ABSTRACT
Coagulase (Coa) and Efb, secreted Staphylococcus aureus proteins, are important virulence factors in staphylococcal infections. Coa interacts with fibrinogen (Fg) and induces the formation of fibrin(ogen) clots through activation of prothrombin. Efb attracts Fg to the bacterial surface and forms a shield to protect the bacteria from phagocytic clearance. This communication describes the use of an array of synthetic peptides to identify variants of a linear Fg binding motif present in Coa and Efb which are responsible for the Fg binding activities of these proteins. This motif represents the first Fg binding motif identified for any microbial protein. We initially located the Fg binding sites to Coa’s C-terminal disordered segment containing tandem repeats by using recombinant fragments of Coa in enzyme-linked immunosorbent assay-type binding experiments. Sequence analyses revealed that this Coa region contained shorter segments with sequences similar to the Fg binding segments in Efb. An alanine scanning approach allowed us to identify the residues in Coa and Efb that are critical for Fg binding and to define the Fg binding motifs in the two proteins. In these motifs, the residues required for Fg binding are largely conserved, and they therefore constitute variants of a common Fg binding motif which binds to Fg with high affinity. Defining a specific motif also allowed us to identify a functional Fg binding register for the Coa repeats that is different from the repeat unit previously proposed.
IMPORTANCE
Staphylococcus aureus infections are a major health problem that affects an estimated 50 million people globally and causes the death of about 20,000 Americans each year. A number of experimental vaccines have been developed during the past years. However, these vaccines have all failed in clinical trials. The ability of S. aureus to form an Fg shield surrounding and protecting bacterial cells from clearance may explain why the vaccines are failing. Furthermore, S. aureus coagulase can induce the formation of a fibrin(ogen) shield in experimental abscess models which surrounds and protects bacteria in the microcolony from clearance. In this study, we identified for the first time a microbial Fg binding motif. Variants of this motif are present in coagulase and Efb. Our results provide a molecular basis for the rational design of inhibitors that could potentially prevent the formation of the obstructing Fg shield.
INTRODUCTION
Fibrinogen (Fg) is a dimeric glycoprotein, with each half composed of three polypeptides: Aα, Bβ, and γ; it is best known for its role in blood coagulation. In this process, thrombin proteolytically converts Fg to fibrin, which then spontaneously assembles into the structural core of the clot (1). However, Fg also plays a critical role in the innate immune defense against pathogens (1, 2). A genetically engineered mouse expressing a mutant form of Fg that is not recognized by the leukocyte integrin αMβ2 showed a profound impediment in clearing Staphylococcus aureus following intraperitoneal inoculation (3). Fg also interacts with the complement system and modulates complement-dependent clearance of bacteria (4, 5).
S. aureus is an opportunistic pathogen capable of causing a variety of infections, from superficial skin infections to life-threatening diseases such as endocarditis, pneumonia, and sepsis (6). It produces a number of virulence factors (VFs) and, surprisingly, many of these VFs bind Fg with high affinity and specificity. Furthermore, studies have shown that the Fg binding activity is critical for the virulence potential of at least some of these VFs (7–11). Recent studies of secreted Fg binding S. aureus VFs have pointed to yet another mechanism of Fg-dependent inhibition of bacterial clearance. Efb, a secreted 16-kDa S. aureus protein, assembles an Fg protective shield around the bacteria, which results in impaired clearance of the organism (7). To achieve this, Efb has to bind to C3b deposited on the surface of the bacterium via its C-terminal folded C3d binding domain, and the disordered N-terminal Fg binding domain (12–14) needs to recruit Fg. Furthermore, Efb can directly interfere with the interactions of neutrophils with Fg (13), interactions that are primarily mediated by the integrin αMβ2. This inhibition is not the consequence of a direct competition between Efb and αMβ2 for the same binding site in Fg (13).
The secreted Fg binding protein coagulase (Coa) and von Willebrand factor binding protein (vWbp) colocalize with fibrin(ogen) during abscess formation in a mouse model of S. aureus infection and form part of a protective structure that borders the uninfected tissue and appears to prevent phagocytes from accessing and clearing bacteria in the center of the abscess (15).
Coa is a “historic” S. aureus protein that is best known for its ability to induce blood coagulation, which enables the classification of the staphylococcal genus into coagulase-positive or -negative species. More recent studies have shown that Coa is a critical VF in staphylococcal diseases (15). Coa is comprised of the D1D2 domain in the N-terminal part and tandem repeats of a 27-residue-long segment in the C-terminal part. Coa induces blood coagulation by activating prothrombin through insertion of the Ile1-Val2 N terminus of the Coa D1D2 domain into the Ile16 pocket of prothrombin, inducing a conformational change and a functional active site in the serine protease (16). The Coa/prothrombin complex then recognizes Fg as a specific substrate and converts this protein into fibrin (17). The crystal structure of the Coa/prothrombin complex revealed that exosite 1 of α-thrombin, which is the Fg recognition site, is blocked by the D2 domain of Coa (16). This observation raises questions concerning the nature of Fg recognition and subsequent cleavage by the Coa/prothrombin complex.
Coa can interact with Fg directly without the aid of prothrombin, and this Fg binding activity was tentatively located to the C-terminal part of Coa (18), where a C-terminal 27-residue-long sequence is tandemly repeated (19). This sequence is relatively conserved, but the number of repeats varies from 5 to 8 in different strains (19). In this study, we have characterized the Fg binding activity of Coa and identified for the first time an Fg binding motif in the Coa repeat region. The Fg binding motif in Coa is structurally and functionally related to the Fg binding sites within Efb, and it binds Fg with surprisingly high affinity. The motif defines a functional register for the Coa repeat region, which is different from the repeat unit previously proposed.
RESULTS
Staphylococcal Coa contains multiple fibrinogen binding sites.
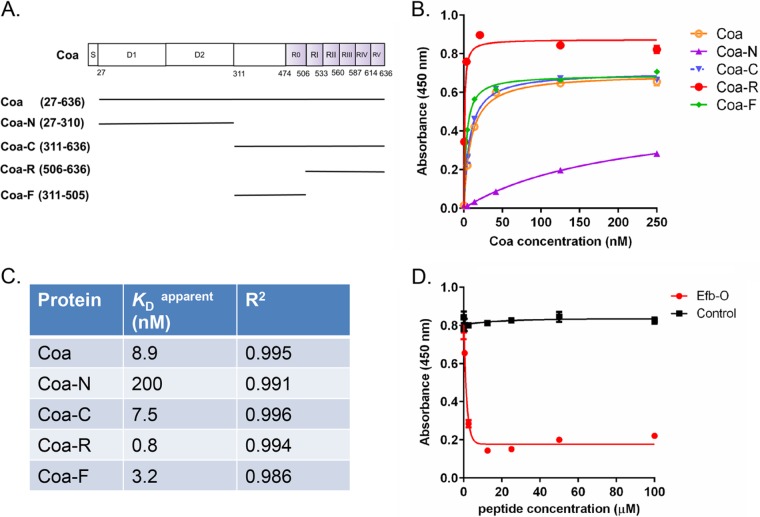
We first constructed a panel of glutathione-S-transferase (GST)–fusion proteins that covered different regions of Coa (Fig. 1A) and examined their Fg binding activities by using enzyme-linked immunosorbent assay (ELISA)-type binding experiments (Fig. 1B). The results confirmed earlier observations (18) that Coa interacts with Fg primarily through the C-terminal part of the protein (Coa-C, residues 311 to 636), which we predicted to be disordered using the DisEMBL program (20). Fg binding to Coa-C was a concentration-dependent process that exhibited saturation kinetics and had an apparent KD (concentration required for half-maximal binding) of 7.5 nM (Fig. 1C). The tandem repeat region of Coa (Coa-R, residues 506 to 636) bound to Fg in a similar way but with a higher affinity (apparent KD, 0.8 nM) than Coa-C (Fig. 1B and C). A recombinant protein corresponding to the segment between D2 and Coa-R was therefore constructed (Coa-F, residues 311 to 505). This protein bound to Fg with an apparent KD of 3.2 nM. The N-terminal D1D2 domain of Coa (Coa-N, residues 27 to 310) harboring the prothrombin binding site also interacted with Fg; however, the affinity observed (apparent KD, 200 nM) was much lower than those recorded for the Coa-C fragments, and therefore Coa-N was not further examined in this study.
FIG 1 .
Staphylococcal Coa contains multiple fibrinogen binding sites. (A) Schematic presentation of the Coa structural organization and fragments generated in this study. S, signal peptide; D1 and D2, prothrombin binding domains; R0 to RV, Fg binding repeats. (B) Concentration-dependent binding of GST-tagged Coa proteins to immobilized Fg. (C) Apparent KD values for Coa proteins binding to Fg, corresponding to concentrations required for half-maximum binding and the goodness of fit (R2) determinations. (D) Concentration-dependent inhibition of Coa protein (4 nM) binding to Fg by peptide Efb-O. Bovine serum albumin was used as a control.
The fibrinogen binding sites in Coa and Efb are functionally related.
Efb is another secreted S. aureus protein where the Fg binding activity has been located to a disordered region in the N-terminal part of the protein (13). We previously identified two related Fg binding segments, which we named Efb-O (residues 68 to 98) and Efb-A (residues 30 to 67). The Efb-O segment has higher affinity for Fg than does Efb-A; however, the two motifs likely bind to the same region in Fg, since Efb-O effectively inhibits Efb-A binding to the host protein (13). Because the Fg binding activities in Efb and Coa are both located in disordered regions and both proteins can induce a protective Fg-containing barrier around staphylococci, we explored the possibility that the Fg binding motifs in these two proteins are functionally related and possibly target the same domain in Fg. To this end, we used a competition ELISA where the binding of Coa to Fg-coated wells was determined in the presence of increasing concentrations of a synthetic peptide version of Efb-O. The peptide Efb-O effectively inhibited Coa binding to Fg (Fig. 1D), suggesting that the Fg binding activities of Coa and Efb are functionally related and that the two proteins likely bind to the same or overlapping sites in Fg.
Residues in EfB-O important for fibrinogen binding.
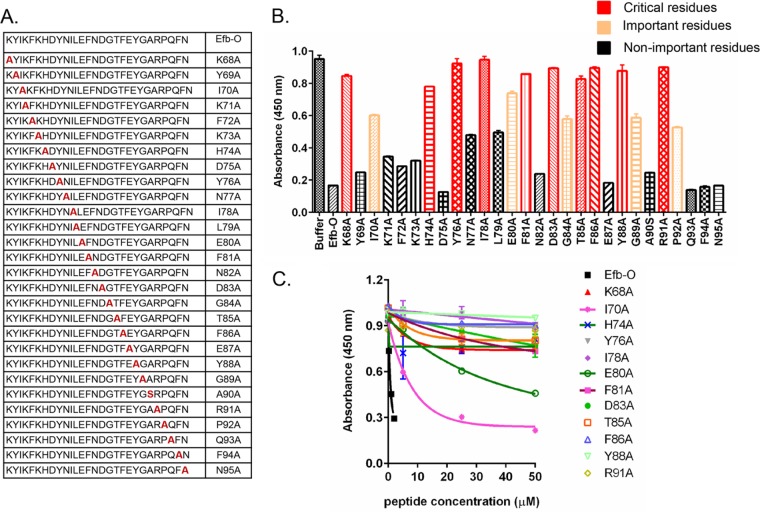
To determine the residues in Efb-O that are important for Fg binding, we used an alanine scanning approach. A panel of Efb-O variant peptides were synthesized where each residue in the sequence was individually replaced with Ala (or Ser when the native amino acid was Ala) (Fig. 2A). We then examined the ability of these synthetic peptides to compete with the binding of recombinant Efb-O (5 nM) to immobilized Fg. The inhibitory activity of the peptides was compared at a fixed concentration (2 µM) for each peptide (Fig. 2B) and at increasing concentrations for selected peptides (Fig. 2C). As the Efb-O sequence is found in a disordered segment of the protein, the peptides are likely to be very flexible in solution and Ala residue substitutions are unlikely to change the conformational freedom of the peptides. Therefore, it is reasonable to assume that a peptide’s relative inhibitory activity corresponds to its relative affinity for Fg and reflects the functional importance of the replaced residue in the Fg binding process.
FIG 2 .
Residues in EfB-O important for fibrinogen binding. (A) The panel of Efb-O variant peptides synthesized. (B) Efb-O variant peptides inhibited Efb-O protein (5 nM) binding to immobilized Fg in ELISA-type competition experiments. Each peptide (2 µM) was mixed with Efb-O protein (5 nM) and incubated in the Fg wells for 1 h. Data for residues critical for Fg binding (absorbance, ≥0.80), residues important for Fg binding (0.52 ≤ absorbance < 0.80), and residues not important for Fg binding (absorbance, <0.52) are shown. (C) Concentration-dependent inhibition of Efb-O (5 nM) binding to immobilized Fg by selected peptides.
As expected, the control parent peptide Efb-O efficiently blocked the corresponding recombinant protein Efb-O from binding to Fg, demonstrating that peptide Efb-O has full Fg binding activity in this assay. Surprisingly, Ala substitution of 15 residues distributed throughout the 25-amino-acid-long Efb-O segment resulted in loss (absorbance, ≥0.80) (Fig. 2B, red bar) or significant reduction (0.52 ≤ absorbance < 0.80) (Fig. 2B, orange bar) in inhibitory activity, suggesting that residues throughout the segment are involved in Fg binding. The 12 peptides with lowest inhibitory activities were further examined at increasing concentrations of the peptides. Of these, only 2 (I70A and E80A) showed a concentration-dependent increase in inhibitory activity, whereas the remaining peptides had essentially no inhibitory activity with all the concentrations tested (Fig. 2C). Thus, residues K68, H74, Y76, I78, F81, D83, T85, F86, Y88, and R91 (Fig. 2B, in red) are critical for binding of Efb-O to Fg, whereas residues I70, E80, G84, G89, and P92 (Fig. 2B, in orange) play some but less important roles in the Fg interaction. Thus, these results define a 25-amino-acid-long Fg binding motif in the Efb-O segment.
Coa-F contains Efb-like fibrinogen binding motifs.
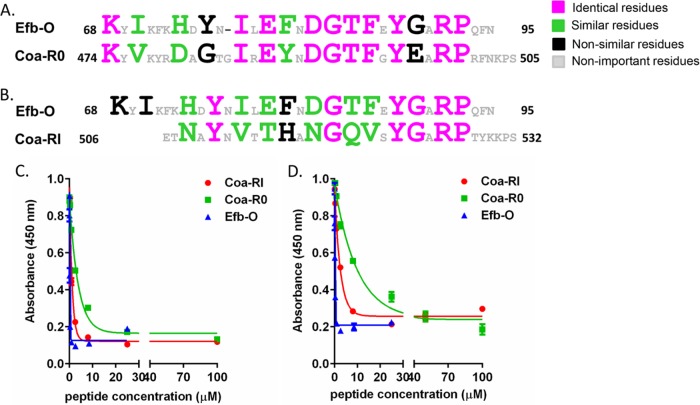
Next, we investigated if sequences similar to the Fg binding motifs in Efb could be found in Coa. In fact, some sequence similarities between Efb and Coa have been previously noticed (21). The C-terminal part of Coa harbors tandem repeats of a 27-residue segment, and this region has been shown to bind Fg (18) (Fig. 1A and B). However, an Fg binding motif has not been identified in the repeat region. An initial BLAST search identified a subsegment corresponding to Coa residues 474 to 505 that showed 56% amino acid identity and 75% similarity to that of the Efb-O motif (see Fig. S1 in the supplemental material). This subsegment, which we named Coa-R0, is mostly located in the N-terminal extension of the previously identified repeat region. Strikingly, of the residues in Efb-O determined to be critical or important for Fg binding (Fig. 2B, residues in red or orange, and 3A, residues in pink, green, or black), all but two are identical (Fig. 3A, residues in pink) or similar (Fig. 3A, residues in green) in Coa-R0, indicating that Coa-R0 likely constitutes an Efb-like Fg binding motif. When the Efb-O sequence and the following Coa sequence (Coa-RI, residues 506 to 532) were overlaid, we found remnants of the Efb motif also in this Coa sequence (Fig. 3B, residues in pink and green). This observation suggests that this Coa segment also may bind Fg.
FIG 3 .
Coa contains Efb-like fibrinogen binding motifs. (A and B) Comparison of amino acid sequences of Efb-O with Coa-R0 (residues 474 to 505) (A) and with Coa-RI (residues 506 to 532) (B). Large letters in Efb-O indicate the residues critical or important for Fg binding. (C and D) Concentration-dependent inhibition of Efb-N protein (residues 30 to 104; 2 nM) (C) and Coa-C protein (residues 311 to 636; 2 nM) (D) binding to Fg by peptides Coa-R0, Coa-RI, and Efb-O.
The Efb-like motifs in Coa bind fibrinogen.
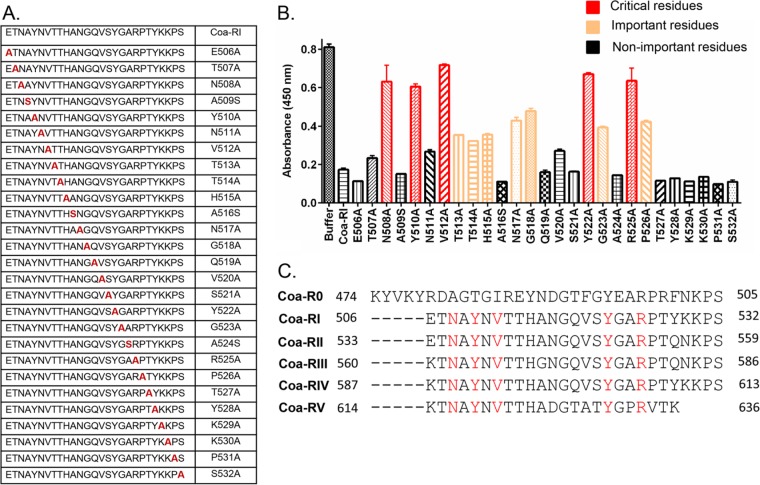
To examine if the Efb-like motifs in Coa bind Fg, we synthesized peptides corresponding to the Coa-R0 and Coa-RI regions and compared their Fg binding activities to that of peptide Efb-O in a competition ELISA. Microtiter wells were coated with Fg, and the binding of the recombinant N-terminal segment of Efb (Efb-N) (Fig. 3C) or Coa-C (Fig. 3D) was quantitated in the presence of increasing concentrations of the different synthetic peptides. As expected, both peptides Coa-R0 and Coa-RI as well as the control peptide Efb-O effectively inhibited the binding of Efb-N and Coa-C to Fg, demonstrating that Coa-R0 and Coa-RI both bind Fg and that they likely target the Efb binding site(s) in Fg. Furthermore, peptide Efb-O was the most potent inhibitor in both assays, and peptide Coa-RI was somewhat more effective than Coa-R0 despite the fact that the Coa-R0 sequence is more similar to that of Efb-O than is Coa-RI. These observations suggest that some of the residues unique to Coa-RI participate in the Fg interaction. To determine what residues in Coa-RI are important for Fg binding, we again used an alanine scanning approach. The peptide panel generated and tested is shown in Fig. 4A. Binding of a fixed concentration (2 nM) of Coa-C protein to immobilized Fg was determined in the presence of a fixed concentration (50 µM) of the individual peptides in the Ala scanning panel (Fig. 4B). Interestingly, the results revealed a similar pattern to that observed for Efb-O, showing that the Ala substitution of over 12 residues distributed throughout the 27-amino-acid-long Coa-RI segment resulted in loss in inhibitory activity (absorbance, ≥0.60) (Fig. 4B, red bar) or a significant reduction in inhibitory activity (0.32 ≤ absorbance < 0.60) (Fig. 4B, orange bar). These results suggest that, similar to Efb-O, residues in almost the entire segment of Coa-RI are involved in Fg binding. The results further showed that 5 peptides, N508A, Y510A, V512A, Y522A, and R525A (Fig. 4B, red bar) essentially lost their ability to inhibit Coa-C binding, indicating that these residues are critical for Coa-RI to bind to Fg with high affinity. Noticeably, these residues are absolutely conserved in all subsequent Coa repeat units (Coa 506 to 636) (Fig. 4C, residues in red), suggesting that these 5 residues, N508, Y510, V512, Y522, and R525, are critical for the Fg binding activity of the repeated motifs in Coa, e.g., Coa-RI through Coa-RV. We further identified that residues T513, T514, H515, N517, G518, G523, and P526 are important for Fg binding (0.32 ≤ absorbance < 0.60) (Fig. 4B, orange bar). By comparing the Efb-O and Coa-RI motifs, we found that most of the critical and important residues are conserved in the two motifs but that residues T513 and H515 are unique to the Coa-RI Fg binding motif (Fig. 3B; see also Fig. 6, below).
FIG 4 .
The residues in Coa-RI important for fibrinogen binding. (A) The panel of Coa-RI variant peptides synthesized. (B) Coa-RI variant peptides (50 µM) inhibited Coa-C (2 nM) binding to immobilized Fg in competition ELISAs. Data for residues critical for Fg binding (absorbance, ≥0.60), residues important for Fg binding (0.32 ≤ absorbance < 0.60), residues not important for Fg binding (absorbance, <0.32) are shown. (C) Proposed Fg binding motifs in the Coa tandem repeats segment. Shown in red are residues critical for Fg binding that are conserved in Coa-RI to Coa-RV.
FIG 6 .
Fg binding motif. (A) Comparison of amino acid sequences of Fg binding sites Efb-A, Efb-O, Coa-R0, and Coa-RI. Red, residues critical for Fg binding; orange, residues important for Fg binding. (B) Amino acid frequencyies in sequences of Efb-A, Efb-O, Coa-R0, and Coa-RI. The size of a letter indicates the frequency of the amino acid in each corresponding position. Red, residues critical for Fg binding in Efb-O or Coa-RI; orange, residues important for Fg binding in Efb-O and Coa-RI.
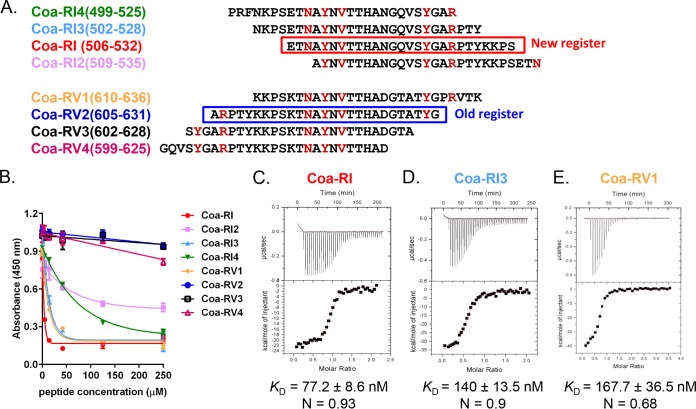
The functional register of the Coa repeat region.
In previous studies, the repeated unit in Coa was proposed to start with residues alanine A497 in S. aureus strain Newman (22, 23). This register was based exclusively on comparisons of Coa sequences from different strains. Initial studies showed that the unit of this register did not bind Fg (Fig. 5, peptide Coa-RV2). To experimentally define a register of the repeats based on their Fg binding function, we synthesized a panel of peptides, 27 residues in length, with each peptide having 22 to 24 residues overlapping with a subsequent peptide and largely spanning repeat I (RI) and repeat V (RV) (Fig. 5A). The Fg binding activities of these peptides were then investigated in a competition ELISA in which the binding of the Coa-C protein to Fg-coated microtiter wells was determined in the presence of increasing concentrations of peptide (Fig. 5B). Peptide Coa-RI (residues 506 to 532) appears to be the most potent inhibitor among these eight peptides, suggesting that Coa-RI has the highest affinity for Fg (Fig. 5B). The order of the five critical residues appears to be required for high-affinity binding to Fg, as peptides Coa-RI3, Coa-RI4, and Coa-RV1, which contain the five critical residues (Fig. 4C and 5A, in red) in the same order as Coa-RI, also are effective inhibitors (and thus potent Fg binders) although somewhat weaker then Coa-RI (Fig. 5B). On the other hand, peptides Coa-RI2, Coa-RV2, and Coa-RV3, as well as Coa-RV4, have significantly reduced or a complete loss of their Fg binding activity compared to that of Coa-RI (Fig. 5B). In these peptides, the order of the five critical residues is altered (Fig. 5A).
FIG 5 .
The functional register within the repeat region. (A) Schematic presentation of the Coa peptides synthesized. Residues in red are those critical for Fg binding; the red box shows the new Fg binding register proposed in this study; the blue box shows the old register based on sequence comparison. (B) Concentration-dependent inhibition of Coa-C (4 nM) protein binding to Fg by Coa peptides. Peptide Coa-RI is the most potent inhibitor. (C to E) Characterization of Coa peptide binding to Fg D-fragment by ITC. Binding isotherms for the interaction of Fg-D with Coa peptide Coa-RI (C), Coa-RI3 (D), and Coa-RV1 (E) were generated by titrating the peptides (~200 µM) into an ITC cell containing 6 µM Fg-D. (Top panels) Heat differences upon injection of Coa peptides. (Bottom panels) Integrated heat of injections. The data were fitted to a single binding site model and binding affinities are expressed as dissociation constants (KD) calculated as the reciprocal of the association constants determined by using Microcal Origin software. N represents the binding ratio. Data in panels C to E are representative of results from 5 independent experiments.
To generate more quantitative binding data for the Coa peptide-Fg interaction, we used isothermal titration calorimetry and titrated the Coa peptides into a solution containing a fixed concentration of Fg-D fragment. Peptide Coa-RI bound to Fg-D fragment with a high affinity (KD = 77.2 ± 8.6 nM) and a binding stoichiometry of 0.93 (Fig. 5C; see also Table S2 in the supplemental material), suggesting that one molecule of Coa-RI bound to one Fg-D fragment. Peptide Coa-RI3 and peptide Coa-RV1 bound the Fg-D fragment with a KD values of 140 ± 13.5 nM and 167.7 ± 36.5 nM, respectively (Fig. 5D and E; see also Table S2). These results corroborated our competition ELISA results (Fig. 5B) and showed that Coa-RI bound Fg-D with higher affinity than Coa-RI3 and Coa-RV1. Collectively, these results suggest that Coa-RI represents the functional repeat unit that interacts with Fg and define the functional (Fg binding) register of the repeat section, as outlined in Fig. 4C.
The fibrinogen binding motif is important for Efb-mediated inhibition of phagocytosis.
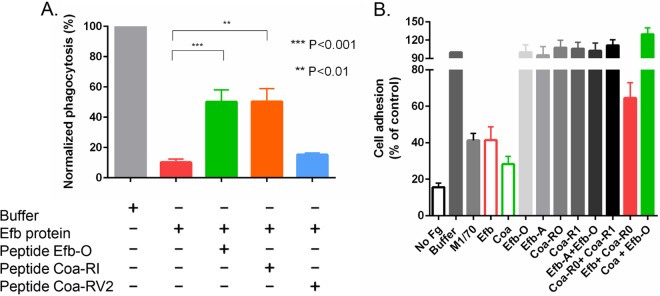
We previously reported that Efb can assemble an antiphagocytic Fg-containing shield tht surrounds bacteria. This activity requires Efb to simultaneously bind bacterium-associated C3b and recruit Fg from the plasma (7). To evaluate if the identified Fg binding motif is involved in this process, phagocytosis of fluorescein isothiocyanate (FITC)-labeled S. aureus by freshly isolated human neutrophils was quantified in the presence of human plasma (the source for complement components and Fg), Efb (0.1 µM), and synthetic peptides (50 µM). As shown earlier (7), the Efb protein efficiently inhibited phagocytosis. Addition of the peptides Efb-O and Coa-RI, which both bind Fg with high affinity, reversed Efb’s inhibition of bacterial phagocytosis, whereas addition of the non-Fg binding peptide Coa-RV2 did not affect phagocytosis. These data suggest that the identified Fg binding motif is critical for Efb’s ability to form the protective Fg shield (Fig. 6).
Coa and Efb use the fibrinogen binding motif to inhibit monocytic cell adherence to fibrinogen.
As Coa and Efb share a similar Fg binding motif and could competitively inhibit each other’s binding to Fg, we explored if Coa, like Efb, can inhibit the adherence of monocytic cells (THP-1) to Fg. THP-1 cells adhere to immobilized Fg primarily through the αMβ2 integrin (also named Mac-1 or CR3) (3). Consequently an antibody against the αM integrin subunit (M1/70) inhibited the adherence of THP-1 cells to immobilized Fg (Fig. 7B). We previously showed that Efb blocks the αMβ2-dependent adherence of neutrophils to Fg (13). Here, as expected, the Efb protein also efficiently inhibited the Fg adherence of THP-1 cells (Fig. 7B). Similar to Efb, Coa, which harbors multiple Fg binding motifs, also inhibited THP-1 cell adherence to an Fg surface. Interestingly, individual synthetic peptides Efb-O or Efb-A, each containing one single Fg binding motif or a combination of both Efb-O and Efb-A peptides, did not inhibit THP-1 cell adherence. The same phenomena were observed using the peptides Coa-R0 and Coa-RI, suggesting that the inhibition of THP-1 cell adherence to Fg requires that an effective inhibitor contains at least two copies of the identified Fg binding motif. A possible mode of action could involve Fg binding of the inhibitor protein via both binding sites, positioning the inhibitor so that it sterically interferes with the integrin-Fg interaction. In support of this model, we found that an excess amount of a single peptide actually reversed the inhibitory effect of Efb or Coa proteins (Fig. 7B). In this experiment, 50 µM peptide Coa-R0 or Efb-O was mixed with Efb (0.2 µM) or Coa (0.5 µM) protein, respectively, in the cell adherence assay. The results showed neutralization of the inhibitory activities of both recombinant Efb and Coa proteins. Collectively, we showed here that the secreted staphylococcal proteins Efb and Coa use the identified Fg binding motif to interact with Fg and inhibit phagocytosis of bacteria and neutrophil adherence to Fg.
FIG 7 .
Efb and Coa use an Fg binding motif to inhibit phagocytosis and monocytic cell adherence. (A) Phagocytosis of FITC-labeled S. aureus cells by human neutrophils in the presence of 5% human plasma and Efb (0.1 µM) in the absence or presence of Fg binding peptide Efb-O or Coa-RI, or with non-Fg binding peptide, Coa-RV2. Peptides were all used at 50 µM. Phagocytosed bacteria were analyzed by flow cytometry. (B) Adherence of THP-1 cells to Fg immobilized in 48-well plates was inhibited by the addition of monoclonal αM antibody M1/70 (20 µg/ml), Efb (0.2 µM), and Coa (0.5 µM) proteins. Addition of a single peptide alone (Efb-O or Efb-A, as well as Coa-RI and Coa-R0, respectively; 0.5 µM each) or a combination of two peptides together (Efb-A + Efb-O or Coa-R0 + Coa-RI), at 0.5 µM each, did not inhibit THP-1 adherence. Preincubation of Efb protein (0.2 µM) with peptide Coa-R0 (50 µM) (Efb + Coa-R0) or Coa protein (0.5 µM) with peptide Efb-O (50 µM) (Coa + Efb-O) reversed the inhibitory activities elicited by Efb or Coa proteins. The presented normalized data were determined by setting the control group (buffer) at 100. In panels A and B, data are mean results ± standard errors; n ≥ 3. **, P < 0.01; ***, P < 0.001 (unpaired two-tailed t test).
DISCUSSION
The pathogenic potential of S. aureus is a consequence of its multitude of VFs that have evolved to interact with a number of host molecules. As a result, S. aureus can survive and thrive at many tissue sites in the host and cause a wide range of diseases. Fg is a surprisingly common target for many of the staphylococcal VFs. The known Fg binding staphylococcal proteins largely fall into two groups: a family of structurally related cell wall-anchored proteins of the MSCRAMM (microbial surface components recognizing adhesive matrix molecules) type, which include ClfA, ClfB, FnbpA, FnbpB, and Bbp/SdrE (24) and a group of secreted smaller proteins (sometimes referred to as the SERAMs [secretable expanded repertoire adhesive molecules]), which include Efb, Coa, vWbp, Emp (extracellular matrix binding protein), and Eap (extracellular adherence protein) (25). The Fg binding sites in the MSCRAMMs are located in a segment of the proteins composed of two IgG folded subdomains that interact with Fg by variants of the multistep “dock, lock, and latch” binding mechanism (26, 27). In this mechanism, a short disordered segment of Fg docks in a trench formed between the two subdomains through beta-complementation to a strand of the second subdomain. This interaction triggers conformational changes in the MSCRAMM that result in subsequent steps.
The Fg-binding SERAMs do not share a domain organization, and the mechanism(s) of Fg binding used by these proteins remains largely unknown. However, these proteins do have some features in common. First, they all interact with multiple ligands, with Fg being the common ligand among them. Second, they all contribute to S. aureus abscess formation in animal infection models (15, 28, 29). Third, intrinsically disordered regions represent a significant part of each protein, and we have previously shown that the Fg binding sites in Efb are located in its disordered region (12). A disordered protein segment is particularly suited for accommodating multiple ligand binding sites, since several interacting motifs can fit in a short segment of the protein and these motifs can be overlapping, because the segment has structural plasticity (30). Furthermore, we observed that there is a rather high frequency of sequence variation in the ligand binding motif within disordered proteins, for instance, the fibronectin binding motif in FnbpA from S. aureus, in BBK32 from Borrelia burgdorferi, and in SfbI from Streptococcus pyogenes (31–35). Sequence divergence makes it especially challenging to identify interactive motifs in these types of segments, particularly if a longer segment of the protein is involved.
Here, we demonstrated that the staphylococcal Coa protein contains multiple Fg binding sites that all are variants of a common motif, and six copies are found in Coa (from Coa-R0 to Coa-RV) from strain Newman. Two copies of this motif are also found in Efb (Efb-O and Efb-A). Using an alanine scanning approach, we identified the residues in the sequences of Efb-O and Coa-RI critical for Fg binding, and we found that these residues are largely conserved. Based on sequence similarities in Coa-R0, Coa-RI, Efb-O, and Efb-A (Fig. 6A), we have defined a core Fg binding motif (Fig. 6B). This core motif defines the Fg binding register of the tandem repeats in Coa, which is different from the previously reported repeating unit. Coa-R0 is slightly longer and more similar to Efb-O, compared to the sequences in Coa-RI to Coa-RV, which in turn are very similar to each other. Coa-RI contains five residues that are critical for Fg binding (Fig. 4B, shown in red), and these have to appear in the right order for high-affinity Fg binding (Fig. 5). These five residues are absolutely conserved in Coa-RI to Coa-RV (Fig. 4C). This core Fg binding motif has several unique characteristics. First, the motif is unusually long (21 to 26 residues) (Fig. 6B) compared to other known and well-characterized interactive motifs (36). Second, many residues throughout the length of the Fg binding motif are engaged in Fg binding, but exchange for similar residues is often tolerated. We have searched for the Efb/Coa Fg binding motif in other eukaryotic and prokaryotic proteins, but so far we have not found any hits, suggesting that the motif evolved in S. aureus rather than being horizontally transferred.
There are some intriguing similarities between the Fg binding motif in Coa/Efb and the fibronectin (Fn) binding motif previously identified in some Fn binding proteins from staphylococcal, streptococcal, and borrelia species (31, 33–35). Both motifs are found in disordered segments of the proteins (12, 37, 38), are long (~26 and ~40 residues, respectively) (13, 33), can bind to their host target with very high affinities (13, 31, 39), occur in several variant forms (31), and appear to have evolved in the microbes, since no counterpart has been found in any potential host.
Efb can protect staphylococci from phagocytosis by inducing the formation of an Fg-containing shield surrounding the bacterial surface (7). Opsonizing antibodies and phagocytes cannot penetrate the shield and access the bacteria. Formation of the Efb-dependent Fg shield associated with the bacteria requires that Efb binds to C3b deposited on the microbial surface. It is likely that Coa can also induce the formation of an Fg-containing protective shield, because its Fg binding motifs are related to those of Efb. However, Coa is not known to interact directly with the bacterial surface but could induce the formation of an Fg shield at some distance from the bacterial surface. Indeed, in the abscess model, Coa facilitates the formation of a fibrin(ogen) structure that surrounds a bacterial microcolony, and Coa has been detected in this structure (15). At these sites, it is likely that at least some of the Fg molecules are converted to fibrin through the action of Coa-activated prothrombin. We recently reported a gene-targeted mouse line that expresses FibAEK, a mutant form of Fg where the Aα chain has been selectively mutated to eliminate thrombin-mediated proteolysis. In this mouse, fibrin formation cannot occur while hemostatic capacity and clotting function are retained (40). The availability of the FibAEK mice provides a great means to better understand and dissect the role of Fg and fibrin in shield formation.
Efb and Coa also inhibit αMβ2-dependent adherence of cells to Fg. This inhibitory activity depends on the presence of at least two Fg binding motifs in the proteins, and a peptide containing a single Fg binding motif can reverse the activity of the inhibitory proteins. We can speculate on the molecular details of this inhibition (see above). Resolution of the crystal structure of a Coa-Fg complex and/or an Efb-Fg complex will provide additional clues and these studies are under way. Our studies presented here show that Coa and Efb play roles in evasion of the host defense and use the identified Fg binding motif to exert an antiphagocytosis function and block the αMβ2-dependent adherence of neutrophils/monocytes to Fg.
The motif identified in this study provides a molecular basis for the interactions of staphylococcal proteins Efb and Coa with Fg and should lead to a better understanding of their roles in the pathogenic processes. Ultimately, this information could lead to novel approaches for the prevention and treatment of staphylococcal infections.
MATERIALS AND METHODS
Ethics statement.
Study participants provided written informed consent in accordance with the Declaration of Helsinki. Our human subject research protocol was reviewed and approved by the Institutional Review Board of Texas A&M University Human Research Protection Program (HRPP; approved protocol IRB2011-0890D).
Bacterial strains, plasmids, and culture conditions.
Escherichia coli XL-1 Blue (Stratagene) was used as the host for plasmid cloning, whereas E. coli BL21 (GE Healthcare) were used for expression of GST-tagged fusion proteins. Chromosomal DNA from S. aureus strain Newman was used to amplify the Coa DNA sequence. E. coli XL-1 Blue and BL21 containing plasmids were grown on LB medium supplemented with ampicillin (100 µg/ml).
Cloning of Coa constructs.
Genomic DNA isolated from S. aureus strain Newman was used as the template for all PCR experiments with the oligonucleotide primers described in Table S1 of the supplemental material. PCR products were digested with BamHI and SalI and ligated into the pGEX-5x-1 vector (GE Healthcare). The ligation mixture was transformed into Escherichia coli XL-1 Blue cells (Stratagene), which were grown on an LB agar plates containing 100 µg/ml ampicillin to select for transformants. Insertions were confirmed by DNA sequencing.
Expression and purification of recombinant proteins.
Methods for expression and purification of recombinant Efb proteins were described earlier (13). Plasmids encoding N-terminal GST-tagged Coa fusion proteins were expressed in E. coli strain BL21. Bacteria were grown overnight at 37°C in LB containing appropriate antibiotics as described above. The overnight cultures were diluted 1:20 into fresh LB medium, and recombinant protein expression was induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside for 2 to 3 h. Bacteria were harvested by centrifugation and lysed using a French press (SLM-Aminco). Soluble proteins were purified through a glutathione-Sepharose 4B column (GE Healthcare) according to the manufacturer’s manual. Purified proteins were dialyzed into Tris-buffered saline (TBS) and stored at −20°C. Protein concentrations were determined via the Bradford assay (Pierce).
Enzyme-linked immunosorbent assays.
Wells of 4HBX microtiter plates (Thermo Scientific) were coated overnight at 4°C with 0.25 µg of human fibrinogen (diluted in phosphate-buffered saline [PBS]; Enzyme Research). After blocking the wells with 3% bovine serum albumin (BSA) in PBS, recombinant proteins were added and the plates were incubated for 1 h. Following incubation with horseradish peroxidase (HRP)-conjugated anti-GST polyclonal antibodies (1:5,000 dilution) for 1 h, the substrate o-phenylenediamine dihydrochloride (Sigma) was added. Bound proteins were quantified by measuring the absorbance at 450 nm in a microplate reader (Thermomax), and apparent KD values and goodness of fit (R2) measures were analyzed by using GraphPad Prism software version 6.05.
In the peptide inhibition assay, various concentrations of Efb or Coa peptides were mixed with a fixed concentration of Coa-GST or Efb-GST fusion protein (5 to 10 nM) in TBS, and the bound GST-fusion proteins were detected through incubation with HRP-conjugated rabbit anti-GST polyclonal antibodies (Abcam; 1:5,000 dilution). All proteins were diluted in TBS containing 1% BSA and 0.05% Tween 20, and the ELISAs were carried out at room temperature.
Preparation of Fg-D fragments.
Fg purchased from Enzyme Research was further purified using size exclusion chromatography (SEC; GE Healthcare). D fragments of Fg were generated by digestion of purified Fg (10 to 15 mg/ml) with 0.5 ml (a 50% slurry) of immobilized trypsin (Sigma) in TBS containing 10 mM CaCl2 for 6 h at 37°C. After centrifugation to remove the trypsin-immobilized resin, Fg-D fragments (~85 kDa) were purified by gel filtration on Sephacryl S-200 medium (GE Healthcare) and analyzed by SDS-PAGE.
Isothermal titration calorimetry.
The interaction between Coa peptides and the soluble, isolated Fg-D fragment was further characterized by isothermal titration calorimetry (ITC) using a VP-ITC microcalorimeter (MicroCal). The ITC cell contained 6 to 10 µM Fg-D fragment, and the syringe contained 100 to 200 µM Coa peptides in TBS (25 mM Tris, 3.0 mM KCl, 140 mM NaCl; pH 7.4). All proteins were filtered through 0.22-µm membranes and degassed for 20 min before use. The titrations were performed at 27°C using a single preliminary injection of 2 to 4 µl of Coa peptide followed by 30 to 40 injections of 5 to 8 µl with an injection speed of 0.5 µl s−1. Injections were spaced over 5-min intervals at a stirring speed of 260 rpm. Raw titration data were fitted to a one-site model of binding using MicroCal Origin version 5.0.
Synthesis of peptides.
The Efb and Coa peptides were purchased from HangHong, Inc. All the peptides were purified using high-performance liquid chromatography and were >95% pure.
Phagocytosis assay.
Phagocytosis of FITC-labeled bacteria (5 × 107/ml) were mixed with human plasma for 2 min at 37°C in the presence of recombinant Efb (0.1 µM) and synthetic peptides (50 µM). Freshly isolated human neutrophils (5 × 106/ml) were added, and phagocytosis was allowed for 15 min at 37°C. Paraformaldehyde was then added to stop the reaction, and phagocytosed bacteria were analyzed by flow cytometry (LSRII apparatus; BD).
Purification of human plasma and neutrophils.
Human plasma was prepared by collecting venous blood from 5 to 10 healthy volunteers into glass vacutainers (BD) containing the anticoagulant lepirudin (50 mg/ml) (41). Plasma was collected after centrifugation for 10 min at 4,000 rpm at 4°C, pooled, and subsequently stored at −80°C. Human neutrophils were isolated fresh from heparinized blood using the Ficoll-Histopaque gradient method (42) and used on the same day.
Fluorescent labeling of bacteria.
For the phagocytosis assay, we used the S. aureus laboratory strain Newman cultured overnight on tryptic soy blood agar (BD) at 37°C. For fluorescent labeling, bacteria were resuspended in PBS and incubated with 0.5 mg/ml FITC (Sigma) for 30 min on ice. Bacteria were washed twice with PBS, resuspended in RPMI medium with human serum albumin, and stored at −20°C until further use.
Cell adherence assay.
A monocytic cell line, THP-1, stably expressing αMβ2, was maintained in RPMI 1640 (Lonza) supplemented with 10% fetal bovine serum (FBS), 2 µM l-glutamine, 100 units/ml penicillin, and 100 µg/ml streptomycin. Prior to use, cells were harvested by centrifugation, washed, and suspended in RPMI 1640–1% human serum albumin (Octapharma). Wells of 48-well plates (Costar) were coated with 200 µl of fibrinogen (10 µg/ml; Enzyme Research) overnight at 4°C followed by 1 h at 37°C before blocking with 1% polyvinylpyrrolidone (PVP; 3,600 kDa; Sigma) for 45 min at 37°C. Subsequently, the cells were seeded at 2 × 105/well in the presence or absence of Coa or Efb recombinant proteins or synthetic peptides and incubated at 37°C for 25 min. Nonadherent cells were removed by washing gently three times with PBS–1% BSA. Adherent cells were quantitated using the CyQuant kit (Invitrogen) according to the manufacturer’s manual.
Statistics.
Statistical analyses were performed using GraphPad Prism 6.0 package, and the differences between groups were analyzed for significance using an unpaired two-tailed t test.
SUPPLEMENTAL MATERIAL
ClustalW alignment of amino acid sequence from Efb-O (Efb residues 68 to 98) and Coa from the Newman strain (Col-Newman). Sequence similarity was identified at Coa residues 474 to 505. Asterisks denote conserved residues, and two dots represent similar residues. Download
Primers for Coa constructs.
Thermodynamic parameters for the interactions of the Fg-D fragment with Coa-RI, Coa-RI3, and Coa-RV1.
ACKNOWLEDGMENTS
This work was supported by NIH grant A1020624 to M.H. and support from the Hamill Foundation to Y.-P.K.
We thank the flow cytometry core facility at IBT, TAMUHSC, for assistance and Arthur Laganowsky and Timothy J. Foster for critical reading of the manuscript.
Footnotes
Citation: Ko Y-P, Kang M, Ganesh VK, Ravirajan D, Li B, Höök M. 2016. Coagulase and Efb of Staphylococcus aureus have a common fibrinogen binding motif. mBio 7(1):e01885-15. doi:10.1128/mBio.01885-15.
REFERENCES
- 1.Mosesson MW, Siebenlist KR, Meh DA. 2001. The structure and biological features of fibrinogen and fibrin. Ann N Y Acad Sci 936:11–30. doi: 10.1111/j.1749-6632.2001.tb03491.x. [DOI] [PubMed] [Google Scholar]
- 2.Laurens N, Koolwijk P, de Maat MPM. 2006. Fibrin structure and wound healing. J Thromb Haemost 4:932–939. doi: 10.1111/j.1538-7836.2006.01861.x. [DOI] [PubMed] [Google Scholar]
- 3.Flick MJ, Du X, Witte DP, Jiroušková M, Soloviev DA, Busuttil SJ, Plow EF, Degen JL. 2004. Leukocyte engagement of fibrin(ogen) via the integrin receptor αMβ2/Mac-1 is critical for host inflammatory response in vivo. J Clin Invest 113:1596–1606. doi: 10.1172/JCI20741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlsson F, Sandin C, Lindahl G. 2005. Human fibrinogen bound to Streptococcus pyogenes M protein inhibits complement deposition via the classical pathway. Mol Microbiol 56:28–39. doi: 10.1111/j.1365-2958.2005.04527.x. [DOI] [PubMed] [Google Scholar]
- 5.Endo Y, Nakazawa N, Iwaki D, Takahashi M, Matsushita M, Fujita T. Interactions of ficolin and mannose-binding lectin with fibrinogen/fibrin augment the lectin complement pathway. J Innate Immun 2:33–42. doi: 10.1159/000227805. [DOI] [PubMed] [Google Scholar]
- 6.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 7.Ko Y, Kuipers A, Freitag CM, Jongerius I, Medina E, van Rooijen WJ, Spaan AN, van Kessel KPM, Höök M, Rooijakkers SHM. 2013. Phagocytosis escape by a Staphylococcus aureus protein that connects complement and coagulation proteins at the bacterial surface. PLoS Pathog 9:e1003816. doi: 10.1371/journal.ppat.1003816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Josefsson E, Higgins J, Foster TJ, Tarkowski A. 2008. Fibrinogen binding sites P336 and Y338 of clumping factor A are crucial for Staphylococcus aureus virulence. PLoS One 3:e2206. doi: 10.1371/journal.pone.0002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miajlovic H, Loughman A, Brennan M, Cox D, Foster TJ. 2007. Both complement- and fibrinogen-dependent mechanisms contribute to platelet aggregation mediated by Staphylococcus aureus clumping factor B. Infect Immun 75:3335–3343. doi: 10.1128/IAI.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNamara C, Zinkernagel AS, Macheboeuf P, Cunningham MW, Nizet V, Ghosh P. 2008. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science 319:1405–1408. doi: 10.1126/science.1154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flick MJ, Du X, Prasad JM, Raghu H, Palumbo JS, Smeds E, Hook M, Degen JL. 2013. Genetic elimination of the binding motif on fibrinogen for the S. aureus virulence factor ClfA improves host survival in septicemia. Blood 121:1783–1794. doi: 10.1182/blood-2012-09-453894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee LYL, Liang X, Hook M, Brown EL. 2004. Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrinogen-binding protein (Efb). J Biol Chem 279:50710–50716. doi: 10.1074/jbc.M408570200. [DOI] [PubMed] [Google Scholar]
- 13.Ko Y-, Liang X, Smith CW, Degen JL, Hook M. 2011. Binding of Efb from Staphylococcus aureus to fibrinogen blocks neutrophil adherence. J Biol Chem 286:9865–9874. doi: 10.1074/jbc.M110.199687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammel M, Sfyroera G, Ricklin D, Magotti P, Lambris JD, Geisbrecht BV. 2007. A structural basis for complement inhibition by Staphylococcus aureus. Nat Immunol 8:430–437. doi: 10.1038/ni1450. [DOI] [PubMed] [Google Scholar]
- 15.Cheng AG, McAdow M, Kim HK, Bae T, Missiakas DM, Schneewind O. 2010. Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog 6:e1001036. doi: 10.1371/journal.ppat.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. 2003. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature 425:535–539. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 17.Panizzi P, Friedrich R, Fuentes-Prior P, Richter K, Bock PE, Bode W. 2006. Fibrinogen substrate recognition by staphylocoagulase (pro)thrombin complexes. J Biol Chem 281:1179–1187. doi: 10.1074/jbc.M507956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDevitt D, Vaudaux P, Foster TJ. 1992. Genetic evidence that bound coagulase of Staphylococcus aureus is not clumping factor. Infect Immun 60:1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phonimdaeng P, O’Reilly M, Nowlan P, Bramley AJ, Foster TJ. 1990. The coagulase of Staphylococcus aureus 8325-4. Sequence analysis and virulence of site-specific coagulase-deficient mutants. Mol Microbiol 4:393–404. doi: 10.1111/j.1365-2958.1990.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 20.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. 2003. Protein disorder prediction: implications for structural proteomics. Structure 11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Bodén MK, Flock J. 1994. Cloning and characterization of a gene for a 19 kDa fibrinogen-binding protein from Staphylococcus aureus. Mol Microbiol 12:599–606. doi: 10.1111/j.1365-2958.1994.tb01046.x. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe S, Ito T, Takeuchi F, Endo M, Okuno E, Hiramatsu K. 2005. Structural comparison of ten serotypes of staphylocoagulases in Staphylococcus aureus. J Bacteriol 187:3698–3707. doi: 10.1128/JB.187.11.3698-3707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaida S, Miyata T, Yoshizawa Y, Kawabata S, Morita T, Igarashi H, Iwanaga S. 1987. Nucleotide sequence of the staphylocoagulase gene: its unique COOH-terminal 8 tandem repeats. J Biochem 102:1177–1186. [DOI] [PubMed] [Google Scholar]
- 24.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chavakis T, Wiechmann K, Preissner KT, Herrmann M. 2005. Staphylococcus aureus interactions with the endothelium: the role of bacterial “secretable expanded repertoire adhesive molecules” (SERAM) in disturbing host defense systems. Thromb Haemost 94:278–285. doi: 10.1160/TH05-05-0306. [DOI] [PubMed] [Google Scholar]
- 26.Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SVL. 2003. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115:217–228. doi: 10.1016/S0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- 27.Ganesh VK, Rivera JJ, Smeds E, Ko Y, Bowden MG, Wann ER, Gurusiddappa S, Fitzgerald JR, Höök M. 2008. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog 4:e1000226. doi: 10.1371/journal.ppat.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jongerius I, von Köckritz-Blickwede M, Horsburgh MJ, Ruyken M, Nizet V, Rooijakkers SHM. 2012. Staphylococcus aureus virulence is enhanced by secreted factors that block innate immune defenses. J Innate Immun 4:301–311. doi: 10.1159/000334604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. 2009. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J 23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. 2005. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J 272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz-Linek U, Werner JM, Pickford AR, Gurusiddappa S, Kim JH, Pilka ES, Briggs JAG, Gough TS, Höök M, Campbell ID, Potts JR. 2003. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature 423:177–181. doi: 10.1038/nature01589. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz-Linek U, Höök M, Potts JR. 2004. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol Microbiol 52:631–641. doi: 10.1111/j.1365-2958.2004.04027.x. [DOI] [PubMed] [Google Scholar]
- 33.Bingham RJ, Rudino-Pinera E, Meenan NAG, Schwarz-Linek U, Turkenburg JP, Hook M, Garman EF, Potts JR. 2008. Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc Natl Acad Sci U S A 105:12254–12258. doi: 10.1073/pnas.0803556105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raibaud S, Schwarz-Linek U, Kim JH, Jenkins HT, Baines ER, Gurusiddappa S, Hook M, Potts JR. 2005. Borrelia burgdorferi binds fibronectin through a tandem beta-zipper, a common mechanism of fibronectin binding in staphylococci, streptococci, and spirochetes. J Biol Chem 280:18803–18809. doi: 10.1074/jbc.M501731200. [DOI] [PubMed] [Google Scholar]
- 35.Talay SR, Valentin-Weigand P, Timmis KN, Chhatwal GS. 1994. Domain structure and conserved epitopes of Sfb protein, the fibronectin-binding adhesin of Streptococcus pyogenes. Mol Microbiol 13:531–539. doi: 10.1111/j.1365-2958.1994.tb00448.x. [DOI] [PubMed] [Google Scholar]
- 36.Ruoslahti E, Pierschbacher MD. 1986. Arg-Gly-Asp: a versatile cell recognition signal. Cell 44:517–518. doi: 10.1016/0092-8674(86)90259-X. [DOI] [PubMed] [Google Scholar]
- 37.Kim JH, Singvall J, Schwarz-Linek U, Johnson BJB, Potts JR, Hook M. 2004. BBK32, a fibronectin binding MSCRAMM from Borrelia burgdorferi, contains a disordered region that undergoes a conformational change on ligand binding. J Biol Chem 279:41706–41714. doi: 10.1074/jbc.M401691200. [DOI] [PubMed] [Google Scholar]
- 38.Penkett CJ, Redfield C, Jones JA, Dodd I, Hubbard J, Smith RA, Smith LJ, Dobson CM. 1998. Structural and dynamical characterization of a biologically active unfolded fibronectin-binding protein from Staphylococcus aureus. Biochemistry (Mosc) 37:17054–17067. [DOI] [PubMed] [Google Scholar]
- 39.Meenan NAG, Visai L, Valtulina V, Schwarz-Linek U, Norris NC, Gurusiddappa S, Hook M, Speziale P, Potts JR. 2007. The tandem beta-zipper model defines high affinity fibronectin-binding repeats within Staphylococcus aureus FnBPA. J Biol Chem 282:25893–25902. doi: 10.1074/jbc.M703063200. [DOI] [PubMed] [Google Scholar]
- 40.Prasad JM, Gorkun OV, Raghu H, Thornton S, Mullins ES, Palumbo JS, Ko Y-, Hook M, David T, Coughlin SR, Degen JL, Flick MJ. 2015. Mice expressing a mutant form of fibrinogen that cannot support fibrin formation exhibit compromised antimicrobial host defense. Blood 126:2047–2058. doi: 10.1182/blood-2015-04-639849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mollnes TE, Brekke OL, Fung M, Fure H, Christiansen D, Bergseth G, Videm V, Lappegard KT, Kohl J, Lambris JD. 2002. Essential role of the C5a receptor in E. coli-induced oxidative burst and phagocytosis revealed by a novel lepirudin-based human whole blood model of inflammation. Blood 100:1869–1877. [PubMed] [Google Scholar]
- 42.Bestebroer J, Poppelier MJJG, Ulfman LH, Lenting PJ, Denis CV, van Kessel KPM, van Strijp JAG, de Haas CJC. 2007. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin-mediated neutrophil rolling. Blood 109:2936–2943. doi: 10.1182/blood-2006-06-015461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ClustalW alignment of amino acid sequence from Efb-O (Efb residues 68 to 98) and Coa from the Newman strain (Col-Newman). Sequence similarity was identified at Coa residues 474 to 505. Asterisks denote conserved residues, and two dots represent similar residues. Download
Primers for Coa constructs.
Thermodynamic parameters for the interactions of the Fg-D fragment with Coa-RI, Coa-RI3, and Coa-RV1.