ABSTRACT
Although progress in Chlamydia genetics has been rapid, genomic modification has previously been limited to point mutations and group II intron insertions which truncate protein products. The bacterium has thus far been intractable to gene deletion or more-complex genomic integrations such as allelic exchange. Herein, we present a novel suicide vector dependent on inducible expression of a chlamydial gene that renders Chlamydia trachomatis fully genetically tractable and permits rapid reverse genetics by fluorescence-reported allelic exchange mutagenesis (FRAEM). We describe the first available system of targeting chlamydial genes for deletion or allelic exchange as well as curing plasmids from C. trachomatis serovar L2. Furthermore, this approach permits the monitoring of mutagenesis by fluorescence microscopy without disturbing bacterial growth, a significant asset when manipulating obligate intracellular organisms. As proof of principle, trpA was successfully deleted and replaced with a sequence encoding both green fluorescent protein (GFP) and β-lactamase. The trpA-deficient strain was unable to grow in indole-containing medium, and this phenotype was reversed by complementation with trpA expressed in trans. To assess reproducibility at alternate sites, FRAEM was repeated for genes encoding type III secretion effectors CTL0063, CTL0064, and CTL0065. In all four cases, stable mutants were recovered one passage after the observation of transformants, and allelic exchange was limited to the specific target gene, as confirmed by whole-genome sequencing. Deleted sequences were not detected by quantitative real-time PCR (qPCR) from isogenic mutant populations. We demonstrate that utilization of the chlamydial suicide vector with FRAEM renders C. trachomatis highly amenable to versatile and efficient genetic manipulation.
IMPORTANCE
The obligate intracellular nature of a variety of infectious bacteria presents a significant obstacle to the development of molecular genetic tools for dissecting pathogenicity. Although progress in chlamydial genetics has been rapid, genomic modification has previously been limited to point mutations and group II intron insertions which truncate protein products. The bacterium has thus far been intractable to gene deletion or more-complex genomic integrations such as allelic exchange. Here, we present a novel suicide vector dependent on inducible expression of a chlamydial gene that renders Chlamydia trachomatis fully genetically tractable and permits rapid reverse genetics by fluorescence-reported allelic exchange mutagenesis (FRAEM). We describe the first available system of targeting chlamydial genes for deletion or allelic exchange as well as curing plasmids from C. trachomatis L2. Furthermore, this approach permits monitoring of mutagenesis by fluorescence microscopy without disturbing bacterial growth, a significant asset when manipulating obligate intracellular organisms.
INTRODUCTION
Understanding the contribution of microbial virulence factors to disease is critical for developing new methods of treating and controlling infection. Unfortunately, the obligate intracellular nature of pathogens such as Chlamydia trachomatis presents a significant obstacle to deconstructing virulence mechanisms. This is of particular significance to chlamydial infection, as it remains the most frequently reported infectious disease in the United States (1). C. trachomatis, responsible for blinding trachoma (serovars A to C) as well as genital infections (serovars D to K and L1 to L3), has been unreceptive to common genetic methods until recently (2). Advances in transformation have enabled introduction and stable maintenance of recombinant vectors, while ethyl methanesulfonate (EMS) treatment and the TargeTron system have successfully produced genomic mutations (3–7). Although progress has been rapid, the tools for chlamydial manipulation lack the versatility of those available for the most genetically amenable systems, such as Escherichia coli. EMS treatment disrupts expression by introducing an in-frame, early termination codon. Since mutations are limited to nucleotide transitions, only specific sites have the potential to disrupt translation. Furthermore, EMS mutagenesis is random and therefore requires laborious screening (4). The TargeTron system disrupts gene expression by introducing a group II intron within the target open reading frame (5). This approach necessitates the use of proprietary algorithms and limits integration to sites evaluated to be efficient. Although both methods are invaluable tools for chlamydial genetics, a system of allelic exchange by homologous recombination would enable genomic deletions while significantly improving the availability of target sites.
Classical reverse genetics by homologous recombination requires the introduction of a recombinant vector with desired modifications, sufficient maintenance for the exchange of nucleotides between the plasmid and genome, and subsequent elimination of the vector from the organism (8–11). Although stable transformation and homologous recombination in Chlamydia have been demonstrated, a method of removing vectors after introduction has not been reported (3, 12). In the case of genetically tractable organisms such as E. coli, transformation vectors commonly require only expression of drug resistance and specific origins of replication for isolation and maintenance. Such plasmids are often lost in the absence of selective pressure. However, chlamydial maintenance requires that the transformation vector include the sequence of the native pL2 plasmid found in most C. trachomatis isolates (3, 12, 13). As a result, Chlamydia bacteria, unlike E. coli, most often retain such plasmids indefinitely, even in the absence of any selective pressures (3). Homologous sequences introduced on these vectors may recombine with the genome, but without a means of removing the original, intact gene now present on the stable plasmid, changes in phenotype will not occur. Expression of popular counterselectable markers such as sacB or ccdB would eliminate all transformed Chlamydia regardless of mutation status (14, 15). Although vectors lacking the pL2 sequence have been successfully introduced into Chlamydia, the brief presence of such constructs after transformation has resulted in only one report of allelic exchange with exogenous DNA—a sequence of 1 kb integrating four nucleotide substitutions restricted to the 16S rRNA region of Chlamydia psittaci, a uniquely compatible recombination event as it produced chlamydial resistance to both kasugamycin and spectinomycin (12). In order to utilize the versatility of homologous recombination for gene deletion and sequence insertion routinely demonstrated in other more genetically tractable bacteria, a mechanism to control plasmid maintenance is critical.
We demonstrate that by regulating expression of pgp6 found on the native pL2 plasmid, we were able to alter the stability of the transformed vector. Using this as a backbone, we targeted and successfully exchanged trpA for a 2.2-kb cassette encoding both β-lactamase and green fluorescent protein (GFP). The presence of the mCherry gene on the vector backbone permitted the real-time observation of successful mutation, as inclusions expressing only green fluorescence emerged from dual-fluorescent transformants. In order to assess the target versatility of this approach, this system of fluorescence-reported allelic exchange mutagenesis (FRAEM) was repeated for ctl0063, ctl0064, and ctl0065. All mutagenesis attempts were successful and specific, as confirmed by whole-genome sequencing. Here, we present FRAEM as a convenient method of chlamydial reverse genetics with the versatility of those used in genetically tractable organisms.
RESULTS
pSU6 maintenance by ATc.
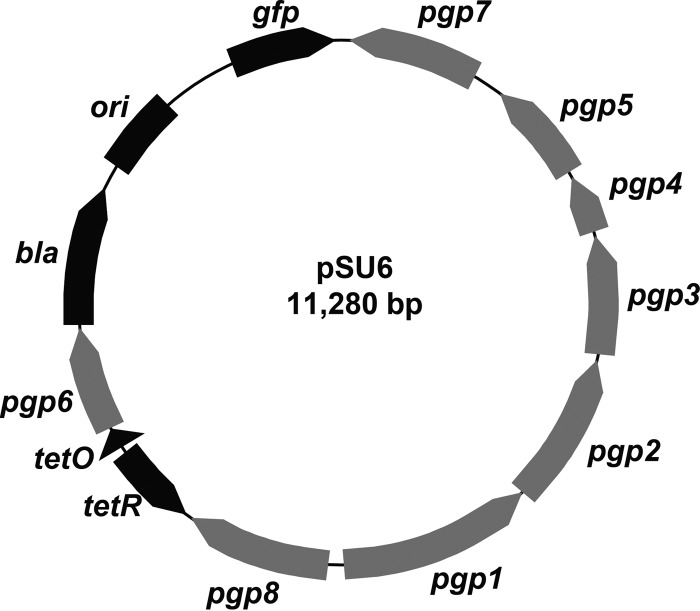
Despite having unknown function, expression of pgp6 was targeted for constructing a plasmid with regulatable stability (16, 17). Of the eight open reading frames present on pL2, only deletion of pgp6 produces transformants unable to maintain the plasmid through multiple rounds of infection. Deletion of other sequences results in no effect (pgp3, pgp4, pgp5, and pgp7), mixed maintenance (pgp8), or no recoverable transformants (pgp1 and pgp2) according to previous reports (16). Although pgp1 and pgp2 may also be viable candidates, the infancy of chlamydial transformation protocols and relatively low success rate compared to E. coli made the absence of transformants a less desirable result than the observable introduction and subsequent loss of pgp6-deficient constructs. Thus, pSU6 was generated from pBOMB4-Tet-mCherry by placing pgp6 under tetracycline (Tet) regulation (Fig. 1), an established system of efficiently regulating gene expression in C. trachomatis (18, 19). The gfp and bla genes were also present on pSU6, providing a fluorescence reporter and β-lactam resistance, respectively, as real-time indicators of the presence of the plasmid. No additional sequences were added, and all other pgp open reading frames and corresponding promoters as present on the native pL2 plasmid were unaltered. All primers used in this work are listed in Table S1 in the supplemental material.
FIG 1 .
pSU6 vector map. Conditional expression of Pgp6 is accomplished by transfer of pgp6 from the endogenous coding position to an engineered locus downstream of a tetracycline-inducible promoter. The remaining pgp genes are expressed under the control of the native promoters in Chlamydia. Constitutively expressed bla and gfp genes provide drug and fluorescence selection capability, respectively.
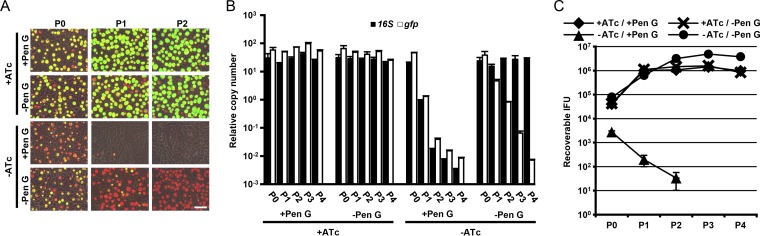
McCoy cell monolayers were infected with C. trachomatis serovar L2 transformed with pSU6 and incubated in medium with or without anhydrotetracycline (ATc) inducer or penicillin G (Pen G) selective pressure (Fig. 2A). In order to observe effects across multiple passages (passage 0 [P0] to passage 2 [P2]), C. trachomatis was harvested, diluted 1,000-fold, and applied to new monolayers every 48 h postinfection (p.i.). Within the first chlamydial developmental cycle (P0), the absence of ATc correlated with a decreased level of observable green fluorescence, regardless of the presence of Pen G. By P2, green fluorescence was not visible in either sample lacking ATc inducer, indicating the loss of pSU6. Furthermore, in the absence of Pen G, chlamydial inclusions were present at comparable numbers regardless of ATc induction, suggesting that the loss of pSU6 was independent of chlamydial development. Visual observations were corroborated by determining the copy numbers of chlamydial genomes and pSU6 by quantitative real-time PCR (qPCR) targeting chlamydial 16S rRNA and gfp DNA sequences, respectively (Fig. 2B). As expected, in the presence of ATc, no significant effect on chlamydial genome or pSU6 copy number was observed through P4, while the absence of inducer resulted in a decrease in detected pSU6 of almost 4 log units, regardless of Pen G.
FIG 2 .
pSU6 maintenance by ATc. McCoy cell monolayers were infected with C. trachomatis L2 carrying pSU6 and incubated in medium in the presence (+) or absence (−) of anhydrotetracycline (ATc) or penicillin G (Pen G). (A) Twenty-four hours p.i., samples were examined for GFP expression (green) and stained with anti-Hsp60 (red) for visualization of L2 (P0). Forty-eight hours p.i., C. trachomatis L2 was harvested from a replicate sample, diluted 1,000-fold, and used to infect a fresh monolayer. Inspection 24 h p.i. and reinfection were repeated for additional time points (P1 and P2). Bar = 50 µm. (B) The relative number of copies of pSU6 and chlamydial genome were determined by qPCR analysis of gfp and the 16S rRNA region, indicating elimination of the vector in the absence of ATc (data are represented as means plus standard deviations [SDs] [error bars]; n = 3). (C) Recoverable IFU from each time point were determined (data are represented as means ± SDs; n = 3). Samples in medium lacking ATc but containing Pen G (−ATc / +Pen G) produced no detectable infectious progeny after P2, suggesting that elimination of pSU6 in the presence of antibiotics provides significant selective pressure.
In order to account for a potentially low homologous recombination rate of bla into the genome, thorough elimination of the suicide vector is essential. In the presence of Pen G, any C. trachomatis retaining pSU6 despite the absence of ATc may outgrow chlamydial mutants which are either produced in low numbers or have significant growth defects. Thus, the recoverable inclusion-forming units (IFU) during each passage were examined (Fig. 2C). All samples in medium with ATc or lacking Pen G produced infectious progeny throughout the experiment. Only the condition with Pen G and without ATc failed to produce detectable infectious progeny after P2, suggesting that the loss of pSU6 in conjunction with drug selection may be sufficient pressure for isolating chlamydial mutants.
Plasmid elimination by transformation with pSU6.
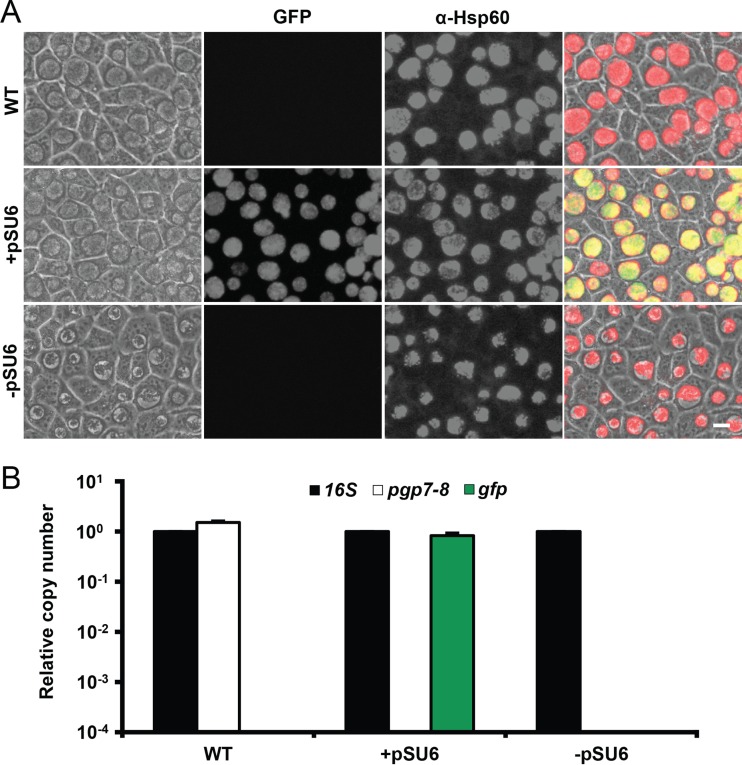
Plasmid elimination with agents such as ethidium bromide and novobiocin has been demonstrated in a variety of organisms including Chlamydia muridarum (20). However, success in curing pL2 from C. trachomatis L2 has not been published. During the process of chlamydial transformation, the loss of all copies of native plasmid in exchange for the introduced vector were previously reported (3). Thus, transformation and elimination of pSU6 as a potential method of curing C. trachomatis L2 was examined. Due to the efficiency with which Chlamydia bacteria laterally transfer plasmids and other genetic material, we attempted to minimize the potential for reacquiring pL2 from nontransformed Chlamydia by isolating pSU6 transformants (+pSU6) by limiting dilutions in the presence of ATc immediately after visual identification (21, 22). C. trachomatis transformed with pSU6 produced inclusions similar in appearance to those produced by wild-type C. trachomatis, with the exception of green fluorescence (Fig. 3A). An additional round of limiting dilutions in the absence of ATc was then applied, resulting in the loss of fluorescence, as well as a unique inclusion morphology with a central aggregate of Chlamydia surrounded by an apparently vacuous lumen lacking visible motion (−pSU6). Given that similar inclusions are produced by the naturally occurring plasmid-free strain L2 (25667R), these features were suggestive of successful plasmid elimination (23). Relative copy numbers of the chlamydial genome, native plasmid, and pSU6 were determined by qPCR of the 16S rRNA region, pgp7-pgp8 junction, and gfp gene, respectively. Since all additional sequences required for construction of pSU6 from pL2 are located between pgp7 and pgp8, oligonucleotide primers surrounding this junction are unable to amplify from pSU6 during the 30-s qPCR extension time. Thus, the pgp7-pgp8 amplicon is generated only from native pL2 plasmid, not pSU6 or any other derivatives described in this work. Given that two rRNA operons are present per L2 genome, wild-type C. trachomatis contained roughly four pL2 plasmids per bacterium (Fig. 3B) (24). Analysis of transformants indicated the loss of native plasmid and the presence of approximately one pSU6 plasmid per genome. Limiting dilutions in the absence of ATc (−pSU6) resulted in samples with undetected levels of either pL2 or pSU6, confirming that pL2 had been successfully cured. This presents a novel method for removal of the pL2 plasmid as well as vectors from C. trachomatis L2. Additionally, this may provide a valuable tool for confirming the loss of phenotype after removal of constructs to corroborate gain of phenotype upon introduction.
FIG 3 .
Plasmid elimination by transformation with pSU6. C. trachomatis was transformed with pSU6 and isolated by limiting dilutions in the presence of ATc immediately upon identification of fluorescent inclusions (+pSU6). Transformants underwent an additional round of limiting dilutions in the absence of ATc (−pSU6). McCoy cell monolayers were infected with each strain. WT, wild type. (A) Forty hours p.i., samples were examined for GFP expression (green), fixed, and probed for chlamydial Hsp60 (red). Elimination of pSU6 resulted in a loss of green fluorescence as well a unique late-inclusion phenotype with a central aggregate of Chlamydia surrounded by an apparently vacuous lumen lacking visible motion, similar to that observed of inclusions produced by infection with the plasmid-free isolate C. trachomatis L2 (25667R) (23). α-Hsp60, anti-Hsp60 antibody. Bar = 10 µm. (B) Relative number of copies of the 16S rRNA region, the native pL2 plasmid (pgp7-pgp8), and gfp were determined by qPCR (data are represented as means plus SDs; n = 3). Neither plasmid was detected in DNA extracted from transformants in the absence of ATc, indicating effective curing.
trpA deletion by FRAEM.
The trpBA operon encodes tryptophan synthase capable of utilizing indole for the synthesis of tryptophan (25). Several members of Chlamydia such as C. trachomatis serovars A and C carry mutations in trpBA and are thus incapable of productive growth in indole-supplemented medium lacking tryptophan (26, 27). Recently, synthesis of TrpB was also disrupted in serovar D by random chemical mutagenesis (4). Due to the thorough characterization of the operon as well as the previous report indicating that mutation produced nonlethal effects, trpA was targeted for deletion as proof of principle. To construct pSUΔtrpA, an amplicon spanning trpA with additional ~3-kb flanking sequences was introduced into pSU6 (Fig. 4). The trpA open reading frame was removed and replaced with an ~2.2-kb cassette consisting of bla and gfp under the regulation of constitutive promoters. Given that the mutagenesis rate was unknown, both antibiotic selection and fluorescence were included in order to discern true homologous recombinants which have exchanged trpA with the cassette from spontaneously resistant C. trachomatis which may develop over time (24). The mCherry gene was included in the backbone of the plasmid, providing a real-time, visual method of separately distinguishing the presence of the integrating cassette (GFP) and the conditional-suicide vector (mCherry). This FRAEM approach enabled the identification of successful mutants by fluorescence microscopy without disturbing chlamydial growth.
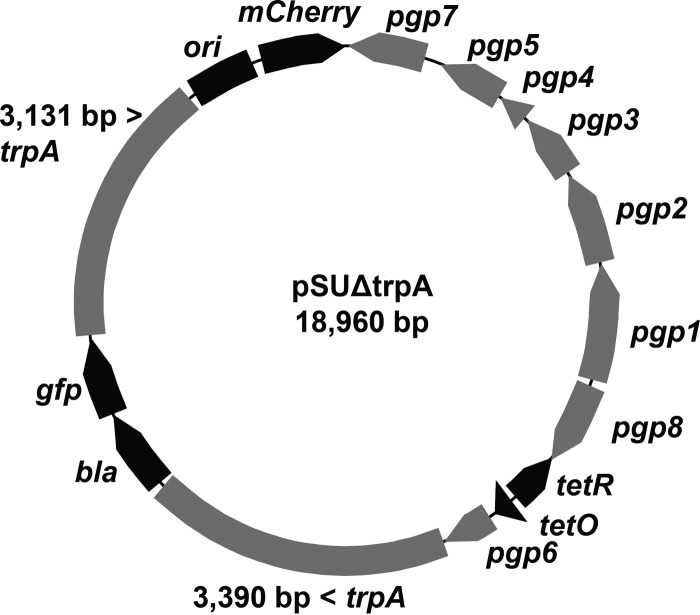
FIG 4 .
pSUΔtrpA vector map. pSU6 was modified for deletion of trpA by insertion of a cassette containing selection markers bla and gfp surrounded by chlamydial DNA corresponding to ca. 3 kb of genomic sequence flanking trpA. The constitutively expressed mCherry gene is included as a fluorescent marker of maintenance of the vector backbone.
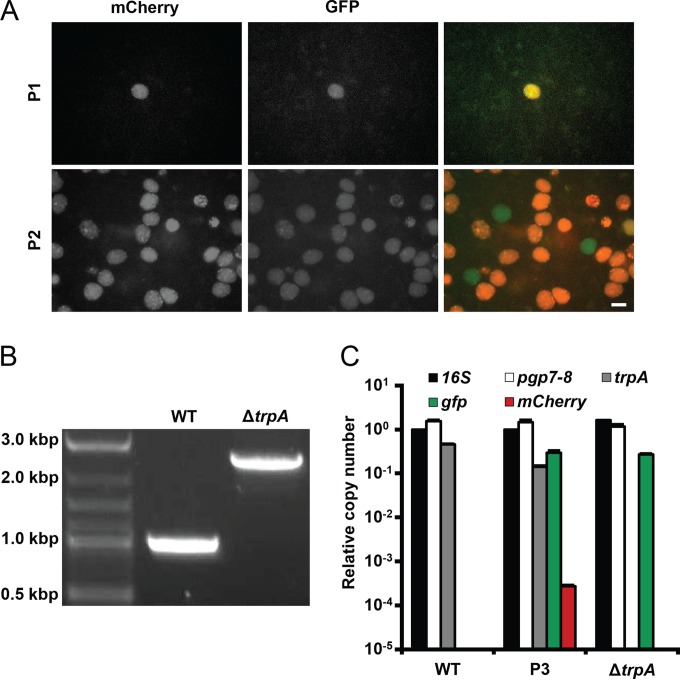
In order to ensure maintenance of the plasmid, C. trachomatis was transformed (P0) and subsequently passaged every 48 h (P1 and P2) in the presence of 50 ng/ml ATc. Four days after transformation (P1, 36 h p.i.), isolated inclusions were observed, each expressing both green and red fluorescence (Fig. 5A). Samples were harvested and used to infect the subsequent passage (P2). Thirty-six hours p.i., a significant increase in the number of inclusions was observed compared to the same time point of P1. While most were both red and green, several dim inclusions expressing only green fluorescence were also identified. The presence of green fluorescence and the absence of red fluorescence were suggestive of successful integration of the cassette and loss of the pSUΔtrpA backbone, respectively. Thus, transformants were separated and expanded by limiting dilutions. DNA was extracted from the ΔtrpA candidate as well as wild-type C. trachomatis and analyzed by PCR for changes in the amplicon with primers surrounding trpA (Fig. 5B). An increase in length of approximately 1.4 kb was observed, as expected for successful recombination (deletion of the 762-bp trpA gene and insertion of the 2,165-bp cassette results in an increase of 1,403 bp). Copy numbers of native pL2 plasmid, trpA, gfp, and the mCherry gene relative to the 16S rRNA region were determined by qPCR for the ΔtrpA candidate, wild-type C. trachomatis, and an additional sample of expanded chlamydial transformants which had not yet undergone limiting dilutions (P3) (Fig. 5C). P2 did not provide sufficient DNA for reliable analysis, so further expansion was required. Neither trpA nor mCherry gene copies were detected in the ΔtrpA candidate, indicating successful deletion of the target gene as well as elimination of the pSUΔtrpA backbone. Exchange of trpA with the cassette and the presence of a single insertion were confirmed by whole-genome sequencing.
FIG 5 .
Deletion of trpA. (A) C. trachomatis was transformed with pSUΔtrpA and passaged every 48 h for two rounds (P1 and P2) in the presence of 50 ng/ml ATc. Green fluorescence and red fluorescence were used as indicators of the presence of the integrating cassette and the vector backbone, respectively. Observation of inclusions expressing exclusively green fluorescence in P2 suggested successful homologous recombination and vector elimination. Bar = 10 µm. (B) DNA extracted from the ΔtrpA candidate was analyzed by PCR with primers surrounding the trpA target site. An increase in amplicon length of approximately 1.4 kb was observed, as expected for successful recombination (deletion of 762-bp trpA and insertion of a 2,165-bp cassette results in an increase of 1,403 bp). (C) Copy numbers of native pL2 plasmid (pgp7-pgp8), trpA, gfp, and the mCherry gene relative to the 16S rRNA region were determined by qPCR for the ΔtrpA candidate, wild-type C. trachomatis, and an additional sample of expanded chlamydial transformants which had not yet undergone limiting dilutions (P3) (data are represented as means plus SDs; n = 3). P2 provided insufficient DNA for reliable analysis. trpA and the mCherry gene were not detected in the ΔtrpA mutant, indicative of successful recombination, while the native pL2 plasmid was detected in all samples.
Contrary to chlamydial transformation with nonrecombining vectors, transformation with pSUΔtrpA did not eliminate the native pL2 plasmid, as indicated by pgp7-pgp8 amplification. The significant disparity between copies of gfp and the mCherry gene in the P3 mixed population suggests rapid cassette integration and elimination of the vector backbone. Although transformants carrying the nonrecombining pSU6 plasmid maintained the vector throughout numerous rounds of expansion, trpA deletion required maintenance of pSUΔtrpA through only one developmental cycle, as indicated by the emergence of green inclusions lacking red fluorescence only one passage after transformation. Thus, it is plausible that the brief presence of the allelic exchange vector is insufficient to cure the native plasmid, unlike the prolonged presence of nonrecombining vectors throughout numerous replication cycles.
Utilization of indole by the ΔtrpA mutant.
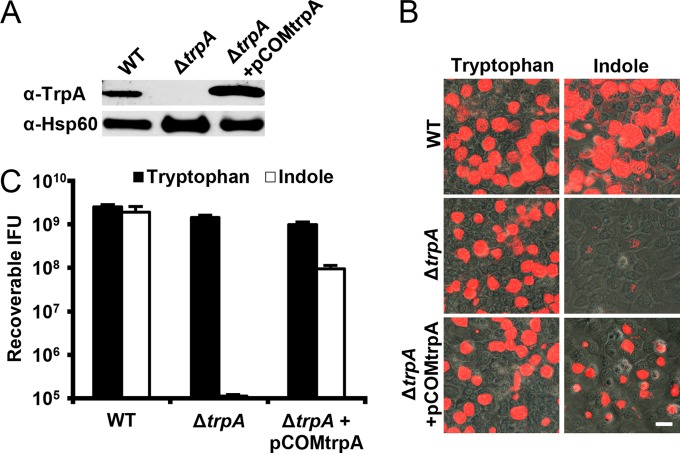
Prior to indole utilization studies, the mutation was complemented with a copy of the trpA open reading frame, as is routine with genetically tractable organisms. Since resistance to β-lactam antibiotics was introduced during homologous recombination, aadA was included in the complementation vector, permitting the use of spectinomycin as the selective pressure for isolating transformants. Expression of complementary trpA was simplified by use of the constitutive Neisseria meningitidis promoter utilized in pGFP::SW2 due to the complex nature of trpBA regulation by trpR (3, 28). Wild-type, the ΔtrpA mutant, and the ΔtrpA mutant complemented with pCOMtrpA were probed for TrpA and Hsp60 (Fig. 6A). TrpA was not detected in the deletion mutant, while complementation restored expression.
FIG 6 .
Utilization of indole by ΔtrpA. (A) Wild-type, the ΔtrpA mutant, and the ΔtrpA mutant with pCOMtrpA were probed for TrpA and Hsp60 as a loading control. Deletion and complementation of trpA eliminated and restored TrpA levels, respectively. (B) HeLa cells infected with wild-type, the ΔtrpA mutant, and the ΔtrpA mutant carrying pCOMtrpA were grown in medium containing either tryptophan or indole. Forty-eight hours p.i., the samples were fixed and stained with Chlamydia-specific antilipopolysaccharide (red). All samples produced inclusions in the presence of tryptophan. However, only wild-type and the ΔtrpA mutant carrying pCOMtrpA developed inclusions in the presence of indole. Bar = 10 µm. (C) Forty-eight hours p.i., C. trachomatis was harvested from replicate samples, and recoverable IFU were determined (data are represented as means plus SDs; n = 3). The detection of infectious progeny from only the wild-type bacteria and the ΔtrpA mutant carrying pCOMtrpA when grown in indole indicated that the deletion of trpA prevented utilization of the indole substrate for chlamydial development.
C. trachomatis was grown in medium containing either tryptophan or indole for direct utilization or conversion by tryptophan synthase, respectively. Infection appeared comparable among all strains when grown in the presence of tryptophan (Fig. 6B). However, the ΔtrpA mutant was unable to develop inclusions when forced to utilize indole. Complementation restored the appearance of inclusions. Forty-eight hours p.i., C. trachomatis was harvested, and recoverable IFU for each strain were determined (Fig. 6C). Wild-type, the ΔtrpA mutant, and the complemented ΔtrpA mutant produced comparable numbers of infectious progeny in the presence of tryptophan. However, with only indole present, the ΔtrpA mutant was incapable of productive growth. While complementation resulted in restored production of infectious progeny, wild-type levels were not achieved. This was not unexpected, as proper, wild-type regulation of trpBA was not restored by complementation with trpA under artificial, constitutive expression.
Target versatility.
Previously reported allelic exchange in C. psittaci provided significant insight into the potential for manipulation of chlamydial genetics. Insertion of point mutations at the 16S rRNA region while using an unstable transformation vector demonstrated the efficiency of homologous recombination in Chlamydia (12). In this work, we attempted to increase the volume of integrated genetic material by regulating the stability of the transformation vector. The ability to insert a complete drug resistance gene would remove requirements restricting targets to unique point mutations which provide antibiotic resistance. The deletion of trpA and insertion of two open reading frames demonstrated that, with the use of a suicide vector, targets need not be localized to specific genomic points, provided sequence homology of the construct and genome are sufficient for recombination. However, the possibility remained that the trpA region of C. trachomatis possesses sequence uniquely susceptible to allelic exchange.
To evaluate the target versatility of this approach, FRAEM, as described for trpA, was repeated for deletion of open reading frames ctl0063, ctl0064, and ctl0065 (966 bp, 1,194 bp, and 1,176 bp, respectively). While little concerning CTL0065 is known, CTL0063 and CTL0064 are type III secretion system effectors translocated into the host cell upon infection (22, 29). As was the case with pSUΔtrpA, transformation of pSUΔctl0063, pSUΔctl0064, and pSUΔctl0065 resulted in deletion of the respective targets. The rate of emergence of green fluorescent inclusions from the dual-fluorescent population was comparable among all transformants and comprised the majority of isolates recovered after limiting dilutions. PCR analysis of DNA extracted from green inclusions with primers surrounding each site indicated the predicted band shifts resulting from exchange of the target sequence for the cassette (Fig. 7A). Each recombination event was specific and independent, as confirmed by whole-genome sequencing of Δctl0063, Δctl0064, and Δctl0065 mutants. Samples were probed for CTL0063, CTL0064, CTL0065, TrpA, and Hsp60 to confirm loss of protein for anticipated targets (Fig. 7B). Although sequencing confirmed that nucleotide manipulation was specific and did not interfere with adjacent open reading frames, homologous recombination at ctl0063 produced a polar effect, resulting in the loss of both CTL0063 and CTL0064. This was not unexpected, as ctl0063 and ctl0064 are cotranscribed (22). The increase in nucleotides separating ctl0064 from the shared promoter of ctl0063 upon insertion of the dual cassette likely disrupted transcription of the downstream open reading frame. Although this poses a limitation unique to manipulating cotranscribed sequences, complementation of deletions remains effective in confirming the cause of phenotypic changes in chlamydial mutants produced by homologous recombination. The presence of target sequences ctl0063, ctl0064, ctl0065, and trpA and the relative levels of the native pL2 plasmid were determined by qPCR (Fig. 7C). Only the specific target sequence of each deletion mutant was not detected, and in all cases, the native pL2 plasmid was maintained, as additionally confirmed by whole-genome sequencing. The similarities shared throughout the process of mutagenizing all four sequences suggest the potential for application of FRAEM to a variety of targets.
FIG 7 .
Target versatility. (A) Δctl0063, Δctl0064, Δctl0065, ΔtrpA, and wild-type C. trachomatis were analyzed by PCR with primers immediately surrounding ctl0063, ctl0064, ctl0065, and trpA. Within each mutant, only the amplicon surrounding the predicted target indicated an increase in size. (B) Δctl0063, Δctl0064, Δctl0065, ΔtrpA, and wild-type C. trachomatis were probed for CTL0063, CTL0064, CTL0065, TrpA, and Hsp60 as a loading control. Due to cotranscription of ctl0063 and ctl0064, the Δctl0063 mutation produced a polar effect on protein levels of CTL0064 (22). Δctl0064, Δctl0065, and ΔtrpA eliminated only the predicted target protein. (C) The number of copies of the 16S rRNA region, the native pL2 plasmid (pgp7-pgp8), ctl0063, ctl0064, ctl0065, and trpA relative to those in the wild-type C. trachomatis were determined by qPCR analysis of Δctl0063, Δctl0064, Δctl0065, ΔtrpA, and wild-type C. trachomatis DNA (data are represented as means plus SDs; n = 3). Only the predicted target sequence of each mutation was not detected, while pL2 was detected at comparable levels from all samples.
DISCUSSION
Gene inactivation via mutagenesis is considered essential for definitively assigning function and fulfilling the molecular Koch’s postulates (30). However, application of this experimental criterion to the study of Chlamydia has proved challenging, as traditional molecular-genetic techniques utilized for genetically tractable organisms such as E. coli have been difficult to adapt to the chlamydial system. In order to circumvent this impasse, novel approaches including chemical mutagenesis by exposure to EMS or gene disruption with group II introns were developed for use with Chlamydia. While these approaches demonstrate tremendous progress in chlamydial genetics, these methods are not without limitations. Aside from the highly laborious screening required of random chemical mutagenesis and the use of proprietary algorithms for the design of group II introns, both methods are restricted to disruption of open reading frames in order to truncate protein products. Neither provides a means to completely delete genes or the flexibility of exchanging sequences for modified constructs granted by traditional molecular-genetic techniques such as allelic exchange.
In this work, we demonstrated that by controlling expression of chlamydial pgp6, we have generated a suicide vector enabling, for the first time, regulation of plasmid stability, the curing of the native pL2 plasmid, targeted genomic sequence deletion and allelic exchange, and the simultaneous insertion of multiple genes into the C. trachomatis L2 genome. This will enable more-nuanced genomic modifications such as region deletions and exchange of genes for those with modified domains as well as promoters. Furthermore, the use of fluorescent markers permits the observation and identification of successfully generated mutants without sacrificing the bacterial population for DNA extraction and screening—a significant bottleneck when attempting to isolate specific mutants from heterogenic communities of obligate intracellular pathogens.
Previously, a variety of viable chlamydial mutants were produced through random chemical mutagenesis (7). In addition to these mutants, here we present the successful deletion of four target genes including two known type III secretion system effectors while maintaining viable progeny. Although this suggests the presence of numerous available targets in C. trachomatis, disruption of essential genes is of particular concern when handling the minimalist chlamydial genome. Genetic manipulations that abolish infectivity are tantamount to lethal mutations. Thus, in addition to targets that interfere with basic replication, any targets that disrupt infectivity or invasion would not be feasible for investigation using EMS or TargeTron systems. Even the assessment that such targets are essential may not be confirmed by these methods as, when targeting such genes, there is no indication of lethality besides the absence of recoverable mutants, an ambiguous result that may be the product of lethal mutation or technical failure. Here, we demonstrate that during FRAEM, successful transformation acts as a precursor to the generation of mutants. In all cases, green fluorescent recombinant C. trachomatis bacteria were consistently identified one passage after the observation of transformants. Therefore, an absence of mutants after multiple rounds of passaging transformants has the potential to act as a method of indicating lethality. This may be confirmed by allelic exchange of the native gene for one constructed under regulation of an inducible promoter. The essential gene may still be characterized by exchange with alleles with specific domain modifications. Additionally, mutations that do not disrupt replication but abolish infectivity would theoretically continue to produce progeny for a single replication cycle. Such green fluorescent, noninfectious elementary bodies would be observable within a mature inclusion and available for examination upon host cell lysis.
As has been previously reported, we also used spectinomycin as a second selective pressure in C. trachomatis (31). The potential for generating isolates with multiple mutations will continue to rise with the number of available selective pressures in Chlamydia. The ability to replace the drug resistance gene within the targeting sequence of the suicide vector grants this approach the flexibility to target additional sites within a previously mutagenized genome. Furthermore, due to the high rate of lateral transfer and genomic recombination, it is likely that mutations of different genes present in distinct chlamydial isolates can be combined into the genomes of progeny by simultaneously applying multiple selective pressures during coinfection. In the future, the isolation of strains with multiple mutations may provide substantive benefits for the dissection of chlamydial functions utilizing multiple or redundant components. The examples of genomic manipulation presented in this work demonstrate the potential of the suicide vector and FRAEM as tools for advancing chlamydial research while establishing the genetic tractability of C. trachomatis.
MATERIALS AND METHODS
Cell cultures and organisms.
C. trachomatis serovar L2 (LGV 434) and derivative mutants were cultivated and examined for indole utilization in HeLa 229 cell monolayers (CCL-2.1; ATCC). McCoy cell monolayers (CRL-1696; ATCC) were used for all additional assays. Except where specified, host cells were routinely maintained at 37°C in an atmosphere of 5% CO2 and 95% humidified air in RPMI 1640 containing 2 mM l-glutamine (Gibco) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (Gibco). Elementary bodies were purified from HeLa cells by centrifugation through MD-76R (diatrizoate meglumine and diatrizoate sodium injection USP; Mallinckrodt Pharmaceuticals) density gradients (DG purified) as previously described (32).
Cloning.
pBOMB4-Tet-mCherry was kindly provided by Ted Hackstadt (Laboratory of Intracellular Parasites, Hamilton, MT) (18). pSU6 was constructed by removing pgp6 from pBOMB4-Tet-mCherry by divergent PCR amplification and religation, followed by exchange of the mCherry gene with pgp6 amplified from C. trachomatis L2 genomic DNA by insertion/deletion PCR as previously described (33). Template for amplification of the dual-fluorescence/drug resistance cassette was assembled by inserting gfp amplified from pBOMB4-Tet-mCherry into pUC19 downstream of bla by insertion/deletion PCR, producing pUC19G. In order to construct the suicide vector backbone for deletion of target genes, gfp in pSU6 was replaced with the mCherry gene amplified from pBOMB4-Tet-mCherry by insertion/deletion PCR, producing pSUmC. aadA amplified from Gateway pDONR223 (Invitrogen) was introduced into the HindIII restriction site of pUC18, resulting in pUC18A. Regions spanning trpA, ctl0063, ctl0064, and ctl0065 with additional ~3-kbp flanking arms were amplified from C. trachomatis L2 genomic DNA and inserted into pUC18A by insertion/deletion PCR. Target gene open reading frames were removed by divergent amplification, and the linearized PCR product was ligated to a 2.2-kbp amplicon spanning bla and gfp from pUC19G in order to produce homologous recombination targeting sequences with cassettes providing both fluorescence and drug resistance. These targeting sequences were fused to pSUmC by insertion/deletion PCR, generating pSUΔtrpA, pSUΔctl0063, pSUΔctl0064, and pSUΔctl0065. pCOMa was constructed by introducing aadA from pDONR223 and the mCherry gene with the Neisseria meningitidis promoter from pGFP::SW2 into pBOMB4-Tet-mCherry by insertion/deletion PCR, thereby removing the anhydrotetracycline (ATc)-inducible mCherry gene and tetR (3). gfp was then replaced with trpA amplified from C. trachomatis L2 genomic DNA by insertion/deletion PCR, producing pCOMtrpA. All primers were custom DNA oligonucleotides from Integrated DNA Technologies (see Table S1 in the supplemental material). Q5 high-fidelity DNA polymerase and Quick Ligation kits (New England Biolabs) were used for all PCR amplifications and ligations, respectively.
Transformation and FRAEM.
C. trachomatis L2 was transformed and isolated as previously described with modifications (22). pCOMtrpA was transformed with 500 µg/ml spectinomycin in lieu of penicillin G sodium salt. During transformation with suicide vector-based constructs, 50 ng/ml ATc was added to the medium to prevent premature loss of plasmid. After successful transformation of gene deletion constructs, the observation of inclusions expressing exclusively green fluorescence was used as a cue to begin isolating clonal populations by limiting dilutions in the absence of drugs, as previously described (22). DNA was extracted from wells containing C. trachomatis-infected monolayers (34), and relative counts of the 16S rRNA region, ctl0063, ctl0064, ctl0065, trpA, mCherry gene, gfp, and the native pL2 plasmid were determined by quantitative real-time PCR using the Bio-Rad CFX96 real-time system, iTaq Universal SYBR green supermix (Bio-Rad), and appropriate primers (see Table S1 in the supplemental material). Traditional PCR was employed to amplify target regions for identification of band shifts (Table S1). C. trachomatis mutants were DG purified, and DNA was extracted and analyzed by whole-genome sequencing (ACGT) (35).
Immunostaining and microscopy.
Western blot analysis of CTL0063, CTL0064, CTL0065, TrpA, and Hsp60 was performed as described previously (4, 22, 29). Anti-TrpA was kindly generated by Grant McClarty (University of Manitoba) and kindly provided by Harlan Caldwell (NIH/NIAID). Progeny counts were determined by staining recoverable IFU as previously described (36). Infected host cells were probed with anti-Hsp60 and anti-LPS for visualization of C. trachomatis. All images were acquired by epifluorescence microscopy using a 40× objective on an Olympus CKX41 inverted microscope equipped with an Olympus DP12 camera.
Indole rescue.
The ability of C. trachomatis strains to utilize indole as a substrate to synthesize tryptophan was performed as previously described (26, 27). Briefly, HeLa cell monolayers were maintained in Dulbecco modified Eagle medium (DMEM) (Gibco). For analysis of chlamydial growth in indole, medium was replaced with custom medium DMEM without l-tryptophan (UCSF Cell Culture Facility) with 100 µM indole (Sigma) 20 h prior to infection. All media were supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (Gibco).
Whole-genome sequencing data accession number.
Sequences reported in this paper have been deposited in the NCBI Sequence Read Archive under accession number PRJNA298309.
SUPPLEMENTAL MATERIAL
Primers used in this work.
ACKNOWLEDGMENTS
We are grateful to Ted Hackstadt and Harlan Caldwell for critically reading the manuscript and kindly providing reagents. We also thank Bill Goldman and Virginia Miller for helpful comments.
This work was supported by a Public Health Service grant from the National Institutes of Health, NIAID (AI065530) to K. A. Fields.
NIAID had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Mueller KE, Wolf K, Fields KA. 2016. Gene deletion by fluorescence-reported allelic exchange mutagenesis in Chlamydia trachomatis. mBio 7(1):e01817-15. doi:10.1128/mBio.01817-15.
REFERENCES
- 1.Centers for Disease Control and Prevention 2014. Sexually transmitted disease surveillance 2013. Division of STD Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 2.Schachter J. 1978. Chlamydial infections. N Engl J Med 298:428–435. doi: 10.1056/NEJM197802232980805. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, Nelson DE, Caldwell HD. 2011. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci U S A 108:7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson CM, Fisher DJ. 2013. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS One 8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen B, Valdivia R. 2014. A chemical mutagenesis approach to identify virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis. Methods Mol Biol 1197:347–358. doi: 10.1007/978-1-4939-1261-2_20. [DOI] [PubMed] [Google Scholar]
- 7.Kokes M, Dunn JD, Granek JA, Nguyen BD, Barker JR, Valdivia RH, Bastidas RJ. 2015. Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe 17:716–725. doi: 10.1016/j.chom.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newland JW, Green BA, Holmes RK. 1984. Transposon-mediated mutagenesis and recombination in Vibrio cholerae. Infect Immun 45:428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selvaraj G, Iyer VN. 1983. Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol 156:1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 11.Skrzypek E, Haddix PL, Plano GV, Straley SC. 1993. New suicide vector for gene replacement in yersiniae and other gram-negative bacteria. Plasmid 29:160–163. doi: 10.1006/plas.1993.1019. [DOI] [PubMed] [Google Scholar]
- 12.Binet R, Maurelli AT. 2009. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc Natl Acad Sci U S A 106:292–297. doi: 10.1073/pnas.0806768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tam JE, Davis CH, Wyrick PB. 1994. Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can J Microbiol 40:583–591. doi: 10.1139/m94-093. [DOI] [PubMed] [Google Scholar]
- 14.Schweizer HP. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol 6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 15.Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. 2013. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun 81:636–644. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong S, Yang Z, Lei L, Shen L, Zhong G. 2013. Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J Bacteriol 195:3819–3826. doi: 10.1128/JB.00511-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickstrum J, Sammons LR, Restivo KN, Hefty PS. 2013. Conditional gene expression in Chlamydia trachomatis using the tet system. PLoS One 8:e76743. doi: 10.1371/journal.pone.0076743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connell CM, Nicks KM. 2006. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology 152:1601–1607. doi: 10.1099/mic.0.28658-0. [DOI] [PubMed] [Google Scholar]
- 21.Demars R, Weinfurter J, Guex E, Lin J, Potucek Y. 2007. Lateral gene transfer in vitro in the intracellular pathogen Chlamydia trachomatis. J Bacteriol 189:991–1003. doi: 10.1128/JB.00845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller KE, Fields KA. 2015. Application of beta-lactamase reporter fusions as an indicator of effector protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PLoS One 10:e0135295. doi: 10.1371/journal.pone.0135295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson JH, Whitmire WM, Crane DD, Wicke L, Virtaneva K, Sturdevant DE, Kupko JJ III, Porcella SF, Martinez-Orengo N, Heinzen RA, Kari L, Caldwell HD. 2008. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun 76:2273–2283. doi: 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binet R, Maurelli AT. 2005. Frequency of spontaneous mutations that confer antibiotic resistance in Chlamydia spp. Antimicrob Agents Chemother 49:2865–2873. doi: 10.1128/AAC.49.7.2865-2873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miles EW. 1995. Tryptophan synthase. Structure, function, and protein engineering. Subcell Biochem 24:207–254. [PubMed] [Google Scholar]
- 26.Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD, McClarty G. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J Biol Chem 277:26893–26903. doi: 10.1074/jbc.M203937200. [DOI] [PubMed] [Google Scholar]
- 27.Caldwell HD, Wood H, Crane D, Bailey R, Jones RB, Mabey D, Maclean I, Mohammed Z, Peeling R, Roshick C, Schachter J, Solomon AW, Stamm WE, Suchland RJ, Taylor L, West SK, Quinn TC, Belland RJ, McClarty G. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest 111:1757–1769. doi: 10.1172/JCI17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akers JC, Tan M. 2006. Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J Bacteriol 188:4236–4243. doi: 10.1128/JB.01660-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hower S, Wolf K, Fields KA. 2009. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol 72:1423–1437. doi: 10.1111/j.1365-2958.2009.06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falkow S. 1988. Molecular Koch’s postulates applied to microbial pathogenicity. Rev Infect Dis 10(Suppl 2):S274–S276. doi: 10.1093/cid/10.Supplement_2.S274. [DOI] [PubMed] [Google Scholar]
- 31.Lowden NM, Yeruva L, Johnson CM, Bowlin AK, Fisher DJ. 2015. Use of aminoglycoside 3′ adenyltransferase as a selection marker for Chlamydia trachomatis intron-mutagenesis and in vivo intron stability. BMC Res Notes 8:570. doi: 10.1186/s13104-015-1542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 31:1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geiser M, Cèbe R, Drewello D, Schmitz R. 2001. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques 31:88–92. [DOI] [PubMed] [Google Scholar]
- 34.Wolf K, Malinverni R. 1999. Effect of azithromycin plus rifampin versus that of azithromycin alone on the eradication of Chlamydia pneumoniae from lung tissue in experimental pneumonitis. Antimicrob Agents Chemother 43:1491–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeffrey BM, Suchland RJ, Quinn KL, Davidson JR, Stamm WE, Rockey DD. 2010. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect Immun 78:2544–2553. doi: 10.1128/IAI.01324-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf K, Fischer E, Hackstadt T. 2000. Ultrastructural analysis of developmental events in Chlamydia pneumoniae-infected cells. Infect Immun 68:2379–2385. doi: 10.1128/IAI.68.4.2379-2385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this work.