ABSTRACT
Oil reservoirs are major sites of methane production and carbon turnover, processes with significant impacts on energy resources and global biogeochemical cycles. We applied a cultivation-independent genomic approach to define microbial community membership and predict roles for specific organisms in biogeochemical transformations in Alaska North Slope oil fields. Produced water samples were collected from six locations between 1,128 m (24 to 27°C) and 2,743 m (80 to 83°C) below the surface. Microbial community complexity decreased with increasing temperature, and the potential to degrade hydrocarbon compounds was most prevalent in the lower-temperature reservoirs. Sulfate availability, rather than sulfate reduction potential, seems to be the limiting factor for sulfide production in some of the reservoirs under investigation. Most microorganisms in the intermediate- and higher-temperature samples were related to previously studied methanogenic and nonmethanogenic archaea and thermophilic bacteria, but one candidate phylum bacterium, a member of the Acetothermia (OP1), was present in Kuparuk sample K3. The greatest numbers of candidate phyla were recovered from the mesothermic reservoir samples SB1 and SB2. We reconstructed a nearly complete genome for an organism from the candidate phylum Parcubacteria (OD1) that was abundant in sample SB1. Consistent with prior findings for members of this lineage, the OD1 genome is small, and metabolic predictions support an obligately anaerobic, fermentation-based lifestyle. At moderate abundance in samples SB1 and SB2 were members of bacteria from other candidate phyla, including Microgenomates (OP11), Atribacteria (OP9), candidate phyla TA06 and WS6, and Marinimicrobia (SAR406). The results presented here elucidate potential roles of organisms in oil reservoir biological processes.
IMPORTANCE
The activities of microorganisms in oil reservoirs impact petroleum resource quality and the global carbon cycle. We show that bacteria belonging to candidate phyla are present in some oil reservoirs and provide the first insights into their potential roles in biogeochemical processes based on several nearly complete genomes.
INTRODUCTION
Study of the deep subsurface has been limited by access to samples suitable for microbial characterization. However, the infrastructure and sampling techniques developed for oil and gas exploration and recovery enable investigations of deep subsurface life. Petroleum reservoirs are unusual environments due to their combinations of very high carbon compound concentrations, elevated temperatures in deeper locations, and long history of separation from surface inputs (although this changes as soon as the reservoir is drilled). The metabolic capacities of individual organisms in these environments, their survival strategies, and intracommunity interactions may influence some chemical and physical characteristics of oil reservoirs. The types of organisms and their roles in hydrocarbon transformations likely vary over the range of different environmental conditions that occur in oil fields. For example, certain microorganisms degrade short-chain hydrocarbons (1–5), converting light to heavy crude oil that is harder to recover. Heavy oils yield less gasoline and diesel fuel and have a more negative impact on the environment during refining (6).
Natural gas from petroleum reservoirs primarily consists of methane, with small amounts of alkanes, carbon dioxide, nitrogen, and hydrogen sulfide. When sulfate or other sulfur compounds are present, sulfidogenic bacteria and archaea can produce hydrogen sulfide (7). Overall, microbial production of H2S leads to petroleum reservoir souring and has significant economic impacts, in part related to worker health and pipeline corrosion (3, 4, 8, 9). From the perspective of oil field management, understanding reservoir microbiology, as well as processes that minimize the activities of H2S-producing bacteria and archaea, may have important long-term economic benefits.
There have been many studies of microbial consortia in oil field environments (10–34). These have used culture-based (16–21), 16S rRNA gene-based culture-independent (10, 20, 22–33), and metagenomic (10, 27, 30, 34) methods. A prior 16S rRNA gene-based PhyloChip study that examined the Alaska North Slope oil field samples studied here identified organisms that may contribute to methane and hydrogen sulfide production and hydrocarbon degradation (32). Although a number of organisms from lineages lacking cultivated representatives were identified, the full diversity and functional capacities of these organisms remained uncertain. Prior metagenomic analyses of microbial community composition from other systems involved extraction and sequencing of genomic DNA of coexisting organisms (10, 30, 34). Two of these investigations applied relatively small-scale (<1 Gbp) DNA sequencing to two samples from an oil reservoir on the Norwegian Continental Shelf. The authors identified sulfate-reducing bacteria, methanogenic archaea, and fermentative bacteria and concluded that genetically similar organisms occurred in both samples, although at different abundance levels. An et al. investigated diverse hydrocarbon-containing samples, and obtained 10 small (<1 Gbp, from various locations) and 2 larger (>1 Gbp, from coal beds) metagenomic datasets, including one small-scale library from a cool (30°C) and shallow (850-m) oil reservoir in Alberta, Canada. The authors found a surprisingly high proportion of genes for enzymes involved in aerobic hydrocarbon metabolism in several samples, while there were more genes for anaerobic hydrocarbon metabolism and methanogenesis in the oil reservoir sample (34). A recent study sequenced fosmid libraries to analyze hydrocarbon degradation pathways in an enrichment culture (35), and an older study (27) used both 16S rRNA gene amplicon sequencing and fosmid library sequencing to investigate produced water from a mesothermic petroleum reservoir.
In this study, we used genome-resolved metagenomic analyses of gigabase-pair-scale sequence datasets for six samples from two North Slope oil fields in Alaska. Compared to 16S rRNA gene profiling, metagenomic analysis provides information about potential microbial physiology. The method also has significantly higher taxonomic resolution, capturing species- and strain-level variants present in natural communities. Furthermore, the approach can detect organisms whose 16S rRNA gene sequences escape detection due, for example, to primer mismatch (36). Our objectives were to identify the organisms in each sample, to compare samples across the range of physical and chemical conditions and to predict metabolic roles based on de novo recovery of draft genomes for the more abundant organisms. Included within the analysis were samples from different depths and temperatures, with or without hydrogen sulfide production (souring), that had or had not been impacted by seawater injection. The results clearly differentiated consortia from different sampling sites, revealed potential metabolic processes, and uncovered potential roles for candidate phyla in biogeochemical transformations in petroleum reservoirs.
RESULTS AND DISCUSSION
Temperature is one of the key factors that shape the community.
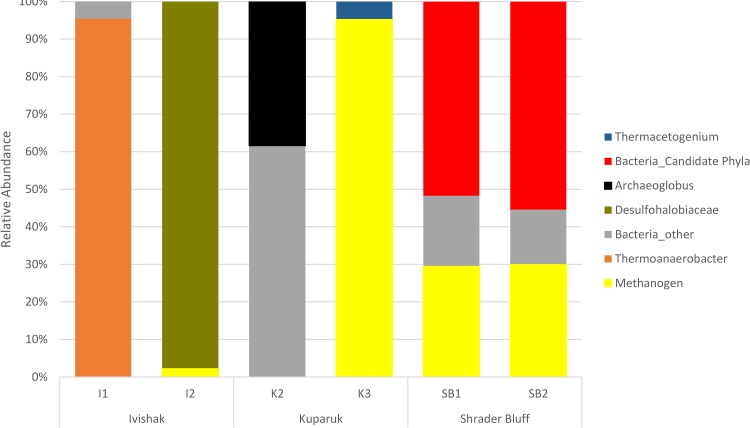
Fifty-seven unique 16S rRNA gene sequences (with a minimum length of 960 bp) were reconstructed using EMIRGE. The results indicate the presence of both bacteria and archaea in samples SB1, SB2, K2, K3, and I2 but only bacteria in I1 (Fig. 1 and 2). The highest-temperature Ivishak samples (I1 and I2) are each dominated by one organism (>95% relative abundance) (Fig. 1; also, see Fig. S1A and B in the supplementl material), Thermoanaerobacter and Desulfonauticus sp. (Desulfohalobiaceae), respectively. The number of nearly full-length sequences recovered by EMIRGE for samples K2 and K3 is low (three to four sequences each). In contrast, many more sequences were recovered from SB1 (28 sequences) and SB2 (18 sequences).
FIG 1 .
Dominant microbial community member groups and their relative abundances based on full-length 16S rRNA gene sequences (>960 bp) reconstructed by using EMIRGE (76). Currently named bacterial phyla were grouped in Bacteria_other (grey), whereas candidate phyla are shown separately (red). The category “methanogen” (yellow) includes several archaeal sequences affiliated with various methanogens.
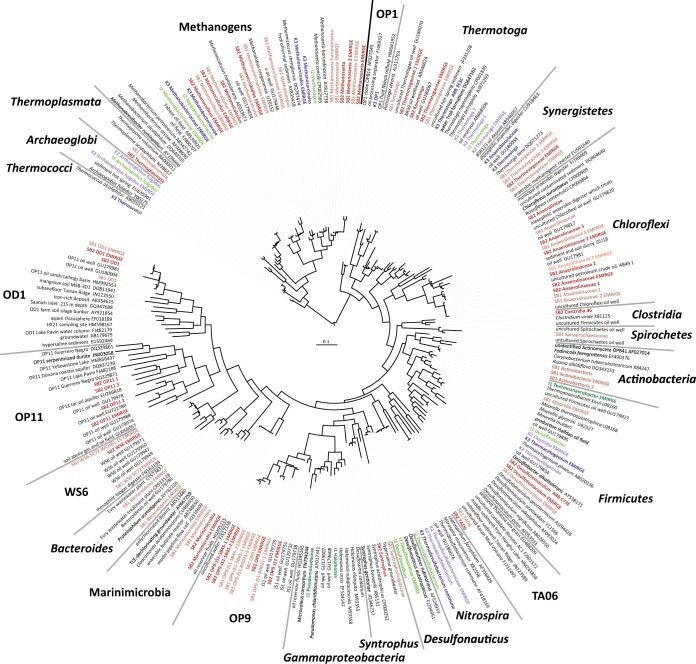
From all of the assembled datasets, we recovered 30 full-length and 30 partial 16S rRNA gene sequences (>300 bp) from bacteria and archaea, including many from organisms in candidate phyla. The sequences from candidate phyla primarily derived from the Schrader Bluff genomic datasets. Specifically, we recovered sequences from Marinimicrobia (SAR406, class AB16), Parcubacteria (OD1), candidate phylum TA06 (37), Atribacteria (OP9), candidate phylum WS6, and Microgenomates (OP11). However, one full-length 16S rRNA gene of Acetothermia (OP1) was recovered from K3. The overall community structures, based on the combined EMIRGE and contig 16S rRNA gene analysis (Fig. 2), are in agreement with PhyloChip results reported previously for the same samples (32). However, the prior study failed to detect archaea in Ivishak samples (I1 and I2) using their standard PCR conditions (primers 4Fa and 1492R, 2 µl template, 30 cycles). Additionally, bacteria in the candidate phyla OP1 (Kuparuk), OP11 (Schrader Bluff), and TA06 (Schrader Bluff) were not reported. There were no sequences classified as OP1 or TA06 in the taxonomy used when probes were designed for the G3 PhyloChip, and there was only one sequence noted as being included in a “former candidate division OP11,” but that operational taxonomic unit (OTU) was not detected in the data set.
FIG 2 .
Phylogenetic tree (constructed using the RAxML method [84] in the ARB software package [85]) illustrating microbial communities represented by 16S rRNA genes. The sequences most closely related to those recovered in this study were included from the Silva database (86). Sequences recovered from Ivishak, Kuparuk, and Schrader Bluff samples were colored in groups of green, red, and purple, respectively (dark green, sample I1; light green, sample I2; dark red, sample K2; light red, sample K3; dark purple, sample SB1; light purple, sample SB2). Closely related reference sequences are in black.
The relative abundances of organisms (calculated based on the fraction of reads for any sample that were binned to specific genomes and normalized based on estimated genome size) also agreed with the observation that as temperature increased, the microbial community appeared to be less complex and dominated by a few organisms (see Fig. S1A to F in the supplemental material).
Inference of major biogeochemical functions and metabolite roles by recovered genomes.
Our analyses focused on 37,933 assembled genome fragments (scaffolds) >2,000 bp in length, a total of 227 Mbp of reconstructed genomic sequences (see Table S2 in the Text S2 file in the supplemental material). We recovered 3 to 50 draft (partial and nearly complete) genomes for bacteria and 0 to 13 draft genomes for archaea per sample (see Table S3 in the Text S2 file in the supplemental material). Subsequent analyses of the potential roles microbial community members played in different geologic formations and souring environments are based on these genomes (although many other scaffolds were assigned to plasmids and phage). Based on homology of conserved proteins, some genomes have very closely related sequences to the genomes in public databases (see Table S4 in the Text S2 file in the supplemental material).
(i) Ivishak Formation.
Samples I1 and I2 were both from 80 to 83°C Ivishak Formation produced water. I1 was not considered soured, whereas I2 was the most soured (200 mg/liter sulfide) of the samples analyzed in this study (see Table S1 in the Text S2 file in the supplemental material).
Consistent with the prediction from the 16S rRNA data (see above), almost 90% of the sequences from Ivishak sample I1 were assigned to two high-quality genomes and one partial genome. This sample was almost entirely dominated by a Thermoanaerobacter organism (Fig. 1; also, see Fig. S1A in the supplemental material). Based on the genome data, we identified the organism as Thermoanaerobacter thermocopriae (see Table S4 in the Text S2 file in the supplemental material). We expected that this highly dominant organism would have metabolic attributes consistent with the geochemistry of this well. The dissimilatory sulfate reduction pathway is absent from the reconstructed genome, consistent with I1 not being soured (see Table S1 in the Text S2 file in the supplemental material).
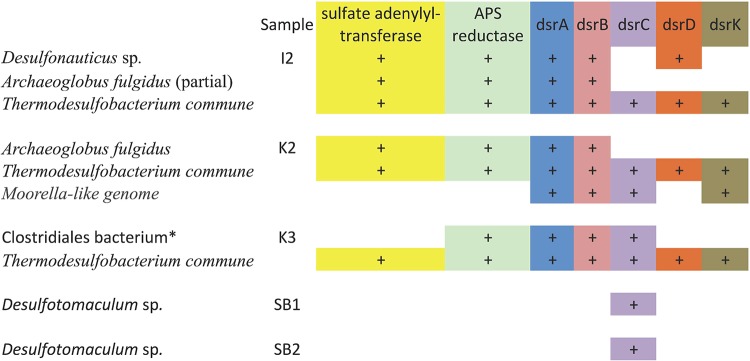
The Ivishak sample I2 also was highly dominated by one organism (Fig. 1; also, see Fig. S1B in the supplemental material), a Desulfonauticus sp. (the 16S rRNA gene is 99% identical to that of Desulfonautics autotrophicus DSM 4206, a thermophilic, sulfate-reducing organism isolated from oil production water [38]). Additionally, both methanogenic and nonmethanogenic archaea were present. Notably, a partial genome for Archaeoglobus fulgidus (not reported previously [32] because only bacteria were profiled for Ivishak samples in the previous study) was recovered. The 1,492-bp 16S rRNA gene from this organism is 99.46% identical to that of Archaeoglobus fulgidus. This soured sample has a longer history of seawater injection than I1, and the sulfate availability of I2 produced water was still high (611 mg/liter) (see Table S1 in the Text S2 file in the supplemental material) (32). There are three major sulfate reducers in this sample (Desulfonauticus, Thermodesulfobacterium, and Archaeoglobus), based on key enzymes in the dissimilatory sulfate reduction pathway predicted in the respective genomes (Fig. 3). Considering that Desulfonauticus is 63.2% in relative abundance and the other two genomes combined comprise <5%, we believe that Desulfonauticus is the principal source of H2S. Desulfonauticus has been isolated and its sequences recovered from oil fields previously (38, 39). It is a thermophilic (D. autotrophicus has a growth optimum of 58°C), halophilic, and sulfidogenic bacterium (38) which uses hydrogen and formate as electron donors [supported by the presence of non-F(420)-reducing hydrogenase and formate dehydrogenase in the binned genome (this study)] and a variety of sulfur compounds as electron acceptors (38). Given the sulfate reduction potential and abundance of sulfate, it makes sense that sample I2 was soured.
FIG 3 .
Key enzymes of the dissimilatory sulfate reduction pathway recovered in draft genomes (from metagenome data) from produced water samples collected from oil reservoirs in the Schrader Bluff Formation (SB1 and SB2), the Kuparuk Formation (K2 and K3), or the Ivishak Formation (I2) of the Alaska North Slope. APS, adenosine-5′-phosphosulfate; dsr, dissimilatory sulfite reductase. The asterisk indicates a genome that is likely for a member of the family Peptococcaceae, based on the marker enzymes (GyrA and ribosomal protein S3 are 71% and 82% identical to those of “Candidatus Desulforudis audaxviator”).
We evaluated genome bins in this sample for genes related to hydrogen cycling. Hydrogenases were found in 10 of 11 genomes recovered from sample I2, though it is generally uncertain whether these are involved in hydrogen production or oxidation. One Thermotoga organism (about 1% in relative abundance) possesses an Fe hydrogenase and therefore is a candidate for hydrogen production in this community.
Since the complete hydrocarbon degradation pathway overlaps other carbon catabolism pathways, we defined hydrocarbon degradation capability based on the presence of at least one of the genes in the activation phase or acting on aromatic substrates (enzymes listed in Table S5 in the Text S2 file in the supplemental material). By this criterion, no hydrocarbon degradation pathways were found in any of the genomes in the two Ivishak samples, consistent with oil from this reservoir being a lighter crude than from the shallower reservoirs and therefore indicating little biodegradation in situ.
A large inventory of glycosyl hydrolases was found in both I1 (Thermoanaerobacter) and I2 (several Thermotoga genomes). Oil reservoirs are not known to contain abundant polysaccharides, but these compounds may have been introduced in the drilling fluids. Cellulose is often used in these fluids to block the fluid leak-off into the rock, and polymers (usually Biozan, containing heteropolysaccharides) are used to increase the viscosity of drilling fluids so that rock cuttings and cellulose can be swept out of the well prior to cementation and oil production. The introduced polysaccharides may have promoted growth of organisms with glycosyl hydrolases (although the wells under study were drilled several years ago). Given that we do not know whether these enzymes are active (since we studied only DNA), it is not certain how much of the drilling influence is still seen today. In the reservoir, these enzymes also could be used for degradation of cell wall materials or may be used to take advantage of dissolved organic matter (DOM), likely brought in by the seawater flooding process. Ocean DOM is known to contain polysaccharides (40, 41), such as xylan from marine algae (42), and sugars, such as galactose and mannose (43). Harvey reported mono- and polysaccharide concentrations in seawater, including one study reporting 70 to 280 µg/liter monosaccharides and 160 to 225 µg/liter polysaccharides in summer months (44). Sakugawa and Handa reported mono-, oligo-, and polysaccharide concentrations of 4 to 100 µg/liter in north Pacific Ocean and Bering Sea water (45). We did not, however, analyze the injection water for sugars in this study. Additionally, genes potentially involved in polysaccharide degradation also exist in ocean microbial population (46). Various marine bacteria have been shown to have such activities (47–50).
(ii) Kuparuk Formation.
The Kuparuk samples (K2 and K3) have relatively low diversity, although they are not highly dominated by a single genotype, as were the Ivishak samples. Fourteen and sixteen partial genomes were reconstructed from these samples, respectively (see Fig. S1C and D in the supplemental material). It is interesting that the dominant organism in sample K2 is Archaeoglobus fulgidus, a well-known archaeon that is capable of H2S production (51) and is commonly found in oil reservoirs (1, 22, 52, 53). Sulfate reduction genes were found in Archaeoglobus fulgidus and Thermodesulfobacterium commune genomes (Fig. 3). Based on the community composition and predicted metabolic potential, we would have expected that the well from which sample K2 was collected would have been soured; however, at 14 mg/liter hydrogen sulfide, this well is not considered soured. The low sulfate concentration (1.4 mg/liter) of the water used to support secondary production (Table 2 in reference 32) is expected to limit the possibility of souring. Although sulfide oxidation, if it occurred in the reservoir, could keep sulfide concentrations low, we did not recover any genomes of known sulfide oxidizers, so it is not a plausible explanation, given the data.
We evaluated other possible electron donors and acceptors that may be used by Archaeoglobus. Its genome content supported the ability to use fatty acids, amino acids, and other small organic acids (51). A previous enrichment culture study demonstrated that Archaeoglobus is capable of oxidizing hydrogen (54), and a recent transcriptomic analysis suggested roles of fatty acid metabolism during growth with H2 (55). We speculate that in K2, Archaeoglobus uses organic acids with H2 as the electron donor, since many of the consortium members have the ability to produce H2. Notable in this regard is Thermococcus sibiricus (56). Additionally, previous studies showed that multiple species of Thermotoga can produce H2 (57, 58), and Fe hydrogenase genes, which are predicted to produce hydrogen in fermentative organisms, were recovered from four Thermotogaceae genomes in sample K2, including from a highly abundant (23%) species. Fe hydrogenase genes also were identified in one Caldanaerobacter genome in this study. While information on genomes alone is insufficient to determine for certain the active hydrogen donor(s), several members of these genera have been shown to produce hydrogen from various fermentable substrates (54, 57), though attempting to measure such substrates was beyond the scope of the current work.
Sample K3 was collected from a low-sulfate and high-sulfide produced fluid. Due to the management of the well, it is not clear what fraction of the sulfide in the produced fluid, if any, is attributable to biological activity, since there was no increase in sulfide of the produced fluid compared to the miscible injectant. Based on recovered sulfate reduction genes, a potential sulfate reducer was Thermodesulfobacterium commune, and a member of a novel genus within the Clostridiales (likely a member of the family Peptococcaceae) also may be able to reduce sulfate (Fig. 3). The reconstructed Clostridiales genome lacks a sulfate adenylyltransferase gene, though it is possible that we did not detect this gene because the genome is partial (this genome was estimated at 74.5% completeness based on the inventory of ribosomal proteins recovered). A Thermococcus organism also could contribute to sulfide production in K3 (see the supplemental material).
The microbial community in K3 was distinct from that in K2 in another way, in that K2 had only nonmethanogenic archaea, whereas the three archaea in K3 were exclusively methanogens. Conversely, bacteria such as Thermotoga, Thermococcus (a member of the Thermodesulfobacterales), Caldanaerobacter, Thermodesulfobacterium commune, and Clostridia were present in both K2 and K3. Since the injection water sources for these two wells are very different, these shared bacteria could be indigenous to the Kuparuk Formation.
Hydrocarbon degradation genes were rare in Kuparuk samples (only one partial benzoyl coenzyme A [benzoyl-CoA] reductase sequence was present in K3). A. fulgidus has been shown to be capable of long-chain alkane degradation (59); however, due to very low homology of the proposed activation enzyme to the known alkylsuccinate synthases or benzylsuccinate synthases, detailed investigation of candidate proteins would be required to support their inclusion in hydrocarbon transformation processes.
Acetate oxidation has been proposed as an important mechanism in anaerobic hydrocarbon degradation in oil reservoirs (2, 5). Piceno et al. suggested that syntrophic acetate-oxidizing and hydrogentrophic methanogenesis processes were prominent in the Kuparuk reservoir (32). They speculated that acetate from degraded carbon might be oxidized to CO2 and H2, with subsequent utilization of H2 and CO2 by hydrogentrophic methanogens, as discussed by Jones et al. (2). We recovered a genome for Thermoacetogenium phaeum in sample K3, an organism reported previously from the Kuparuk Formation (22, 32). This is a thermophilic, syntrophic acetate oxidizer, perhaps associated with hydrogentrophic methanogens (60). Genes for previously described major enzymes in T. phaeum grown syntrophically on acetate (61) (CO dehydrogenase, formate dehydrogenase, hydrogenase, and formyl-hydrofolate ligase) were identified in the T. phaeum genome bin.
Sample K3 was from an area that at the time of sampling had a higher CO2 concentration (measured as moles percent in the gas phase in equilibrium with the oil and water at standard temperature and pressure) than other parts in the same formation. The elevated CO2 concentration was a result of this area having been swept with miscible injectant (containing 25 mol% CO2). The gas phase CO2 was much higher in K3 (11 to 12.5 mol%, in 2013) than in K2 (~0.5 mol%). This condition may provide an additional opportunity for organisms that are able to fix CO2 with concomitant acetate production. A nearly complete genome of Acetothermia (OP1) was recovered from sample K3 (Table 1). The 1,534-bp 16S rRNA gene for this organism is 99% identical to a sequence (1,538 bp) recovered from a nonflooded, high-temperature petroleum reservoir (25), but the previous study authors did not reconstruct a genome. The first genome of this phylum was recovered from a subsurface thermophilic microbial mat community, and it was predicted to have an acetogenic lifestyle based on the Wood-Ljungdahl pathway (62), which removes hydrogen that accumulates during biodegradation, and assimilates CO2 to produce acetate (63, 64). The 16S rRNA gene and RecA protein sequences of the new OP1 genome recovered from K3 were 86% and 71% identical to that of the prior draft genome, respectively. We evaluated autotrophic CO2 fixation pathways, i.e., reductive tricarboxylic acid cycle and the Wood-Ljungdahl pathway. The new OP1 genome has genes for 2-oxoglutarate synthase and ATP citrate lyase (alpha subunit) but lacks a fumarate reductase gene. While we found several enzymes in Wood-Ljungdahl pathway, one key component, CO dehydrogenase/acetyl-CoA synthase, was not detected in the current assembly, which still has many gaps.
TABLE 1 .
OP1 (Acetothermia) draft genome statistics and major metabolic functional pathway genes recovered in the genome
| Characteristic | Result |
|---|---|
| No. of contigs | 22 |
| Total sequence length (bp) | 1,816,458 |
| G+C content | 64.3% |
| 16S rRNA gene | 1,525 bp, 99.6% identical to OP1, class OPB14 |
| Ribosomal proteins | 46/55 |
| Single copy genes | 48/51 |
| Carbon utilization | Glycosyl hydrolases |
| Hydrogenases | Membrane bound (MBH 2) |
| Dissimilatory sulfate reduction | Not found |
| Sulfur oxidation | Not found |
| Nitrate-nitrite reduction | Not found |
OP1 has many glycosyl hydrolase genes. Most of these enzymes (except one endoglucanase) were not found in the previous OP1 genome (Candidatus “Acetothermum autotrophicum”), suggesting significant differences in metabolic capacity between the two genomes. Since both genomes have gaps, discussion here is limited to the sequences available. OP1 has a group 4 NiFe membrane-bound hydrogenase (MBH). This type of MBH could produce hydrogen using reduced ferrodoxin (Fd) from carbohydrate fermentation or oxidize hydrogen, pumping protons via electron transfer to various quinones and cytochromes. We found no evidence of aerobic metabolism (no cytochrome oxidase), or alternative pathways for energy production. For instance, OP1 lacks genes for dissimilatory sulfate reduction and nitrate or nitrite reduction. This organism also appears to lack flagellum-based motility and has no lipopolysaccharide biosynthetic genes, suggesting that it does not have a Gram-negative cell envelope.
(iii) Schrader Bluff Formation
The ability to degrade hydrocarbons was more prominent in organisms of the mesothermic Schrader Bluff samples (SB1 and SB2) than in other samples studied here. This is consistent with previous hydrocarbon profiles showing that Schrader Bluff oil was the most degraded of the oils from the Alaska North Slope samples (32). Hydrocarbon degradation genes were identified in Desulfotomaculum and in another partial genome in Clostridiales (Clostridiales_45_118_partial in Table 2). These genomes have the key enzyme required for the first step in anaerobic hydrocarbon degradation (alkylsuccinate synthase [ASS] or benylsuccinate synthase [BSS]) and the requisite activase. Additional genomes contain incomplete BSS subunits or only enzymes involved in downstream steps or steps in degrading aromatic compounds (Table 2). It is not certain whether the partial gene structures are due to the lack of intact operons or the incompleteness of the genomes.
TABLE 2 .
Anaerobic hydrocarbon degradation genes recovered from binned genomes (metagenome analysis) from produced water samples collected from oil reservoirs in the Schrader Bluff Formation (SB1 and SB2) or the Kuparuk Formation (K3) of the Alaska North Slopea
| Draft genome | Sample | Anaerobic hydrocarbon degradation gene product(s) | Recovered genome size (Mbp) | Estimated genome coverage based on rRNA gene retrieval (%) |
|---|---|---|---|---|
| Clostridia_62_21 | K3 | Benzoyl-CoA reductase | 1.3 | 74.5 |
| Bacterium_34_27_partial | SB1 | Tungsten-dependent benzoyl-CoA reductase | 0.82 | 5.5 |
| Clostridia_33_59 | SB1 | Benzylsuccinate synthase (gamma subunit); glycyl- radical enzyme activating protein | 0.5 | 65.5 |
| Clostridia_51_5 | SB1 | Benzylsuccinate synthase (alpha, gamma subunits); | 0.64 | 18.2 |
| Desulfotomaculum_46_80d | SB1 | Alkylsuccinate synthase; benzoyl-CoA reductase; (R)-benzylsuccinyl-CoA dehydrogenase; glycyl-radical enzyme-activating protein | 2.25 | 80.0 |
| Desulfotomaculum_46_296d | SB1 | Benzylsuccinate synthase (alpha, beta, gamma subunits); glycyl-radical enzyme activating protein; benzoyl-CoA reductase; (R)-benzylsuccinyl- CoA dehydrogenase | 2.33 | 81.8 |
| Firmicute_34_26_partial | SB1 | Benzylsuccinate synthase (gamma subunit); glycyl- radical enzyme activating protein | 1.2 | 38.2 |
| Firmicute_40_6_partial | SB1 | Benzylsuccinate synthase (gamma subunit) | 1.19 | 14.5 |
| Mesotoga_prima_46_7 | SB1 | Benzoyl-CoA reductase | 1.71 | 54.5 |
| OP9_34_73_partial | SB1 | Benzylsuccinate synthase (gamma subunit); glycyl- radical enzyme activating protein | 0.22 | 47.3 |
| OP9_34_128 | SB1 | Alkylsuccinate synthase (AssA); glycyl-radical enzyme-activating protein | 0.9 | 76.4 |
| Syntrophobacterales_55_5 _plusb | SB1 | Benzoyl-CoA reductase; benzoate-CoA ligase | 35/55+ | |
| Unbinnedb | SB1 | Alkylsuccinate synthase; benzoyl-CoA reductase | ||
| Chloroflexi_43_5_mixb | SB2 | 4-Hydroxybenzoate-CoA ligase | 42/55+ | |
| OP9_34_191_partial | SB2 | Benzylsuccinate synthase (gamma subunit); glycyl- radical enzyme activating protein | 0.63 | 67.3 |
| Clostridiales_45_118_partiald | SB2 | Benzylsuccinate synthase (alpha, beta, gamma subunits); glycyl-radical enzyme activating protein; benzoyl-CoA reductase; (R)-benzylsuccinyl- CoA dehydrogenase | 2.34 | 10.9 |
| OP9_34_868 | SB2 | Benzylsuccinate synthase (gamma subunit); glycyl- radical enzyme activating protein | 0.2 | 36.4 |
| OP9-like_34_37 | SB2 | Benzylsuccinate synthase (alpha, gamma subunits); glycyl-radical enzyme activating protein | 0.91 | 38.2 |
| Syntrophobacterales_51_5_partial | SB2 | Benzoyl-CoA reductase | 1.11 | 60 |
| Unbinnedc | SB2 | Alkylsuccinate synthase; glycyl-radical enzyme activating protein benzylsuccinate synthase |
Genome names have the format taxonomy_percent G+C content_calculated genome coverage.
These bins contains multiple marker genes, indicating 1 to 3 genomes in the same family or genus.
Contig that was not binned to any draft genomes.
Predicted hydrocarbon-degrading bacteria.
Schrader Bluff produced water samples were not soured, and sulfate concentrations were below detection levels (see Table S1 in the Text S2 file in the supplemental material). From the metagenomic data reported here, we did not recover a sulfate reduction pathway in any of the Schrader Bluff genome bins. Therefore, neither chemical nor biological factors indicated souring of this reservoir. Compared to published Desulfotomaculum cluster 1 sequences (65), the Desulfotomaculum 16S rRNA genes from SB1 and SB2 are similar, clustering with Ii (see Fig. S2 in the supplemental material). The current genome-based findings are consistent with the role proposed previously (32) for the Schrader Bluff Desulfotomaculum organisms, where syntrophic growth rather than sulfate reduction likely explains the prominence of members of this genus in this nonsoured reservoir. We propose that the microbial community has adapted to this low-sulfate environment by relying on fermentation of organic substrates (e.g., propionate, reported in produced water from the Schrader Bluff Formation [SB] [26]). To make this energetically feasible, the hydrogen concentrations must be kept low. The hydrogen- and formate-consuming methanogens in the community likely make a synthophic lifestyle favorable.
The presence of many methanogens, including acetoclastic methanogens, is consistent with the detection of biogenic methane based on stable isotope data (32). In SB1 and SB2, both hydrogentrophic and acetotrophic methanogens are present, but the latter dominate. The same species, Methanosaeta harundinacea (gyrA genes are 100% identical), is relatively abundant in both SB1 and SB2. Methanosaeta is likely to produce methane from acetate (acetoclastic methanogenesis) using the acetyl-CoA synthetase pathway (66, 67). This is consistent with the previous assertion that acetogenic methanogenesis is more prominent in the Schrader Bluff than in the Kuparuk Formation (32).
Several organisms may be syntrophs. We recovered partial Syntrophobacteriales genomes from both SB1 and SB2. Members of this order have been studied for aromatic compound degradation and syntrophic metabolism (60). The benzoyl-CoA reductase genes associated with this genome support this function. We also recovered some components proposed to be essential for syntrophic metabolism: formate hydrogenlyase and heterodisulfide reductase (HdrA), which is part of the HdrABC electron transfer complex (37). We infer that Syntrophobacteriales organisms in oil field environments degrade aromatic compounds in syntrophy with hydrogentrophic methanogens (contributing to syntrophic H2 and formate generation). Additionally, genomes of Proteiniphilum acetatigens were reconstructed from SB1 (and a genome was assigned to Proteiniphilum but not to the species Proteiniphilum acetatigens in SB2). The possible function for this member is utilizing protein substrates from cellular debris and producing acetate and CO2 (68).
In contrast to the samples from Ivishak and Kuparuk, where bacteria from candidate phyla comprised 0 to 0.4%, bacteria from candidate phyla are well represented in the SB1 and SB2 microbial communities, in terms of both variety and abundance (Fig. 1; also, see Fig. S1E and F in the supplemental material). Candidate phylum OP9 (Atribacteria [69]) comprises 30% of the community in SB2 and 12% in SB1. Parcubacteria (OD1) represented 9.5% and 6% of communities in SB1 and SB2, respectively. Bacteria from several other candidate phyla, including TA06, WS6, and Microgenomates (OP11), were also sampled.
We reconstructed Marinimicrobia (SAR406) genomes from both Schrader Bluff samples (identified based on full-length 16S rRNA genes). The primary water source for Schrader Bluff wells was a mixture of water from other oil wells in the Kuparuk and Schrader Bluff formations and from other Cretaceous era marine sandstone formations. To our knowledge, the SB1 and SB2 samples are not influenced by modern-day seawater, suggesting that Marinimicrobia genomes are indigenous to the subsurface ecosystem. The reconstructed genomes from candidate phyla (see the supplemental material) indicate that some of them may be involved with carbon and hydrogen cycling. For example, Marinimicrobia may produce hydrogen (Fe hydrogenase present), and both WS6 and OP11 have predicted fermentative lifestyles, based on the lack of electron transfer chain components and incomplete TCA cycles.
Some archaeal and bacterial genome bins contained nitrogenase genes. However, methanogenic archaea are predicted to have the largest share of nitrogen fixation genes in organisms present in the SB1 and SB2 communities (Table 3), in terms of both diversity (4 archaea versus 1 bacterium) and relative abundance. This finding is in remarkable contrast to those for other ecosystems, where such genes are typically primarily associated with bacteria (70, 71). I2 and K3 samples also contain three organisms with genomes that contain nitrogen fixation genes. All three genomes belong to the order Methanobacteriales. Nitrogen fixation genes were not recovered for any organism in samples I1 and K2.
TABLE 3 .
Nitrogen fixation genes present in draft genomes (from metagenome data) from produced water samples collected from oil reservoirs in the Schrader Bluff formation (SB1 and SB2), the Kuparuk formation (K3), or the Ivishak Formation (I2) of the Alaska North Slopea
| Genome | Sample | Nitrogen fixation gene(s) | Affiliation |
|---|---|---|---|
| Methanosaeta_harundinacea_57_489 | SB1 | nifH (nitrogenase Fe alpha subunit), nifK (MoFe beta subunit), nifE (nitrogenase MoFe cofactor biosynthesis protein) | Methanogenic Archaea |
| Methanocalculus_52_23 | SB1 | nifH, nifK | Methanogenic Archaea |
| Methanoculleus_marisnigri_60_61_partial | SB1 | nifH, nifK | Methanogenic Archaea |
| Desulfotomaculum_46_80 | SB1 | nifH, nifK, nifE, nifB (nitrogenase cofactor biosynthesis protein) | Bacteria |
| Methanobacteriales_53_19_partial | SB2 | nifH, nifK, | Methanogenic Archaea |
| Methanoculleus_60_29 | SB2 | nifH, nifK, nifE, anfO (nitrogenase iron-iron accessory protein) | Methanogenic Archaea |
| Methanosaeta_haundinacea_56_747 | SB2 | nifH | Methanogenic Archaea |
| Clostridia_45_118_partial | SB2 | nifH, nifK, nifE, nifB | Bacteria |
| Methanothermobacter_50_10 | I2 | nifH, nifK | Methanogenic Archaea |
| Methanobacteriaceae_41_258_partial | I2 | nifH, nifK | Methanogenic Archaea |
| Methanobacteria_50_154 | K3 | nifH, nifK, nifE, glnB (nitrogen metabolism regulatory protein) | Methanogenic Archaea |
Genome names have the format taxonomy_percent G+C content_calculated genome coverage.
Summary.
In this study, we analyzed petroleum reservoir produced water samples collected from six production wells, from different depths, temperatures, and H2S concentrations using a genome-resolved metagenomic approach. These samples had previously been investigated using 16S rRNA gene-based PhyloChip analyses. The PhyloChip data for bacteria, the 16S rRNA genes reconstructed in metagenomes (Fig. 3), and genome phylogeny, based on other single copy gene information (Fig. S1A to F), are in overall agreement. However, the genome-resolved approach has higher taxonomic resolution and enables detailed metabolic predictions. Generally, the richness of the microbial communities decreased as temperatures increased, with 44 to 60 recovered genomes from the mesothermic reservoir yet only 3 genomes reconstructed from the highest temperature reservoir. The microbial communities are much more diverse and evenly distributed in the mesothermic reservoir (Fig. S1A to F).
Clostridiales are likely the major contributors to hydrocarbon degradation in the low temperature Schrader Bluff oil reservoir. Whether some candidate phyla also played a role in this process is uncertain. We clearly demonstrated the existence of the dissimilatory sulfate reduction pathways, regardless of the souring status of the well. We assembled several nearly complete genomes from candidate phyla bacteria and provided insights into their ecosystem roles. Nitrogen fixation potential is predicted to be largely associated with methanogens. The conclusions from this study provide valuable insights into functional roles individual organisms, including those from candidate phyla, may play in these petroleum reservoirs.
MATERIALS AND METHODS
Sample descriptions.
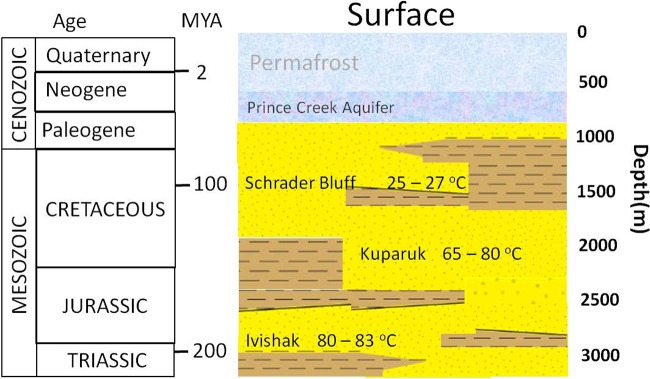
Sample collection methods, DNA extraction, and chemical analyses have been described previously (32). Briefly, six produced water samples were collected from Milne Point and Prudhoe Bay oil field subsurface reservoirs in the Alaska North Slope (ANS) in September 2011 and in various months of 2013. The samples were collected from three geologic formations (Fig. 4). Two samples (SB1 and SB2) were collected in September 2011 from the Schrader Bluff Formation, which formed as a marine deposit during the Late Cretaceous period (72). Throughout this formation, the temperature is estimated to range between 24 and 27°C at depths of 1,200 to 1,400 m below the surface. Two samples, K2 and K3, were obtained in May 2013 from the Kuparuk Formation, a marine shelf siliciclastic sandstone deposit that formed in the Early Cretaceous period (73). Within this formation, the temperatures range between 47 and 70°C at depths between 1,785 and 2,150 m below the surface. Two samples, I1 and I2, were collected from the Ivishak Formation, a marine siliciclastic sedimentary unit formed in the Early Triassic period (74). The Ivishak Formation hosts the deepest and the hottest reservoir studied here. The temperature range in this formation is between 80 and 83°C at depths of 2,750 to 3,100 m.
FIG 4 .
Schematic illustration of relative positions of geologic formations containing the oil reservoirs sampled in the Alaska North Slope and temperature ranges associated with the formations. This illustration was adapted from reference 83.
Over the period of reservoir oil production from the ANS, water is commonly pumped into the reservoir to enhance oil recovery. The water used for this purpose may be taken from the ocean, obtained from a subsurface aquifer, or recycled from other wells. Both aquifer water and recycled water, but not seawater, have been injected into the Schrader Bluff Formation reservoir. No sulfide was detected in the two samples from this reservoir. Production from the K2 sample well in the Kuparuk Formation reservoir has been supported only with aquifer water, whereas that from the K3 sample well has a complex history, with some use of recycled seawater. Production from the Ivishak Formation reservoir sample well I1 has only recently been supported with seawater, whereas I2 has a long history of seawater injection. Elevated hydrogen sulfide concentrations were associated with I2 (~200 mg/liter) (32), and by industry standards, this reservoir region is soured. Hydrogen sulfide concentrations for K2 and I1 wells were below the level associated with reservoir souring (17.5 and 14 mg/liter, respectively). The H2S level was ~130 mg/liter in the produced water of K3. Since the initial report (32), however, it has been established that this area of the formation has been swept with alternating slugs of produced water (relatively low sulfate) and miscible injectant (usually solvents/gases injected to enhance oil recovery); in this case, the injectant contained ~130 mg/liter sulfide. Therefore, this sample is not regarded as biologically soured. Chemical properties of the produced water are provided in Table S1 in the Text S2 file in the supplemental material.
Metagenome sequencing and analysis.
Produced reservoir water (125, 80, 3,125, 4,190, 5,000, and 2,935 ml for SB1, SB2, K2, K3, I1, and I2, respectively) was filtered through a 0.2-µm filter. Genomic DNA was extracted from cells retained on this filter. Genomic DNA was extracted using a modification of the method of Miller et al. (75) and further purified with a MoBio UltraSoil DNA extraction kit (MoBio, Carlsbad, CA) as described previously (32). DNA from SB1 and SB2 was sheared using a Covaris instrument (Woburn, MA), and then SPRI (Agencourt Ampure) beads (Beckman Coulter, Brea, CA) were used to size-select fragments in the range of 400 bp. The size and quality of DNA were examined using Bioanalyzer HS DNA assay (Agilent Technologies, Santa Clara, CA). Genomic DNA from the other samples was sent directly to the Yale Center for Genomic Analysis for library construction. Illumina kits were used to prepare libraries for either MiSeq (2011 samples) or multiplexed HiSeq (2013 samples) sequencing runs per the standard protocols performed at the Yale Center for Genomic Analysis. Compared to the HiSeq option (read length at about 150 bp), the MiSeq option provided longer reads (~250 bp) for greater ease of assembly but shallower sequencing depth (see the discussion in the supplemental material).
Data analysis was initiated by removing adapter sequences using Cutadapt (https://pypi.python.org/pypi/cutadapt/1.2.1), and sequences were trimmed for quality using Trimmomatic (http://www.usadellab.org/cms/index.php?page=trimmomatic) (parameters: leading, 3; trailing, 3; sliding window, 4; quality score, ≥15; minimum read length of 60 bases). The dominant species (based on reconstructed full-length 16S rRNA gene) in each sample were identified using EMIRGE (76). The trimmed reads were assembled using IDBA-UD (77). Using the ggkBase (http://ggkbase.berkeley.edu) online binning tools, assembled genome fragments were assigned to draft genomes of origin (binned) based on GC content, read coverage [calculated as (read count × read length)/sequence length], and phylogenetic profiles. The genome bins were refined based on tetranucleotide sequence information analyzed using an emergent self-organizing map (ESOM) (78), with refinement of some bins using organism abundance pattern data. The genome of one organism was manually curated in Geneious (v7.0.6) to improve the accuracy of the contigs. Gene predictions were made using Meta-Prodigal (79), and functional predictions were made using a standard annotation pipeline, including amino acid similarity searches against UniRef90 (80) and KEGG (81, 82). Functional annotations were searched across the data set in ggKbase to predict the metabolic repertoire of specific organisms. Relative abundance was calculated as percentage of all reads assigned to a genome, with a correction based on estimated genome completeness (see the supplemental materials and methods). The unassigned reads were accounted for in the total reads.
Nucleotide sequence accession numbers.
This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession numbers LDZP00000000 to LDZU00000000 and LGEN00000000 to LGHI00000000. The version described in this paper is version LDZP00000000 to LDZU00000000 and LGEN00000000 to LGHI00000000. The raw reads have been deposited at DDBJ/EMBL/GenBank under the accession number SRP057267. The project description and related metadata are accessible through BioProject number PRJNA278302.
SUPPLEMENTAL MATERIAL
Supplemental methods and results. Download
File containing Tables S1 to S5. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample I1. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample I2. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample K2. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample K3. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample SB1. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample SB2. Download
Phylogenetic tree of Desulfotomaculum subcluster. Download
ACKNOWLEDGMENTS
Francine Reid, Craig Graff, and Jiabin Han collected samples, and Francine Reid assisted in sample processing.
This work was supported by a subcontract from the University of California, Berkeley, Energy Biosciences Institute to Lawrence Berkeley National Laboratory under its U.S. Department of Energy contract DE-AC02-05CH11231. The ggkbase is supported by grant DE-SC0004918 (Systems Biology Knowledge Base Focus Area) from the Department of Energy.
Funding Statement
This work was supported by a subcontract from the University of California at Berkeley, Energy Biosciences Institute to Lawrence Berkeley National Laboratory under its U.S. Department of Energy contract DE-AC02-05CH11231. The ggkbase is supported by grant DE-SC0004918 (Systems Biology Knowledge Base Focus Area) from Department of Eenergy.
Footnotes
Citation Hu P, Tom L, Singh A, Thomas BC, Baker BJ, Piceno YM, Andersen GL, Banfield JF. 2016. Genome-resolved metagenomic analysis reveals roles for candidate phyla and other microbial community members in biogeochemical transformations in oil reservoirs. mBio 7(1):e01669-15. doi:10.1128/mBio.01669-15.
REFERENCES
- 1.Head IM, Jones DM, Larter SR. 2003. Biological activity in the deep subsurface and the origin of heavy oil. Nature 426:344–352. doi: 10.1038/nature02134. [DOI] [PubMed] [Google Scholar]
- 2.Jones DM, Head IM, Gray ND, Adams JJ, Rowan AK, Aitken CM, Bennett B, Huang H, Brown A, Bowler BF, Oldenburg T, Erdmann M, Larter SR. 2008. Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451:176–U176. doi: 10.1038/Nature06484. [DOI] [PubMed] [Google Scholar]
- 3.Aitken CM, Jones DM, Larter SR. 2004. Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs. Nature 431:291–294. doi: 10.1038/Nature02922. [DOI] [PubMed] [Google Scholar]
- 4.Huang H, Larter S. 2005. Biodegradation of petroleum in subsurface geological reservoirs, p 91–121. In Ollivier B, Magot M (ed), Petroleum biology. ASM Press, Washington, DC. [Google Scholar]
- 5.Head IM, Larter SR, Gray ND, Sherry A, Adams JJ, Aitken CM, Jones DM, Rowan AK, Huang H, Roling WFM. 2010. Hydrocarbon degradation in petroleum reservoirs, p 3097–3109. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Spring-Verlag, Berlin, Germany. [Google Scholar]
- 6.Connan J. 1984. Biodegradation of crude oils in reservoirs, p 299–335. In Brooks J, Welte J (ed), Advances in petroleum geochemistry, vol. 1 Academic Press, London, United Kingdom. [Google Scholar]
- 7.Pereira IA, Ramos AR, Grein F, Marques MC, da Silva SM, Venceslau SS. 2011. A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front Microbiol 2:69. doi: 10.3389/Fmicb.2011.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gieg LM, Jack TR, Foght JM. 2011. Biological souring and mitigation in oil reservoirs. Appl Microbiol Biotechnol 92:263–282. doi: 10.1007/s00253-011-3542-6. [DOI] [PubMed] [Google Scholar]
- 9.Liang R, Grizzle RS, Duncan KE, McInerney MJ, Suflita JM. 2014. Roles of thermophilic thiosulfate-reducing bacteria and methanogenic archaea in the biocorrosion of oil pipelines. Front Microbiol 5:89. doi: 10.3389/fmicb.2014.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewin A, Johansen J, Wentzel A, Kotlar HK, Drabløs F, Valla S. 2014. The microbial communities in two apparently physically separated deep subsurface oil reservoirs show extensive DNA sequence similarities. Environ Microbiol 16:545–558. doi: 10.1111/1462-2920.12181. [DOI] [PubMed] [Google Scholar]
- 11.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkes RJ, Cragg BA, Wellsbury P. 2000. Recent studies on bacterial populations and processes in subseafloor sediments: a review. Hydrogeol J 8:11–28. doi: 10.1007/Pl00010971. [DOI] [Google Scholar]
- 13.D’Hondt S, Rutherford S, Spivack AJ. 2002. Metabolic activity of subsurface life in deep-sea sediments. Science 295:2067–2070. doi: 10.1126/science.1064878. [DOI] [PubMed] [Google Scholar]
- 14.Jørgensen BB. 2011. Deep subseafloor microbial cells on physiological standby. Proc Natl Acad Sci U S A 108:18193–18194. doi: 10.1073/pnas.1115421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. 2012. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci U S A 109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gieg LM, Davidova IA, Duncan KE, Suflita JM. 2010. Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oilfields. Environ Microbiol 12:3074–3086. doi: 10.1111/j.1462-2920.2010.02282.x. [DOI] [PubMed] [Google Scholar]
- 17.Mueller RF, Nielsen PH. 1996. Characterization of thermophilic consortia from two souring oil reservoirs. Appl Environ Microbiol 62:3083–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsen RK, Beeder J, Thorstenson T, Torsvik T. 1996. Distribution of thermophilic marine sulfate reducers in North Sea oil field waters and oil reservoirs. Appl Environ Microbiol 62:1793–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsen RK, Torsvik T, Lien T. 1996. Desulfotomaculum thermocisternum sp. nov., a sulfate reducer isolated from a hot North Sea oil reservoir. Int J Syst Bacteriol 46:397–402. doi: 10.1099/00207713-46-2-397. [DOI] [Google Scholar]
- 20.Orphan VJ, Taylor LT, Hafenbradl D, Delong EF. 2000. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl Environ Microbiol 66:700–711. doi: 10.1128/Aem.66.2.700-711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berdugo-Clavijo C, Gieg LM. 2014. Conversion of crude oil to methane by a microbial consortium enriched from oil reservoir production waters. Front Microbiol 5:197. doi: 10.3389/Fmicb.2014.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duncan KE, Gieg LM, Parisi VA, Tanner RS, Tringe SG, Bristow J, Suflita JM. 2009. Biocorrosive thermophilic microbial communities in Alaskan north slope oil facilities. Environ Sci Technol 43:7977–7984. doi: 10.1021/Es9013932. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa R, Toyama K, Miyanaga K, Tanji Y. 2014. Identification of crude-oil components and microorganisms that cause souring under anaerobic conditions. Appl Microbiol Biotechnol 98:1853–1861. doi: 10.1007/S00253-013-5107-3. [DOI] [PubMed] [Google Scholar]
- 24.Kleinsteuber S, Schleinitz KM, Vogt C. 2012. Key players and team play: anaerobic microbial communities in hydrocarbon-contaminated aquifers. Appl Microbiol Biotechnol 94:851–873. doi: 10.1007/s00253-012-4025-0. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi H, Endo K, Sakata S, Mayumi D, Kawaguchi H, Ikarashi M, Miyagawa Y, Maeda H, Sato K. 2012. Phylogenetic diversity of microbial communities associated with the crude-oil, large-insoluble-particle and formation-water components of the reservoir fluid from a non-flooded high-temperature petroleum reservoir. J Biosci Bioeng 113:204–210. doi: 10.1016/J.Jbiosc.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Okoro C, Smith S, Chiejina L, Lumactud R, An D, Park HS, Voordouw J, Lomans BP, Voordouw G. 2014. Comparison of microbial communities involved in souring and corrosion in offshore and onshore oil production facilities in Nigeria. J Ind Microbiol Biotechnol 41:665–678. doi: 10.1007/S10295-014-1401-Z. [DOI] [PubMed] [Google Scholar]
- 27.Pham VD, Hnatow LL, Zhang S, Fallon RD, Jackson SC, Tomb JF, DeLong EF, Keeler SJ. 2009. Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ Microbiol 11:176–187. doi: 10.1111/j.1462-2920.2008.01751.x. [DOI] [PubMed] [Google Scholar]
- 28.Voordouw G, Armstrong SM, Reimer MF, Fouts B, Telang AJ, Shen Y, Gevertz D. 1996. Characterization of 16S rRNA genes from oil field microbial communities indicates the presence of a variety of sulfate-reducing, fermentative, and sulfide-oxidizing bacteria. Appl Environ Microbiol 62:1623–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang LY, Ke WJ, Sun XB, Liu JF, Gu JD, Mu BZ. 2014. Comparison of bacterial community in aqueous and oil phases of water-flooded petroleum reservoirs using pyrosequencing and clone library approaches. Appl Microbiol Biotechnol 98:4209–4221. doi: 10.1007/S00253-013-5472-Y. [DOI] [PubMed] [Google Scholar]
- 30.Kotlar HK, Lewin A, Johansen J, Throne-Holst M, Haverkamp T, Markussen S, Winnberg A, Ringrose P, Aakvik T, Ryeng E, Jakobsen K, Drabløs F, Valla S. 2011. High coverage sequencing of DNA from microorganisms living in an oil reservoir 2.5 kilometres subsurface. Env Microbiol Rep 3:674–681. doi: 10.1111/J.1758-2229.2011.00279.X. [DOI] [PubMed] [Google Scholar]
- 31.Dahle H, Garshol F, Madsen M, Birkeland NK. 2008. Microbial community structure analysis of produced water from a high-temperature North Sea oil-field. Anton Leeuw. Int J G93:37–49. doi: 10.1007/S10482-007-9177-Z. [DOI] [PubMed] [Google Scholar]
- 32.Piceno YM, Reid FC, Tom LM, Conrad ME, Bill M, Hubbard CG, Fouke BW, Graff CJ, Han J, Stringfellow WT, Hanlon JS, Hu P, Hazen TC, Andersen GL. 2014. Temperature and injection water source influence microbial community structure in four Alaskan north slope hydrocarbon reservoirs. Front Microbiol 5:409. doi: 10.3389/fmicb.2014.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang F, She YH, Chai LJ, Banat IM, Zhang XT, Shu FC, Wang ZL, Yu LJ, Hou DJ. 2012. Microbial diversity in long-term water-flooded oil reservoirs with different in situ temperatures in China. Sci Rep 2:760. doi: 10.1038/Srep00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An D, Caffrey SM, Soh J, Agrawal A, Brown D, Budwill K, Dong X, Dunfield PF, Foght J, Gieg LM, Hallam SJ, Hanson NW, He Z, Jack TR, Klassen J, Konwar KM, Kuatsjah E, Li C, Larter S, Leopatra V, Nesbo CL, Oldenburg T, Page AP, Ramos-Padron E, Rochman FF, Saidi-Mehrabad A, Sensen CW, Sipahimalani P, Song YC, Wilson S, Wolbring G, Wong ML, Voordouw G. 2013. Metagenomics of hydrocarbon resource environments indicates aerobic Taxa and genes to be unexpectedly common. Environ Sci Technol 47:10708–10717. doi: 10.1021/Es4020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sierra-García IN, Alvarez J, de Vasconcellos SP, de Souza A, dos Santos EV, de Oliveira VM. 2014. New hydrocarbon degradation pathways in the microbial metagenome from Brazilian petroleum reservoirs. PLoS One 9:e90087. doi: 10.1371/journal.pone.0090087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 37.Nobu MK, Narihiro T, Hideyuki T, Qiu YL, Sekiguchi Y, Woyke T, Goodwin L, Davenport KW, Kamagata Y, Liu WT. 2014. The genome of Syntrophorhabdus aromaticivorans strain UI provides new insights for syntrophic aromatic compound metabolism and electron flow. Environ Microbiol. 10.1111/1462-2920.12444. [DOI] [PubMed] [Google Scholar]
- 38.Mayilraj S, Kaksonen AH, Cord-Ruwisch R, Schumann P, Spröer C, Tindall BJ, Spring S. 2009. Desulfonauticus autotrophicus sp. nov., a novel thermophilic sulfate-reducing bacterium isolated from oil-production water and emended description of the genus Desulfonauticus. Extremophiles 13:247–255. doi: 10.1007/s00792-008-0212-4. [DOI] [PubMed] [Google Scholar]
- 39.Li D, Greenfield P, Rosewarne CP, Midgley DJ. 2013. Draft genome sequence of Thermoanaerobacter sp. strain A7A, reconstructed from a metagenome obtained from a high-temperature hydrocarbon reservoir in the Bass Strait, Australia. Genome Announc 1:e00701-13. doi: 10.1128/genomeA.00701-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azam F. 2010. Microbial control of oceanic carbon flux: the plot thickens, p 7–8. In Jiao N, Azam F, Sanders S (ed), Microbial carbon pump in the ocean. AAAS, Washington, DC. [Google Scholar]
- 41.Kujawinski EB. 2011. The impact of microbial metabolism on marine dissolved organic matter. Annu Rev Mar Sci 3:567–599. doi: 10.1146/annurev-marine-120308-081003. [DOI] [PubMed] [Google Scholar]
- 42.Iriki Y, Suzuki T, Nisizawa K, Miwa T. 1960. Xylan of siphonaceous green algae. Nature 187:82–83. doi: 10.1038/187082a0. [DOI] [PubMed] [Google Scholar]
- 43.Mopper K, Dawson R, Liebezeit G, Ittekkot V. 1980. The monosaccharide spectra of natural waters. Mar Chem 10:55–66. doi: 10.1016/0304-4203(80)90058-4. [DOI] [Google Scholar]
- 44.Harvey GR. 1983. Dissolved carbohydrates in the New York bight and the variability of marine organic matter. Mar Chem 12:333–339. doi: 10.1016/0304-4203(83)90060-9. [DOI] [Google Scholar]
- 45.Sakugawa H, Handa N. 1985. Chemical studies on dissolved carbohydrates in the water samples collected from the North Pacific and Bering Sea. Oceanol Acta 8:185–196. [Google Scholar]
- 46.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM. 2006. Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 47.Arnosti C. 2000. Substrate specificity in polysaccharide hydrolysis: contrasts between bottom water and sediments. Limnol Oceanogr 45:1112–1119. doi: 10.4319/lo.2000.45.5.1112. [DOI] [Google Scholar]
- 48.Arnosti C. 2008. Functional differences between Arctic seawater and sedimentary microbial communities: contrasts in microbial hydrolysis of complex substrates. FEMS Microbiol Ecol 66:343–351. doi: 10.1111/j.1574-6941.2008.00587.x. [DOI] [PubMed] [Google Scholar]
- 49.Kwon YK, Kim JJ, Kim JH, Jeon SM, Ye BR, Jang J, Heo SJ, Park SC, Kang DH, Oh C. 2012. Draft genome sequence of the xylan-degrading marine bacterium strain S124, representing a novel species of the genus Oceanicola. J Bacteriol 194:6325. doi: 10.1128/JB.01614-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziervogel K, Arnosti C. 2008. Polysaccharide hydrolysis in aggregates and free enzyme activity in aggregate-free seawater from the north-eastern Gulf of Mexico. Environ Microbiol 10:289–299. doi: 10.1111/j.1462-2920.2007.01451.x. [DOI] [PubMed] [Google Scholar]
- 51.Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, Richardson DL, Kerlavage AR, Graham DE, Kyrpides NC, Fleischmann RD, Quackenbush J, Lee NH, Sutton GG, Gill S, Kirkness EF, Dougherty BA, McKenney K, Adams MD, Loftus B, Peterson S, Reich CI, McNeil LK, Badger JH, Glodek A, Zhou LX, Overbeek R, Gocayne JD, Weidman JF, McDonald L, Utterback T, Cotton MD, Spriggs T, Artiach P, Kaine BP, Sykes SM, Sadow PW, DAndrea KP, Bowman C, Fujii C, Garland SA, Mason TM, Olsen GJ, Fraser CM, Smith HO, Woese CR, Venter JC. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 52.Beeder J, Nilsen RK, Rosnes JT, Torsvik T, Lien T. 1994. Archaeoglobus fulgidus isolated from hot north sea oil field waters. Appl Environ Microbiol 60:1227–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stetter KO, Huber R, Blöchl E, Kurr M, Eden RD, Fielder M, Cash H, Vance I. 1993. Hyperthermophilic archaea are thriving in deep north sea and Alaskan oil reservoirs. Nature 365:743–745. doi: 10.1038/365743a0. [DOI] [Google Scholar]
- 54.Fardeau M, Goulhen F, Bruschi M, Khelifi N, Cayol J, Ignatiadis I, Guyot F, Ollivier B. 2009. Archaeoglobus fulgidus and Thermotoga elfii, thermophilic isolates from deep geothermal water of the Paris Basin. Geomicrobiol J 26:119–130. doi: 10.1080/01490450802674970. [DOI] [Google Scholar]
- 55.Hocking WP, Stokke R, Roalkvam I, Steen IH. 2014. Identification of key components in the energy metabolism of the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus by transcriptome analyses. Front Microbiol 5:95. doi: 10.3389/Fmicb.2014.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mardanov AV, Ravin NV, Svetlitchnyi VA, Beletsky AV, Miroshnichenko ML, Bonch-Osmolovskaya EA, Skryabin KG. 2009. Metabolic versatility and indigenous origin of the archaeon Thermococcus sibiricus, isolated from a Siberian oil reservoir, as revealed by genome analysis. Appl Environ Microbiol 75:4580–4588. doi: 10.1128/AEM.00718-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balk M, Weijma J, Stams AJ. 2002. Thermotoga lettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int J Syst Evol Microbiol 52:1361–1368. doi: 10.1099/Ijs.0.02165-0. [DOI] [PubMed] [Google Scholar]
- 58.D’Ippolito G, Dipasquale L, Vella FM, Romano I, Gambacorta A, Cutignano A, Fontana A. 2010. Hydrogen metabolism in the extreme thermophile Thermotoga neapolitana. Int J Hydrogen Energ 35:2290–2295. doi: 10.1016/J.Ijhydene.2009.12.044. [DOI] [Google Scholar]
- 59.Khelifi N, Amin Ali O, Roche P, Grossi V, Brochier-Armanet C, Valette O, Ollivier B, Dolla A, Hirschler-Réa A. 2014. Anaerobic oxidation of long-chain n-alkanes by the hyperthermophilic sulfate-reducing archaeon, Archaeoglobus fulgidus. ISME J 8:2153–2166. doi: 10.1038/ismej.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sieber JR, McInerney MJ, Gunsalus RP. 2012. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 66:429–452. doi: 10.1146/Annurev-Micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 61.Hattori S, Kamagata Y, Hanada S, Shoun H. 2000. Thermacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int J Syst Evol Microbiol 50:1601–1609. doi: 10.1099/00207713-50-4-1601. [DOI] [PubMed] [Google Scholar]
- 62.Takami H, Noguchi H, Takaki Y, Uchiyama I, Toyoda A, Nishi S, Chee GJ, Arai W, Nunoura T, Itoh T, Hattori M, Takai K. 2012. A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PLoS One 7:e30559. doi: 10.1371/journal.pone.0030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ragsdale SW. 1997. The eastern and western branches of the Wood/Ljungdahl pathway: how the east and west were won. Biofactors 6:3–11. doi: 10.1002/biof.5520060102. [DOI] [PubMed] [Google Scholar]
- 64.Drake HL, Kusel K, Matthies C. 2002. Ecological consequences of the phylogenetic and physiological diversities of acetogens. Antonie von Leeuwenhoek G81:203–213. doi: 10.1023/A:1020514617738. [DOI] [PubMed] [Google Scholar]
- 65.Imachi H, Sekiguchi Y, Kamagata Y, Loy A, Qiu YL, Hugenholtz P, Kimura N, Wagner M, Ohashi A, Harada H. 2006. Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl Environ Microbiol 72:2080–2091. doi: 10.1128/Aem.72.3.2080-2091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fournier GP, Gogarten JP. 2008. Evolution of acetoclastic methanogenesis in Methanosarcina via horizontal gene transfer from cellulolytic Clostridia. J Bacteriol 190:1124–1127. doi: 10.1128/JB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White D. 2000. The physiology and biochemistry of prokaryotes, 2nd ed. Oxford University Press, New York, NY. [Google Scholar]
- 68.Chen S, Dong X. 2005. Proteiniphilum acetatigenes gen. nov., sp. nov., from a UASB reactor treating brewery wastewater. Int J Syst Evol Microbiol 55:2257–2261. doi: 10.1099/Ijs.0.63807-0. [DOI] [PubMed] [Google Scholar]
- 69.Dodsworth JA, Blainey PC, Murugapiran SK, Swingley WD, Ross CA, Tringe SG, Chain PSG, Scholz MB, Lo CC, Raymond J, Quake SR, Hedlund BP. 2013. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nat Commun 4:1854 doi: 10.1038/Ncomms2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raymond J, Siefert JL, Staples CR, Blankenship RE. 2004. The natural history of nitrogen fixation. Mol Biol Evol 21:541–554. doi: 10.1093/Molbev.Msh047. [DOI] [PubMed] [Google Scholar]
- 71.Reed SC, Cleveland CC, Townsend AR. 2011. Functional ecology of free-living nitrogen fixation: a contemporary perspective. Annu Rev Ecol Evol S42:489–512. doi: 10.1146/Annurev-Ecolsys-102710-145034. [DOI] [Google Scholar]
- 72.Flores RM, Myers MD, Houseknecht DW, Stricker GD, Brizzolara DW, Ryherd TJ, Takahashi KI. 2007. Stratigraphy and facies of Cretaceous Schrader Bluff and Prince Creek formations in Colville River Bluffs, North Slope, Alaska. U.S. Geological Survey Professional Paper, 1748. U.S. Geological Survey, Washington, DC. [Google Scholar]
- 73.Carman GJ, Hardwick P. 1983. Geology and regional setting of Kuparuk oil field, Alaska. AAPG Bull 67:1014–1031. [Google Scholar]
- 74.Tye RS, Bhattacharya JP, Lorsong JA, Sindelar ST, Knock DG, Puls DD, Levinson RA. 1999. Geology and stratigraphy of fluvio-deltaic deposits in the Ivishak formation: applications for development of Prudhoe Bay field Alaska. AAPG Bull 83:1588–1623. [Google Scholar]
- 75.Miller DN, Bryant JE, Madsen EL, Ghiorse WC. 1999. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol 65:4715–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller CS, Baker BJ, Thomas BC, Singer SW, Banfield JF. 2011. EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol 12:R44. doi: 10.1186/Gb-2011-12-5-R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peng Y, Leung HC, Yiu SM, Chin FY. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 78.Dick GJ, Andersson AF, Baker BJ, Simmons SL, Yelton AP, Banfield JF. 2009. Community-wide analysis of microbial genome sequence signatures. Genome Biol 10:R85. doi: 10.1186/Gb-2009-10-8-R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. 2012. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28:2223–2230. doi: 10.1093/bioinformatics/bts429. [DOI] [PubMed] [Google Scholar]
- 80.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. 2007. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 81.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. 1999. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Housenecht DW, Bird KJ. 2005. Oil and gas resources of the Arctic Alaska petroleum province. U.S. Geological Survey Professional Paper 1732-A. U.S. Geological Survey, Washington, DC: http://pubs.usgs.gov/pp/pp1732/pp1732a/. [Google Scholar]
- 84.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 85.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods and results. Download
File containing Tables S1 to S5. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample I1. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample I2. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample K2. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample K3. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample SB1. Download
Rank abundance curve of petroleum reservoir microbial consortia, sample SB2. Download
Phylogenetic tree of Desulfotomaculum subcluster. Download