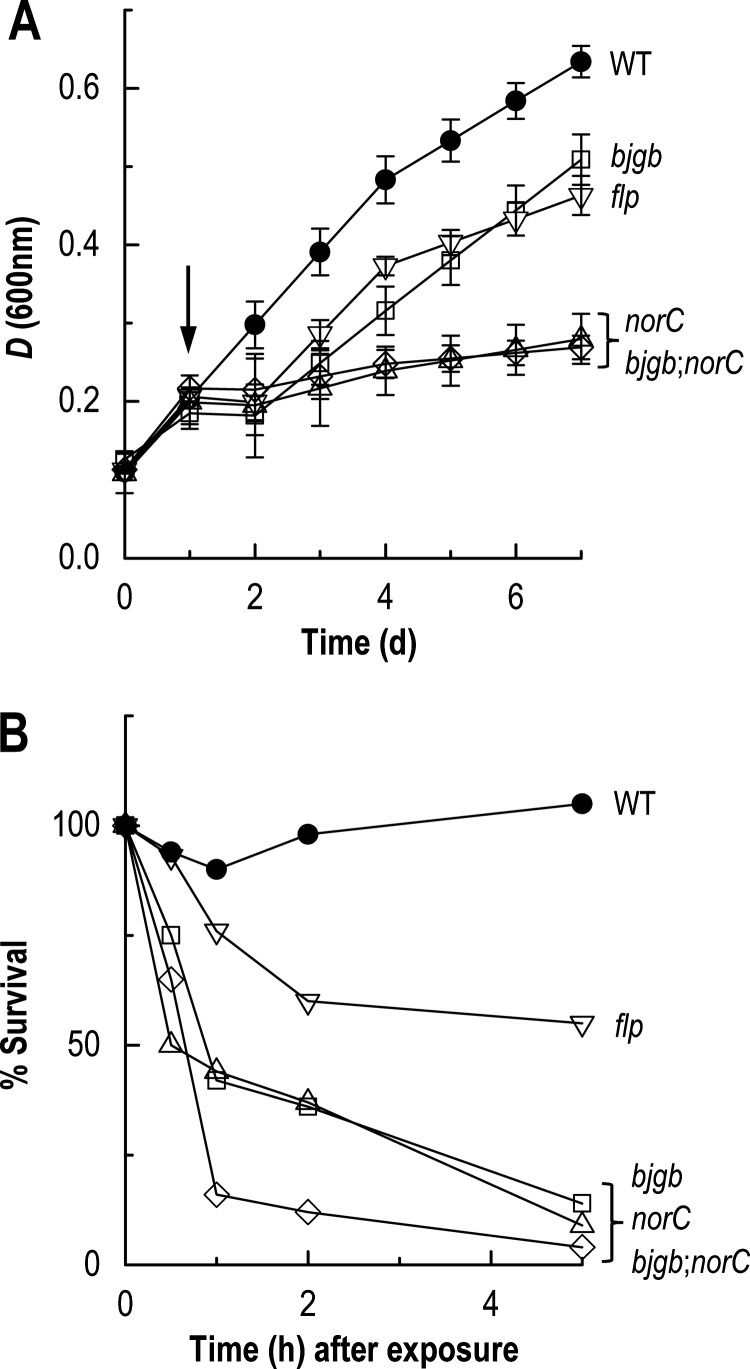
A marked difference in growth between the bjgb mutant [μmax (app) ∼ 0.02 h−1, maximum D (at 600 nm)=0.45±0.02] and WT strains [μmax (app) ∼ 0.04 h−1, maximum D (at 600 nm)=0.61±0.03] was observed under anaerobic conditions, in minimal medium with NO3− as N-source (Figure 2B; Supplementary Table S3). By contrast, growth of the bjgb mutant and WT strains was similar under aerobic conditions (Figure 2A; Supplementary Table S3). These observations suggest that Bjgb has a key role in vivo for NO3− assimilation under anaerobic conditions, but not during aerobic growth. Anaerobic NO3− reduction is known to generate the potent cytotoxin NO, which requires NO-detoxification and nitrosative stress defence systems for bacterial survival [43,44]. To investigate the role of Bjgb in NO-metabolism, the nitrosative stress agent SNP was added (at 1 mM final concentration) to microaerobic B. japonicum cultures following growth in minimal medium with L-glutamate (BG) as sole N-source. Growth of WT cells was not significantly perturbed, whereas addition of SNP resulted in transient growth arrest of bjgb and flp strains that was restored after 24 h (Figure 5A). Perhaps most significantly, a norC or a bjgb;norC double mutant showed a substantially longer period of growth inhibition of approximately 7 days following addition of SNP to cultures (Figure 5A). The effect of SNP on cell viability was also assayed by performing viable cell counts on samples taken at intervals spanning a 5-h period following addition of SNP to cultures. Although WT cell viability was not significantly affected, addition of SNP caused a ∼60% decrease in cell survival for norC or bjgb cultures after 2 h (Figure 5B). The most prominent effect was observed with the bjgb;norC double mutant, which was the most sensitive to nitrosative stress. Here, approximately 80% of cells were killed within 1–2 h following SNP exposure (Figure 5B). Furthermore, the addition of SNP provoked a ∼40% decrease in flp viability after 2 h incubation. These results revealed the importance of Bjgb and Flp for protection against nitrosative stress in B. japonicum under free-living conditions. NO is a product of SNP breakdown and a similar sensitivity of bjgb or flp mutants to NO was observed using spermine NONOate as an NO-generating compound (result not shown). Importantly, complementation with pDB4014 (harbouring a functional plasmid-borne copy of bjgb) allowed the bjgb mutant to grow anaerobically with NO3− to near WT levels (Supplementary Table S3). This confirmed that the growth phenotype observed for the bjgb mutant was not caused by a downstream effect on flp gene expression.
Figure 5. Growth inhibition curves (A) and cell viability assays (B) for B. japonicum WT (●), bjgb (□), flp (▽), norC (△) and bjgb;norC (◇) strains in response to nitrosative stress induced by addition of SNP.
For growth inhibition curves (A), B. japonicum strains were cultured microaerobically in BG minimal medium and 1 mM SNP was added after 1 d (as indicated by the arrow) and cell viability was measured from 1 to 5 h after exposure (B). The results presented are the mean of three biological replicates.
NO formed during NO3− assimilation induces nor gene expression
To further investigate the role of Bjgb and Flp in NO metabolism, the ability of B. japonicum bjgb and flp strains to consume NO was analysed. Here, cells were incubated in BSN3 medium, with 2% initial O2 and NO consumption rates were determined using an NO-electrode (Supplementary Figure S3). A ∼2.5-fold increase in NO consumption was observed in the bjgb mutant compared with the WT strain (Table 3). This increase was not observed in the flp mutant, which showed NO consumption rates marginally lower than that observed in WT cells (Table 3; Supplementary Figure S3). NO consumption in the norC or the bjgb;norC mutants was approximately 1.6- and 1.7-fold lower respectively, compared with that observed in WT cells (Table 3; Supplementary Figure S3). The presence of residual activity in the bjgb;norC implies that under our experimental conditions, another enzyme(s) or perhaps a chemical process may be involved in NO consumption. The ability of bjgb cells to produce N2O following incubation in BSN3 medium with 2% initial O2 was also investigated. The bjgb mutant produced approximately 2.5-fold more N2O than WT cells. By contrast, the level of N2O produced by the flp mutant was comparable to WT (Table 3). Given that N2O production was not detected for either the norC or the bjgb;norC mutants, this suggested the NorCB enzyme was the main source of N2O in vivo.
Table 3. NO consumption activity and N2O levels for B. japonicum WT, bjgb, flp, norC and bjgb;norC strains cultured in BSN3 minimal medium under 2% (v/v) initial O2.
Data are expressed as the means±S.D. from at least two different cultures assayed in triplicate; n.d., not detectable.
| B. japonicum str. | Genotype | NO consumption activity (nmol·h−1·mg·protein−1) | N2O (mM) |
|---|---|---|---|
| USDA110 | WT | 155±29 | 1.04±0.26 |
| 4001 | bjgb | 384±65 | 2.34±0.16 |
| 4002 | flp | 101±17 | 0.88±0.03 |
| GRC131 | norC | 97±14 | n.d. |
| GRC131-4001 | bjgb;norC | 92±18 | n.d. |
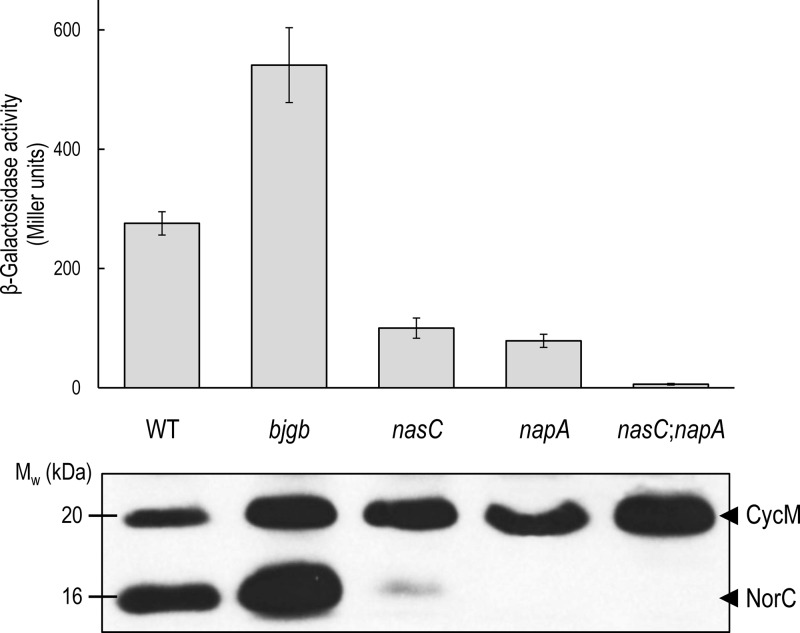
To test whether the higher levels of NO consumption and N2O production observed by the bjgb mutant were due to an induction of NorCB expression, norC transcription and relative abundance of NorC in membrane extracts were analysed, using a norC-lacZ transcriptional fusion and haem staining SDS-PAGE respectively. Firstly, a ∼2-fold increase in norC-lacZ expression was observed in the bjgb mutant compared with WT (Figure 6). Given that the norC promoter is highly sensitive to N-oxides, including NO [31], an induction of β-galactosidase activity implies that Bjgb may act as a net sink for NO in WT cells. By contrast, β-galactosidase activity of the norC-lacZ transcriptional fusion was similar for both the nasC and the napA mutants, being approximately 3-fold lower compared with WT levels (Figure 6). Activity of the norC-lacZ transcriptional fusion was essentially abolished in the nasC;napA double mutant, implying that NO3− reduction by NasC or NapA was the source of NO required for norC-lacZ expression (Figure 6).
Figure 6. Expression of B. japonicum nor genes during NO3−-dependent growth.
β-Galactosidase expression levels for the norC-lacZ transcriptional fusion in the WT, bjgb, nasC, napA and nasC;napA strains grown in BSN3 minimal medium containing 2% initial O2 (v/v) and NO3− as sole N-source. Haem-staining SDS-PAGE analysis of membrane fractions from B. japonicum strains is inset below. Each lane contains ∼20 μg of total protein for the strains described. Haem-staining bands for previously identified c-type cytochromes, CycM and NorC, are indicated.
SDS-PAGE analysis of membranes (that were normalized for total protein) by haem staining was used as a qualitative assay for expression of the NorC cytochrome. In bjgb cells, NorC levels were significantly increased relative to WT (Figure 6 inset; compare lanes 1 and 2). However, a clear decrease in NorC expression was observed in the nasC mutant compared with WT (Figure 6 inset; compare lanes 1 and 3). Furthermore, haem staining failed to detect NorC expression in membranes prepared from either the napA or the nasC;napA mutant (Figure 6 inset; compare lane 1 with lane 4 or 5).
DISCUSSION
Defining the key components and transcriptional architecture of NO3− and NO2− assimilation in B. japoniucum
A series of molecular genetics studies have established that genes encoded at two distinct loci, blr2806–09 and bll4571–73 of the B. japonicum genome (http://genome.kazusa.or.jp/rhizobase/), encode structural and regulatory components of a combined assimilatory NO3− reductase and NO detoxification system (Figure 1). RT-PCR experiments demonstrate that the narK-bjgb-flp-nasC genes (present at blr2806–09 respectively) constitute a transcriptional unit. However, three putative genes (blr2803–05) predicted to encode a NO3− transport system (similar to NrtABC, reviewed in [45]) and that lie immediately upstream of the narK operon are transcribed from a different promoter. The nasTS-nirA gene cluster (present at bll4571–73 respectively) lies some 2 Mb from the narK operon in the genome and encodes a NO3−/NO2− responsive two-component regulatory system, NasS-NasT [22] and a putative ferredoxin-dependent NO2− reductase (NirA).
A role for the bjgb (blr2807) gene product in NO detoxification has been described [3,20], but the functions of other putative proteins encoded within the narK operon and biochemical components for the assimilatory NO3− reductase pathway in B. japonicum were unknown. In the present work, we have demonstrated that the assimilatory NO3− reductase (we rename herein as NasC) is encoded by blr2809 and is essential for NO3−-dependent growth. The second core cytoplasmic enzyme component of the NO3− assimilation pathway is NirA, which is required for growth on either NO3− or NO2− as sole N-source. Consistent with our findings, it has recently been demonstrated that NirA (encoded by bll4571) is required for utilization of NO3− or NO2− as sole N-source in B. japonicum [46]. NO3−-dependent induction of nasC (as part of the narK operon) and nirA expression is mediated by the two-component regulator NasS-NasT, an observation that is consistent with the role of this system in other α-proteobacteria [21].
Phenotypic analyses of a mutant lacking Flp (encoded by blr2808) suggest that Flp mediates electron transfer to NasC, but not to NirA. Consecutive genes from the same operon encode Flp and NasC, but lie in a different genetic locus to bll4571 (nirA). This genetic organization may explain the requirement of Flp for NO3− assimilation but not for NO2− assimilation, which instead is ferredoxin dependent (Figure 7). In contrast with B. japonicum, in P. denitrificans the regulatory and structural elements for a cytoplasmic NO3−/NO2− reductase system comprise a large gene cluster, nasTSABGHC [23]. The absence of a nasG homologue in either the narK operon or the nirA cluster in B. japonicum is notable. NasG may mediate electron flux to both the NO3− reductase and the NO2− reductase in other bacteria to prevent accumulation of excess NO2− by NO3− reduction in the cytoplasmic compartment [23,25]. Instead, for B. japonicum, genes encoding systems for NO2− transport and NO-detoxification are present within the operon encoding the NO3− reductase, which generates NO2−.
Figure 7. Proposed biochemical pathway for NO3−-assimilation and NO-detoxification (red), alongside well-characterized systems for dissimilatory NO3− respiration (blue) in B. japonicum.
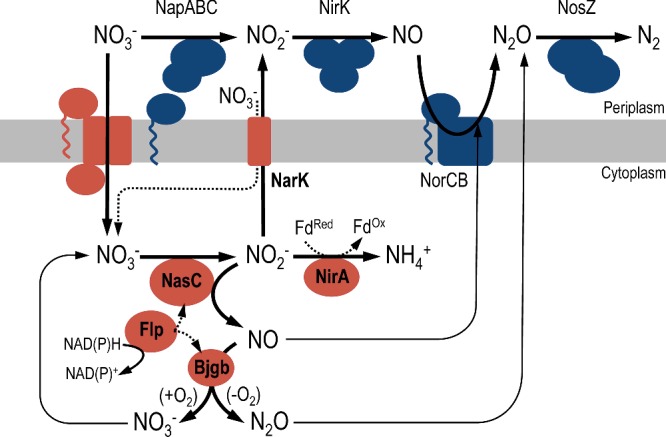
Assimilatory reduction of NO3− to NH4+ is performed by sequential action of the NO3− reductase NasC and ferredoxin (Fd)-dependent NO2− reductase NirA. Electrons from NAD(P)H are supplied to NasC and also Bjgb by Flp. During assimilatory NO3− reduction, cytoplasmic NO2− may accumulate and be further reduced, by NasC, to generate cytotoxic NO. NarK can counteract accumulation of NO2− by exporting it to the periplasm. Alternatively, Bjgb may detoxify the NO, formed by adventitious reduction in cytosolic NO2−, to NO3− or N2O in the presence or absence of O2 respectively. Expression of NorCB is up-regulated during NO3− assimilation and this respiratory system may assist Bjgb to limit accumulation of NO and maintain cell viability.
Sequence comparison of blr2806 with homologous proteins from diverse bacterial phyla suggests that this gene encodes an MFS-type NO3−/NO2− antiporter with similarity to E. coli NarK. The capacity of a B. japonicum narK mutant to accumulate NO2− inside the cell demonstrates the involvement of NarK in NO2− export. Further, phenotypic analyses reveal that NarK is not the main system for cytoplasmic NO3− import, as narK cells were still able to grow on NO3−. Instead, the narK mutant showed enhanced growth compared to WT cells with NO3− as sole N-source, either under aerobic or anaerobic conditions. These observations imply that NarK acts to lower cytoplasmic NO2− levels by exporting NO2− to the periplasm and this process may involve corresponding import of NO3− (Figure 7) [40]. In this respect, it is significant that B. japonicum NarK performs a very different role to the MFS-type NO3−/NO2− transporter NasA, which supplies NO3− to the cytoplasmic NO3−/NO2− reductase pathway in other α-proteobacteria [23]. Instead, by counteracting NO2− accumulation, the B. japonicum NarK protein may thus represent a first level of protection to mitigate the production of cytotoxic NO, by adventitious reduction of NO2− within the cytoplasm [43]. However, as a consequence, in WT cells NarK may also lower substrate availability for NirA and thus limit growth on NO3−.
Deletion of blr2803–05 that bioinformatics analyses had predicted to collectively encode an NrtABC family transporter, did not affect the ability of B. japonicum to assimilate NO3− as sole N-source. Therefore, the main route(s) for assimilatory NO3− import remains to be established. Although blr2803–05 are not required for NO3− assimilation, there are other NtrABC-like candidates present on the chromosome (e.g. bll5732–34) that may facilitate NO3− import to the cytoplasm.
A modular detoxification system for NO generated during NO3− assimilation
In general, Nas systems have a high degree of structural plasticity, yet most contain proteins for transport and reduction of NO3− and NO2− [23,25,45,47,48]. In the present work, a novel NO3− assimilation system that also includes proteins for NO-detoxification is reported. The narK-bjgb-flp-nasC operon in B. japonicum encodes the sdHb Bjgb [3,20], which is homologous to the N-terminal haem-containing domain of E. coli FHb (Hmp) as well as the sdHbs from Vitreoscilla stercoraria (Vgb) and Campilobacter jejuni (Cgb) [20].
Deletion of bjgb had a strong negative affect on O2-limited growth with NO3− as sole N-source, relative to WT, which implies a role for Bjgb in protecting B. japonicum cells from nitrosative stress. Importantly, in the absence of Bjgb, NO3− respiring cells were also highly sensitive to exogenous NO. Since growth of the bjgb mutant was not affected under aerobic conditions, the role of Bjgb may be restricted to anaerobic NO3−-dependent growth. However, our data suggest that the contribution of Bjgb to N2O production in vivo is low. These observations are consistent with studies performed in E. coli, which reveal Hmp can reduce NO to N2O under anaerobic conditions, but with a much lower rate compared with the activity of the FlRd NorV [44]. Furthermore, expression of the respiratory NorCB is significantly up-regulated in the bjgb mutant, relative to WT (see Figure 6), in response to increased intracellular NO levels that arise during NO3−-dependent growth. This result suggests that increased NorCB expression may counteract accumulation of cytotoxic NO and may partially compensate for the absence of the cytoplasmic Bjgb NO-detoxification system to maintain cell viability, albeit with a detrimental impact on anaerobic growth. Consequently, the bulk of the N2O produced by the bjgb mutant can be attributed to NorCB activity, which is increased by ∼2-fold relative to WT levels.
In E. coli Hmp, the FAD prosthetic group within the C-terminal NADH-reductase domain provides electrons from NAD(P)H that are required to reduce the NO-bound haem active site and complete the catalytic cycle. Aside from NO dioxygenation, Hmp has also been shown to perform slower reduction of NO to N2O under anoxic conditions, which operates at approximately 1% of the rate observed for aerobic dioxygenase activity [49–52]. In the case of Cgb (an sdHb family protein that like Bjgb lacks the reductase domain present in the FHb Hmp), the electron–donor protein remains to be identified. However, recent heterologous expression studies of Cgb in E. coli have reported a minor role for the NADH-(flavo)rubredoxin oxidoreductase NorW [53]. In B. japonicum, the enhanced sensitivity of the flp mutant to chemical NO-donors suggests that Flp may supply electrons from NAD(P)H that are required for Bjgb activity (Figure 7).
Sources of NO: NasC and NapA activity is responsible for elevated NorCB expression
In eukaryotes, NO synthase (NOS) enzymes have been well described as the main NO-forming pathway for cell signalling and anti-microbial host defence [54]. By contrast, NO-formation in prokaryotes has been considered a by-product of denitrification, anaerobic ammonium oxidation and other related respiratory pathways [55–58]. However, NO is now increasingly recognized as a key substrate for ‘non-respiratory’ pathways in bacteria, e.g. those that protect against nitrosative stress and the link between NO-detoxification and pathogenicity has been the focus of several studies (reviewed by Maia and Moura [56,59]). The biochemical basis for NO-formation during anaerobic bacterial respiration has been shown to result from enzymic reduction of the pseudo-substrate NO2− by the respiratory membrane-bound NO3− reductase, Nar [43,60,61]. Furthermore, a small contribution (less than 3%) has been attributed to the periplasmic enzyme, Nap [43,61]. In the context of this present study, the potential contribution of cytoplasmic NO2− reduction to NO formation, by NasC, during NO3−/NO2− assimilation has not yet been investigated.
In the denitrifying endosymbiotic bacterium B. japonicum, reduction of NO2− by the periplasmic copper-dependent NO2− reductase NirK is the main NO-forming process, which occurs during anaerobic NO3− respiration [1,42]. Many studies have proposed that NO activates transcription of nor genes and that this control is mediated by regulatory proteins designated NNR/NnrR and DNR (reviewed by Spiro [18,62]). In the present study, we demonstrate that cells lacking the periplasmic respiratory NO3− reductase NapA, where NO synthesis from denitrification is blocked, results in very low expression of NorCB. Perhaps our most important finding was that, in addition to NapA, the assimilatory NO3− reductase (NasC) is also responsible for generating NO, as induction of NorCB was significantly lowered and completely abolished, relative to WT, in the nasC and nasC;napA mutant strains respectively (Figure 6). Therefore, the importance of NasC not only in NO3− assimilation but also in NO production has been demonstrated.
Co-expression of bjgb, flp and nasC that constitute a combined NO3− assimilation/NO-detoxification system may represent a novel method by which bacteria maintain cytoplasmic NO homeostasis and protect against nitrosative stress imposed during NO3−-dependent growth, where pathways for both respiratory denitrification and NO3−/NO2− assimilation are active (Figure 7). Although co-regulation between similar NO-forming and consuming systems has been proposed in Aspergillus nidulans [63], to our knowledge, this is the first time where this mechanism has been reported in bacteria. Finally, should production of NO exceed concentrations that can be contained by Bjgb-Flp, a ‘safety’ mechanism exists to enhance expression of NorCB to drive reduction of excess NO to N2O.
Acknowledgments
We thank Dr Hans-Martin Fischer (Institute of Microbiology, ETH Zürich, Switzerland) for providing the pSUP3535 plasmid.
Abbreviations
- Bjgb
Bradyrhizobium japonicum haemoglobin
- FHb
flavohaemoglobin
- Flp
flavoprotein
- FlRd
flavorubredoxin
- MFS
major facilitator superfamily
- MV-NIR
methyl viologen-dependent nitrite reductase
- MV-NR
methyl viologen-dependent nitrate reductase
- NapABC
periplasmic respiratory quinol/nitrate oxidoreductase
- NarK
nitrate/nitrite transporter
- Nas
assimilatory nitrate reductase
- NirA
assimilatory nitrite reductase
- NirK
copper-dependent respiratory nitrite reductase
- NorCB
cytochrome-c nitric oxide reductase
- NosZ
nitrous oxide reductase
- PSY
peptone-salts-yeast extract
- sdHb
single-domain haemoglobin
- SNP
sodium nitroprusside
- WT
wild-type
- μmax (app)
apparent maximum growth rate
AUTHOR CONTRIBUTION
Juan Cabrera, Ana Salas, María Torres, Eulogio Bedmar, David Richardson, Andrew Gates and María Delgado designed the research. Juan Cabrera, Ana Salas, María Torres, Andrew Gates and María Delgado performed the research. Juan Cabrera, Ana Salas, David Richardson, Andrew Gates and María Delgado analysed the data. Andrew Gates and María Delgado wrote the manuscript.
FUNDING
This work was supported by European Regional Development Fund (ERDF) co-financed grants from Ministerio de Economía y Competitividad, Spain [grant numbers: AGL2010-18607 and AGL2013-45087-R (to M.J.D.)]; the Junta de Andalucía [grant number PE2012-AGR1968 (to E.J.B.)]; the Biotechnology and Biological Sciences Research Council [grant number BB/M00256X/1 (to A.J.G.)]; and the Royal Society International Exchanges Programme, U.K. [grant number IE140222 (to A.J.G. and M.J.D.)]. J.J.C. was supported by a fellowship from the Consejo Superior de Investigaciones Científicas (CSIC) JAE programme. D.J.R. is a Royal Society Wolfson Foundation Merit Award holder.
References
- 1.Bedmar E.J., Robles E.F., Delgado M.J. The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum. Biochem. Soc. Trans. 2005;33:141–144. doi: 10.1042/BST0330141. [DOI] [PubMed] [Google Scholar]
- 2.Delgado M.J., Casella S., Bedmar E.J. Denitrification in rhizobia-legume symbiosis. In: Bothe H., Ferguson S.J., Newton W.E., editors. Biology of the Nitrogen Cycle. Amsterdam: Elsevier; 2007. pp. 57–66. [Google Scholar]
- 3.Sánchez C., Cabrera J.J., Gates A.J., Bedmar E.J., Richardson D.J., Delgado M.J. Nitric oxide detoxification in the rhizobia-legume symbiosis. Biochem. Soc. Trans. 2011;39:184–188. doi: 10.1042/BST0390184. [DOI] [PubMed] [Google Scholar]
- 4.Bedmar E.J., Bueno E., Correa D., Torres M.J., Delgado M.J., Mesa S. Ecology of denitrification in soils and plant-associated bacteria. In: González M.B.R., Gonzalez-López J., editors. Beneficial Plant-Microbial Interactions: Ecology and Applications. Florida: CRC Press; 2013. pp. 164–182. [Google Scholar]
- 5.Horchani F., Prévot M., Boscari A., Evangelisti E., Meilhoc E., Bruand C., Raymond P., Boncompagni E., Aschi-Smiti S., Puppo A., Brouquisse R. Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 2011;155:1023–1036. doi: 10.1104/pp.110.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inaba S., Ikenishi F., Itakura M., Kikuchi M., Eda S., Chiba N., Katsuyama C., Suwa Y., Mitsui H., Minamisawa K. N2O emission from degraded soybean nodules depends on denitrification by Bradyrhizobium japonicum and other microbes in the rhizosphere. Microbes Environ. 2012;27:470–476. doi: 10.1264/jsme2.ME12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meakin G.E., Bueno E., Jepson B., Bedmar E.J., Richardson D.J., Delgado M.J. The contribution of bacteroidal nitrate and nitrite reduction to the formation of nitrosylleghaemoglobin complexes in soybean root nodules. Microbiology. 2007;153:411–419. doi: 10.1099/mic.0.2006/000059-0. [DOI] [PubMed] [Google Scholar]
- 8.Sánchez C., Gates A.J., Meakin G.E., Uchiumi T., Girard L., Richardson D.J., Bedmar E.J., Delgado M.J. Production of nitric oxide and nitrosylleghemoglobin complexes in soybean nodules in response to flooding. Mol. Plant Microbe Interact. 2010;23:702–711. doi: 10.1094/MPMI-23-5-0702. [DOI] [PubMed] [Google Scholar]
- 9.Bates B.C., Kundzewicz S., Wu J., Palutikof P. Technical Paper of the Intergovernmental Panel on Climate Change. Geneva: IPCC Secretariat; 2008. Climate change and water; p. 210. [Google Scholar]
- 10.Crutzen P.J., Mosier A.R., Smith K.A., Winiwarter W. N2O release from agro-biofuel production negates global warming reduction by replacing fossil fuels. Atmos. Chem. Phys. 2008;8:389–395. doi: 10.5194/acp-8-389-2008. [DOI] [Google Scholar]
- 11.Ravishankara A.R., Daniel J.S., Portmann R.W. Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- 12.Kato K., Kanahama K., Kanayama Y. Involvement of nitric oxide in the inhibition of nitrogenase activity by nitrate in Lotus root nodules. J. Plant Physiol. 2010;167:238–241. doi: 10.1016/j.jplph.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Meakin G.E., Jepson B.J., Richardson D.J., Bedmar E.J., Delgado M.J. The role of Bradyrhizobium japonicum nitric oxide reductase in nitric oxide detoxification in soya bean root nodules. Biochem. Soc. Trans. 2006;34:195–196. doi: 10.1042/BST0340195. [DOI] [PubMed] [Google Scholar]
- 14.Mills P.C., Rowley G., Spiro S., Hinton J.C., Richardson D.J. A combination of cytochrome c nitrite reductase (NrfA) and flavorubredoxin (NorV) protects Salmonella enterica serovar Typhimurium against killing by NO in anoxic environments. Microbiology. 2008;154:1218–1228. doi: 10.1099/mic.0.2007/014290-0. [DOI] [PubMed] [Google Scholar]
- 15.Pittman M.S., Elvers K.T., Lee L., Jones M.A., Poole R.K., Park S.F., Kelly D.J. Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol. Microbiol. 2007;63:575–590. doi: 10.1111/j.1365-2958.2006.05532.x. [DOI] [PubMed] [Google Scholar]
- 16.Poole R.K. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 2005;33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 17.Pullan S.T., Monk C.E., Lee L., Poole R.K. Microbial responses to nitric oxide and nitrosative stress: growth, “omic,” and physiological methods. Methods Enzymol. 2008;437:499–519. doi: 10.1016/S0076-6879(07)37025-0. [DOI] [PubMed] [Google Scholar]
- 18.Spiro S. Nitric oxide metabolism: physiology and regulatory mechanisms. In: Moir J.W.B., editor. Nitrogen Cycling in Bacteria: Molecular Analysis. Norfolk: Caister Academic Press; 2011. pp. 177–196. [Google Scholar]
- 19.Kaneko T., Nakamura Y., Sato S., Minamisawa K., Uchiumi T., Sasamoto S., Watanabe A., Idesawa K., Iriguchi M., Kawashima K., et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002;9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 20.Cabrera J.J., Sánchez C., Gates A.J., Bedmar E.J., Mesa S., Richardson D.J., Delgado M.J. The nitric oxide response in plant-associated endosymbiotic bacteria. Biochem. Soc. Trans. 2011;39:1880–1885. doi: 10.1042/BST20110732. [DOI] [PubMed] [Google Scholar]
- 21.Luque-Almagro V.M., Lyall V.J., Ferguson S.J., Roldán M.D., Richardson D.J., Gates A.J. Nitrogen oxyanion-dependent dissociation of a two-component complex that regulates bacterial nitrate assimilation. J. Biol. Chem. 2013;288:29692–29702. doi: 10.1074/jbc.M113.459032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sánchez C., Itakura M., Okubo T., Matsumoto T., Yoshikawa H., Gotoh A., Hidaka M., Uchida T., Minamisawa K. The nitrate-sensing NasST system regulates nitrous oxide reductase and periplasmic nitrate reductase in Bradyrhizobium japonicum. Environ. Microbiol. 2014;16:3263–3274. doi: 10.1111/1462-2920.12546. [DOI] [PubMed] [Google Scholar]
- 23.Gates A.J., Luque-Almagro V.M., Goddard A.D., Ferguson S.J., Roldán M.D., Richardson D.J. A composite biochemical system for bacterial nitrate and nitrite assimilation as exemplified by Paracoccus denitrificans. Biochem. J. 2011;435:743–753. doi: 10.1042/BJ20101920. [DOI] [PubMed] [Google Scholar]
- 24.Pino C., Olmo-Mira F., Cabello P., Martínez-Luque M., Castillo F., Roldán M.D., Moreno-Vivián C. The assimilatory nitrate reduction system of the phototrophic bacterium Rhodobacter capsulatus E1F1. Biochem. Soc. Trans. 2006;34:127–129. doi: 10.1042/BST0340127. [DOI] [PubMed] [Google Scholar]
- 25.Luque-Almagro V.M., Gates A.J., Moreno-Vivián C., Ferguson S.J., Richardson D.J., Roldán M.D. Bacterial nitrate assimilation: gene distribution and regulation. Biochem. Soc. Trans. 2011;39:1838–1843. doi: 10.1042/BST20110688. [DOI] [PubMed] [Google Scholar]
- 26.Delgado M.J., Bonnard N., Tresierra-Ayala A., Bedmar E.J., Muller P. The Bradyrhizobium japonicum napEDABC genes encoding the periplasmic nitrate reductase are essential for nitrate respiration. Microbiology. 2003;149:3395–3403. doi: 10.1099/mic.0.26620-0. [DOI] [PubMed] [Google Scholar]
- 27.Robles E.F., Sanchez C., Bonnard N., Delgado M.J., Bedmar E.J. The Bradyrhizobium japonicum napEDABC genes are controlled by the FixLJ-FixK2-NnrR regulatory cascade. Biochem. Soc. Trans. 2006;34:108–110. doi: 10.1042/BST0340108. [DOI] [PubMed] [Google Scholar]
- 28.Regensburger B., Hennecke H. RNA polymerase from Rhizobium japonicum. Arch. Microbiol. 1983;135:103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 29.Bergersen F.J. A treatise on dinitrogen fixation. In: Hardy R.W., Silver W., editors. Biology: Section III. New York: John Wiley & Sons; 1977. pp. 519–556. [Google Scholar]
- 30.Torres M.J., Argandoña M., Vargas C., Bedmar E.J., Fischer H.M., Mesa S., Delgado M.J. The global response regulator RegR controls expression of denitrification genes in Bradyrhizobium japonicum. PLoS One. 2014;9:e99011. doi: 10.1371/journal.pone.0099011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torres M.J., Bueno E., Mesa S., Bedmar E.J., Delgado M.J. Emerging complexity in the denitrification regulatory network of Bradyrhizobium japonicum. Biochem. Soc. Trans. 2011;39:284–288. doi: 10.1042/BST0390284. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Russell D. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor; 2001. [Google Scholar]
- 33.Simon R., Priefer U., Pühler A. Vector plasmids for in-vivo and in-vitro manipulation of gram-negative bacteria. In: Pühler A., editor. Molecular genetics of the bacteria-plant interaction. Berlin: Springer-Verlag; 1983. pp. 98–106. [DOI] [Google Scholar]
- 34.Mesa S., Velasco L., Manzanera M.E., Delgado M.J., Bedmar E.J. Characterization of the norCBQD genes, encoding nitric oxide reductase, in the nitrogen fixing bacterium Bradyrhizobium japonicum. Microbiology. 2002;148:3553–3560. doi: 10.1099/00221287-148-11-3553. [DOI] [PubMed] [Google Scholar]
- 35.Egelhoff T.T., Fisher R.F., Jacobs T.W., Mulligan J.T., Long S.R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA. 1985;4:241–248. doi: 10.1089/dna.1985.4.241. [DOI] [PubMed] [Google Scholar]
- 36.Hauser F., Pessi G., Friberg M., Weber C., Rusca N., Lindemann A., Fischer H.M., Hennecke H. Dissection of the Bradyrhizobium japonicum NifA+σ54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol. Genet. Genomics. 2007;278:255–271. doi: 10.1007/s00438-007-0246-9. [DOI] [PubMed] [Google Scholar]
- 37.Miller J.H. Experiments in Molecular Genetics. New York: Cold Spring Harbor; 1972. [Google Scholar]
- 38.Vargas C., McEwan A.G., Downie J.A. Detection of c-type cytochromes using enhanced chemiluminescence. Anal. Biochem. 1993;209:323–326. doi: 10.1006/abio.1993.1127. [DOI] [PubMed] [Google Scholar]
- 39.Nicholas D.J.D., Nason A. Determination of nitrate and nitrite. In: Colowick S.P., Kaplan N.O., editors. Methods in Enzymology. New York: Academic Press; 1957. pp. 981–984. [DOI] [Google Scholar]
- 40.Fukuda M., Takeda H., Kato H.E., Doki S., Ito K., Maturana A.D., Ishitani R., Nureki O. Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK. Nat. Commun. 2015;6:7097. doi: 10.1038/ncomms8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goddard A.D., Moir J.W., Richardson D.J., Ferguson S.J. Interdependence of two NarK domains in a fused nitrate/nitrite transporter. Mol. Microbiol. 2008;70:667–681. doi: 10.1111/j.1365-2958.2008.06436.x. [DOI] [PubMed] [Google Scholar]
- 42.Velasco L., Mesa S., Delgado M.J., Bedmar E.J. Characterization of the nirK gene encoding the respiratory, Cu-containing nitrite reductase of Bradyrhizobium japonicum. Biochim. Biophys. Acta. 2001;1521:130–134. doi: 10.1016/S0167-4781(01)00279-2. [DOI] [PubMed] [Google Scholar]
- 43.Rowley G., Hensen D., Felgate H., Arkenberg A., Appia-Ayme C., Prior K., Harrington C., Field S.J., Butt J.N., Baggs E., Richardson D.J. Resolving the contributions of the membrane-bound and periplasmic nitrate reductase systems to nitric oxide and nitrous oxide production in Salmonella enterica serovar Typhimurium. Biochem. J. 2012;441:755–762. doi: 10.1042/BJ20110971. [DOI] [PubMed] [Google Scholar]
- 44.Vine C.E., Cole J.A. Nitrosative stress in Escherichia coli: reduction of nitric oxide. Biochem. Soc. Trans. 2011;39:213–215. doi: 10.1042/BST0390213. [DOI] [PubMed] [Google Scholar]
- 45.Moreno-Vivián C., Flores E. Nitrate assimilation in bacteria. In: Bothe H., Ferguson S.J., Newton W.E., editors. Biology of the Nitrogen Cycle. Amsterdam: Elsevier; 2007. pp. 263–292. [DOI] [Google Scholar]
- 46.Franck W.L., Qiu J., Lee H.I., Chang W.S., Stacey G. DNA microarray-based identification of genes regulated by NtrC in Bradyrhizobium japonicum. Appl. Environ. Microbiol. 2015;81:5299–5308. doi: 10.1128/AEM.00609-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin J.T., Stewart V. Nitrate assimilation by bacteria. Adv. Microb. Physiol. 1998;39:379. doi: 10.1016/s0065-2911(08)60014-4. [DOI] [PubMed] [Google Scholar]
- 48.Moreno-Vivián C., Luque-Almagro V.M., Cabello P., Roldán M.D., Castillo F. Transport and assimilation of inorganic nitrogen in bacteria. In: Moir J.W.B., editor. Nitrogen Cycling in Bacteria. Norfolk: Caister Academic Press; 2011. pp. 101–122. [Google Scholar]
- 49.Angelo M., Hausladen A., Singel D.J., Stamler J.S. Interactions of NO with hemoglobin: from microbes to man. Methods Enzymol. 2008;436:131–168. doi: 10.1016/S0076-6879(08)36008-X. [DOI] [PubMed] [Google Scholar]
- 50.Gardner P.R. Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases. J. Inorg. Biochem. 2005;99:247–266. doi: 10.1016/j.jinorgbio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Hernández-Urzúa E., Mills C.E., White G.P., Contreras-Zentella M.L., Escamilla E., Vasudevan S.G., Membrillo-Hernández J., Poole R.K. Flavohemoglobin Hmp, but not its individual domains, confers protection from respiratory inhibition by nitric oxide in Escherichia coli. J. Biol. Chem. 2003;278:34975–34982. doi: 10.1074/jbc.M303629200. [DOI] [PubMed] [Google Scholar]
- 52.Kim S.O., Orii Y., Lloyd D., Hughes M.N., Poole R.K. Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 1999;445:389–394. doi: 10.1016/S0014-5793(99)00157-X. [DOI] [PubMed] [Google Scholar]
- 53.Tinajero-Trejo M., Vreugdenhil A., Sedelnikova S.E., Davidge K.S., Poole R.K. Nitric oxide reactivities of the two globins of the foodborne pathogen Campylobacter jejuni: roles in protection from nitrosative stress and analysis of potential reductants. Nitric Oxide. 2013;34:65–75. doi: 10.1016/j.niox.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Alderton W.K., Cooper C.E., Knowles R.G. Nitric oxide synthases: structure, function and inhibition. Biochem. J. 2001;357:593–615. doi: 10.1042/bj3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kartal B., van Niftrik L., Keltjens J.T., Op den Camp H.J., Jetten M.S. Anammox-growth physiology, cell biology, and metabolism. Adv. Microb. Physiol. 2012;60:211–262. doi: 10.1016/B978-0-12-398264-3.00003-6. [DOI] [PubMed] [Google Scholar]
- 56.Maia L.B., Moura J.J. How biology handles nitrite. Chem. Rev. 2014;114:5273–5357. doi: 10.1021/cr400518y. [DOI] [PubMed] [Google Scholar]
- 57.Rinaldo S., Cutruzzolá F. Nitrate reductases in denitrification. In: Bothe H., Ferguson S.J., Newton W.E., editors. Biology of the Nitrogen Cycle. Amsterdam: Elsevier Science; 2007. pp. 37–55. [DOI] [Google Scholar]
- 58.Zumft W.G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maia L.B., Moura J.J. Nitrite reduction by molybdoenzymes: a new class of nitric oxide-forming nitrite reductases. J. Biol. Inorg. Chem. 2015;20:403–433. doi: 10.1007/s00775-014-1234-2. [DOI] [PubMed] [Google Scholar]
- 60.Gilberthorpe N.J., Poole R.K. Nitric oxide homeostasis in Salmonella typhimurium: roles of respiratory nitrate reductase and flavohemoglobin. J. Biol. Chem. 2008;283:11146–11154. doi: 10.1074/jbc.M708019200. [DOI] [PubMed] [Google Scholar]
- 61.Vine C.E., Purewal S.K., Cole J.A. NsrR-dependent method for detecting nitric oxide accumulation in the Escherichia coli cytoplasm and enzymes involved in NO production. FEMS Microbiol. Lett. 2011;325:108–114. doi: 10.1111/j.1574-6968.2011.02385.x. [DOI] [PubMed] [Google Scholar]
- 62.Spiro S. Nitrous oxide production and consumption: regulation of gene expression by gas-sensitive transcription factors. Philos. Trans. R Soc. Lond. B Biol. Sci. 2012;367:1213–1225. doi: 10.1098/rstb.2011.0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schinko T., Berger H., Lee W., Gallmetzer A., Pirker K., Pachlinger R., Buchner I., Reichenauer T., Guldener U., Strauss J. Transcriptome analysis of nitrate assimilation in Aspergillus nidulans reveals connections to nitric oxide metabolism. Mol. Microbiol. 2010;78:720–738. doi: 10.1111/j.1365-2958.2010.07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]