ABSTRACT
Herpesviruses must contend with host cell epigenetic silencing responses acting on their genomes upon entry into the host cell nucleus. In this study, we confirmed that unchromatinized herpes simplex virus 1 (HSV-1) genomes enter primary human foreskin fibroblasts and are rapidly subjected to assembly of nucleosomes and association with repressive heterochromatin modifications such as histone 3 (H3) lysine 9-trimethylation (H3K9me3) and lysine 27-trimethylation (H3K27me3) during the first 1 to 2 h postinfection. Kinetic analysis of the modulation of nucleosomes and heterochromatin modifications over the course of lytic infection demonstrates a progressive removal that coincided with initiation of viral gene expression. We obtained evidence for three phases of heterochromatin removal from an early gene promoter: an initial removal of histones and heterochromatin not dependent on ICP0, a second ICP0-dependent round of removal of H3K9me3 that is independent of viral DNA synthesis, and a third phase of H3K27me3 removal that is dependent on ICP0 and viral DNA synthesis. The presence of ICP0 in transfected cells is also sufficient to promote removal of histones and H3K9me3 modifications of cotransfected genes. Overall, these results show that ICP0 promotes histone removal, a reduction of H3K9me3 modifications, and a later indirect reduction of H3K27me3 modifications following viral early gene expression and DNA synthesis. Therefore, HSV ICP0 promotes the reversal of host epigenetic silencing mechanisms by several mechanisms.
IMPORTANCE
The human pathogen herpes simplex virus (HSV) has evolved multiple strategies to counteract host-mediated epigenetic silencing during productive infection. However, the mechanisms by which viral and cellular effectors contribute to these processes are not well defined. The results from this study demonstrate that HSV counteracts host epigenetic repression in a dynamic stepwise process to remove histone 3 (H3) and subsequently target specific heterochromatin modifications in two distinct waves. This provides the first evidence of a stepwise reversal of host epigenetic silencing by viral proteins. This work also suggests that targets capable of disrupting the kinetics of epigenetic regulation could serve as potential antiviral therapeutic agents.
INTRODUCTION
The assembly of histones onto incoming naked DNA by the recipient cell appears to have evolved in eukaryotic cells as an intrinsic response to silence foreign DNA through compaction into repressive heterochromatin (1, 2), which serves to silence the foreign DNA and prevent the expressed gene products from affecting the host cell. Chromatin has a dynamic structure that serves an important role in regulating gene expression. Chromatin structure can be modified by histone chaperones facilitating the selective addition and removal of nucleosome components, chromatin remodelers modulating nucleosome density and positioning, or a variety of chromatin-modifying enzymes directing the addition or removal of specific covalent modifications to or from histone tails. These factors are necessary to establish and maintain distinct forms of chromatin that can be interpreted by chromatin “readers” to impact transcription of genes. Chromatin that is densely compacted with regularly spaced nucleosomes through association with heterochromatin protein 1 (HP1) and covalent modifications at histone tails such as histone 3 (H3) lysine 9-trimethylation (H3K9me3) and lysine 27-trimethylation (H3K27me3) is not accessible to RNA polymerase II and transcription factors and thus is epigenetically silenced (3, 4). Conversely, chromatin that is less densely packed with nucleosomes and/or enriched in modifications such as H3 lysine 9-acetylation (H3K9ac) or lysine 4-trimethylation (H3K4me3) is accessible to RNA polymerase II and transcription factors for transcription (5). Histone-mediated interactions also regulate many processes, including transcriptional response to signaling (6–8), mRNA splicing (9), DNA repair (10), and positioning of chromatin within the three-dimensional nuclear space (11).
During productive infection, herpes simplex virus (HSV) genes are expressed in an ordered cascade of immediate-early (IE), early (E), and late (L) genes (12, 13), in a process dependent on viral and cellular transcriptional machinery (14, 15). This was originally thought to be the result of a series of gene activation steps (13), but more recent studies have shown that reversal of epigenetic silencing is also involved in the sequential activation of viral gene expression (16–18).
The double-stranded DNA genome of HSV-1 enters the host cell free of histones (19–22). Histones are known to be loaded rapidly on HSV-1 E and L promoters and then removed (17, 19). Histone H3K9me3 is also present at high levels at early times that decrease on the IE ICP0 gene promoter (23), consistent with removal of heterochromatin on IE genes. Acetylation of H3 on viral IE and E gene promoters increases during infection, consistent with the addition of euchromatin markers (24). The changes in IE gene chromatin require VP16 and associated host proteins (25, 26).
Each round of lytic infection must contend with the cellular chromatinization response to prevent silencing, initiate the ordered cascade of lytic gene expression, and facilitate production of infectious progeny. Previous studies have demonstrated that, as lytic infection of HSV-1 progresses, viral promoters associate with unstable nucleosomes that are enriched for histones bearing activating modifications (17, 27–29). Epigenetic changes can occur as both cause and consequence of transcription, and their specific roles and relative levels of importance to viral infection have generated some debate (30, 31). It has been demonstrated that efficient productive HSV-1 infection requires cellular chromatin modifiers such as the SNF2H chromatin remodeler for IE viral gene expression (32), while the CHD3 chromatin remodeler has been implicated in initial repression of gene expression (33). Additional factors such as the histone chaperone human antisilencing factor1a (Asf1a) and the exchange of histone variants have also been implicated in epigenetic regulation of HSV-1 (34, 35).
Viral proteins, including VP16 and ICP0, have also been implicated in chromatin regulation critical for lytic infection. The VP16 tegument protein activates IE gene expression through interaction with cellular factors HCF-1 and Oct-1 and the recruitment of chromatin methylation modifiers, including SETD1A methyltransferase, LSD1 (an H3K9 di- or monodemethylase), and JMJD2 (an H3K9me3 demethylase) (23). In addition, VP16 can recruit nucleosome remodelers such as BRG1 and BRM, as well as histone acetyltransferases (HATs) such as p300 and CBP, to viral IE promoters (25, 31). Accordingly, in the absence of VP16, IE gene promoters are associated with increased histone accumulation, and E gene promoters are associated with reduced histone acetylation (25).
The HSV IE ICP0 protein, a multifunctional E3 ubiquitin ligase and potent gene transactivator in infected (36, 37) and cotransfected (38–41) cells, is required for expression of early proteins such as ICP8 (36, 37, 42). During lytic infection of HeLa cells, ICP0 was found to promote the acetylation of viral chromatin and to reduce total histone accumulation on the viral genome (17). This argues that one mechanism by which ICP0 promotes viral gene expression is through regulation of chromatin on the viral genome by limiting histone association and/or promoting association with active euchromatin. ICP0 promotes the acetylation of viral chromatin by preventing the removal of histone acetylations by binding to CoREST and disrupting histone deacetylase 1 (HDAC1) binding to the HDAC1/CoREST/LSD1/REST (HCLR) repressor complex, ultimately causing relocalization of HCLR components to the cytoplasm later in infection (16, 43–45). During infection, stabilization and recruitment of CLOCK H3/H4 acetyltransferases facilitate expression of viral genes, and CLOCK overexpression can compensate for defects in ICP0-deficient viruses (46, 47). ICP0 also promotes the degradation of nuclear domain 10 (ND10) components, namely, PML, Sp100, and Daxx, which are thought to be involved in epigenetic silencing of the viral genome (48). However, the specific mechanism of ND10 silencing of the HSV genome remains to be defined. An additional restriction factor, IFI16, can promote accumulation of silencing H3K9me3 modifications on the viral genome and other foreign unchromatinized DNA (49); however, ICP0 promotes the degradation of IFI16 during infection with HSV-1 (50). These studies highlight the importance and diversity of targets of ICP0 in lytic infection associated with its ability to prevent chromatin-mediated silencing.
Previous studies of HSV-1 chromatin during lytic infection have focused mainly on IE gene promoters. Thus, studies have identified H3K9me3 heterochromatin associated with viral genomes during initial acute infection (26, 51) but excluded from replication compartments (52). Additionally, a reduction of H3K9me3 heterochromatin at the ICP0 promoter coincided with the expression of IE genes (26). However, little was known about the effects of heterochromatin on E viral promoters. In this study, we examined the kinetics of histone association and heterochromatin modifications H3K9me3 and H3K27me3 on an E gene promoter during lytic infection of primary human foreskin fibroblasts (HFFs). Additionally, we evaluated the role of the ICP0 protein by studies of two ICP0-deficient viruses that either fail to transcribe the ICP0 gene or express only a truncated ICP0 protein. We found that, during lytic infection, viral genomes were initially associated with repressive heterochromatin modifications followed by a stepwise restructuring of chromatin at viral gene promoters. The ICP0-null viruses were unable to effectively reverse host-mediated epigenetic silencing, indicating that ICP0, either directly or indirectly, impacts the level of associated histones and heterochromatin markers to facilitate viral gene expression.
RESULTS
Kinetic analysis of HSV-1 chromatin reveals early association with histones and heterochromatin modifications during lytic infection.
To examine the kinetics of chromatin association with an HSV E gene promoter, we infected HFF cells at 3 PFU/cell with wild-type (WT) KOS strain HSV-1 and collected cells at every hour postinfection (hpi) for 12 h. Cells were subsequently processed, and chromatin was analyzed by immunoprecipitation with antibodies specific for H3, H3K9me3, or H3K27me3. We performed chromatin immunoprecipitation (ChIP) analysis using quantitative PCR (qPCR) with specific primers to measure the extent of association with histones and specific histone modifications at the viral ICP8 E gene promoter normalized to the proportion of cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene DNA immunoprecipitated from the same reaction.
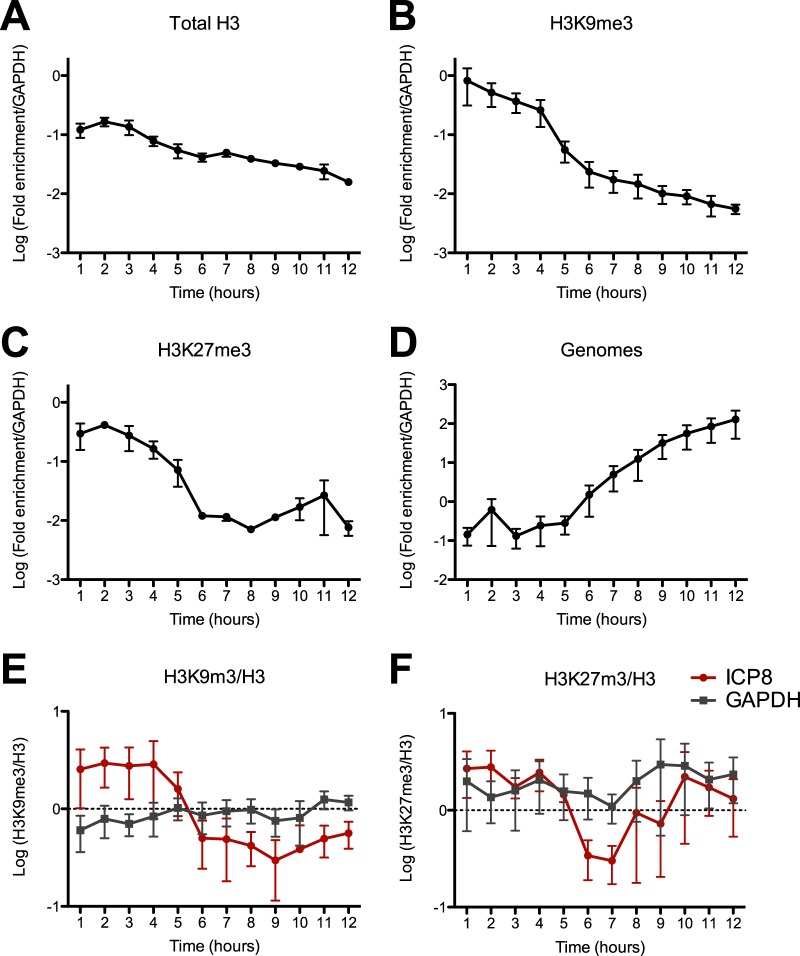
Over 12 h of infection, the levels of total histone and heterochromatin modifications remained constant at the cellular GAPDH gene promoter (not shown) but decreased dramatically at the viral ICP8 promoter (Fig. 1). We observed that the ICP8 gene promoter accumulated the highest levels of H3 and of heterochromatin modifications H3K9me3 and H3K27me3 at 1 to 2 hpi (Fig. 1A to C). Overall, relative to the initial peak of accumulation, the ICP8 promoter showed a 10-fold reduction in the levels of total H3 and a 50- to 100-fold reduction in the H3K9me3 and H3K27me3 heterochromatin modifications, respectively, during the following 10 h. To identify the time periods when chromatin levels changed most dramatically, we calculated the fold change for each hour relative to the previous hour (see Fig. S1 in the supplemental material). We observed a significant reduction in total H3 before 4 hpi, in H3K9me3 at 3 to 7 hpi, and H3K27me3 at 5 hpi (see Fig. S1). Interestingly, while the absolute levels of total H3, H3K9me3, and H3K27me3 changed rapidly during the first 4 hpi, the relative proportion of H3 bearing K9me3 or K27me3 remained constant (Fig. 1E and F). This resulted in a reduced proportion of histones bearing H3K9me3 or H3K27me3 modifications at the ICP8 promoter relative to cellular GAPDH gene sequences by 6 hpi.
FIG 1 .
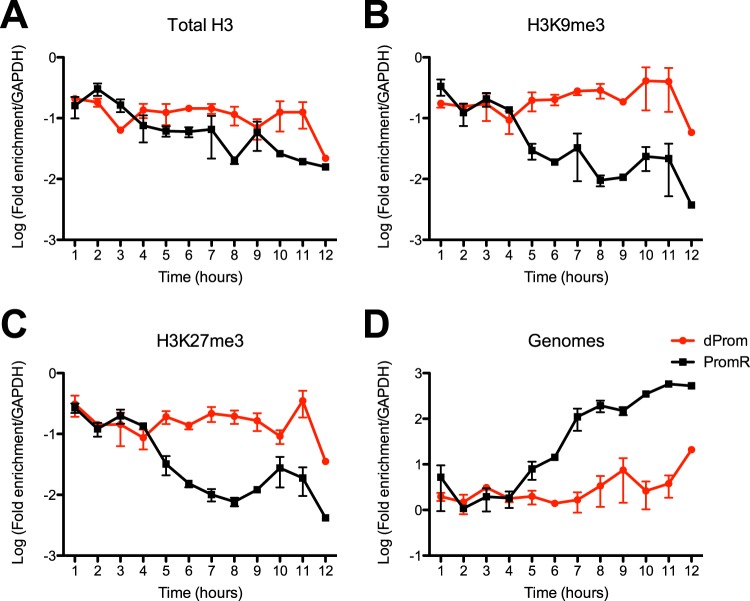
Kinetic analyses of chromatin association with HSV-1 during lytic infection. (A to C) Chromatin immunoprecipitation assay of histones and heterochromatin on the ICP8 promoter of WT HSV-1 KOS was performed for 12 h following infection of human foreskin fibroblast cells at 3 PFU/cell. ChIP was carried out using antibodies specific for total H3 (A), H3K9me3 (B), and H3K27me3 (C). The amounts of input and percentage of immunoprecipitated DNA were measured by quantitative PCR using primers specific for the ICP8 promoter and cellular GAPDH pseudogene. ChIP data are presented as the fold enrichment of immunoprecipitated ICP8 DNA relative to GAPDH gene DNA. (D) Chromatin inputs were used to calculate relative genome copy numbers. (E and F) The relative proportions of H3 bearing heterochromatin modifications were calculated from the percentage of DNA immunoprecipitated with antibodies specific for (E) H3K9me3 or (F) H3K27me3 modifications normalized to the percentage of DNA immunoprecipitated with an antibody specific to total H3. The mean values and standard errors of the means of results of at least three independent experiments are shown.
Removal of nucleosomes and H3K9me3 heterochromatin modifications occurred independently of viral DNA synthesis.
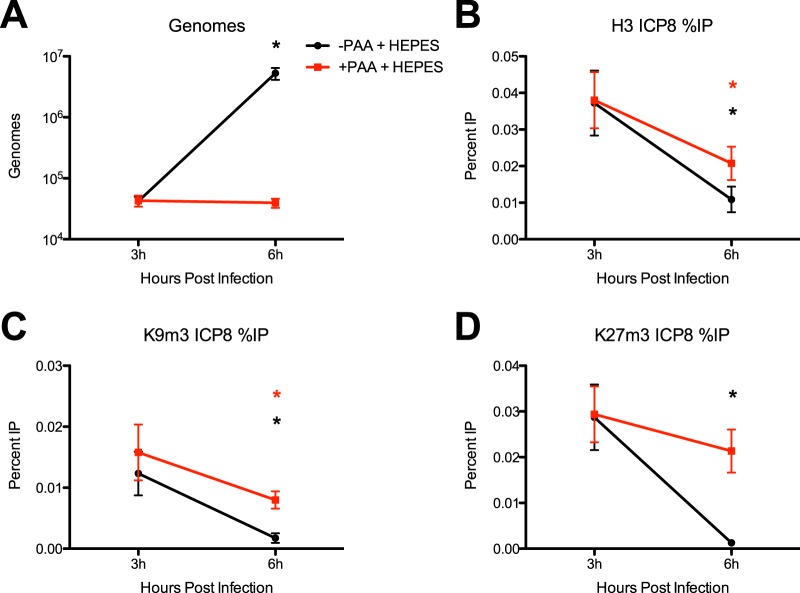
The time period of the most prominent heterochromatin removal on the ICP8 promoter relative to total H3 removal occurred after 4 hpi, coinciding with the initiation of viral DNA synthesis (Fig. 1D). Therefore, we wanted to determine whether viral DNA synthesis or potential dilution of histones and heterochromatin modification by newly synthesized genomes or both were responsible for their apparent reduction. We infected HFF cells at 3 PFU/cell with WT HSV-1 in the presence or absence of a viral DNA polymerase inhibitor, sodium phosphonoacetate (PAA). Control-treated infections demonstrated a significant increase in viral genomes between 3 and 6 hpi, while PAA-treated cells showed no increases in viral genome copy number (Fig. 2A). We performed ChIP analysis on samples isolated at 3 and 6 hpi with antibodies specific for H3, H3K9me3, and H3K27me3 as described above. We detected significant decreases in the levels of total H3, H3K9me3, and H3K27me3 in control cells between 3 and 6 hpi (Fig. 2B to D). The levels of H3 and H3K9me3 (Fig. 2B and C) decreased significantly in PAA-treated cells; however, H3K27me3 levels did not change significantly between 3 and 6 hpi (Fig. 2D) in the PAA-treated cells. These results argued that removal of histones and H3K9me3 occurred independently of viral DNA synthesis. Surprisingly, however, the H3K27me3 modification was not removed in cells treated with PAA, indicating that removal of H3K27me3 was dependent on viral DNA synthesis or the participation of a viral L gene product.
FIG 2 .
Effect of viral DNA synthesis on removal of histones and chromatin modifications. HFF cells were treated with the viral polymerase inhibitor PAA or mock treated and infected with WT HSV-1 at 3 PFU/cell to evaluate if histone removal was due to vDNA synthesis. Cell lysates were prepared at 3 and 6 hpi. (A) Relative viral genome copy numbers were determined by qPCR of viral ICP8 DNA normalized to cellular GAPDH gene DNA. (B to D) ChIP assays were carried out with antibodies specific for total H3 (B), H3K9me3 (C), and H3K27me3 (D). The fraction of ICP8 DNA immunoprecipitated was measured by qPCR, and data are shown as the means and standard errors of the means for results from at least three independent experiments. Samples with mean values that varied significantly from 3 to 6 hpi (P < 0.05, paired Student’s t test) are indicated (*).
Construction and analysis of HSV-1 ICP0 mutant viruses.
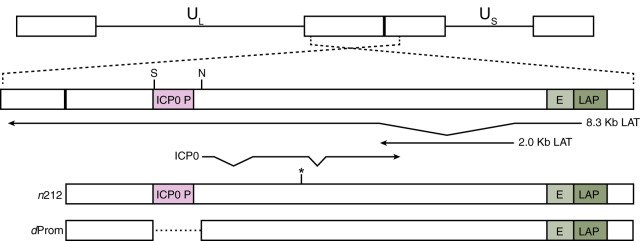
We wanted to determine whether ICP0 was facilitating the dynamic reversal in chromatin association during lytic infection to reduce chromatin silencing and promote viral gene expression. We tested two independently constructed ICP0 mutant HSV-1 strains, n212 and dProm (Fig. 3). The n212 virus (53) contains a nonsense mutation in codon 212 that results in expression of a truncated ICP0. We constructed the n212R rescued virus as the ICP0-positive (ICP0+) control virus. The dProm virus contained a 711-bp deletion of the ICP0 transcription start site and upstream promoter region that eliminated transcription of the ICP0 gene (Fig. 4A). Construction of the dProm virus and of the cognate rescued virus PromR is described in Materials and Methods. Characterization of these viruses confirmed reduced viral gene expression and replication by the ICP0− viruses compared to the rescued viruses in HFF cells (see Fig. S2 in the supplemental material), characteristic of ICP0-null mutant viruses.
FIG 3 .
HSV-1 genomic map of the ICP0 region. The unique short regions (US) and unique long regions (UL) of HSV-1 flanked by inverted repeats are shown here. A view of the ICP0 region, which encodes ICP0, the antisense primary 8.3-kb LAT transcript, and the stable 2-kb LAT intron, is magnified. The dProm mutant virus has a 711-bp deletion between the NcoI (N) and StuI (S) sites depicted on this map. The n212 mutant contains a nonsense mutation in codon 212 in exon 2 of the ICP0 transcript (indicated by a star).
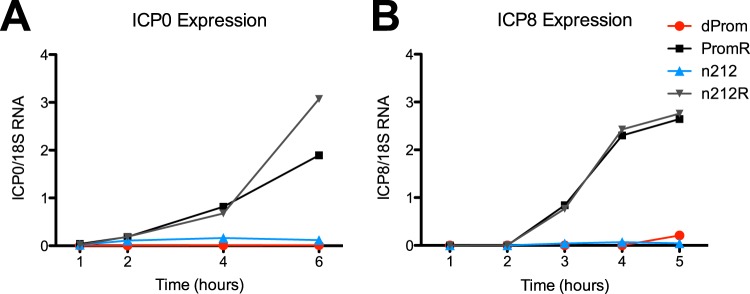
FIG 4 .
Viral transcript accumulation during lytic infection. HFF cells were infected at 3 PFU/cell with the PromR or n212R ICP0+ viruses or with the dProm or n212 ICP0-null mutant viruses. Total RNA was isolated from cell lysates and quantified using quantitative reverse transcription-PCR (qRT-PCR). Viral transcripts are expressed normalized to cellular 18S rRNA.
ICP0 mutant HSV-1 strains show no removal of nucleosomes and heterochromatin.
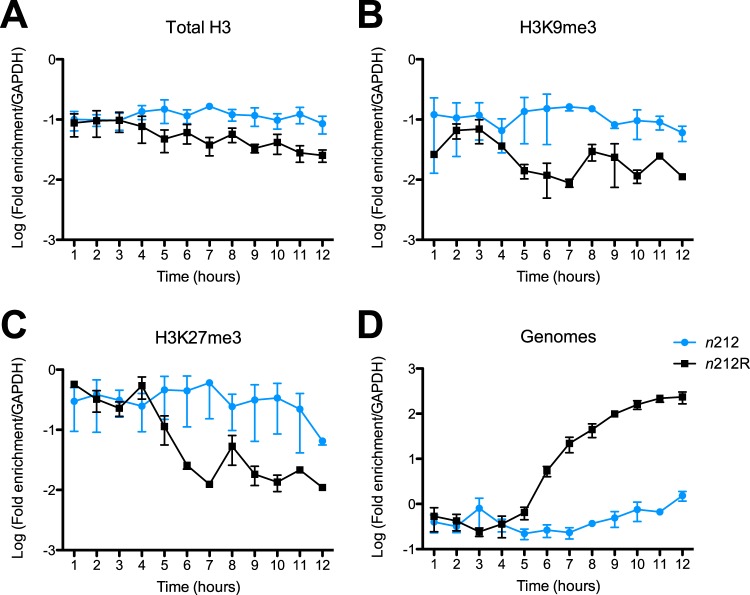
To study the kinetics and structure of viral chromatin in ICP0 mutant-infected cells, we infected HFF cells with n212 or dProm virus as well as with the corresponding rescued viruses at 3 PFU/cell. Viral replication and genome accumulation were quantified by measurement of levels of viral DNA normalized to the GAPDH gene. The ICP0 mutant viruses demonstrated reduced and delayed viral DNA synthesis (Fig. 5D and 6D) and reduced ICP8 RNA expression (Fig. 4B). ChIP analysis was carried out to measure the histone modifications of total H3 and of H3K9me3 and H3K27me3 associated with the HSV-1 genome. Immunoprecipitated DNA levels were measured by qPCR and expressed as the fraction of the ICP8 gene promoter sequences that were immunoprecipitated normalized to the fraction of the GAPDH gene promoter sequences that were immunoprecipitated from the same reaction. We observed that the n212R and PromR ICP0+ viruses demonstrated kinetics of initial histone and heterochromatin association and subsequent removal similar to those observed previously with the WT KOS strain virus (Fig. 5A to C and Fig. 6A to C). The n212 and dProm ICP0− mutant HSV-1 strains accumulated histones and heterochromatin modifications to levels similar to those seen with ICP0+ viral strains at 1 to 2 hpi, but these associations were maintained throughout the 12 hpi. While the trends in data were reproducible, the small sample size and the scatter resulted in statistically insignificant changes. Therefore, we combined the values for samples from 1 to 4 hpi, 5 to 8 hpi, and 9 to 12 hpi and tested for significance using a two-tailed Wilcoxon ranked-sum test. Combined analysis of the data from the first 4 hpi did not show statistically significant differences between ICP0+ and ICP0− viruses. However, from 5 to 8 hpi, the ICP0 mutant viruses were unable to effectively remove histones and repressive histone modifications, resulting in a significant (P < 0.05) difference from infection with ICP0+ viruses. The trend was upheld at late times postinfection but was not statistically significant in dProm and PromR infections, which was the result of high variability of the samples taken at 12 hpi, likely resulting from cell death. Overall, these results indicated that ICP0 promotes removal of histones and heterochromatin during the progression of lytic infection and facilitates viral early gene transcription and DNA replication.
FIG 5 .
Kinetic analyses of histones and heterochromatin on the ICP8 promoter in an ICP0 mutant dProm virus relative to its rescued virus PromR during lytic infection. (A to C) HFF cells were infected with dProm or PromR viruses at 3 PFU/cell. Cells were collected hourly for 12 h, and chromatin was processed for immunoprecipitation with antibodies specific for total H3 (A), H3K9me3 (B), or H3K27me3 (C). ChIP results are presented as the proportion of the ICP8 promoter DNA immunoprecipitated relative to cellular GAPDH gene DNA measured by qPCR. (D) Relative viral genome copy numbers were calculated from input chromatin measurement of viral ICP8 DNA normalized to cellular GAPDH gene DNA. The means and standard errors of the means of results of at least three independent experiments are shown.
FIG 6 .
Histone and heterochromatin association on an ICP0 mutant n212 virus compared to the rescued n212R virus during lytic infection. HFF cells were infected with n212 or n212R viruses at 3 PFU/cell. (A to C) Cells were collected hourly for 12 h, and chromatin was processed for immunoprecipitation with antibodies specific for total H3 (A), H3K9me3 (B), and H3K27me3 (C). ChIP results are presented as the proportion of the ICP8 promoter DNA immunoprecipitated relative to cellular GAPDH gene DNA measured by qPCR. (D) Relative viral genome copy numbers were calculated from input chromatin measurement of ICP8 DNA normalized to GAPDH gene DNA. The means and standard errors of the means of results of at least three independent experiments are shown.
ICP0 is sufficient for heterochromatin reduction on a cotransfected ICP8 gene promoter.
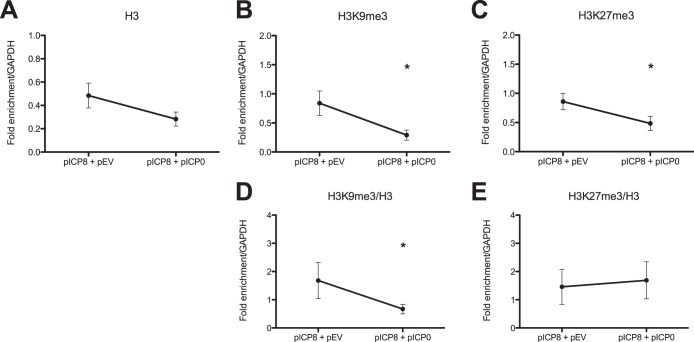
ICP0 mutant viruses are defective for viral gene expression during infection of primary HFF cells, so the effect of ICP0 on viral chromatin could be an indirect effect caused by reduced expression of another viral gene product with chromatin modulatory activity or reduced viral DNA synthesis. However, ICP0 expressed alone in transfected cells is capable of transactivating expression of a cotransfected gene (38, 54). Therefore, we wanted to determine whether ICP0 was sufficient to modulate chromatin on the ICP8 promoter in the absence of other viral gene products, as a potential mechanism of transactivation. We cotransfected 293T cells with a plasmid expressing ICP8-green fluorescent protein (GFP) from the native ICP8 promoter and either an empty vector plasmid or a plasmid expressing ICP0. As observed previously (38), cotransfection with the ICP0-encoding plasmid promoted expression of ICP8-GFP (results not shown). We performed ChIP assays to determine occupancy of histones and heterochromatin modifications on the ICP8 promoter and found that ICP0 expression resulted in significantly reduced levels of H3K9me3 and H3K27me3 (Fig. 7B and C). Total H3 occupancy demonstrated a similar trend, but the results were not statistically significant (Fig. 7A). Normalization of H3K9me3 to total H3 showed a significant reduction when ICP0 was expressed (Fig. 7D), but when H3K27me3 was normalized to total H3, ICP0 did not cause a reduction (Fig. 7E). Therefore, ICP0 appeared to cause a specific reduction in H3K9me3 levels but not in H3K27me3 levels. Therefore, we concluded that ICP0 was sufficient to reduce certain heterochromatin modifications at the ICP8 promoter sequences.
FIG 7 .
ICP0 is sufficient to reduce chromatin and heterochromatin on transfected ICP8 gene plasmid. Cells (293T) were cotransfected with pICP8-GFP plasmid and either an empty vector control (pEV) plasmid or the pICP0 plasmid. (A to C) After 48 h, cells were harvested and processed for ChIP with antibodies specific for total H3 (A), H3K9me3 (B), and H3K27me3 (C). ChIP results were measured by qPCR as the proportion of the immunoprecipitated ICP8 promoter DNA relative to cellular GAPDH gene DNA. The relative proportions of H3 bearing (D) H3K9me3 and (E) H3K27me3 heterochromatin modifications are also presented. Results are shown as the means and standard errors of the means of results of at least three independent experiments. Samples with mean values that varied significantly (P < 0.05, paired Student’s t test) are indicated (*).
DISCUSSION
Epigenetic regulation is a dynamic process that progresses in stages during lytic infection.
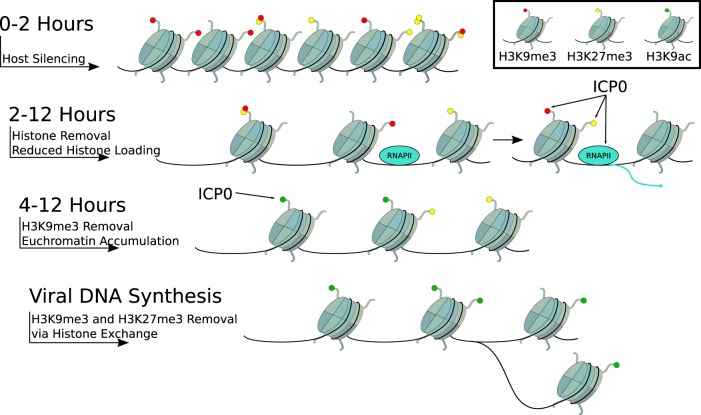
Previous studies have shown that histones are loaded and removed rapidly on HSV-1 E and L promoters and that heterochromatin was removed from IE gene promoters (2). However, the kinetics of heterochromatin association with early gene promoters has not been investigated. In this study, we examined the kinetics of total nucleosome and heterochromatin association on a prototype HSV-1 early gene promoter during the lytic infection of primary HFFs (Fig. 8) and found that the accumulation of histones and heterochromatin modifications H3K9me3 and H3K27me3 peaked during the first 2 hpi. Interestingly, the proportion of histones bearing the H3K9me3 modification was initially higher on the viral ICP8 promoter than on the cellular GAPDH gene. Therefore, the cellular epigenetic silencing response may preferentially assemble histones bearing heterochromatin modifications or rapidly recruit histone methyltransferases. At 2 to 4 hpi, total H3 and heterochromatin levels declined rapidly; however, the ratio of histone to heterochromatin modification remained constant. This argues that the initial antisilencing response by HSV-1 targets nucleosomes indiscriminately, potentially by recruiting histone chaperones or chromatin remodelers. This could expose critical regulatory sequences and initiation sites for the assembly of RNA polymerase II to poise viral genes for transcriptional activation. In the next phase (from 4 to 6 hpi), histone and heterochromatin removal continued, resulting in lower total histone association at the ICP8 promoter region relative to the GAPDH gene sequences. However, while H3 removal slowed, the reduction of H3K9me3 and H3K27me3 heterochromatin modifications increased. Kinetic studies showed parallel losses of H3K9me3 and H3K27me3, but inhibition of viral DNA revealed a differential inhibition of H3K27 removal, demonstrating a functional separation of the two events. Therefore, by 6 hpi, the histones that remained bound to the ICP8 region had a lower proportion of heterochromatin-modified H3 relative to GAPDH gene sequences. This argues that, after 4 hpi, heterochromatin is preferentially removed, indicating either (i) the selective removal of histones bearing heterochromatin modifications or (ii) the recruitment of specific demethylases to remove methyl groups from the remaining histones. Additionally, the dependence of H3K27me3 removal on viral DNA synthesis suggests that histone exchange during DNA replication could be an important mechanism for heterochromatin removal. Interestingly, the timing of heterochromatin reduction coincided with the derepression of E viral gene transcription. This argues that histone and heterochromatin reduction is an important mechanism by which HSV promotes E gene expression and the progression of lytic infection and could mediate the transition from IE to E viral gene expression (46, 55). While transcription could lead to chromatin removal, previous studies (17) have shown that chromatin removal on the ICP8 gene can occur in the absence of transcription.
FIG 8 .
Model of progressive epigenetic regulation of the HSV-1 genome during lytic infection. HSV-1 enters the host cell nucleus and is subjected to host cell repression through the assembly of silenced heterochromatin during the first 2 hpi. After 2 h, HSV-1 reduces epigenetic repression by removing histones to reduce total histone occupancy and heterochromatin levels while maintaining a stable proportion of histones bearing heterochromatin modifications. RNA polymerase II (RNAPII) is recruited and initiates transcription of viral early genes. After 4 h, H3K9me3 and H3K27me3 heterochromatin modifications are specifically targeted for removal, resulting in a lower proportion of histones at viral promoters bearing heterochromatin modifications relative to cellular sequences. H3K27me3 removal is dependent on viral DNA synthesis. ICP0 promotes viral gene transcription, removal of histones and heterochromatin, and accumulation of euchromatic histone modifications.
Removal of H3K9 trimethylation.
The removal of the H3K9me3 heterochromatin marker from 3 to 7 h is dependent on ICP0 and independent of viral DNA synthesis. Because this was also apparent in transfected cells, this appears to be an intrinsic function of ICP0. There is no evidence that ICP0 is a demethylase, so it must work through cellular functions. It will be important to determine if ICP0 requires other demethylases such as LSD1 to promote these chromatin changes. IFI16, a cellular antiviral protein which promotes the H3K9me3 modification on histones on the HSV IE ICP4 gene promoter, is degraded by ICP0 (49). Possible mechanisms for the effect of ICP0 on histone acetylation are known, but it is not clear how ICP0 affects histone methylation. Furthermore, it is not known how ICP0 exerts its effects on the viral genome. It is conceivable that ICP0 targets specific host proteins that are bound to or localized near viral DNA, such as PML (41) or IFI16 (50). In any event, it appears that H3K9me3 exerts a silencing function on the early ICP8 promoter and that ICP0 counters that effect. This likely represents the checkpoint that characterized the immediate early to early gene expression transition (56).
Reduction of H3K9 trimethylation on IE gene promoters in HFFs is known to require LSD1 (KDM1A) and KDM4A–D (23). KDM3A is required for ICP8 expression in U2OS cells (42). Further studies are needed to define the host factors needed for H3K9me3 removal in HFF cells and the mechanism(s) by which ICP0 promotes these factors.
Removal of H3K27 trimethylation.
The removal of the H3K27me3 modification is initially accomplished through nucleosome removal and is dependent on viral DNA synthesis from 4 to 6 hpi. Because ICP0 is required for expression of the E proteins involved in viral DNA synthesis, at least part of the requirement for ICP0 in promoting H3K27me3 removal is indirect. The requirement for viral DNA synthesis could be due to viral DNA synthesis diluting out the histone association, or a late viral gene product could promote the demethylation. Removal of H3K27me3 could be part of the mechanism by which viral DNA replication promotes late gene expression. Thus, the removal of H3K27me3 could be related to activation of viral late genes upon viral DNA replication. ICP8 is expressed in the absence of viral DNA synthesis; thus, the H3K27me3 modification cannot exert a complete silencing effect because it remains on the ICP8 promoter until the genome is replicated. Interestingly, the KDM6A H3K27me2/3 demethylase is required for optimal ICP8-GFP expression in U2OS cells (42). Further studies are needed to determine the host factors needed for H3K27me3 removal in HFF cells. In eukaryotic cells, H3K9me3 is generally associated with constitutive heterochromatin and permanent repression, while H3K27me3 is associated with facultative heterochromatin that is characterized by repression with periodic activation, as seen in developmental genes and in bivalent domains in stem cells (57, 58). Further studies to define the complete range of histone modifications on viral chromatin on the ICP8 gene promoter are needed to understand the full function of the H3K27me3 histone modification.
These results argue that the H3K9 trimethylation and H3K27 trimethylation modifications are independent events. We have recently observed that during latent infection that there is an apparent inverse relationship between the H3K27me3 and H3K9me3 modifications (59). These results in combination provide further evidence for the complex nature of posttranslational modifications of histones and their role in epigenetics. The elucidation of the mechanisms by which the three stages of HSV chromatin modification defined here, namely, general removal of chromatin, removal of H3K9me3, and removal of H3K27me3, should identify additional targets for intervention against HSV infection.
MATERIALS AND METHODS
Plasmids, cells, and viruses.
Primary HFF, HeLa, Vero, U2OS, and 293T cells were obtained from the American Type Culture Collection (Manassas, VA). The n212 ICP0 nonsense mutant virus contains a nonsense mutation in codon 212 (53). For this study, we constructed a corresponding ICP0+ rescued virus, n212R, by homologous recombination of a full-length ICP0 gene in linearized pCIΔLAT.full plasmid with infectious n212 viral DNA. The dProm mutant virus and the PromR rescued virus were constructed by homologous recombination with ICP0-null 7134 infectious viral DNA (53) together with the pCIΔLAT.fulldProm ICP0 promoter mutant plasmid and the pCIΔLAT.full WT full-length ICP0 plasmid, respectively. See the supplemental material for more details. We verified that the rescued viruses followed infection kinetics similar to those seen with WT HSV-1 KOS, which was also titrated on U2OS cells.
Infections.
HFF cells were infected with viruses at 3 PFU/cell. For ChIP experiments testing the effects of inhibiting viral DNA synthesis, sodium phosphonoacetate (PAA) was added to the medium at 200 µg/ml along with 10 mM HEPES at the time of infection and maintained in the medium after infection until the cells were harvested at the indicated time points (17, 60, 61). See Text S1 (supplemental Materials and Methods) for more details.
Cotransfection of ICP0 and ICP8-GFP genes.
HEK 293T cells were seeded in 6-well plates to ensure <50% confluence on the day of transfection. Cells were cotransfected with 0.4 µg of pICP8-GFP (42) and either 0.4 µg of pICP0 (49) or 0.4 µg of an empty vector plasmid (pEV) using Effectene (Qiagen) according to the manufacturer’s instructions. At 48 h after transfection, cells were harvested and processed for ChIP assays or for quantification of RNA expression.
Chromatin immunoprecipitation.
ChIP experiments were carried out as previously described (17, 24, 62, 63), with some modifications. See Text S1 (supplemental Materials and Methods) for detailed descriptions of chromatin immunoprecipitation and quantification of viral gene expression.
SUPPLEMENTAL MATERIAL
Supplemental Materials and Methods. Download
Kinetic analyses of association of H3 and heterochromatin markers on WT HSV-1. The fold change at each hour was calculated relative to the previous hour for total H3, H3K9me3, and H3K27me3. We statistically evaluated the hours at which there was a significant fold change relative to a hypothetical value of 1.0 for no change, assuming equal probabilities of encountering significant change at every hour. Intervals of significant change (P < 0.05, one sample t test) are indicated (*). Download
Analysis of ICP0 mutant viruses. HFF cells were infected with n212, n212R, dProm, or PromR HSV-1 strains at 3 PFU/ml. (A) Whole-cell lysates were collected at 2, 4, 6, 8, and 12 hpi and analyzed by Western blotting for ICP8, ICP27, and GAPDH protein expression. (B) HFF or Vero cells were infected at 1 PFU/ml for 24 h. Infected cells in their overlay medium were collected, and the lysate was titrated on U2OS cells to determine viral yields. Download
ACKNOWLEDGMENTS
We thank Jeho Shin for technical assistance and Patrick T. Waters for assistance with the manuscript.
This research was supported by NIH grant AI098681 to D.M.K.
Footnotes
Citation Lee JS, Raja P, Knipe DM. 2016. Herpesviral ICP0 protein promotes two waves of heterochromatin removal on an early viral promoter during lytic infection. mBio 7(1):e02007-15. doi:10.1128/mBio.02007-15.
REFERENCES
- 1.Cereghini S, Yaniv M. 1984. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J 3:1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knipe DM, Lieberman PM, Jung JU, McBride AA, Morris KV, Ott M, Margolis D, Nieto A, Nevels M, Parks RJ, Kristie TM. 2013. Snapshots: chromatin control of viral infection. Virology 435:141–156. doi: 10.1016/j.virol.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greer EL, Shi Y. 2012. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 5.Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 6.Badeaux AI, Shi Y. 2013. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol 14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hübner MR, Eckersley-Maslin MA, Spector DL. 2013. Chromatin organization and transcriptional regulation. Curr Opin Genet Dev 23:89–95. doi: 10.1016/j.gde.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suganuma T, Workman JL. 2013. Chromatin and signaling. Curr Opin Cell Biol 25:322–326. doi: 10.1016/j.ceb.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price B, D’Andrea A. 2013. Chromatin remodeling at DNA double-strand breaks. Cell 152:1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dion V, Gasser S. 2013. Chromatin movement in the maintenance of genome stability. Cell 152:1355–1364. doi: 10.1016/j.cell.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Honess RW, Roizman B. 1974. Regulation of herpesvirus macro-molecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol 14:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honess RW, Roizman B. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci U S A 72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alwine JC, Steinhart WL, Hill CW. 1974. Transcription of herpes simplex type 1 DNA in nuclei isolated from infected HEp-2 and KB cells. Virology 60:302–307. doi: 10.1016/0042-6822(74)90390-0. [DOI] [PubMed] [Google Scholar]
- 15.Costanzo F, Campadelli-Fiume G, Foa-Tomasi L, Cassai E. 1977. Evidence that herpes simplex virus DNA is transcribed by cellular RNA polymerase B. J Virol 21:996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu H, Liang Y, Mandel G, Roizman B. 2005. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci U S A 102:7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cliffe AR, Knipe DM. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J Virol 82:12030–12038. doi: 10.1128/JVI.01575-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roizman B. 2011. The checkpoints of viral gene expression in productive and latent infection: the role of the HDAC/CoREST/LSD1/REST repressor complex. J Virol 85:7474–7482. doi: 10.1128/JVI.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh J, Fraser NW. 2008. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J Virol 82:3530–3537. doi: 10.1128/JVI.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen GH, Ponce de Leon M, Diggelmann H, Lawrence WC, Vernon SK, Eisenberg RJ. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol 34:521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pignatti PF, Cassai E. 1980. Analysis of herpes simplex virus nucleoprotein complexes extracted from infected cells. J Virol 36:816–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 23.Liang Y, Vogel JL, Arbuckle JH, Rai G, Jadhav A, Simeonov A, Maloney DJ, Kristie TM. 2013. Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci Transl Med 5:167ra165. doi: 10.1126/scitranslmed.3005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol 83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrera FJ, Triezenberg SJ. 2004. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J Virol 78:9689–9696. doi: 10.1128/JVI.78.18.9689-9696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med 15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lacasse JJ, Schang LM. 2012. Herpes simplex virus 1 DNA is in unstable nucleosomes throughout the lytic infection cycle, and the instability of the nucleosomes is independent of DNA replication. J Virol 86:11287–11300. doi: 10.1128/JVI.01468-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kent JR, Zeng P-, Atanasiu D, Gardner J, Fraser NW, Berger SL. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J Virol 78:10178–10186. doi: 10.1128/JVI.78.18.10178-10186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J, Kent JR, Placek B, Whelan KA, Hollow CM, Zeng P-, Fraser NW, Berger SL. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J Virol 80:5740–5746. doi: 10.1128/JVI.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henikoff S, Shilatifard A. 2011. Histone modification: cause or cog? Trends Genet 27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Kutluay SB, Triezenberg SJ. 2009. Role of chromatin during herpesvirus infections. Biochim Biophys Acta 1790:456–466. doi: 10.1016/j.bbagen.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryant KF, Colgrove RC, Knipe DM. 2011. Cellular SNF2H chromatin-remodeling factor promotes herpes simplex virus 1 immediate-early gene expression and replication. mBio 2:e00330-10. doi: 10.1128/mBio.00330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arbuckle JH, Kristie TM. 2014. Epigenetic repression of herpes simplex virus infection by the nucleosome remodeler CHD3. mBio 5:e01027-13. doi: 10.1128/mBio.01027-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh J, Ruskoski N, Fraser NW. 2012. Chromatin assembly on herpes simplex virus 1 DNA early during a lytic infection is Asf1a dependent. J Virol 86:12313–12321. doi: 10.1128/JVI.01570-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Placek BJ, Huang J, Kent JR, Dorsey J, Rice L, Fraser NW, Berger SL. 2009. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J Virol 83:1416–1421. doi: 10.1128/JVI.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stow ND, Stow EC. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol 67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 37.Sacks WR, Schaffer PA. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol 61:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinlan MP, Knipe DM. 1985. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol 5:957–963. doi: 10.1128/MCB.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Hare P, Hayward GS. 1985. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol 53:751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gelman IH, Silverstein S. 1985. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci U S A 82:5265–5269. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Everett RD. 1984. Transactivation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J 3:3135–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh HS, Bryant KF, Nieland TJF, Mazumder A, Bagul M, Bathe M, Root DE, Knipe DM. 2014. A targeted RNA interference screen reveals novel epigenetic factors that regulate herpesviral gene expression. mBio 5:e01086-13. doi: 10.1128/mBio.01086-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu H, Roizman B. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci U S A 104:17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou G, Te D, Roizman B. 2010. The CoREST/REST repressor is both necessary and inimical for expression of herpes simplex virus genes. mBio 2:e00313-10. doi: 10.1128/mBio.00313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou G, Du T, Roizman B. 2013. The role of the CoREST/REST repressor complex in herpes simplex virus 1 productive infection and in latency. Viruses 5:1208–1218. doi: 10.3390/v5051208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalamvoki M, Roizman B. 2010. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci U S A 107:17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalamvoki M, Roizman B. 2011. The histone acetyltransferase CLOCK is an essential component of the herpes simplex virus 1 transcriptome that includes TFIID, ICP4, ICP27, and ICP22. J Virol 85:9472–9477. doi: 10.1128/JVI.00876-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glass M, Everett RD. 2013. Components of promyelocytic leukemia nuclear bodies (ND10) act cooperatively to repress herpesvirus infection. J Virol 87:2174–2185. doi: 10.1128/JVI.02950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orzalli MH, Conwell SE, Berrios C, Decaprio JA, Knipe DM. 2013. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci U S A 110:E4492–E4501. doi: 10.1073/pnas.1316194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orzalli MH, DeLuca NA, Knipe DM. 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A 109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narayanan A, Ruyechan WT, Kristie TM. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci U S A 104:10835–10840. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silva L, Cliffe A, Chang L, Knipe DM. 2008. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog 4:e1000071. doi: 10.1371/journal.ppat.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai WZ, Schaffer PA. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol 63:4579–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su L, Knipe DM. 1987. Mapping of the transcriptional initiation site of the herpes simplex virus type 1 ICP8 gene in infected and transfected cells. J Virol 61:615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roizman B, Zhou G. 2015. The 3 facets of regulation of herpes simplex virus gene expression: A critical inquiry. Virology 479–480:562–567. doi: 10.1016/j.virol.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roizman B, Zhou G, Du T. 2011. Checkpoints in productive and latent infections with herpes simplex virus 1: Conceptualization of the issues. J Neurovirol 17:512–517. doi: 10.1007/s13365-011-0058-x. [DOI] [PubMed] [Google Scholar]
- 57.Barski A, Cuddapah S, Cui K, Roh T, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Trojer P, Reinberg D. 2007. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell 28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 59.Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, Foster TP, Bosch-Marce M, Raja P, Lee JS, Bernstein DI, Krause PR, Knipe DM, Kristie TM. 2014. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci Transl Med 6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shipkowitz NL, Bower RR, Appell RN, Nordeen CW, Overby LR, Roderick WR, Schleicher JB, Von Esch AM. 1973. Suppression of herpes simplex virus infection by phosphonoacetic acid. Appl Microbiol 26:264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jofre JT, Schaffer PA, Parris DS. 1977. Genetics of resistance to phosphonoacetic acid in strain KOS of herpes simplex virus type 1. J Virol 23:833–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Solomon MJ, Larsen PL, Varshavsky A. 1988. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53:937–947. doi: 10.1016/S0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 63.Wang Q-Y, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A 102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods. Download
Kinetic analyses of association of H3 and heterochromatin markers on WT HSV-1. The fold change at each hour was calculated relative to the previous hour for total H3, H3K9me3, and H3K27me3. We statistically evaluated the hours at which there was a significant fold change relative to a hypothetical value of 1.0 for no change, assuming equal probabilities of encountering significant change at every hour. Intervals of significant change (P < 0.05, one sample t test) are indicated (*). Download
Analysis of ICP0 mutant viruses. HFF cells were infected with n212, n212R, dProm, or PromR HSV-1 strains at 3 PFU/ml. (A) Whole-cell lysates were collected at 2, 4, 6, 8, and 12 hpi and analyzed by Western blotting for ICP8, ICP27, and GAPDH protein expression. (B) HFF or Vero cells were infected at 1 PFU/ml for 24 h. Infected cells in their overlay medium were collected, and the lysate was titrated on U2OS cells to determine viral yields. Download