Abstract
AIM: To assess the clinical significance of Hepatitis B virus (HBV) DNA localization in the liver tissue of patients with positive HBsAg and negative viremia.
METHODS: HBV virological parameters of 33 HBsAg positive chronic hepatitis patients, including seromarkers and HBV DNA amplification in both sera and liver biopsies, were evaluated.
RESULTS: Ten patients had negative viremia and positive HBV DNA in their liver biopsies. Most of them had HBeAg-negative/HBeAb-positive chronic hepatitis. Their liver biochemical and histopathological profiles were different from the viremic patients. Their disease pattern was designated as “hepatitis B in situ”.
CONCLUSION: Hepatitis B in situ is a consequential entity which can be missed in clinical practice. It is a new clinical pattern of chronic HBV infection that considers HBV in liver biopsy and adds a new indication for antiviral therapy.
Keywords: Hepatitis B in situ, Antiviral therapy, HBV DNA, Chronic hepatitis
INTRODUCTION
Hepatitis B virus is a virus that infects 350 million people worldwide with a clinical spectrum of acute hepatitis, the healthy carrier state, cirrhosis, and hepatocellular carcinoma. The outcome of infection is the result of complicated viral-host interactions. As in other infections with non-cytopathic viruses, the immune response is thought to play a crucial role in disease pathogenesis. However, there are increasing evidences that varieties of viral mechanisms, some depending on the function of virally encoded proteins, have a profound impact on the infected hepatocytes, the liver microenvironment and host antiviral responses[1].
Occult HBV infection is characterized by undetectable HBsAg. Serum HBV DNA level is usually less than 104 copies/mL in these patients. Diagnosis requires a sensitive HBV-DNA assay[2]. To our knowledge, no data are available about the amount of HBV genomes in the liver of patients with chronic HBV infection. However, it has been reported that cases with suppressed HBV activity, despite the very low level of viremia, maintain a relatively high amount of intrahepatic viral genomes. This virus reservoir is likely involved in HBV reactivation, which is usually observed after stopping lamivudine treatment[3].
The detection of HBV genomes in nested PCR-based assays appears to be the most reliable methods for monitoring infection and assessing response to antiviral treatment[4]. The majority of patients with HBsAg in serum have HBV DNA present in serum as well. Some patients have HBsAg but no HBV DNA in serum. These are chronic carriers with normal transaminases[5]. On the other hand, HBV DNA can be detected in serum and liver by PCR in patients who have lost their HBsAg. In addition, hepatic HBV DNA can be detected in cirrhotic liver in the absence of serum HBV DNA[5,6]. HBV DNA can be treated as a sign of HBV activity[7], without ignoring the facts that HBV DNA levels fluctuate in patients with chronic HBV infection and that the correlation between serum HBV DNA levels and histologic activity is poor[8].
In this study, we evaluated the clinical significance of the localization of HBV in liver tissue of chronic HBV patients with negative viremia and elevated ALT.
MATERIALS AND METHODS
This prospective study included chronic liver disease patients who attended the out-patient clinic of the Endemic Medicine Department, Cairo University during the period between September 2002 and July 2003. All patients with data suggestive of liver affection were evaluated, and those with positive HBsAg and elevated ALT were selected as the candidates for this study.
Exclusion criteria were HCV infection, advanced liver disease, alcohol consumption, autoimmune hepatitis, drug-induced hepatitis, hepatic schistosomiasis, evidence of acute hepatitis in the last 6 mo, presence of HDVAb, hepatocellular carcinoma, and advanced co-morbid conditions such as cardiac or renal disease.
Selected patients were subjected to clinical evaluation and laboratory investigations including viral serologic tests using enzyme immunoassay technique applying the Abbott System®. These assays included the detection of HBsAg, HBsAb, HBcAb (total), HBeAg, HBeAb, and HCVAb (total) by 3rd generation ELISA assay (Sorin®, Italy). Abdominal ultrasonography and liver biopsy were performed. The modified HAI score was used to stage and grade liver disease[9]. For the detection of hepatic total HBV DNA (integration plus CCC form), part of the liver sample was incubated at 56 °C for 3 h with 270 µL of phosphate buffer saline containing 160 µg of proteinase K (Promega, Madison, WI, USA) and 30 µL of 10 buffer. Serum HBV DNA was extracted by incubation at 56 °C for 3 h with 270 µL of serum plus 30 µL of 10×buffer containing 160 µg proteinase K. DNA extraction was purified by phenol:chloroform (300 µL, 1:1 V/V) extraction, ethanol/0.2 mol/L sodium acetate precipitation. Qualitative PCR testing for HBV in serum and liver tissue was carried out using nested PCR technique[10]. Total genomic DNA was extracted from serum or liver tissue using phenol/chloroform extraction after proteinase K digestion. Purified DNA (1–2 µg) was utilized as a template in the polymerase chain reaction using two rounds of amplification with nested primers as follows: Outer primers: B1 5’ AAGGTCTTACATAAGAGGAC 3’ (nt 1 644–1 663), B2 5’ CTAACATTGAGATTCCCGAGATTGAGA 3’ (nt 2 458–2 432); Inner primers: B3 5’ GGCTGTAGGCATAAATTGGTC-TG 3’ (nt 1 781–1 803), B4 5’ TTGCCTGAGTGCAGTA-TGGT 3’ (nt 2 075–2 056).
The PCR reactions were performed in 50 µL reaction volume containing 10 µmol/L dNTPs, 50 pmol from each primer, and 2 U of Taq. The amplification protocol for the two nested rounds consisted of 35 cycles, each cycle contained a denaturation at 94 °C for 30 s, annealing at 52 °C for 1 min, and extension at 72 °C for 2 min, followed by a single cycle with final extension at 72 °C for 7 min. Positive samples infected with HBV DNA produced a amplified product of 295 bp in length.
Statistical analysis
Patients’ data were tabulated, and then processed using the software SPSS for Windows 9 version 10.0®. Quantitative variables were expressed as mean±SD and compared using Student’s t test. Qualitative variables were compared using Fisher’s exact test. A P<0.05 was considered statistically significant.
RESULTS
Out of the 670 patients with chronic liver disease, 44 were seropositive for HBsAg. Eleven of the forty-four HBsAg seropositive patients were excluded because of normal liver enzymes. The study was then conducted on the remaining 33 patients (26 males and 7 females; mean age 32.8±12.0 years, range 18–60 years). Fifteen patients were originally from urban areas and 18 from rural areas (10 from Nile Delta, and 8 from Upper Egypt).
According to the modified HAI grading and staging, histopathological findings are shown in Figure 1. Ground glass pattern of the hepatocytes was detected in 21 patients, steatosis was found in 14 patients, while cholestasis was detected in two cases.
Figure 1.
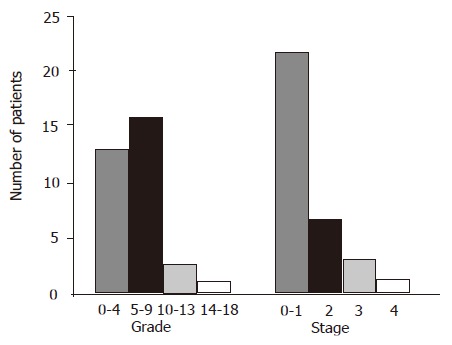
Histopathological grading and staging
HBV markers were spread into Poynard’s table of classification of HBV infected patients[10]. Seventeen patients were fit with the “chronic carrier, pre-core mutant type”, and six with the “chronic carrier, wild type” definitions. Ten patients remained uncategorized, showing a unique pattern (Table 1).
Table 1.
Characteristics of patients uncategorized in Poynard (2000) classification
| HbsAg | HbsAb | HbeAg | HbeAb | HBcAb IgG | HBcAb IgM | DNA serum | DNA liver | Symptom | ALT | HAI/fibrosis | No. of patients | |
| Uncategorized | + | +/– | –/+ | +/– | + | – | – | + | +/– | + | + | 10 |
These 10 patients (group 1) were compared to the remaining 23 chronic hepatitis B patients (group 2) as shown in Tables 2 and 3. No statistically significant differences were observed regarding the liver biochemical or histological parameters. In this study, we found five possible permutations of the HBV seromarkers, which were grouped according to the HBV DNA results (Table 4). Among the HBcAb-negative patients, three patients were also negative for HBeAg and HBeAb. HBV DNA was detectable in six patients with positive HBsAb (four with detectable viral DNA in both serum and liver tissue, while two with detectable viral DNA only in liver tissue). Although four of the HBeAg-negative cases had higher HAI score (10–18) compared to none of the HBeAg-positive chronic hepatitis cases, the two groups showed no significant difference in either biochemical or histological parameters.
Table 2.
Hepatic biochemical tests in group I vs group II patients
| Group I (n = 10) | Group II (n = 23) | P values | |
| Serum bilirubin (mg/dL) | 1.36±1.31 | 1.44±2.1 | 0.9 |
| AST (IU/L) | 63.9±24.1 | 82.0±51.3 | 0.3 |
| ALT (IU/L) | 62.6±27.4 | 0.2 | |
| Serum albumin (g/dL) | 3.9±0.50 | 4.0±0.57 | 0.4 |
| INR | 1.25±0.22 | 1.23±0.14 | 0.7 |
Table 3.
Hepatic histopathologic parameters in group I vs group II patients
| Group I (n = 10) | Group II (n = 23) | P values | ||
| Grading | 0–9 | 8 | 20 | 0.4 |
| 10–18 | 2 | 3 | ||
| Staging | 0–2 | 9 | 19 | 0.9 |
| 3–6 | 1 | 4 |
Table 4.
HBV seromarker pattern according to HBV DNA testing
| Marker pattern | n |
HBV DNA status |
|||
| +ve serum | –ve serum | +ve serum | –ve serum | ||
| +ve liver | –ve liver | –ve liver | +ve liver | ||
| cAb+eAg–eAb+ | 18 | 7 | 1 | 3 | 7 |
| cAb+eAg+eAb– | 7 | 5 | – | 1 | 1 |
| cAb–eAg–/+eAb–/+ | 5 | 2 | 2 | 1 | – |
| cAb+eAg–eAb– | 2 | 1 | – | – | 1 |
| cAb+eAg+eAb+ | 1 | – | – | – | 1 |
| Total | 33 | 15 | 3 | 5 | 10 |
cAb: hepatitis B core antibody; eAg: hepatitis B envelope (e) antigen; eAb: hepatitis B antibody against envelope (e) antigen.
DISCUSSION
HBV infection varies widely from high (>8%, e.g., Africa, Asia, and the Western Pacific) to intermediate (2–7%, e.g., Southern and Eastern Europe) to low (<2%, e.g., Western Europe, North America, and Australia) prevalence areas. Egypt lies in the intermediate prevalence region[10]. HBV DNA is a good marker of the level of viremia and can be correlated with serum transaminase levels. The majority of patients with HBsAg in serum have HBV DNA. However, some patients are positive for HBV DNA in serum even in the absence of HBsAg, while some others have HBsAg but not HBV DNA in serum[5]. The presence of HBV genome is associated with ongoing necroinflammation[8]. In the presence of HBeAg, the diagnosis of replicating chronic HBV can be made whatever the viral load is. In contrast, the interpretation of HBV DNA quantification is difficult in HBeAg-negative/HBeAb-positive patients (pre-core mutant HBV) who generally have lower replication levels than HBeAg-positive patients. Active HBV replication is associated with a significant risk of progression to chronic hepatitis B complications[11]. The presence of HBV DNA is the single most important indication to start antiviral therapy[12]. This work aimed at evaluating the clinical significance of localizing HBV DNA in the liver in case of negative viremia. Patients with HBsAg and elevated ALT were selected among patients who presented at the outpatient clinic of the Tropical Medicine Department, suffering from a variety of chronic liver diseases.
Of 670 patients, the prevalence of HBsAg-positive patients among those with chronic liver disease was 6.6% (44/670). This figure represented both the carriage rate and the infection rate among chronic liver disease patients, whatever the causes were. The prevalence of HBV chronic hepatitis among chronic liver disease patients was 4.9% (33/670). The prevalences of HBV infection in Egypt have been found to be variable among different studies, ranging from 3.2% to 21%[13-19], depending on the number of patients. The higher figures studied the prevalence among cirrhotic patients, the lowest figures were found among school children in certain villages. Thirty-three patients eligible for the present study were considered to have chronic hepatitis B. NASH was not totally excluded. Fourteen liver biopsies showed prominent steatosis. The prevalence of steatosis in Egypt is expected to be quite high. It is known that 35% of the Egyptians are considered obese[20]. Moreover, high comorbid factors are recorded among Egyptians, such as diabetes, pollution, etc. It is worth mentioning that none of our 33 patients had a BMI >29 kg/m2, and we had two diabetic patients.
In this study, the histopathological results showed that most of our patients had relatively mild disease, with an HAI score between 0 and 9, and a fibrosis score between 0 and 2. Conversely, the HBsAb-positive patients had a higher HAI score as compared with the HBsAb-negative patients. This might be explained by a stronger reaction to HBV infection in the former subgroup. The co-existence of both HBsAg and HBsAb has been previously described, and it is thought that HBsAb in such a situation is unable to neutralize the virus[21]. It has also been postulated that infection with multiple types of HBV is another explanation for this finding[22]. Most of our patients (78.8%) were HBeAg-negative chronic hepatitis B. It is known that the core promoter mutants are predominant in Egypt[23]. Compared to the HBeAg-negative chronic hepatitis B, the HBeAg-positive patients had a milder disease histopathologically, which was in agreement with previous reports[23-25]. We observed that the detection of HBV viremia was more in HBeAg-positive patients, which is expected, because of the formerly documented strong correlation between HBV DNA and HBeAg[26]. Characteristically, five patients (15.2%) were negative for both HBeAg and HBeAb, and five patients were negative for HBcAb. Three patients were negative for all seromarkers except HBsAg. These unusual patterns of serological profile can be explained either by mutations or viral DNA integration. Absence of cytotoxic T-lymphocyte (CTL) recognition of epitopes in HBeAg, HBsAg, and other viral proteins contributes to viral persistence. Failure of antigen presentation to CD4 helper T cells, failure of CD4 proliferating responses, immune modulation by secreted proteins, viral mutation, and integration of viral DNA into the cellular genome are all mechanisms involved in viral persistence[27]. Three patients had undetectable HBV DNA in either serum or liver tissue. This might be explained by an enhanced immunological response to HBV DNA, and extraordinarily low viral DNA copy number[28], or by the integration phenomenon affecting the primer orientation used in PCR technique[29]. Another explanation stands on HBV infection being a dynamic process, so that some of these patients might have been in the immune clearance phase[23]. Five patients had negative HBV DNA in the liver tissue, but positive in serum. This can be explained by the liver being a vast size organ, and hence, substantial viremia can result even if each cell produces only a few numbers of virions per day. This hypothesis has been proposed by Branch et al[31] on HCV infection. In our opinion, it can also be applied on HBV infection if the viral genome is present in a very low level to be detected by biopsy. Ten patients (30.3%) did not fulfill any of the clinical criteria identified in literature. They could not classically fit in the Poynard’s 2002 classification. They had one common serological profile, which is, detection of HBV DNA in the liver tissues but not in the sera (negative viremia). The disease burden as evidenced by hepatic biochemical profile and the spectrum of HAI and fibrosis was not different in this group of patients from those with the classic chronic HBV infection. This group has been named in this study as “hepatitis B in situ”. Seven out of the eighteen patients, showing a classic unequivocal pattern of HBeAg-negative chronic hepatitis, had hepatitis B in situ. This implies that the reliance on mere testing of serum HBV DNA would have missed viral replication in such condition. On the other hand, HBeAg-positive hepatitis showed viremia in most cases.
In conclusion, a significant number of patients with chronic hepatitis B fail to show viremia, yet HBV DNA can be detected in their liver tissues. Hepatitis B in situ is a term used to describe the detection of total HBV DNA (Integrated plus CCC form) in liver tissue of patients with positive HBsAg and negative viremia. It is particularly frequent in the HBeAg-negative variety of chronic hepatitis B. The bearing of “hepatitis B in situ” on the liver is no less than the viremic form; serious sequelae are thus liable to take place. The recognition of this form of the disease may have its impact on the choice of therapy, since such patients should not be denied the chance to receive antiviral treatment.
Footnotes
Science Editor Kumar M and Guo SY Language Editor Elsevier HK
References
- 1.Rapicetta M, Ferrari C, Levrero M. Viral determinants and host immune responses in the pathogenesis of HBV infection. J Med Virol. 2002;67:454–457. doi: 10.1002/jmv.10096. [DOI] [PubMed] [Google Scholar]
- 2.Fong TL, Di Bisceglie AM, Gerber MA, Waggoner JG, Hoofnagle JH. Persistence of hepatitis B virus DNA in the liver after loss of HBsAg in chronic hepatitis B. Hepatology. 1993;18:1313–1318. [PubMed] [Google Scholar]
- 3.Cacciola I, Pollicino T, Squadrito G, Cerenzia G, Villari D, de Franchis R, Santantonio T, Brancatelli S, Colucci G, Raimondo G. Quantification of intrahepatic hepatitis B virus (HBV) DNA in patients with chronic HBV infection. Hepatology. 2000;31:507–512. doi: 10.1002/hep.510310235. [DOI] [PubMed] [Google Scholar]
- 4.Pawlotsky JM, Bastie A, Hézode C, Lonjon I, Darthuy F, Rémiré J, Dhumeaux D. Routine detection and quantification of hepatitis B virus DNA in clinical laboratories: performance of three commercial assays. J Virol Methods. 2000;85:11–21. doi: 10.1016/s0166-0934(99)00149-4. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko S, Miller RH, Di Bisceglie AM, Feinstone SM, Hoofnagle JH, Purcell RH. Detection of hepatitis B virus DNA in serum by polymerase chain reaction. Application for clinical diagnosis. Gastroenterology. 1990;99:799–804. doi: 10.1016/0016-5085(90)90971-3. [DOI] [PubMed] [Google Scholar]
- 6.Lee CZ, Huang GT, Yang PM, Sheu JC, Lai MY, Chen DS. Correlation of HBV DNA levels in serum and liver of chronic hepatitis B patients with cirrhosis. Liver. 2002;22:130–135. doi: 10.1034/j.1600-0676.2002.01525.x. [DOI] [PubMed] [Google Scholar]
- 7.Meng Q, Wong C, Rangachari A, Tamatsukuri S, Sasaki M, Fiss E, Cheng L, Ramankutty T, Clarke D, Yawata H, et al. Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA, and human immunodeficiency virus type 1 RNA. J Clin Microbiol. 2001;39:2937–2945. doi: 10.1128/JCM.39.8.2937-2945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu CJ, Lok AS. Clinical utility in quantifying serum HBV DNA levels using PCR assays. J Hepatol. 2002;36:549–551. doi: 10.1016/s0168-8278(02)00039-9. [DOI] [PubMed] [Google Scholar]
- 9.Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, Denk H, Desmet V, Korb G, MacSween RN. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 10.De Castro L, Niel C, Gomes SA. Low frequency of mutations in the core promoter and precore regions of hepatitis B virus in anti-HBe positive Brazilian carriers. BMC Microbiol. 2001;1:10. doi: 10.1186/1471-2180-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poynard T. Hepatitis B. In Hepatitis B and C: management and treatment. 1st ed. Mobipocket ebooks; 2002. pp. 63 http: //www.ebookmall.com/ebook/88983–ebook.htm. [Google Scholar]
- 12.Pawlotsky JM. Molecular diagnosis of viral hepatitis. Gastroenterology. 2002;122:1554–1568. doi: 10.1053/gast.2002.33428. [DOI] [PubMed] [Google Scholar]
- 13.Tsai NC. Practical management of chronic hepatitis B infection. Semin Liver Dis. 2004;24 Suppl 1:71–76. doi: 10.1055/s-2004-828681. [DOI] [PubMed] [Google Scholar]
- 14.Sherif MM, Abou-Aita BA, Abou-Elew MH, el-Kafrawi AO. Hepatitis B virus infection in upper and lower Egypt. J Med Virol. 1985;15:129–135. doi: 10.1002/jmv.1890150205. [DOI] [PubMed] [Google Scholar]
- 15.Kamel MA, Miller FD, el Masry AG, Zakaria S, Khattab M, Essmat G, Ghaffar YA. The epidemiology of Schistosoma mansoni, hepatitis B and hepatitis C infection in Egypt. Ann Trop Med Parasitol. 1994;88:501–509. doi: 10.1080/00034983.1994.11812897. [DOI] [PubMed] [Google Scholar]
- 16.Attia MA. Prevalence of hepatitis B and C in Egypt and Africa. Antivir Ther. 1998;3:1–9. [PubMed] [Google Scholar]
- 17.André F. Hepatitis B epidemiology in Asia, the Middle East and Africa. Vaccine. 2000;18 Suppl 1:S20–S22. doi: 10.1016/s0264-410x(99)00456-9. [DOI] [PubMed] [Google Scholar]
- 18.Angelico M, Renganathan E, Gandin C, Fathy M, Profili MC, Refai W, De Santis A, Nagi A, Amin G, Capocaccia L, et al. Chronic liver disease in the Alexandria governorate, Egypt: contribution of schistosomiasis and hepatitis virus infections. J Hepatol. 1997;26:236–243. doi: 10.1016/s0168-8278(97)80036-0. [DOI] [PubMed] [Google Scholar]
- 19.Zaki A, Bassili A, Amin G, Aref T, Kandil M, Abou Basha LM. Morbidity of schistosomiasis mansoni in rural Alexandria, Egypt. J Egypt Soc Parasitol. 2003;33:695–710. [PubMed] [Google Scholar]
- 20.Reda AA, Arafa MA, Youssry AA, Wandan EH, Ab de Ati M, Daebees H. Epidemiologic evaluation of the immunity against hepatitis B in Alexandria, Egypt. Eur J Epidemiol. 2003;18:1007–1011. doi: 10.1023/a:1025805817101. [DOI] [PubMed] [Google Scholar]
- 21. Available from: http: //www.tribuneindia.com/2000/20000917/world.htm#6.
- 22.Park LC, Maruyama T, Kamiya N, Goto M, Kuma H, Hamasaki N. Mutation detection in the drug-resistant hepatitis B virus polymerase gene using nanostructured reverse micelles. Anal Sci. 2004;20:1609–1611. doi: 10.2116/analsci.20.1609. [DOI] [PubMed] [Google Scholar]
- 23.Saudy N, Sugauchi F, Tanaka Y, Suzuki S, Aal AA, Zaid MA, Agha S, Mizokami M. Genotypes and phylogenetic characterization of hepatitis B and delta viruses in Egypt. J Med Virol. 2003;70:529–536. doi: 10.1002/jmv.10427. [DOI] [PubMed] [Google Scholar]
- 24.Fattovich G. Natural history and prognosis of hepatitis B. Semin Liver Dis. 2003;23:47–58. doi: 10.1055/s-2003-37590. [DOI] [PubMed] [Google Scholar]
- 25.Hadziyannis SJ, Vassilopoulos D. Hepatitis B e antigen-negative chronic hepatitis B. Hepatology. 2001;34:617–624. doi: 10.1053/jhep.2001.27834. [DOI] [PubMed] [Google Scholar]
- 26.Yalcin K, Degertekin H, Yildiz F, Celik Y. Markers of disease activity in chronic hepatitis B virus infection. Clin Invest Med. 2003;26:27–34. [PubMed] [Google Scholar]
- 27.Zhang L, Zhu J, Ma Y, Zhang H. Nucleic acid quantifying assay of hepatitis B virus. Zhonghua GanZangBing ZaZhi. 2002;10:49–51. [PubMed] [Google Scholar]
- 28.Carman WF, Thomas HC, Zuckermann AJ. Molecular variants of hepatitis B. In: viral hepatitis. Zuckermann A, Thomas HC, eds. 2nd ed. Churchill Livingstone. 1998;Section III:141–172. [Google Scholar]
- 29.Kuhns M, McNamara A, Mason A, Campbell C, Perrillo R. Serum and liver hepatitis B virus DNA in chronic hepatitis B after sustained loss of surface antigen. Gastroenterology. 1992;103:1649–1656. doi: 10.1016/0016-5085(92)91191-6. [DOI] [PubMed] [Google Scholar]
- 30.Balderas-Renteria I, Munoz-Espinosa LE, Dector-Carrillo MA, Martinez-Martinez FJ, Barrera-Saldana HA. Detection of hepatitis B virus in seropositive and seronegative patients with chronic liver disease using DNA amplification by PCR. Arch Med Res. 2002;33:566–571. doi: 10.1016/s0188-4409(02)00391-0. [DOI] [PubMed] [Google Scholar]
- 31.Branch AD, Seeff LB. Afterword: HCV in the decades ahead. Semin Liver Dis. 2000;20:233–237. [PubMed] [Google Scholar]


