ABSTRACT
Our fundamental knowledge of the protein-sorting pathways required for plant cell-to-cell trafficking and communication via the intercellular connections termed plasmodesmata has been severely limited by the paucity of plasmodesmal targeting sequences that have been identified to date. To address this limitation, we have identified the plasmodesmal localization signal (PLS) in the Tobacco mosaic virus (TMV) cell-to-cell-movement protein (MP), which has emerged as the paradigm for dissecting the molecular details of cell-to-cell transport through plasmodesmata. We report here the identification of a bona fide functional TMV MP PLS, which encompasses amino acid residues between positions 1 and 50, with residues Val-4 and Phe-14 potentially representing critical sites for PLS function that most likely affect protein conformation or protein interactions. We then demonstrated that this PLS is both necessary and sufficient for protein targeting to plasmodesmata. Importantly, as TMV MP traffics to plasmodesmata by a mechanism that is distinct from those of the three plant cell proteins in which PLSs have been reported, our findings provide important new insights to expand our understanding of protein-sorting pathways to plasmodesmata.
IMPORTANCE
The science of virology began with the discovery of Tobacco mosaic virus (TMV). Since then, TMV has served as an experimental and conceptual model for studies of viruses and dissection of virus-host interactions. Indeed, the TMV cell-to-cell-movement protein (MP) has emerged as the paradigm for dissecting the molecular details of cell-to-cell transport through the plant intercellular connections termed plasmodesmata. However, one of the most fundamental and key functional features of TMV MP, its putative plasmodesmal localization signal (PLS), has not been identified. Here, we fill this gap in our knowledge and identify the TMV MP PLS.
INTRODUCTION
The science of virology began with the discovery of Tobacco mosaic virus (TMV). Since then, TMV has served as an experimental and conceptual paradigm for studies of viruses and dissection of virus-host interactions, truly becoming the ‘virus of many “firsts” ’ (1). For example, TMV was the first virus to be chemically purified and visualized, its RNA was the first viral genome proven sufficient for infectivity, and the TMV coat protein was the first viral protein sequenced. Importantly, it was seminal studies of TMV that led to the discovery of a virus-encoded 30-kDa cell-to-cell-movement protein (MP), which is essential for plant virus spread between host cells. Thus, TMV MP has emerged as the molecular tool of choice for dissecting the details of cell-to-cell transport through plant intercellular connections, the plasmodesmata: it has been shown to target to plasmodesmata, increase plasmodesmal permeability, and traffic through the plasmodesmal channel into neighboring cells (reviewed in reference 1).
Despite intensive studies of TMV MP in the 35 years since its discovery (2), one of the most important and fundamental functional features of this protein, its putative plasmodesmal localization signal (PLS), has yet to be identified. Our prediction of such a signal sequence is based on the concept that the sorting of virtually all proteins to their correct locations within or outside the cell requires targeting sequences that are specific for each destination. In particular, cytosolic synthesized proteins require organelle-specific targeting sequences to be sorted to the proper organelle, from the endoplasmic reticulum (ER), to chloroplasts or mitochondria, to the nucleus. For example, protein import through nuclear pores is mediated by nuclear localization signals (NLSs) within the transported proteins (3–5). That no PLSs have been identified for any virus-encoded cell-to-cell MP, including the TMV MP, has been a significant impediment not only to studies of viral infection, but also to our understanding of fundamental protein-sorting pathways involved in intercellular transport and communication between plant cells. Only three protein sequences for plasmodesmal targeting have been reported, and all of them are for endogenous proteins rather than for viral proteins. The first two are found in plant transcription factors: one is represented by a specific homeobox domain of KN1 (6), a transcription factor that normally moves unidirectionally from the inner cell layers of the leaf to the epidermis (7) and of its KNOX homologs (8), and the second by intercellular trafficking (IT) motifs of Dof transcription factors (9). The third sequence comprises a plant transmembrane domain of the PDLP1 plasmodesmata-resident type I membrane protein (10). Because viral MPs traffic by a mechanism distinct from that employed by transcription factors (7) and because TMV MP does not contain a transmembrane domain (11), the putative PLS sequence in TMV MP must be distinct from these two known targeting sequences. We report here the identification of a functional TMV MP PLS sequence and show that it is necessary and sufficient for protein targeting to plasmodesmata. We also uncoupled TMV MP targeting to plasmodesmata from MP cell-to-cell transport, thereby demonstrating that the identified MP PLS is a bona fide targeting sequence that is not involved in subsequent protein movement into and through the plasmodesmal channel.
RESULTS
The amino terminus of TMV MP contains a functional PLS.
We defined the putative PLS of TMV MP as the shortest sequence both necessary and sufficient for protein localization to plasmodesmata. To identify this sequence, we fused a series of consecutive fragments of TMV MP to cyan fluorescent protein (CFP), which served as both a generic cargo protein and a subcellular localization marker. Protein subcellular localization was visualized by laser scanning confocal microscopy. Localization at plasmodesmata was identified by the diagnostic punctate pattern (12–18) and confirmed by colocalization with a known plasmodesmal marker, PDCB1 (19).
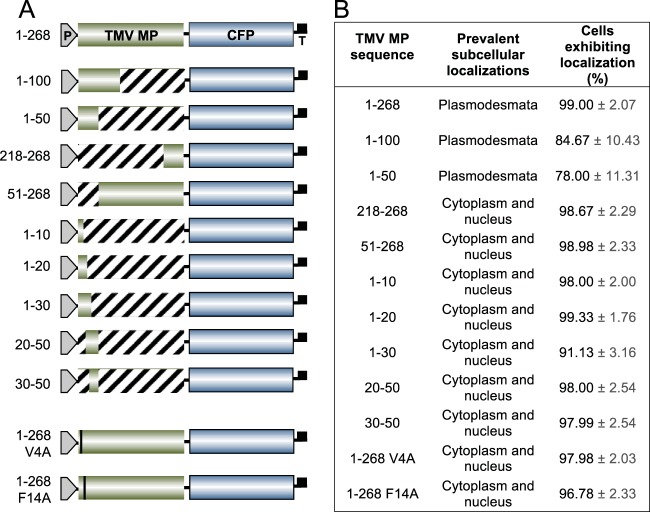
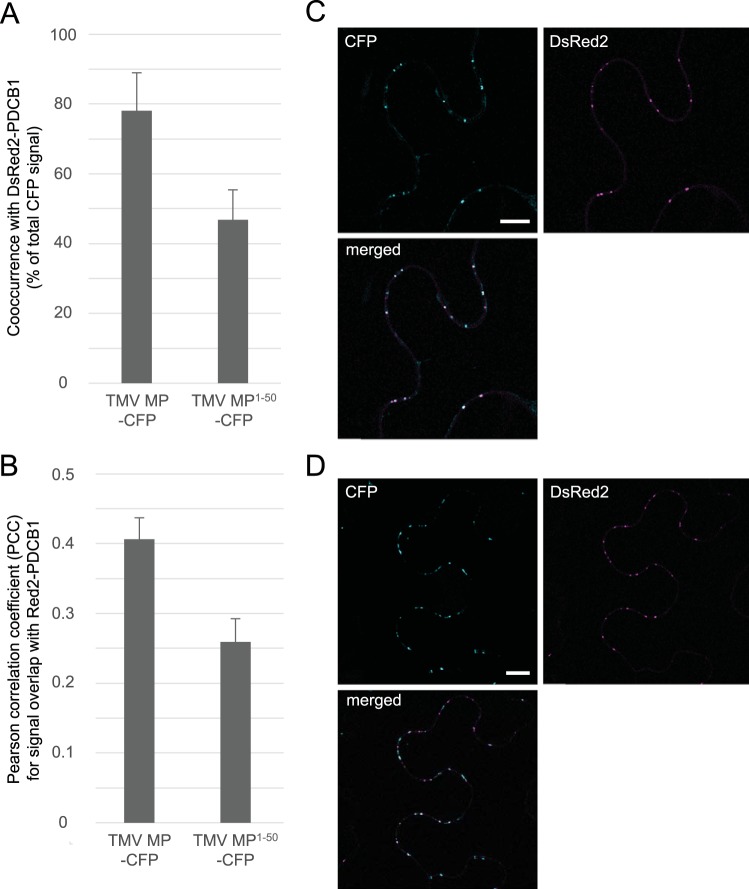
Figure 1A summarizes the series of constructs analyzed. We coexpressed each of these with two reference constructs, Discosoma sp. red fluorescent protein 2 (DsRed2)-tagged PCDB1, to identify plasmodesmata, and free DsRed2, which, due to its small size, partitions between the cell cytoplasm and nucleus (see, e.g., reference 20) and thus served as a marker for nucleocytoplasmic localization. As shown in Fig. 2 and 3, CFP-tagged full-length TMV MP (MP-CFP) displayed the typical plasmodesmata-specific punctate accumulation, co-occurring with DsRed2-tagged PDCB1 at plasmodesmata 78.1% ± 10.9% of the time (Pearson correlation coefficient [PCC], 0.41 ± 0.03).
FIG 1 .
Summary of CFP-fused TMV MP constructs used in this study and their ability to direct CFP to plasmodesmata. (A) Fusion constructs. TMV MP sequences, green boxes; CFP, blue boxes; P, promoter; T, terminator. Numbers on the left indicate the amino acid residues included in each TMV MP fragment. (B) Subcellular localization of each fusion construct. Localization of TMV MP-CFP fusion constructs to plasmodesmata or nuclei was assessed on the basis of colocalization with PDCB1-DsRed2 or free DsRed2, respectively. The percentage of cells expressing each construct is shown, based on counting 100 expressing cells per construct in three independent experiments. Data are means ± standard errors (SE).
FIG 2 .
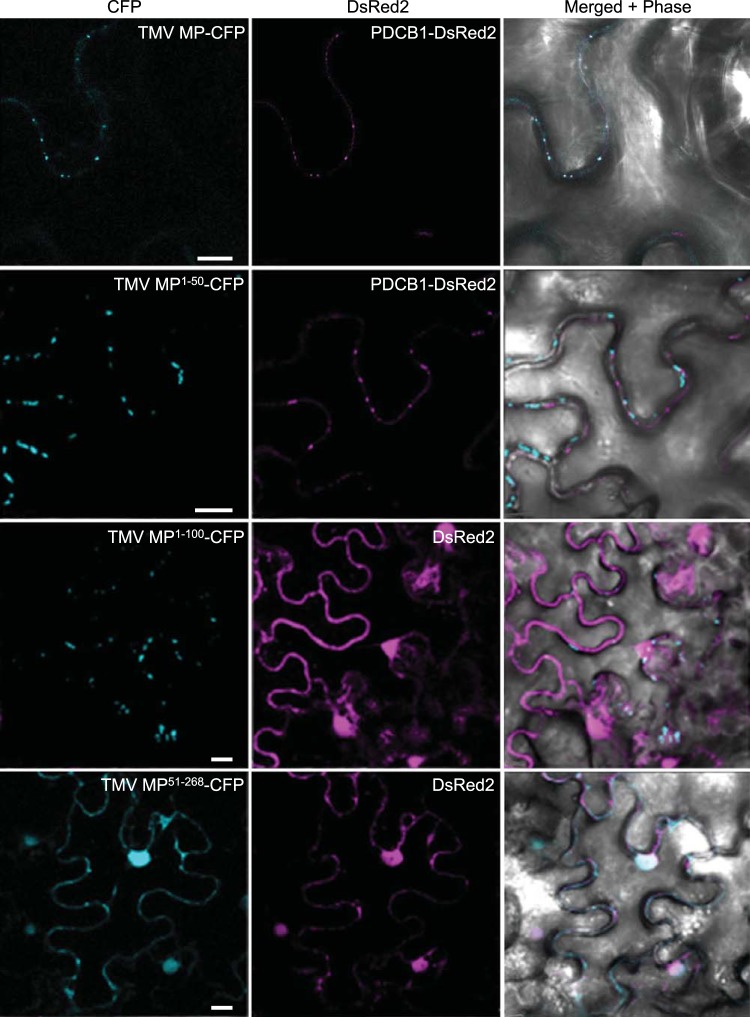
Subcellular localization of TMV MP, MP1–50, and MP51–268. PDCB1-DsRed2 and free DsRed2 were coexpressed as markers for plasmodesmata and nuclei, respectively. CFP signal is blue; DsRed2 signal is red; plastid autofluorescence was filtered out. Images are single confocal sections. Scale bars, 10 µm.
When the first 100 amino acid residues of TMV MP were fused to the CFP cargo, the resulting TMV MP(1)–100-CFP protein accumulated at plasmodesmata in most of the expressing cells (Fig. 1B). The fusion of the amino-terminal 50 residues of TMV MP to CFP (MP1–50-CFP) also predominantly targeted to plasmodesmata, colocalizing with PDCB1-DsRed2 but not with free DsRed2 (Fig. 1B and 2). The frequency of TMV MP1–50-CFP plasmodesmal localization was comparable to that of TMV MP1–100-CFP, being 78% and 85% of all expressing cells, respectively (Fig. 1B). The efficiency of plasmodesmal localization of TMV MP1–50-CFP, as determined on the basis of integrated density measurements of the plasmodesmal-associated fluorescent signal (21), also was at least equivalent to that of TMV MP-CFP (data not shown). As shown in Fig. 3, our quantification of the plasmodesmal localization of TMV MP1–50-CFP showed that TMV MP1–50-CFP co-occurred with DsRed2-tagged PDCB1 at plasmodesmata 46.8% ± 8.6% of the time (PCC, 0.26 ± 0.03).
FIG 3 .
TMV MP and TMV MP1–50 localize to plasmodesmata. (A and B) TMV MP-CFP co-occurred with PDCB1-DsRed2 at plasmodesmata 78.1% ± 10.9% of the time (PCC 0.41 ± 0.03); TMV MP1–50-CFP co-occurred with PDCB1-DsRed2 at plasmodesmata 46.8% ± 8.6% of the time (PCC 0.26 ± 0.03). Data were determined on the basis of analyzing 32 expressing cells in each of three independent experiments and are expressed as means ± SE. (C) Representative single confocal sections of coexpressed TMV MP-CFP and PDCB1-DsRed2. (D) Representative single confocal sections of coexpressed TMV MP1–50-CFP and PDCB1-DsRed2. CFP signal is blue; DsRed2 signal is red; plastid autofluorescence was filtered out. Scale bars, 10 µm.
Collectively, these results showed that the amino-terminal 50 amino acid residues of TMV MP were sufficient to confer plasmodesmal localization. However, was this TMV MP fragment also necessary for efficient targeting, or could the remaining TMV MP sequence between residues 51 and 268 still accumulate at plasmodesmata? Figures 1B and 2 show that TMV MP51–268-CFP did not target to plasmodesmata. Rather, similarly to the coexpressed free DsRed2 marker, it partitioned between the cytoplasm and the nucleus in 99% of expressing cells, although very low basal levels of about 1% to 2% appeared to localize at plasmodesmata. This plasmodesmal association may likely be due to residual membrane association, for example, via its hydrophobic regions (11). Likewise, shorter carboxyl-terminal-proximal TMV MP fragments, such as TMV MP218–268-CFP, which contained the carboxyl-terminal 50 amino acid residues, did not target to plasmodesmata (Fig. 1B).
We next examined whether the entire TMV MP1–50 sequence was required for plasmodesmal targeting by subdividing it into five overlapping fragments, namely, amino acid residues between positions 1 and 10, 1 and 20, 1 and 30, 20 and 50, and 30 and 50 (Fig. 1B). We assayed the subcellular localization of each fragment fused to the CFP cargo. None of these fragments were able to promote significant localization of CFP to plasmodesmata, remaining nucleocytoplasmic in 91% to 99% of expressing cells (Fig. 1B). Collectively, our data suggest that the TMV MP1–50 sequence likely contains the major PLS, which is both necessary and sufficient for efficient plasmodesmal targeting.
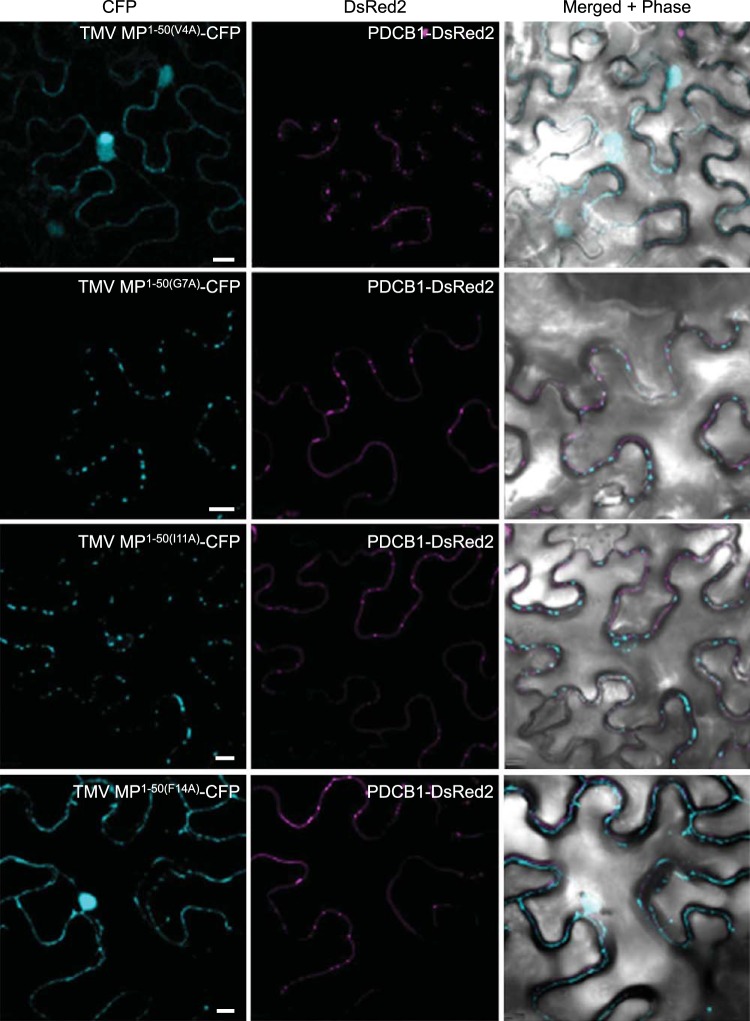
To better understand the potential effects of specific MP residues on plasmodesmal targeting activity we analyzed the TMV MP PLS sequence (MP1–50; Fig. 4A) by alanine-scanning mutagenesis (22). Figure 4B summarizes the series of CFP-tagged single-alanine-substitution mutants of TMV MP1–50 that we tested and their respective subcellular localization patterns, and Fig. 5 illustrates the subcellular localization of selected TMV MP1–50 mutants coexpressed with the PDCB1-DsRed2 plasmodesmal marker. Most alanine substitutions did not affect plasmodesmal localization (Fig. 4B). For example, the G7A and I11A mutants exhibited localization patterns similar to those of PDCB1 (Fig. 5) and TMV MP1–50 (Fig. 2). Thus, whereas the entire TMV MP PLS, comprising the TMV MP1–50 sequence, was required for efficient plasmodesmal targeting, many of its amino acid residues could be mutated, at least individually, without obviously affecting targeting. However, the two V4A and F14A point mutations abolished PLS activity, producing stable CFP fusion proteins with a nucleocytoplasmic localization pattern (Fig. 4B and 5). Fitting with this, when each of these mutations was introduced into the full-length TMV MP, the resulting mutants also lost their ability to target to plasmodesmata (Fig. 1). Thus, the residues Val-4 and Phe-14 potentially represent critical sites for PLS function that most likely affect active protein conformation or protein-protein interactions. We then examined the degree of conservation of Val-4 and Phe-14 among MPs of 10 Tobamoviruses, i.e., TVCV (Turnip vein-clearing virus), YoMV (Youcai mosaic virus), RMV (Ribgrass mosaic virus), SHMV (Sunn-hemp mosaic virus), ORSV (Odontoglossum ringspot virus), TMGMV (Tobacco mild green mosaic virus), PMMV-J (Pepper mild mottle virus, strain Japan), TMOB (Tobamovirus Ob), and ToML (Tomato mosaic virus, strain L) (accession numbers Q88921, Q66221, Q9QDI8, P03585, P22590, P18338, P89658, Q83485, P69513, and P03583, respectively), using the Universal Protein Resource (UniProt; http://www.uniprot.org). Whereas Val-4 was conserved in MPs of TVCV, SHMV, PMMV-J, TOML, and TMV, the Phe-14 residue was conserved among all of the tested tobamoviruses.
FIG 4 .
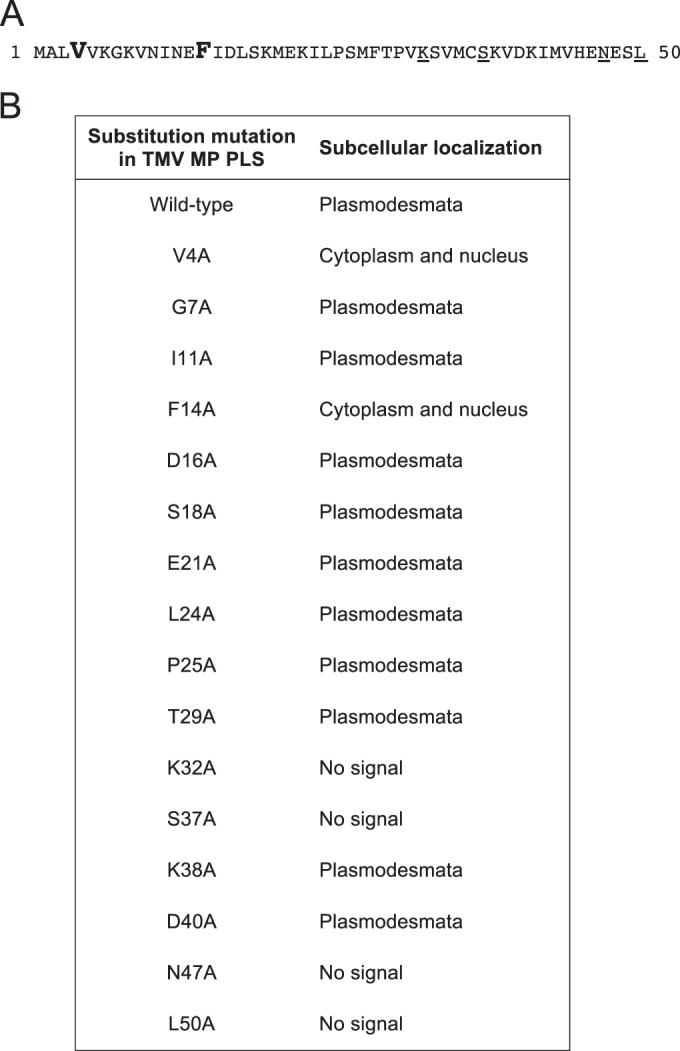
Val-4 and Phe-14 are critical for TMV MP PLS activity. (A) Amino acid sequence of the TMV MP PLS (GenBank accession no. CAD48854.1). Residues critical for PLS activity are in large boldface, and residues important for protein stability are underlined. (B) Effects of alanine-scanning mutations on the subcellular localization of CFP-tagged TMV MP PLS. Localization was assessed on the basis of the wild-type TMV MP PLS. Mutants that exhibited the same subcellular localization as the native TMV MP PLS (coexpressed with PDCB1-DsRed2) were considered to be plasmodesmal.
FIG 5 .
Subcellular localization of selected TMV MP1–50 PLS alanine-scanning mutants. PDCB1-DsRed2 was coexpressed as a marker for plasmodesmata. CFP signal is blue; DsRed2 signal is red; plastid autofluorescence was filtered out. Images are single confocal sections. Scale bars, 10 µm.
Finally, four of the tested mutations, K32A, S37A, N47A, and L50A, did not produce a detectable fluorescent signal (Fig. 4B), most likely due to the fusion protein being unstable or reduced in expression or in autofluorescence.
Bona fide PLS: uncoupling plasmodesmal targeting and entry.
A true PLS would be expected to direct cargo to its specific destination, the plasmodesmata, but not necessarily to participate in subsequent events such as entering and traversing the plasmodesmal channel. We therefore tested the abilities of TMV MP1–50 to promote both entry of its cargo into plasmodesmata and transport through plasmodesmata to a neighboring cell(s).
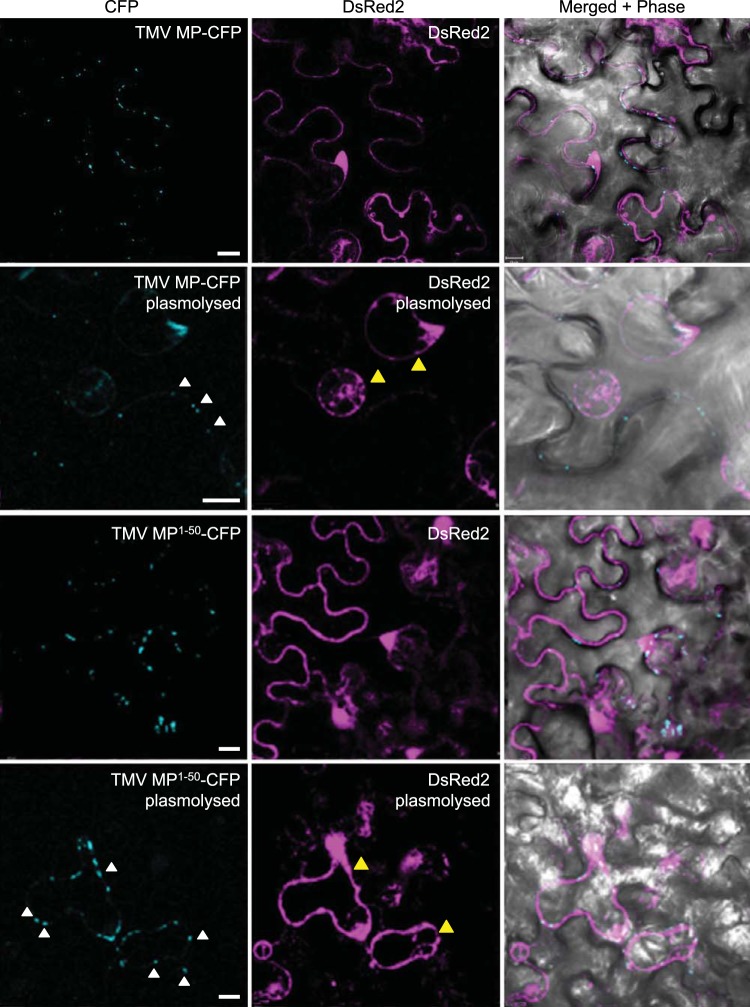
To investigate plasmodesmal entry, we used plasmolysis, which distinguishes protein accumulation within cell wall-associated compartments, such as plasmodesmata, from that within intracellular and membrane compartments (23). Figure 6 shows that full-length TMV MP-CFP formed the characteristic plasmodesmal puncta at the cell periphery under physiological osmotic conditions. Upon plasmolysis, TMV MP-CFP retained this same localization pattern at the cell periphery (Fig. 6, white arrowheads) and did not localize to the displaced membrane and cytoplasm as visualized by free DsRed2 (Fig. 6, yellow arrowheads). This indicated that TMV MP-CFP remained associated with plasmodesmata inside the cell wall. Thus, as expected, TMV MP, which is known to accumulate within the central regions of plasmodesmata (24), indeed targeted to and entered these channels. Under normal osmotic conditions, TMV MP1–50-CFP also localized in the plasmodesmata-specific punctate pattern at the cell wall. However, in contrast to TMV MP-CFP, MP1–50-CFP in plasmolyzed cells retracted internally (Fig. 6, white arrowheads) together with the displaced cellular contents (Fig. 6, yellow arrowheads). Nevertheless, this displaced TMV MP1–50-CFP still retained its punctate pattern of accumulation, which also remained peripheral, diagnostic of the displaced plasma membrane and its residual plasmodesmal components. Notably, the TMV MP-CFP puncta appeared to be more compact than those of TMV MP1–50-CFP, most likely due to TMV MP entering and accumulating within the plasmodesmal channels. Taken together, these data indicate that the TMV MP PLS targets to and docks at the outer orifice of plasmodesma in a relatively stable manner, without entering the channel itself.
FIG 6 .
Subcellular localization of TMV MP and TMV MP1–50 in plasmolyzed cells. Free DsRed2 was coexpressed to show the location of the cytoplasm. CFP signal is blue; DsRed2 signal is red; plastid autofluorescence was filtered out. In plasmolyzed cells, the location of plasmodesmal CFP signals are illustrated by white arrowheads, and the location of retracted intracellular DsRed2 signals are illustrated by yellow arrowheads. Images are single confocal sections. Scale bars, 10 µm.
To examine whether TMV MP1–50-CFP can move its cargo cell to cell through plasmodesmata, we used agroinfiltration to transiently coexpress TMV MP1–50-CFP or full-length TMV MP-CFP, each with free DsRed2 as a marker for the initial infiltrated cell, and compared the dynamics of TMV MP1–50-CFP versus MP-CFP intercellular trafficking. Figure 7 shows that, as expected (18, 25), TMV MP moved from cell to cell, with 70% to 75% of the examined fields showing TMV MP-CFP trafficking from the initial expressing cell into neighboring cells. In contrast, TMV MP1–50 did not move between cells: 94% to 98% of the fields examined showed that MP1–50-CFP remained confined to a single cell (Fig. 7B). Thus, our results effectively uncouple plasmodesmal targeting per se from plasmodesmal entry and transit for a viral MP, thereby identifying a bona fide PLS in TMV MP.
FIG 7 .
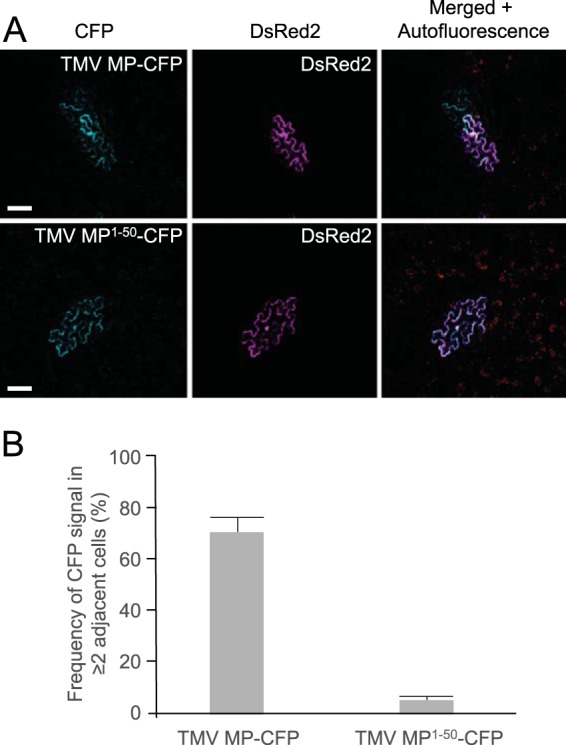
Cell-to-cell movement of TMV MP and TMV MP1–50. (A) Representative images for TMV MP and TMV MP1–50 cell-to-cell movement, with free DsRed2 coexpressed to label the initial infiltrated cell. CFP signal is blue; DsRed2 signal is red. Plastid autofluorescence either was filtered out or is shown, as indicated. Cells that contain both CFP and DsRed2 signals represent infiltrated cells, and those that contain only a CFP signal are cells into which movement had occurred. Images are single confocal sections. Scale bars, 50 µm. (B) Cell-to-cell movement was quantified on the basis of examining 50 fields for the presence of CFP in two or more neighboring cells, only one of which expressed both CFP and DsRed2. Results shown for each construct are from three independent experiments. Data are means ± SE.
DISCUSSION
Cell-to-cell transport through plasmodesmata is mediated by specific virus-encoded MPs and represents a critical event in the interactions between plant viruses and their plant hosts. Yet, a key aspect of this transport, namely, the identity of a signal that specifically targets an MP to plasmodesmata, has eluded discovery. Here, we have identified this functional sequence in the MP encoded by TMV, a paradigm for plant virus cell-to-cell movement (26), and termed it a plasmodesmata localization signal (PLS). This PLS was both necessary and sufficient for targeting an unrelated cargo protein (CFP) to plasmodesmata, and it represented the major such targeting signal within TMV MP. Importantly, TMV MP PLS acted as a bona fide targeting signal. It was required for the specific docking of cargo proteins at plasmodesmata, but did not act on the subsequent steps of cargo entering into and moving through plasmodesmata. Instead, TMV MP PLS most likely directed its cargo to, and promoted its stable association with, the plasma membrane face of the plasmodesmal channel, an association that survived plasmolysis and membrane displacement. This conclusion is consistent with our finding that while both TMV MP and MP1–50 localized to plasmodesmata, the PCC colocalization overlap with PDCB1-DsRed2 was only 0.26 ± 0.03 for TMV MP1–50-CFP, which targets to, but does not enter plasmodesmata, compared to 0.41 ± 0.03 for TMV MP-CFP, which resides within plasmodesmata, as does PDCB1 (19, 24). Additionally, these proteins may simply be targeted to distinct, albeit overlapping, subsets of plasmodesmata.
The PLS comprises the first 50 amino-terminal residues of TMV MP, and our findings suggest that this fragment represents the minimum length of the efficiently functioning signal. In the absence of this amino-terminal PLS, we could not detect that the remaining portion of TMV MP (MP51–268) directed CFP to plasmodesmata. Thus, MP1–50 most likely represents the major PLS of TMV MP. The location of the PLS sequence is consistent with early data indicating that mutations within the amino terminus of TMV MP impair plasmodesmal localization, although that study did not delineate the targeting sequence or address its sufficiency for targeting (27). Clearly, however, we cannot exclude the possibility that there are additional domains in the rest of the MP sequence (i.e., TMV MP51–268) that could contribute to the stability of the targeting.
Within the PLS sequence, our mutational analyses identified only 2 amino acid residues that were clearly critical for plasmodesmal targeting activity, Val-4 and Phe-14. Mutation of either of these residues singly to alanine mistargeted MP1–50-CFP to the nucleocytoplasmic compartment. This suggests that Val-4 and Phe-14 may be essential to allow correct MP folding to present the PLS sequence to the cellular protein-sorting machinery; indeed, Val-4 and Phe-14 were conserved in MPs from 5 and 10 different Tobamoviruses, respectively. Our mutational analyses also suggested that Lys-32, Ser-37, Gln-47, and Leu-50 may be important for the stability of the TMV MP PLS.
Interestingly, while there are regions of secondary structure that are predicted to be conserved between TMV MP1–50 and the equivalent regions of other tobamovirus MPs, there is <50% conservation among their primary sequences (28 and data not shown), suggesting that the TMV MP PLS may be virus specific. In this context, it is striking that the TMV MP PLS is also different from those few targeting domains identified in cellular proteins that reside within plasmodesmata, such as PDCB1, or that move through plasmodesmata, such as KN1/KNOX/Dof transcription factors (6, 8, 10). This sequence diversity among PLSs is reminiscent of the wide spectrum of poorly related NLSs that function in specific importin alpha-dependent nuclear import pathways (5). Thus, our identification of this viral PLS in TMV MP not only is important to better understand TMV MP function and TMV biology, but it also lays a foundation for systematically unraveling the specific plasmodesmal transport pathway that initiates with this signal.
MATERIALS AND METHODS
Plants.
Nicotiana benthamiana plants were grown in soil in an environment-controlled chamber at 20 to 25°C under long day conditions of 16 h of light at ~75 µmol photons m−2 s−1 and 8 h of dark.
Constructs.
TMV MP-CFP was expressed from pSAT1-TMV MP-CFP (29). For TMV MP1–50-CFP, pSAT1-TMV MP-CFP was amplified using primers MP50REcoRI and CFPFEcoRI, digested with DpnI and EcoRI, and self-ligated. TMV MP1–100-CFP was generated using the same strategy with primers MP100REcoRI and CFPFEcoRI.
For TMV MP1–10-CFP, pSAT1-TMV MP-CFP was amplified using primers CFPFEcoRI and MP10REcoRI, digested with DpnI and EcoRI, and self-ligated. TMV MP1–20-CFP and TMV MP1–30-CFP were generated using the same strategy with primer pairs CFPFEcoRI/MP20REcoRI and CFPFEcoRI/MP30REcoRI, respectively.
For TMV MP20–50-CFP, pSAT1-TMV MP1–50-CFP was amplified using primers MP20FBamHI and MPRBamHI, digested with DpnI and BamHI, and self-ligated. For TMV MP30–50-CFP, pSAT1-TMV MP1–50-CFP was amplified using primers MP30FBglII and MPRBamHI, digested with DpnI and BglII, and self-ligated.
For TMV MP51–268-CFP and TMV MP218–268-CFP, the corresponding coding sequences of MP were amplified from pSAT1-TMV MP-CFP using primer pairs MP51FBglII/MP268REcoRI and MP218FBglII/MP268REcoRI, respectively, and cloned into the BglII and EcoRI sites of pSAT1-TMV MP-CFP.
The entire expression cassettes from all pSAT1-based constructs were excised with AscI and inserted into the same site of the binary vector pPZP-RCS2 (20). All PCR primer pairs used in this study are listed in Table S1 in the supplemental material. All constructs were verified by DNA sequencing.
Alanine scanning.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. For TMV MP1–50(V4A)-CFP and TMV MP1–50(F14A)-CFP, pSAT1-TMV MP1–50-CFP was amplified using primer pairs 1-50ASMP4F/1-50ASMP4R and 1-50ASMP14F/1-50ASMP14R, respectively, which contained the desired mutations. The same primers were used to amplify pSAT1-TMV MP-CFP for production of full-length TMV MP(V4A)-CFP and TMV MP(F14A)-CFP. Nonmutated DNA was removed by digestion with DpnI. Other alanine-scanning mutants of TMV MP1–50 were generated using the same strategy and their corresponding primers are listed in Table S1 in the supplemental material.
Agroinfiltration.
Binary plasmids expressing proteins of interest were introduced into Agrobacterium strain GV3101 (30), grown overnight at 25°C, diluted to a cell density A600 value of 0.5 or as indicated, and infiltrated with a 1-ml needleless syringe as previously described (31, 32) into N. benthamiana leaves immediately above the cotyledon. For reproducibility and optimal plasmodesmal localization, the plants must be less than 4 weeks old and at the 4-leaf-to-6-leaf stage, with the diameter of the largest leaves approximately 4 to 6 cm. At least 10 plants were used per experimental condition, and all the experiments were repeated three times.
Confocal microscopy.
Images were collected using a Zeiss LSM 5 Pascal laser scanning confocal microscope. A 458-nm line and a 488-nm line from an argon ion laser were used to excite CFP and green fluorescent protein (GFP), respectively, and a 543-nm line and a 587-nm line from a helium-neon ion laser were used to excite DsRed2 and monomeric red fluorescent protein (mRFP), respectively. All settings for image acquisition, i.e., all laser intensity and photomultiplier tube (PMT) settings, were preserved between experiments. On average, 100 to 120 cells were examined for each experiment.
To measure levels of protein accumulation, all confocal images were collected using exactly the same setting references. Images were imported into Adobe Photoshop, and the signal of nonspecific protein aggregations was eliminated. Then, the integrated signal density at plasmodesmata, i.e., at punctae at the plasma membrane, was quantified using ImageJ software (version 1.49; NIH) with the “analyze particles” command (http://imagej.nih.gov/ij/). Detected particle size was set to 3 to 50 pixels2 and the threshold value adjusted to 50 as previously described (21).
Quantification of protein colocalization.
Agrobacterium cultures at a cell density A600 value of 0.5 containing the construct that expressed TMV MP-CFP or TMV MP1–50-CFP were mixed with Agrobacterium cultures at a cell density A600 value of 0.1 containing the construct that expressed PDCB1-DsRed2 and were coinfiltrated into intact N. benthamiana leaf epidermis. Between 30 and 32 cells per experiment were imaged 24 to 36 h after infiltration. Co-occurrence and PCC values were calculated using Zen software (Version 5.5.0.452; Zeiss) and the “colocalization” tool with the default setting. Calculation area was selected to have CFP and/or DsRed2 signals of similar intensities at the regions of interest, i.e., plasmodesmal puncta (21), and to avoid nonspecific protein aggregates.
Plasmolysis.
Leaf sections were excised, incubated in 5% NaCl as previously described (33) until epidermal cells were visibly plasmolyzed, and examined by confocal microscopy.
Cell-to-cell movement.
Agrobacterium cultures at a cell density A600 value of 0.1 containing the construct that expressed TMV MP-CFP or TMV MP1–50-CFP were mixed with Agrobacterium cultures at a cell density A600 value of 0.05 containing the reference construct that expressed free DsRed2 and were coinfiltrated into intact N. benthamiana leaf epidermis. The transformed tissues were analyzed by confocal microscopy 36 to 48 h after infiltration. Cells that contained both CFP and DsRed2 signals were scored as infiltrated cells, and the adjacent cells that contained only a CFP signal were scored as those that exhibited movement.
SUPPLEMENTAL MATERIAL
List of PCR primers used for generation of constructs for expression of different TMV MP mutants. (a) Letters R and F in primer names indicate reverse and forward primers, respectively. (b) Capital letters in primer sequences indicate EcoRI (GAATTC), BamHI (GGATCC), or BglI (AGATCT) sites or the translation initiation site (ATG).
ACKNOWLEDGMENTS
The work in the laboratory of V.C. is supported by grants from NIH, NSF, USDA/NIFA, BARD, and BSF to V.C., and the laboratory of S.G.L. is supported by the NIH and funds from the Department of Plant Pathology and Plant-Microbe Biology to S.G.L.
Footnotes
Citation Yuan C, Lazarowitz SG, Citovsky V. 2016. Identification of a functional plasmodesmal localization signal in a plant viral cell-to-cell-movement protein. mBio 7(1):e02052-15. doi:10.1128/mBio.02052-15.
REFERENCES
- 1.Creager ANH, Scholthof KBG, Citovsky V, Scholthof HB. 1999. Tobacco mosaic virus: pioneering research for a century. Plant Cell 11:301–308. doi: 10.1105/tpc.11.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishiguchi M, Motoyoshi F, Oshima M. 1980. Further investigation of a temperature-sensitive strain of Tobacco mosaic virus: its behaviour in tomato leaf epidermis. J Gen Virol 46:497–500. doi: 10.1099/0022-1317-46-2-497. [DOI] [Google Scholar]
- 3.Nigg EA, Baeuerle PA, Lührmann R. 1991. Nuclear import-export: in search of signals and mechanisms. Cell 66:15–22. doi: 10.1016/0092-8674(91)90135-L. [DOI] [PubMed] [Google Scholar]
- 4.Nigg EA. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 5.Freitas N, Cunha C. 2009. Mechanisms and signals for the nuclear import of proteins. Curr Genomics 10:550–557. doi: 10.2174/138920209789503941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JY, Rim Y, Wang J, Jackson D. 2005. A novel cell-to-cell trafficking assay indicates that the Knox homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev 19:788–793. doi: 10.1101/gad.332805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JY, Yuan Z, Jackson D. 2003. Developmental regulation and significance of Knox protein trafficking in Arabidopsis. Development 130:4351–4362. doi: 10.1242/dev.00618. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Jackson D, Kim JY. 2014. Identification of evolutionarily conserved amino acid residues in homeodomain of Knox proteins for intercellular trafficking. Plant Signal Behav 9:e28355. doi: 10.4161/psb.28355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Ahmad M, Rim Y, Lucas WJ, Kim JY. 2013. Evolutionary and molecular analysis of dof transcription factors identified a conserved motif for intercellular protein trafficking. New Phytol 198:1250–1260. doi: 10.1111/nph.12223. [DOI] [PubMed] [Google Scholar]
- 10.Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ. 2008. Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 6:e7. doi: 10.1371/journal.pbio.0060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiró A, Martínez-Gil L, Tamborero S, Pallás V, Sánchez-Navarro JA, Mingarro I. 2014. The Tobacco mosaic virus movement protein associates with but does not integrate into biological membranes. J Virol 88:3016–3026. doi: 10.1128/JVI.03648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M. 2000. Function of microtubules in intercellular transport of plant virus RNA. Nat Cell Biol 2:826–832. doi: 10.1038/35041072. [DOI] [PubMed] [Google Scholar]
- 13.Heinlein M, Epel BL, Padgett HS, Beachy RN. 1995. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270:1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- 14.Oparka KJ, Prior DA, Santa Cruz S, Padgett HS, Beachy RN. 1997. Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV). Plant J 12:781–789. doi: 10.1046/j.1365-313X.1997.12040781.x. [DOI] [PubMed] [Google Scholar]
- 15.Crawford KM, Zambryski PC. 2001. Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol 125:1802–1812. doi: 10.1104/pp.125.4.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotlizky G, Katz A, van der Laak J, Boyko V, Lapidot M, Beachy RN, Heinlein M, Epel BL. 2001. A dysfunctional movement protein of Tobacco mosaic virus interferes with targeting of wild-type movement protein to microtubules. Mol Plant Microbe Interact 14:895–904. doi: 10.1094/MPMI.2001.14.7.895. [DOI] [PubMed] [Google Scholar]
- 17.Roberts IM, Boevink P, Roberts AG, Sauer N, Reichel C, Oparka KJ. 2001. Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma 218:31–44. doi: 10.1007/BF01288358. [DOI] [PubMed] [Google Scholar]
- 18.Ueki S, Lacroix B, Krichevsky A, Lazarowitz SG, Citovsky V. 2009. Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat Protoc 4:71–77. doi: 10.1038/nprot.2008.217. [DOI] [PubMed] [Google Scholar]
- 19.Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ. 2009. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21:581–594. doi: 10.1105/tpc.108.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V. 2005. pSAT vectors: a modular series of plasmids for fluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57:503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- 21.Levy A, Zheng JY, Lazarowitz SG. 2015. Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr Biol 25:2018–2025. doi: 10.1016/j.cub.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giesman-Cookmeyer D, Lommel SA. 1993. Alanine scanning mutagenesis of a plant virus movement protein identifies three functional domains. Plant Cell 5:973–982. doi: 10.1105/tpc.5.8.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, Rhee SY, Raikhel NV, Citovsky V. 2004. High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135:25–38. doi: 10.1104/pp.104.040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding B, Haudenshield JS, Hull RJ, Wolf S, Beachy RN, Lucas WJ. 1992. Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell 4:915–928. doi: 10.1105/tpc.4.8.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueki S, Spektor R, Natale DM, Citovsky V. 2010. ANK, a host cytoplasmic receptor for the Tobacco mosaic virus cell-to-cell movement protein, facilitates intercellular transport through plasmodesmata. PLoS Pathog 6:e1001201. doi: 10.1371/journal.ppat.1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee Y, Tzfira T, Chen MH, Waigmann E, Citovsky V. 2000. Cell-to-cell movement of tobacco mosaic virus: enigmas and explanations. Mol Plant Pathol 1:33–39. doi: 10.1046/j.1364-3703.2000.00005.x. [DOI] [PubMed] [Google Scholar]
- 27.Kahn TW, Lapidot M, Heinlein M, Reichel C, Cooper B, Gafny R, Beachy RN. 1998. Domains of the TMV movement protein involved in subcellular localization. Plant J 15:15–25. doi: 10.1046/j.1365-313X.1998.00172.x. [DOI] [PubMed] [Google Scholar]
- 28.Levy A, Zheng JY, Lazarowitz SG. 2013. The tobamovirus Turnip Vein Clearing Virus 30-kilodalton movement protein localizes to novel nuclear filaments to enhance virus infection. J Virol 87:6428–6440. doi: 10.1128/JVI.03390-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen MH, Tian GW, Gafni Y, Citovsky V. 2005. Effects of calreticulin on viral cell-to-cell movement. Plant Physiol 138:1866–1876. doi: 10.1104/pp.105.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzfira T, Jensen CS, Wangxia W, Zuker A, Altman A, Vainstein A. 1997. Transgenic Populus: a step-by-step protocol for its Agrobacterium-mediated transformation. Plant Mol Biol Rep 15:219–235. doi: 10.1023/A:1007484917759. [DOI] [Google Scholar]
- 31.Kapila J, De Rycke R, Van Montagu M, Angenon G. 1997. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci 122:101–108. doi: 10.1016/S0168-9452(96)04541-4. [DOI] [Google Scholar]
- 32.Wroblewski T, Tomczak A, Michelmore R. 2005. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol J 3:259–273. doi: 10.1111/j.1467-7652.2005.00123.x. [DOI] [PubMed] [Google Scholar]
- 33.Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, Schroeder JI, Kangasjärvi J. 2008. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of PCR primers used for generation of constructs for expression of different TMV MP mutants. (a) Letters R and F in primer names indicate reverse and forward primers, respectively. (b) Capital letters in primer sequences indicate EcoRI (GAATTC), BamHI (GGATCC), or BglI (AGATCT) sites or the translation initiation site (ATG).