Summary
Microbiota–mediated effects on the host immune response facilitate pathogen colonization resistance. However, it is unclear whether and how the host immune response can regulate the microbiota to mediate colonization resistance. Id2, an essential transcriptional regulator for the development of innate lymphoid cell (ILC) progenitors, remains highly expressed in differentiated ILCs with unknown function. Using conditionally deficient mice that delete Id2 in differentiated ILC3s, we observed that these mutant mice exhibited greatly impaired gut colonization resistance against Citrobacter rodentium. Utilizing gnotobiotic hosts, we showed that the Id2-dependent early colonization resistance was mediated by interleukin-22 (IL-22) regulation of microbiota. In addition to regulating development, Id2 maintained homeostasis of ILC3s, and controlled IL-22 production through an aryl hydrocarbon receptor (Ahr) and IL-23 receptor pathway. Thus, ILC3s can mediate immune surveillance that constantly maintains proper microbiota to mediate early colonization resistance through an Id2–dependent regulation of IL-22.
Introduction
Pathogen colonization resistance is dependent on direct inhibition by host microbiota (van der Waaij et al., 1971). Microbiota can also regulate the host immune response to mediate indirect pathogen colonization resistance (Buffie and Pamer, 2013). In contrast, various host genetic factors including immune factors also contribute to the varying level of individual susceptibility to pathogen infections (Chapman and Hill, 2012; Wlodarska et al., 2014). However, it is unclear whether and how such host genetic factors contribute to pathogen colonization resistance through shaping of the microbiota. Citrobacter rodentium (C. rodentium) is a natural mouse intestinal pathogen that mimics human Enterohaemorrhagic E. coli and Enteropathogenic E. coli, which cause severe diarrhea (Crim et al., 2014; Ochoa et al., 2008). Both innate and adaptive immune components, including IL-22 producing innate lymphoid cells (ILCs), CD4+ T cells, B cells, and C. rodentium-specific antibodies, are essential for controlling and eradicating the infection (Bry and Brenner, 2004; Bry et al., 2006; Guo et al., 2014; Zheng et al., 2008). Gut flora has been shown to be important in protection against infection. C3H/HeOuJ mice suffer 100% mortality after C. rodentium-induced colitis (Vallance et al., 2003), whereas microbiota transfer from C57BL/6 mice led to a complete rescue of C3H/HeOuJ from death (Ghosh et al., 2011).
IL-22 is induced by IL-23 through the IL23R–STAT3 pathway in the early phase of infection and is essential for host defense against C. rodentium infection (Guo et al., 2014; Zheng et al., 2008). The major function of IL-22 is to promote mucosal epithelial cell survival and proliferation, and to trigger the secretion of antimicrobial peptides, such as RegIIIγ (Pickert et al., 2009). Previous studies also show that exogenous RegIIIγ can partially rescue IL-22 deficient mice from death (Zheng et al., 2008). Interestingly, in vitro assay suggests that RegIIIγ can only kill some Gram-positive bacteria but not the Gram–negative bacteria C. rodentium (Cash et al., 2006). Therefore, it is still unknown how IL-22–induced RegIIIγ controls C. rodentium infection. Multiple studies also show that IL-22 can shape the gut microflora, which contributes to protection or exacerbation of inflammatory bowel disease or infections (Behnsen et al., 2014; Qiu et al., 2013; Zelante et al., 2013). However, it is not known whether IL-22 shapes the microbiota to mediate early C. rodentium colonization resistance.
Group 3 innate lymphoid cells (ILC3s) are the major producer of IL-22 in the naive gut (Guo et al., 2014; Qiu et al., 2011). Innate lymphoid cells (ILCs) are newly defined immune cells that protect the host from various infections and include group 1 ILCs, group 2 ILCs and RORγt+ ILC3s (including CD4+LTi, NCR− ILC3s and NCR+ILC3s) (Spits et al., 2013). To date, the developmental and functional program of ILC3s is known to involve the transcription factors, such as RORγt (Eberl and Littman, 2003; Eberl et al., 2004), Ahr (Kiss et al., 2011; Lee et al., 2011; Qiu et al., 2011), and STAT3 (Guo et al., 2014). Recent data suggest that NCR+ILC3s (NKp46+ RORγt+ ILCs) may originate from NCR−ILC3s (Rankin et al., 2013; Vonarbourg et al., 2010). IL7R signaling is critical for the survival of ILC3s, but it also maintains RORγt expression in mature NCR+ILC3s (Schmutz et al., 2009; Vonarbourg et al., 2010).
E proteins belong to bHLH transcription factor family that contains a basic DNA-binding region and a helix–loop–helix (HLH) dimerization domain. They can form homodimers or heterodimers with other HLH proteins and function as transcription activators or repressors. Inhibitor of DNA binding (ID) proteins are HLH proteins that lack a basic region and can prevent E proteins from binding to DNA. Both E and ID proteins play important roles in the lymphoid cell development (Kee, 2009). In particular, Id2 is thought to be required for the development of the ILC precursor since Id2−/− mice lack all the currently known ILCs, including group 1, 2 and 3 ILCs (Boos et al., 2007; Hoyler et al., 2012). Id2 is continuously and highly expressed in all ILCs, including differentiated ILCs (Hoyler et al., 2012). However, the function of Id2 in these well-differentiated ILCs is still unclear, as Id2−/− mice lack ILCs from the earliest identifiable stage. Here, through conditional deletion of Id2 after RORγt expression in the ILC3 lineage, we demonstrated that continuous Id2 expression is required for the homeostasis and function of ILC3. Using this system we showed that ILC3s were essential for regulating the microbiota to mediate early colonization resistance against intestinal pathogen.
Results
Id2 is continuously expressed in intestinal innate lymphoid cells
To test whether Id2 could function in differentiated ILCs, we first analyzed Id2 protein expression in different ILCs population with Id2gfp/+ mice. As previously reported (Cherrier et al., 2012; Hoyler et al., 2012), Id2 was expressed in differentiated ILCs, including NK, ILC1, ILC2 and ILC3 (Figure S1A). Id2 was not homogeneously expressed in the different subsets of ILC3s. NCR+ILC3 expressed higher concentrations of Id2 compared with LTi and NCR−ILC3 (Figure S1B). Since NCR+ILC3 could be differentiated from NCR−ILC3 (Rankin et al., 2013; Vonarbourg et al., 2010), our findings raise the possibility that Id2 may continue to play a role in the development and function of ILC3 after their formation.
To study the requirements for Id2 in the homeostasis and function of differentiated ILC3s, Id2-floxed mice were crossed with Rorc-cre transgenic mice to achieve specific deletion of Id2 after RORγt expression in ILC3s (RorccreId2fl/fl). Because RORγt is transiently expressed at high levels at the double positive stage of T cell development, RorccreId2fl/fl mice not only lack Id2 expression in RORγt+ ILC3s, but also in most αβ T cells (Figure S1C).
Id2 is essential for early colonization resistance and protection against C. rodentium infection
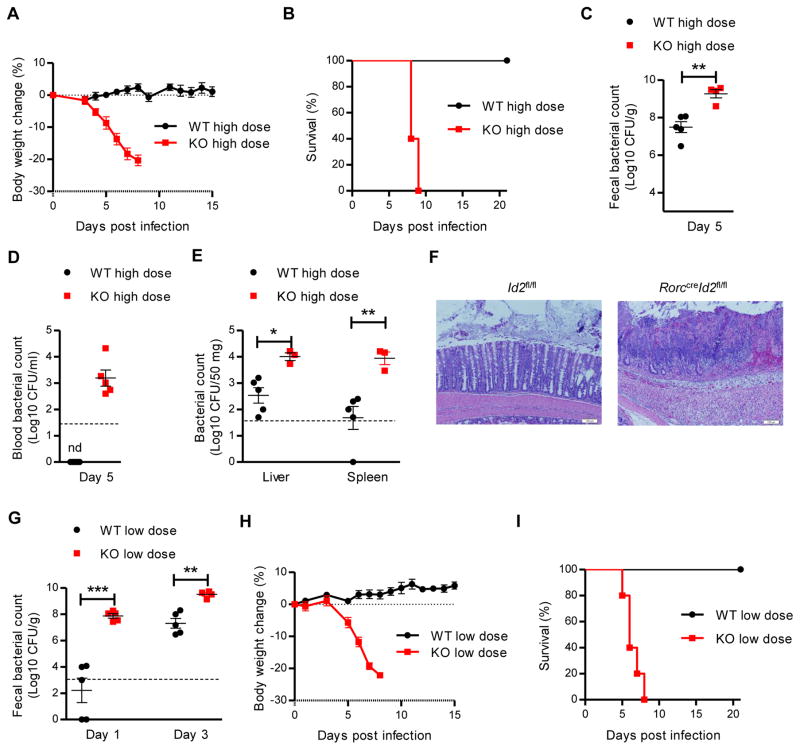
Previous studies have shown that ILC3s are essential for host protection against C. rodentium infection (Guo et al., 2014; Qiu et al., 2011). We next investigated the importance of Id2 for ILC3 function in this infection model. After high doses of C. rodentium infection, RorccreId2fl/fl mice rapidly lost body weight and died around day 10, whereas no weight loss or death was observed in their littermate control Id2fl/fl mice (Figure 1A and 1B). Consistent with the increased morbidity and mortality, RorccreId2fl/fl mice had 10–100 times higher bacterial titers in the feces compared to Id2fl/fl mice at day 5 post-infection (Figure 1C). Systemic dissemination of C. rodentium in RorccreId2fl/fl mice was also observed by increased bacterial titers in the blood, liver and spleen (Figure 1D and 1E). The RorccreId2fl/fl mice also exhibited severe diarrhea, in ammation, and colon pathology upon C. rodentium challenge (data not shown and Figure 1F). Collectively, these data demonstrate that continued Id2 expression in RORγt+ cells is required for host defense against C. rodentium infection.
Figure 1. Id2 is essential to mediate the colonization resistance and protection against C. rodentium infection.
(A–F) 7 weeks old RorccreId2fl/fl (KO, n=5) and their littermate wild type Id2fl/fl mice (WT, n=5) were orally inoculated with high dose (2 × 109 CFU) of C. rodentium. Body weight change (A) and survival rates (B) are shown. Fecal and blood C. rodentium titers at indicated day post infection (C, D) and C. rodentium titers from spleen and liver homogenate cultures at day 8 post infection (E)are shown. Dash line, limit of detection. (F) Histological analysis of representative colons from WT and KO mice at day 8 after infection. Scale bars, 100 μm.
(G–I) 7 weeks old KO (n=5) and their littermate WT (n=5) mice were orally inoculated with low dose (5 × 106 CFU) of C. rodentium. (G) Fecal C. rodentium titers at indicated day post infection are shown. Body weight change (H) and survival rates (I) are shown.
Each dot represents one individual mouse (C, D, E and G). Error bars represent SEM. *P<0.05, **P<0.01, ***P<0.001 (Student’s t-test). nd, nondetectable. Data are representative of three independent experiments (A–D, G–I) or two independent experiments (E, F). See also Figure S1.
Since disease signs, including diarrhea and body weight loss, appeared in Id2 deficient mice before day 5 post infection, we hypothesized that an Id2–dependent mechanism influenced the intestinal environment to limit early colonization even before the innate response was initiated. To test our hypothesis, we challenged both RorccreId2fl/fl and Id2fl/fl littermate mice with a low dose of C. rodentium to determine whether RorccreId2fl/fl mice were more readily colonized. Interestingly, there was significantly more colonization of C. rodentium in the RorccreId2fl/fl mice in the first few days, even as early as day 1, after infection compared with Id2fl/fl mice (Figure 1G). Moreover, C. rodentium could be detected in the whole intestine of RorccreId2fl/fl mice even at day 1 post infection, compared with only in the cecum of Id2fl/fl mice (Figure S1D). As in the high dose infection, RorccreId2fl/fl mice infected with a low dose of C. rodentium had severe diarrhea, rapidly lost body weight and died around day 10 whereas Id2fl/fl mice were not affected (Figure 1H and 1I). Together, our data indicate that Id2 is essential in RORγt+ cells for host defense against a mucosal bacterial pathogen and is required for maintenance of early colonization resistance.
Since it takes a few days for an effective innate immune response to control pathogen, the impaired early colonization resistance against C. rodentium suggested there was a preexisting defect in the RorccreId2fl/fl mice. Previous studies have shown that either increased inflammation or reduced mucus layer could result in increased colonization of intestinal pathogen (Bergstrom et al., 2010; Wlodarska et al., 2011). However, colonic mRNA analysis showed that there was no increased expression of pro-inflammatory cytokines or reduction of mucin in RorccreId2fl/fl mice (Figure S1E and S1F). Moreover, HE and PAS staining showed that there were no obvious alteration of histopathology, mucus layer and goblet cells (Figure S1G). Together, these data suggest that the increased C. rodentium colonization in our Id2 deficient mice is unlikely due to changed colonic inflammation or mucus environment.
Id2 dependent microbiota controls colonization resistance against C. rodentium infection
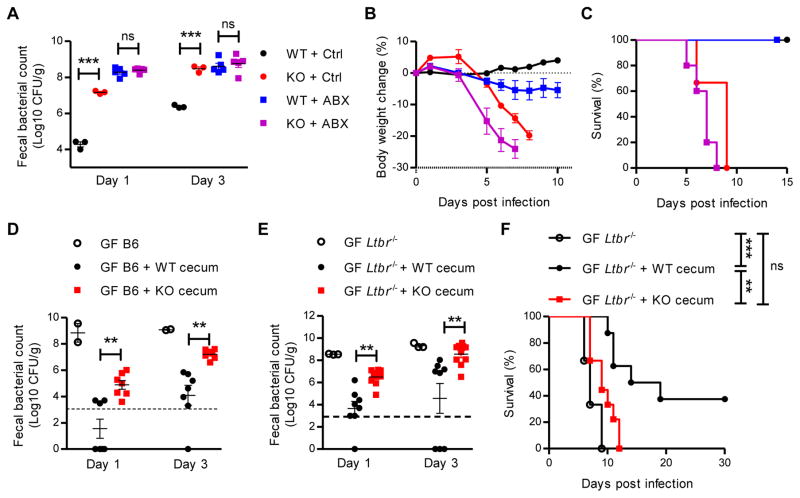
Host microbiota has been recognized as a direct mediator for pathogen colonization resistance (Buffie and Pamer, 2013). To test whether Id2 regulated early C. rodentium colonization through the microbiota, we first treated both Id2fl/fl and RorccreId2fl/fl mice with antibiotics for 1 week, then infected these mice with a low dose of C. rodentium after 1 day of rest. As shown in Figure 2A, fecal C. rodentium titers in antibiotic treated RorccreId2fl/fl mice were much higher than in Id2fl/fl mice at day 1 and 3 post infection. However, bacterial titers were increased in both the Id2fl/fl and RorccreId2fl/fl mice after antibiotic treatment during first few days after infection (Figure 2A). In addition, both Id2fl/fl and RorccreId2fl/fl antibiotic treated mice rapidly lost more body weight than untreated mice (Figure 2B). However, only a little systemic dissemination of C. rodentium was observed in the blood and liver of Id2fl/fl antibiotic treated mice, despite high pathogen titers in the feces (Figure 2A, S2A and S2B). Together, these results suggest that the microbiota are critical for early colonization resistance against C. rodentium, but with limited effect on systemic dissemination of the pathogen.
Figure 2. Id2 dependent microbiota mediates the colonization resistance against C. rodentium infection.
(A–C) 7 weeks old RorccreId2fl/fl (KO) and their littermate Id2fl/fl mice (WT) were treated with either antibiotic (n=5) or control (n=3) in drinking water for one week. One day later, WT and KO mice were orally inoculated with 5 × 106 CFU of C. rodentium. (A) Fecal C. rodentium titers at indicated day post infection. Body weight change (B) and survival rates (C) are shown. Data are representative of two independent experiments.
(D–F) Germ free (GF) wild type B6 mice (D) or GF Ltbr−/− mice (E, F) were colonized with the microbiota from either WT or KO littermate mice by gavage of cecal material. One day later, these mice were orally inoculated with 5 × 106 CFU (D) or 1 × 107 CFU (E, F) of C. rodentium and fecal C. rodentium titers were examined at indicated day post infection (D–E). (F) Survival rates were monitored every day. Data are pooled from three independent experiments.
Each dot represents one individual mouse (A, D, E). Dash line, limit of detection. Error bars represent SEM. *P<0.05, **P<0.01, ***P<0.001; ns, no significant difference (Student’s t-test). See also Figure S2.
Our data led us to hypothesize Id2 controlled the microbiota to maintain colonization resistance. To test this hypothesis we reconstituted gnotobiotic C57BL/6 mice with cecal content from Id2fl/fl or RorccreId2fl/fl mice by oral inoculation. One day later, the reconstituted mice were challenged with a low dose of C. rodentium. As shown in Figure 2D, germ free mice that did not receive a microbiota transplantation exhibited very high fecal C. rodentium titers even at day 1, while the mice reconstituted with Id2fl/fl microbiota showed markedly reduced C. rodentium titers. Importantly, germ free mice inoculated with RorccreId2fl/fl cecal content showed higher C. rodentium titers than Id2fl/fl mice (Figure 2D). These data demonstrate that the Id2 dependent microbiota controls colonization resistance against C. rodentium.
Microbiota can also indirectly mediate colonization resistance through regulation of host immune response (Buffie and Pamer, 2013). Reconstitution of microbiota in germ free mice will induce the host immune response, which may result in increased indirect colonization resistance against C. rodentium. We examined whether the different microbiota from Id2fl/fl and RorccreId2fl/fl mice induce different host immune response of germ free mice. As shown in Figure S2C, reconstitution of Id2fl/fl and RorccreId2fl/fl microflora in germ free mice induced similar amount of expression of pro-inflammatory cytokines and antimicrobial proteins. Together, these data indicate that the early colonization resistance mediated by Id2 dependent microbiota might not be through the regulation of host immune response.
Ltbr−/− mice have multiple defects in innate and adaptive immune responses, including reduced IL-22 and antibody production, which are both essential for the host protection against C. rodentium infection (Kang et al., 2002; Spahn et al., 2004; Tumanov et al., 2011; Wang et al., 2010). To further determine whether the Id2 dependent microbiota could directly mediate colonization resistance, we use Ltbr−/− germ free mice as the recipient. As observed in the C57BL/6 germ free recipient mice, the Ltbr−/− germ free mice repopulated with Id2fl/fl microbiota showed reduced C. rodentium colonization, whereas RorccreId2fl/fl microbiota repopulated Ltbr−/− mice showed more C. rodentium colonization (Figure 2E). Moreover, Id2fl/fl microbiota transplantation improved the survival of Ltbr−/− mice after C. rodentium challenge, while Ltbr−/− germ free mice transferred with RorccreId2fl/fl cecal content showed a similar mortality to the untreated Ltbr−/− germ free mice (Figure 2F). All together, these data indicate that microbiota mediate colonization resistance, which is dependent on continued expression of Id2 in RORγt+ cells.
To further understand how Id2 dependent microbiota regulated the pathogen colonization, we examined the microbiome by bacterial 16S rRNA gene pyrosequencing. Analysis of 16S rRNA genes revealed that although there were no obvious difference of the bacterial diversity and compositions at the phyla level between Id2fl/fl and RorccreId2fl/fl mice (Figure S2D and S2E), we observed several operational taxonomic units (OTUs) were over- and underpresented in RorccreId2fl/fl mice (Figure S2F). Furthermore, quantitative PCR with primers specific for different bacteria demonstrated that Segmented filamentous bacteria (SFB) was overgrown in RorccreId2fl/fl mice (Figure S2G), which has been shown could induce the development of Th17 cells and are regulated by the ILC3s (Ivanov et al., 2009; Qiu et al., 2013). All together, our data suggest that Id2 in in RORγt+ cells regulates the intestinal microbiota.
Id2 mediates the colonization resistance against C. rodentium through IL-22 dependent regulation of microbiota
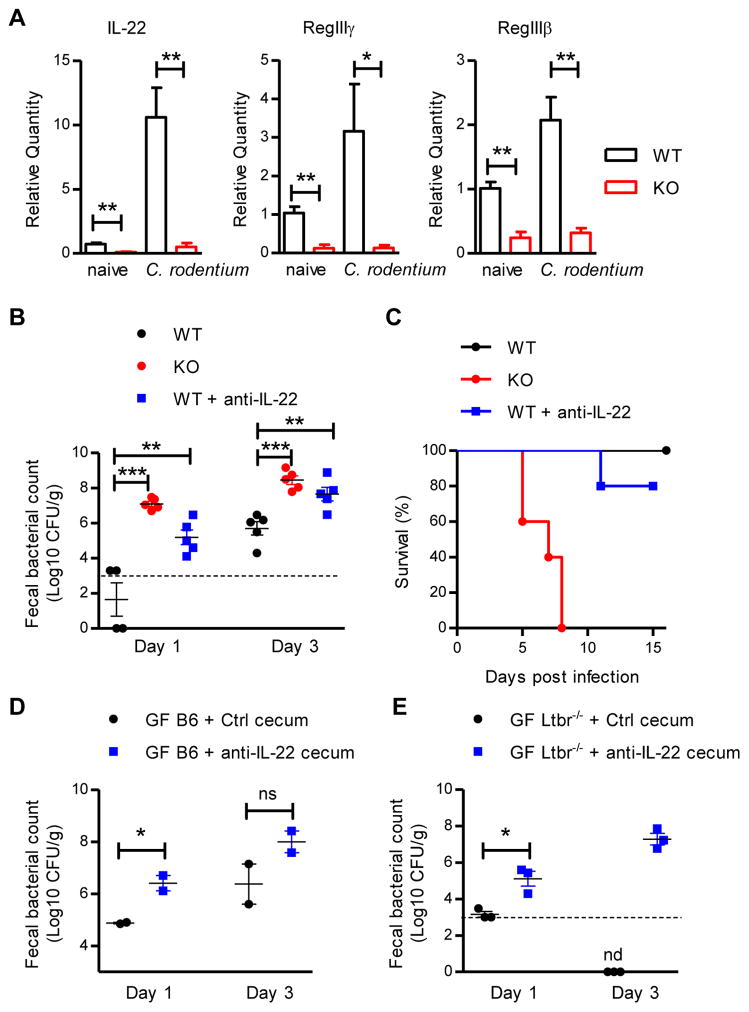
IL-22, mainly produced by RORγt+ cells, is not only required for protection against pathogen infection, but also regulates the homeostasis of microflora in the intestine (Qiu et al., 2013; Qiu et al., 2011). We tested whether Id2 mediated colonization resistance through IL-22 dependent regulation of the microbiota. Id2fl/fl and RorccreId2fl/fl mice were infected with C. rodentium and the expression of Il22 mRNA and mRNA for two antimicrobial proteins dependent on IL-22, RegIIIγ, RegIIIβ, were examined in both naive and infected colon tissues. As shown in Figure 3A, compared with Id2fl/fl mice, RorccreId2fl/fl mice showed significant reduction of Il22, Reg3g and Reg3b mRNA in both naive and infected states, indicating that continued Id2 expression in RORγt+ cells was essential for the IL-22 production in the intestine. To test whether IL-22 regulated the microbiota to mediate early colonization resistance, Id2fl/fl mice were treated with anti-IL-22 neutralization antibody at weaning and challenged with C. rodentium when they were 7 weeks old. Similar to the RorccreId2fl/fl mice, there was increased fecal C. rodentium amounts and decreased survival in anti-IL-22 treated Id2fl/fl mice (Figure 3B and 3C), indicating that blocking IL-22 function in early life destroyed colonization resistance in adulthood. To further determine whether the early colonization resistance was mediated by IL-22 dependent microbiota, but not C. rodentium induced immune response, Id2fl/fl mice were treated with anti-IL-22 antibody 7 days before or at the same day of infection. As shown in Figure S3A–S3C, although only early anti-IL-22 treatment resulted in increased colonization of C. rodentium in feces, both anti-IL-22 treated groups revealed increased C. rodentium CFU in blood, spleen and liver. Our earlier data in this study showed that antibiotic treated microbiota dramatically reduced the early colonization resistance against C. rodentium, but not the systemic dissemination (Figure S3A and S3B). Thus, our data indicate that IL-22 not only mediates the colonization resistance against intestinal pathogen through regulation of microbiota, but also controls the systemic dissemination of pathogen, which is less microbiota dependent. Consistent with this, RorccreStat3fl/fl mice, which lack IL-22 production from RORγt+ cells, also exhibited impaired early colonization resistance (Figure S3D). To further test the requirement for IL-22 to maintain a protective microbiota, we transplanted germ free C57BL/6 and Ltbr−/− mice with microflora from the anti-IL-22 or control treated mice and challenged them with C. rodentium. As observed with germ free mice repopulated with microbiota from RorccreId2fl/fl mice, microbiota from anti-IL-22 treated mice failed to prevent colonization by C. rodentium (Figure 3D and 3E). Collectively, our results suggest that Id2 mediates colonization resistance against C. rodentium through IL-22 dependent regulation of microbiota.
Figure 3. Id2 mediates the colonization resistance against C. rodentium through IL-22 dependent regulation of microbiota.
(A) RorccreId2fl/fl (KO) and their littermate Id2fl/fl (WT) mice were infected with 2 × 109 CFU of C. rodentium. The mRNA expression of IL-22, RegIIIγ and RegIIIβ antimicrobial proteins in the colon of naive or 5 days infected mice were measured by real-time PCR. Data are representative of two independent experiments (n = 3 to 5 per group; mean ± s.e.m.).
(B, C) WT mice were injected intraperitoneally with either anti-IL-22 antibody (8E11.9, 100 μg per mouse per week, n=5) or mouse IgG control (n=5) at 3, 4, 5 weeks old. Two weeks later, 7 weeks old WT and KO mice were orally infected with low dose (5 × 106 CFU) of C. rodentium. (B) Fecal C. rodentium titers at indicated day post infection. (C) Survival rates are shown. Dash line, limit of detection. Data are representative of two independent experiments.
(D) Germ free (GF) wild type B6 mice or
(E) GF Ltbr−/− mice were colonized with the microbiota from either anti-IL-22 or mouse IgG treated WT mice by gavage of cecal material. One day later, these mice were orally inoculated with low dose (5 × 106 CFU, D; 1 × 107 CFU, E) of C. rodentium and fecal C. rodentium titers were examined at indicated day post infection. Data are representative of two (D) or three (E) independent experiments.
Each dot represents one individual mouse (B, D, E). Error bars represent SEM. **P<0.01, ***P<0.001; ns, no significant difference (Student’s t-test). nd, nondetectable. See also Figure S3.
IL-22 producing innate lymphoid cells are necessary and sufficient to mediate the colonization resistance against C. rodentium
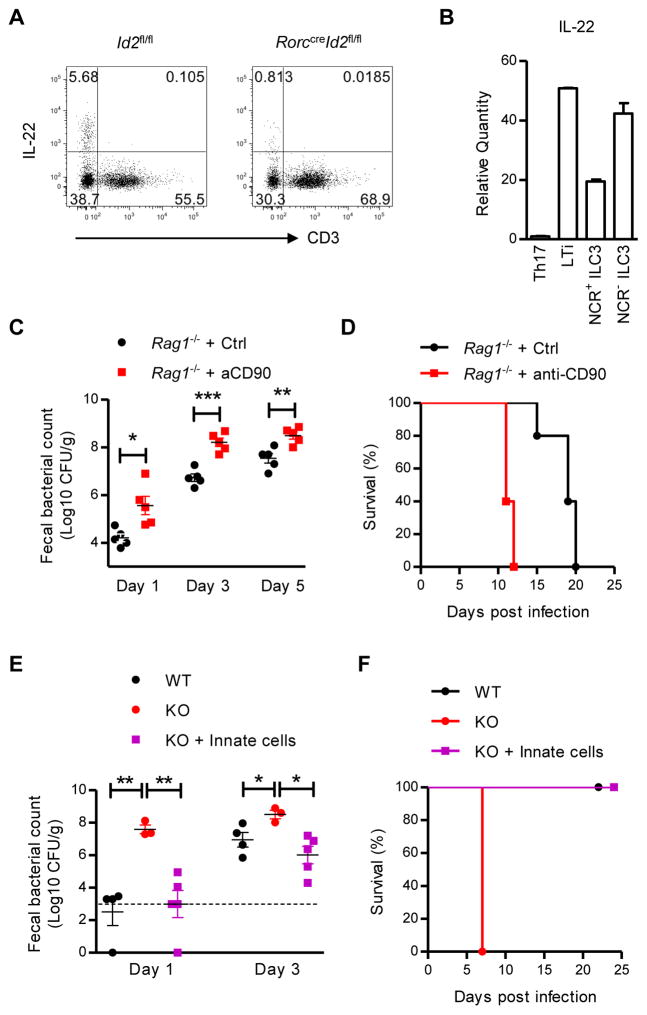
Although IL-22 can be produced by both ILC3s and T cells, we observed that ILC3s are the main producer of IL-22 in the naive state (Figure 4A and 4B) and that innate IL-22 production is markedly reduced in Id2 deficient mice (Figure 4A). This IL-22 expression profile suggests that ILC3s may be responsible for colonization resistance. To test our hypothesis, ILCs were depleted with anti-CD90 antibody in Rag1−/− mice one week before infection. Compared with control treated mice, ILC depleted Rag1−/− mice showed increased early C. rodentium colonization (Figure 4C) and decreased life span post infection (Figure 4D). Furthermore, innate cells from the intestine of Rag1−/− mice were isolated and transferred into RorccreId2fl/fl mice at weaning. Consistent with our hypothesis, innate cells restored the expression of IL-22 and the colonization resistance against C. rodentium in RorccreId2fl/fl mice and completely rescued the RorccreId2fl/fl mice from the death (Figure 4E, 4F and S4A). Together, these results demonstrated that IL-22 producing ILCs mediate the early colonization resistance against an intestinal pathogen.
Figure 4. IL-22 producing innate lymphoid cells mediates the colonization resistance against C. rodentium.
(A) IL-22 expression in CD3− and CD3+ cells were analyzed by intracellular cytokine staining. Intestinal LPLs were isolated from the colons of naive Id2fl/fl (WT) or RorccreId2fl/fl (KO) mice, were stimulated with IL-23 (25 ng/ml) for 4 hours and gated in Thy1+ lymphocytes. Data are representative of at least five independent experiments. (B) RORγt+ ILC3s and T helper cells were purified by flow cytometric sorting from intestinal LPLs of Rorcgfp/+ mice. The mRNA expression of IL-22 was measured by real-time PCR. Data are representative of two independent experiments (mean ± s.e.m. of triplicate samples of real-time PCR).
(C, D) Rag1−/− mice were injected intraperitoneally with either anti-CD90 antibody (30H12, 100 μg per mouse each time) or Rat IgG control at day -10 and day -5 before infection for depletion of ILCs (n=5). Ten days later, mice were orally infected with 1 × 107 CFU of C. rodentium. (C) Fecal C. rodentium titers at indicated day post infection. (D) Survival rates are shown. Data are representative of two independent experiments. Each dot represents one individual mouse (C).
(E, F) KO mice were injected with innate cells at weaning. Four weeks later, WT and KO mice were orally infected with 5 × 106 CFU of C. rodentium. (E) Fecal C. rodentium titers at indicated day post infection. (F) Survival rates are shown. Dash line, limit of detection. Data are representative of two independent experiments.
Error bars represent SEM. *P<0.05, **P<0.01, ***P<0.001 (Student’s t-test). See also Figure S4.
Continued expression of Id2 is required for the development and maintenance of group 3 ILCs
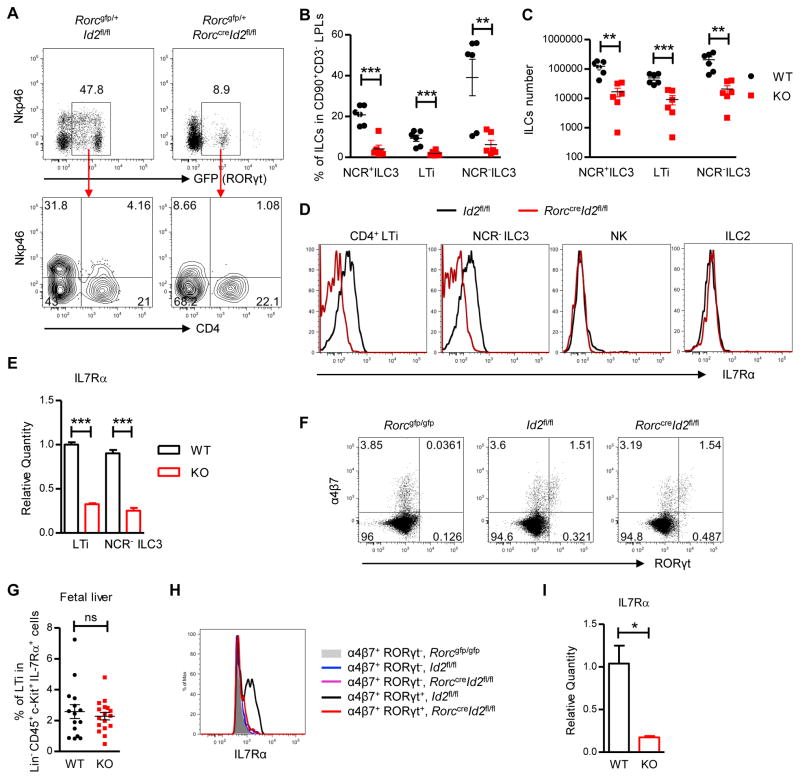
Since Id2 expression in ILC3s is essential for IL-22 dependent regulation of microbiota against pathogen colonization, we next determined how Id2 regulated IL22 production by ILC3s, and whether Id2 regulated ILC3s’ homeostasis or functionality. Id2 is required for the development of all ILC precursors, thus first we tested whether Id2 was still important for the further development of ILC3s. Id2fl/fl and RorccreId2fl/fl mice were crossed with Rorcgfp/+ mice and ILC3s were examined in the intestinal LPLs. RORγt expressing ILC3s were dramatically reduced in both the large and small intestine in Rorcgfp/+RorccreId2fl/fl mice, and the remaining ILC3s are mostly CD4+ LTi cells and NCR− ILC3s (Figure 5A–C and S5A–B). Previous studies have shown that NCR+ ILC3s can develop from NCR− ILC3s, and that IL7R signaling is required for this further development as well as for the survival and proliferation of ILCs (Schmutz et al., 2009; Vonarbourg et al., 2010). We found that Id2 deficient ILC3s has diminished IL7Rα expression at both the protein and mRNA level (Figure 5D and 5E).
Figure 5. Continued expression of Id2 is required for the further development and maintenance of group 3 ILCs.
(A, D, E) Small intestinal LPLs were isolated from 4 weeks old Rorcgfp/+Id2fl/fl (WT) and Rorcgfp/+RorccreId2fl/fl (KO) mice and gated in live CD90+ CD3− ILCs. (A) Different populations of ILCs were analyzed by flow cytometry. Numbers adjacent to outlined areas indicate percent cells in each gate. (D) The expression of IL-7Rα on different ILCs was analyzed by flow cytometry. Data are representative of three independent experiments. NCR+ILC3, Nkp46+CD4−RORγt+ILCs; LTi, Nkp46−CD4+RORγt+ILCs; NCR−ILC3, Nkp46−CD4−RORγt+ ILCs; NK, Nkp46+RORγt− ILCs; ILC2, RORγt−Nkp46−ILCs.
(B, C) Percentage of different ILC3s in the CD90+ CD3− ILCs, as well as the absolute numbers of RORγt+ ILC3s in the small intestine of Id2fl/fl (WT) and RorccreId2fl/fl (KO) mice are shown. Each dot represents one individual mouse. Data are pooled from two independent experiments (mean ± s.e.m.).
(E) LTi and NCR− ILC3 were purified by flow cytometric sorting. The mRNA expression of IL-7Rα was measured by real-time PCR. Data are representative of two independent experiments (mean ± s.e.m. of triplicate samples of real-time PCR).
(F, G) LTi cells in fetal liver were analyzed by flow cytometry. E14-E15 fetuses were isolated and genotyped by PCR. Fetal liver cells were isolated from Rorcgfp/gfp, Id2fl/fl and RorccreId2fl/fl fetus and gated in Lineage−CD45+c-Kit+IL7Rα+ ILCs. Numbers adjacent to outlined areas indicate percent cells in each gate. Data are representative of four independent experiments. (G) Percentage of LTi cells (α4β7+RORγt+) in the Lineage−CD45+c-Kit+IL7Rα+ cells were shown. Each dot represents one individual mouse. Data are pooled from four independent experiments (mean ± s.e.m.).
(H) The expression of IL-7Rα on different fetal liver cells was analysis by flow cytometry. The fetal liver cells were gated in Lineage−CD45+c-Kit+IL7Rα+ cells first and then gated in either α4β 7+RORγt+ LTi cells or α4β7+RORγt− LTi precursor cells. Data are representative of four independent experiments.
(I) Fetal LTi were purified by flow cytometric sorting from fetal liver of Rorcgfp/+Id2fl/fl and Rorcgfp/+RorccreId2fl/fl mice. The mRNA expression of IL-7Rα was measured by real-time PCR. Data are representative of two independent experiments (mean ± s.e.m. of triplicate samples of real-time PCR).
**P<0.01, ***P<0.001; ns, no significant difference (Student’s t-test). See also Figure S5.
To further determine the role of Id2 in ILC3 maintenance, both WT and RorccreId2fl/fl ILC3s were sorted from CD45.1+ C57BL/6 and CD45.2+ RorccreId2fl/fl mice and injected into Rag2−/− Il2γc−/− mice at 1:1 ratio. The gut LPL were isolated 4 weeks later and flow cytometry analysis revealed that WT ILC3s were 4 folds more than RorccreId2fl/fl cells (Figure S5C). Moreover, we found that Id2 deficient ILC3 exhibited increased apoptosis and reduced Bcl2 expression, which is considered as an important anti-apoptotic protein (data not shown and Figure S5D). Since Id2 is required for the development of all the ILCs and early defect of development may also result in dramatic reduction of adult ILC3s (Cherrier et al., 2012), we further isolated the fetal liver cells to examine whether Id2 was required for the development rather than the maintenance of ILC3s. As shown in Figure 5F and 5G, there was no reduction of fetal LTi cells in the liver of RorccreId2fl/fl mice. However, the same as the adult ILC3s, Id2 deficient fetal ILC3s also exhibited reduced expression of IL7Rα (Figure 5H and 5I). Together, our results clearly demonstrate that Id2 expression continues to be required in ILC3 after expression of RORγt for proper expression of IL7Rα and ILC3 maintenance.
Id2 regulates IL-22 production by group 3 ILCs through IL-23R pathway
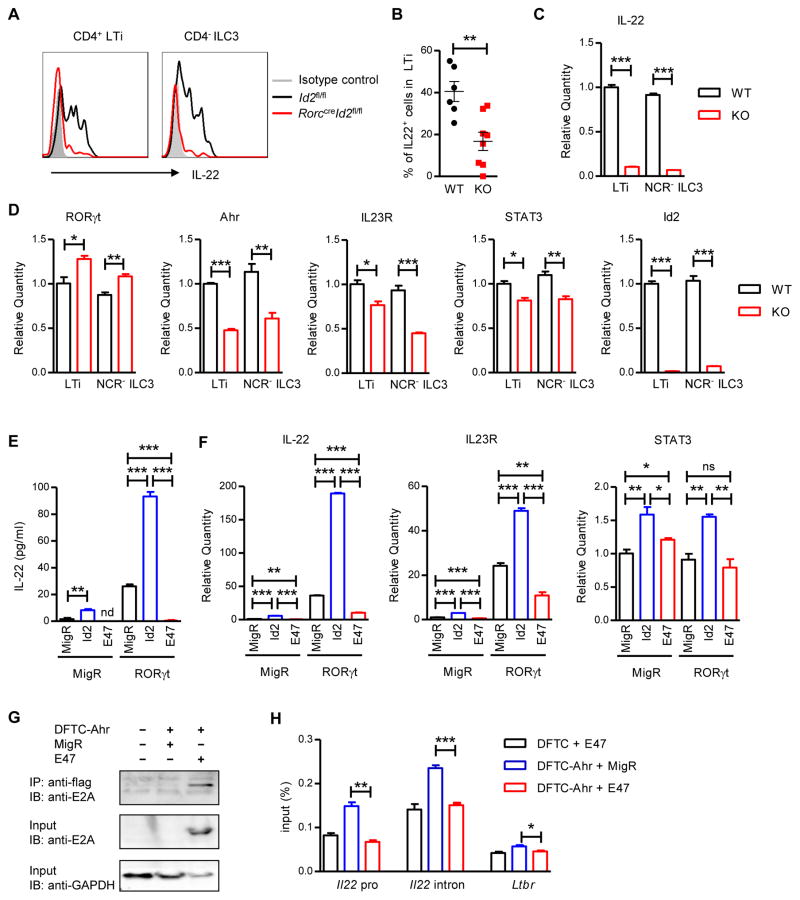
Multiple transcription factors, such as RORγt, Ahr and GATA-3 regulate ILC3 development as well as ILC3 functions (Qiu et al., 2011; Serafini et al., 2014). To further test whether Id2 also regulated the function of ILC3s, both Id2fl/fl and RorccreId2fl/fl LPLs were isolated from the intestine and IL-22 production was examined. Interestingly, the residual Id2 deficient ILC3s were unable to produce IL-22 after IL-23 stimulation (Figure 6A and 6B). This is also confirmed by the marked reduction of IL-22 production in LTi and NCR− ILC3s sorted from Rorcgfp/+RorccreId2fl/fl mice compared with Rorcgfp/+Id2fl/fl ILC3s (Figure 6C and S6A). Together, these data suggest that Id2 also plays an essential role in regulating the function of ILC3s.
Figure 6. Id2 regulates IL-22 production by group 3 ILCs through IL-23R pathway.
(A) Intestinal LPLs were isolated from naive Id2fl/fl (WT) or RorccreId2fl/fl (KO) mice and stimulated with IL-23 (25 ng/ml) for 4 hours. IL-22 expression in CD3−CD90highCD45lowCD4+ LTi or CD3−CD90highCD45lowCD4− ILC3s was analyzed by intracellular cytokine staining followed by flow cytometry. Data are representative of three independent experiments.
(B) Percentages of IL-22 producing cell in LTi cells are shown. Colonic LPLs were isolated from C. rodentium infected WT or KO mice at day 5 post infection, restimulated with IL-23 and gated in CD3−CD90highCD45lowCD4+ LTi cells. Each dot represents one individual mouse. Data are pooled from two independent experiments (mean ± s.e.m.).
(C, D) LTi and NCR− ILC3 were purified by flow cytometric sorting from intestinal LPLs of Rorcgfp/+Id2fl/fl or Rorcgfp/+RorccreId2fl/fl mice and lysed directly for RNA extraction. The mRNA expression of IL-22 (C), RORγt, Ahr, IL23R and STAT3 (D) were measured by real-time PCR. Data are representative of two independent experiments (C, D; mean ± s.e.m. of triplicate samples of real-time PCR).
(E, F) EL4 cells were infected with Id2, E47 expression or control retrovirus with or without RORγt expression retrovirus and stimulated with IL-23 for 2 days. (E) The production of IL-22 in the cell culture supernatant was measured by ELISA. (F) The mRNA expression of IL-22, IL23R and STAT3 were measured by real-time PCR. Error bars represent SEM of triplicate samples. Data are representative of two independent experiments.
(G) E2A physically interacted with Ahr. EL4 cells were stably infected with the indicated expression constructs. Whole cell extracts were immunoprecipitated with anti-flag beads and subsequently immunoblotted with anti-E2A antibodies. Data are representative of three independent experiments.
(H) Empty MigR or E47 were coexpressed by retroviral transduction in EL4 cell lines stably expressing either DFTC or DFTC-Ahr. Ahr binding at the Il22 locus was detected by ChIP assay. *P<0.05, **P<0.01, ***P<0.001; ns, no significant difference (Student’s t-test). nd, nondetectable. See also Figure S6.
Previous studies showed that IL-23 can interact with the IL-23 receptor (IL23R) and activate STAT3, RORγt, , Ahr and STAT3 can bind to the Il22 locus and directly promote IL-22 production (Guo et al., 2014; Qiu et al., 2011). To determine how Id2 regulated ILC3s producing IL-22, we first examined the expression of these transcription factors and cytokine receptor. LTi and NCR− ILC3s were purified from both Rorcgfp/+ and Rorcgfp/+RorccreId2fl/fl mice. Consistent with a previous study (Zhang et al., 2014), Id2 deficient LTi cells showed increased RORγt expression. The expression of Ahr was reduced in both Id2 deficient resting LTi and NCR− ILC3s, but not IL-23 activated LTi cells. Interestingly, the expression of IL23R and STAT3 were reduced in both Id2 deficient LTi and NCR− ILC3s (Figure 6D and S6A). Moreover, Id2 deficient fetal liver LTi cells also showed significant reduction of IL-22 and IL-23 receptor (Figure S6B), suggesting that early Id2 regulation of IL-22 may be through IL23R–STAT3 pathway.
ID proteins are transcription factors that inhibit the function of the E protein transcription factors by preventing them from binding to DNA. Previous studies have shown that Id2 regulates the development of NK and LTi cells through suppression of E2A (Boos et al., 2007). To test whether Id2 also regulated IL-22 production through suppression of E2A, we infected EL4 cells with either Id2 or E47 (one E protein encoded by E2A gene) expressing retrovirus and examined IL-22 production. Since RORγt is the master regulator for IL-22 production, we also infected the EL4 cells with or without RORγt expressing retrovirus. As shown in Figure 6E and 6F, compared with empty MigR retrovirus control group, Id2 retrovirus infected EL4 cells produced more IL-22, while the E47 group showed reduced IL-22 production with or without RORγt expression. Moreover, increased expression of Id2 also causes increased IL23R and STAT3 expression in EL4 cells, while over-expression of E47 inhibited IL23R expression. Together, our data indicate that E2A may regulate IL-22 production through suppression of the IL23R pathway, while Id2 promotes IL-22 production through suppression of E2A activity.
Next we wanted to determine how E2A inhibited IL-22 production. Previous studies have shown that Ahr deficient ILC3s, like Id2 deficient ILC3s, showed reduced IL7Rα and IL23R expression (Qiu et al., 2011). In addition, Ahr is also a member of the bHLH transcription factor family. Although there was slight reduction of Ahr expression in Id2 deficient ILC3s, we also considered the possibility that E2A could interact with Ahr to prevent its transcriptional activity, resulting in a reduction of IL-22 in Id2 deficient ILC3s. Indeed, using EL4 cells expressing the double flag-tagged Ahr (DFTC-Ahr) with or without E47, we detected an interaction between Ahr and E2A by coimmunoprecipitation (Figure 6G). Furthermore, a chromatin immunoprecipitation (ChIP) assay revealed that E2A could suppress Ahr binding to the Il22 locus in EL4 cell lines (Figure 6H). Together, these data suggest that E2A may regulate IL-22 production by directly binding to Ahr and preventing its transcription activity at Il22 locus.
Discussion
Host sequential responses from innate and adaptive immune cells are essential for late stage colonization resistance and clearance of pathogens. Here, our study demonstrates that microbiota is important for early colonization resistance ahead of innate and adaptive responses. Fecal microbiota transplantation has been successfully used in some patients with Clostridium difficile infection (Austin et al., 2014). However, since the stability of the gut microbiota is dependent on many host and environment factors (Lozupone et al., 2012), it remains unclear how a stable healthy microbial community should be introduced to prevent recurrent infection. Our results further demonstrate that baseline IL-22 production by ILC3s can regulate gut microbial homeostasis promoting pathogen colonization resistance. Therefore, innate immunity contributes to colonization resistance not only by rapid responses after invasion but also by maintaining the proper microbiota that limit colonization before invasion. Our study suggests that a combination of microbiota transplantation with immune molecule treatment may restore a stable microbial community to prevent intestinal pathogen colonization.
Commensals have been shown to utilize several different mechanisms to directly mediate colonization resistance against pathogen, including competition for niches and nutrients, altering host environmental conditions (for example, pH), producing bacteriocins, and affecting pathogen virulence by O2 consumption and production of specific metabolites such as the short-chain fatty acid (Kamada et al., 2013). Previous studies have shown that E. coli can directly compete with C. rodentium for nutritional resources and help the host to clear the pathogen (Kamada et al., 2012). Repopulation of Segmented filamentous bacterium (SFB) partially protects the Jackson B6 mice from C. rodentium infection (Ivanov et al., 2009). However, we detected very few E. coli in the mice housed in our SPF facility and the SFB level was greatly increased in conditional Id2 deficient mice, which is consistent with other IL-22 deficient mice (Qiu et al., 2013; Upadhyay et al., 2012). Thus, IL-22 dependent colonization resistance is not mediated through E. coli and SFB. It remains to be determined which commensals are regulated by innate IL-22 for colonization resistance and how commensals suppress C. rodentium colonization, which may leads to discovery of novel therapeutic probiotics and prebiotics.
Notch, Id2, and RORγt sequentially orchestrate the development of ILC3s (Cherrier et al., 2012). However, the function of the highly expressed Id2 in differentiated ILCs is unclear. Our data clearly demonstrate that the maintenance of Id2 expression is required for the further development and function of ILC3s. Consistent with the role of Id2 on IL-7Rαp regulation in CD4+ thymocytes (Jones-Mason et al., 2012), we found that Id2 controls IL-7Rα expression in ILC3s but how Id2 regulates IL-7Rα remains to be determined. Since IL7R signaling is also essential for the homeostasis of the other ILCs (Hoyler et al., 2012), our results suggest Id2 may globally and constantly regulate the ILC lineage from progenitor to effector ILCs.
After IL-23 stimulation, STAT3, Ahr and RORγt can be recruited to the Il22 locus to promote IL-22 production. It has been shown that Ahr requires interaction with RORγt to bind to the Il22 locus. Without RORγt, there is only a little recruitment of Ahr to Il22 locus (Guo et al., 2014; Qiu et al., 2011). Consistently, our data confirm that Ahr weakly binds to Il22 locus without over-expressing RORγt. However, E2A physically interacted with Ahr and completely blocked the binding of Ahr to the Il22 locus. Our studies suggest that two bHLH transcription factors, E2A and Ahr, may form a heterodimer that either fails to bind DNA or has a DNA binding specificity different with the Ahr–RORγt dimer. Therefore, an essential role of Id2 may be to liberate Ahr from E2A and allow Ahr bind to other transcriptional activators, such as RORγt. Whether E2A also interrupts the interaction between Ahr and RORγt remain to be examined. We also showed that Id2 was required for expression of IL7Rα and IL23R. Since Ahr deficient ILC3s also exhibit impaired expression of these two cytokine receptors (Qiu et al., 2011), E2A may also suppress their expression through its interaction with Ahr; however, an complete understanding of the molecular mechanism by which E2A affects gene expression in ILC3s requires further investigation.
Taken together, our studies demonstrated several important findings. (1) Id2 is essential not only for ILC lineage specification but also for the maintenance and further development of RORγt+ILC3s. (2) The Id2–E2A interaction regulates IL-22 production function in ILC3 through the Ahr and IL23R pathway. (3) IL-22 producing ILC3s are essential for the maintenance of the proper microbiota to mediate early pathogen colonization resistance. Proper addition of immune molecule treatment to the fecal microbiota transplantation may help the host to reestablish a more stable and healthier microbial community in patients with recurrent gut infection.
Materials and Methods
Mice
C57BL/6 and Rag1−/− mice were purchased from Harland Teklad. Rag2−/− Il2γc−/−, Rorcgfp/+ (Eberl et al., 2004) and Id2gfp/+ mice (Rawlins et al., 2009) were purchased from The Jackson Lab. RorccreId2fl/fl mice were generated by crossing Id2-floxed mice (Niola et al., 2012) with Rorc-cre transgenic mice (Eberl and Littman, 2004). All mice are on C57BL/6 background. Germ free Ltbr−/− mice were rederived in Taconic. All the germ free mice are maintained in the gnotobiotic facility at the University of Chicago. Animal care and use were in accordance with institutional and National Institutes of Health guidelines and all studies were approved by the Animal Care and Use Committee of the University of Chicago.
Infection with Citrobacter rodentium and Treatment
Mice were orally gavaged with C. rodentium strain DBS100 (ATCC 51459; American Type Culture Collection) and body weight, survival, CFU counts, tissue histology, PAS staining were assessed as previously described (Tumanov et al., 2011; Wang et al., 2010). Where indicated, mice were treated antibiotics (1 g/L Ampicillin, 1 g/L neomycin, 1 g/L metronidazole, 0.5 g/L vancomycin) in drinking water for 1 week, then infected with C. rodentium after 1 day’s rest. Where indicated, mice were injected intraperitoneally with either anti-IL-22 antibody (8E11.9) or mouse IgG1 as isotype control (100 μg per mouse) by the age of 3, 4 and 5 weeks or indicated day. Then the mice were infected with C. rodentium at 7 weeks old. For depletion of innate lymphoid cells, Rag1−/− mice were injected intraperitoneally with anti-CD90 antibody (30H12, 100 μg per mouse) or Rat IgG 10 days before C. rodentium infection. Where indicated, innate cells were isolated from the intestine of Rag1−/− mice and transferred by i.v. injection (1 × 106 innate cells or 2× 105 purified CD45lowCD90high ILC3s per mouse) into 3–4 weeks old RorccreId2fl/fl mice.
Germ free experiments
C57BL/6 or Ltbr−/− germ-free mice were transferred to specific pathogen free environment and immediately gavaged with fresh cecal contents from Id2fl/fl and RorccreId2fl/fl littermate donors, or anti-IL-22 antibody and isotype control treated mice. One day later, these microflora reconstituted mice were infected with C. rodentium (Ahern et al., 2014).
Isolation of Intestinal LPLs and Fetal Liver Cells
The intestinal lamina propria leukocytes were isolated by Lamina Propria Dissociation Kit (Miltenyl Biotec) according to the manufacturer’s recommendations. Fetal liver were dissociated by mechanical shearing in PBS containing 0.5% bovine serum and then filtered through a 70-μM mesh.
Flow Cytometry, Antibodies and ELISA
Antibodies against lineage marker, CD3, CD4, CD45, CD90, Nkp46, CD117, CD127, α4β7, RORγt and Streptavidin-APC were purchased from BioLegend or eBioscience. Anti-IL-22 antibody was a gift from Genentech. For nuclear staining, cells were fixed and permeabilized using a Transcription Factor Staining Buffer Set (eBioscience). For cytokine production, cells were stimulated ex vivo by IL-23 (25 ng/ml, R&D) for 4 hrs. IC Fixation Buffer and Permeabilization Buffer (eBioscience) were used for intracellular cytokine staining. Flow cytometry was performed on LSR-Fortessa (BD Biosciences) instruments and analyzed with FlowJo software (Tree Star Inc.). ILC3s were sorted from the intestine LPLs and fetal liver cells on a FACS Aria III instrument (BD Bioscience). IL-22 in supernatants was measured by ELISA according to the manufacturer’s recommendations (R&D Systems).
Quantitative Real-Time RT-PCR
RNA isolation and Real-time PCR was performed as previously described (Guo et al., 2014) with different primer sets (Table S1).
Retroviral Transduction of Cell Lines
The MigR, Id2, E47 and RORγt expression retrovirus were made as previously described (Qiu et al., 2011). EL4 cells were infected with2 ml of virus supernatant (MigR, Id2 or E47) and 48 hours later, the GFP+ infected cells were sorted on Avalon Cell Sorter (Propel Labs). The sorted EL4 cells were then infected with either MigR or RORγt expression retrovirus. Cell culture supernatant and cell RNA were analyzed by ELISA and Realtime PCR separately.
Co-immunoprecipitation and Chromatin Immunoprecipitation
EL4 cells were infected with DFTC (double flag epitope tagged)–Ahr retroviral vector and selected in the presence of 7 μg/ml puromycin. Ahr stably expressing EL4 cells were then infected either MigR or E47 retrovirus. The GFP+ infected cells were sorted and the whole cell lysate supernatant was immunoprecipitated with EZview™ Red anti-flag affinity gel (Sigma). Western blotting was performed with anti-E2A rabbit antibody (Thermo Scientific) and anti-GAPDH antibody (Sigma). ChIP assays with EL4 cells were performed as previously described (Qiu et al., 2011).
Statistical Methods
Statistical analysis was performed by two-tailed Student’s t test using GraphPad Prism 5.0 program. Data from such experiments are presented as mean values ± SEM; p < 0.05 was considered significant. For survival curves, statistics were done using the log rank (Mantel-Cox) test.
Highlights.
Id2 is essential to mediate the colonization resistance against C. rodentium.
Continued expression of Id2 is required for the homeostasis of ILC3s.
Id2-E2A interaction regulates ILC3 production of IL-22 through Ahr and IL23R pathway.
IL-22 from ILC3s controls the colonization resistance through regulating microbiota.
Acknowledgments
We are grateful to D. Littman (New York University, NY) for Rorccre mice; L. Zhou (Northwestern University, IL) for Ahr plasmid; C. R. Nagler (The University of Chicago, IL) for germ free mice; A. T. Stefka (The University of Chicago, IL) for the analysis of bacterial 16s rRNA gene pyrosequencing; W. Ouyang (Genentech, CA) for anti-IL-22 antibody. The work was supported by the National Institutes of Health grants (AI106352 to B.L.K.; DK080736, DK095962, DK100427 to Y.X.F.).
References
- Ahern PP, Faith JJ, Gordon JI. Mining the Human Gut Microbiota for Effector Strains that Shape the Immune System. Immunity. 2014;40:815–823. doi: 10.1016/j.immuni.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M, Mellow M, Tierney WM. Fecal microbiota transplantation in the treatment of Clostridium difficile infections. Am J Med. 2014;127:479–483. doi: 10.1016/j.amjmed.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, Raffatellu M. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bry L, Brenner MB. Critical role of T cell-dependent serum antibody, but not the gut-associated lymphoid tissue, for surviving acute mucosal infection with Citrobacter rodentium, an attaching and effacing pathogen. J Immunol. 2004;172:433–441. doi: 10.4049/jimmunol.172.1.433. [DOI] [PubMed] [Google Scholar]
- Bry L, Brigl M, Brenner MB. CD4+-T-cell effector functions and costimulatory requirements essential for surviving mucosal infection with Citrobacter rodentium. Infect Immun. 2006;74:673–681. doi: 10.1128/IAI.74.1.673-681.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013 doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet. 2012;13:175–188. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORgammat sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crim SM, Iwamoto M, Huang JY, Griffin PM, Gilliss D, Cronquist AB, Cartter M, Tobin-D’Angelo M, Blythe D, Smith K, et al. Incidence and trends of infection with pathogens transmitted commonly through food--Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006–2013. MMWR Morb Mortal Wkly Rep. 2014;63:328–332. [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Littman DR. The role of the nuclear hormone receptor RORgammat in the development of lymph nodes and Peyer’s patches. Immunol Rev. 2003;195:81–90. doi: 10.1034/j.1600-065x.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- Eberl G, Littman DR. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Dai C, Brown K, Rajendiran E, Makarenko S, Baker J, Ma C, Halder S, Montero M, Ionescu VA, et al. Colonic microbiota alters host susceptibility to infectious colitis by modulating inflammation, redox status, and ion transporter gene expression. Am J Physiol Gastrointest Liver Physiol. 2011;301:G39–49. doi: 10.1152/ajpgi.00509.2010. [DOI] [PubMed] [Google Scholar]
- Guo X, Qiu J, Tu T, Yang X, Deng L, Anders RA, Zhou L, Fu YX. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity. 2014;40:25–39. doi: 10.1016/j.immuni.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Mason ME, Zhao X, Kappes D, Lasorella A, Iavarone A, Zhuang Y. E protein transcription factors are required for the development of CD4(+) lineage T cells. Immunity. 2012;36:348–361. doi: 10.1016/j.immuni.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nunez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Kang HS, Chin RK, Wang Y, Yu P, Wang J, Newell KA, Fu YX. Signaling via LTbetaR on the lamina propria stromal cells of the gut is required for IgA production. Nat Immunol. 2002;3:576–582. doi: 10.1038/ni795. [DOI] [PubMed] [Google Scholar]
- Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2011;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niola F, Zhao X, Singh D, Castano A, Sullivan R, Lauria M, Nam HS, Zhuang Y, Benezra R, Di Bernardo D, et al. Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nat Cell Biol. 2012;14:477–487. doi: 10.1038/ncb2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa TJ, Barletta F, Contreras C, Mercado E. New insights into the epidemiology of enteropathogenic Escherichia coli infection. Trans R Soc Trop Med Hyg. 2008;102:852–856. doi: 10.1016/j.trstmh.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 Innate Lymphoid Cells Inhibit T-Cell-Mediated Intestinal Inflammation through Aryl Hydrocarbon Receptor Signaling and Regulation of Microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2011;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, Mielke LA, McKenzie AN, Carotta S, Nutt SL, Belz GT. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz S, Bosco N, Chappaz S, Boyman O, Acha-Orbea H, Ceredig R, Rolink AG, Finke D. Cutting edge: IL-7 regulates the peripheral pool of adult ROR gamma+ lymphoid tissue inducer cells. J Immunol. 2009;183:2217–2221. doi: 10.4049/jimmunol.0802911. [DOI] [PubMed] [Google Scholar]
- Serafini N, Klein Wolterink RG, Satoh-Takayama N, Xu W, Vosshenrich CA, Hendriks RW, Di Santo JP. Gata3 drives development of RORgammat+ group 3 innate lymphoid cells. J Exp Med. 2014;211:199–208. doi: 10.1084/jem.20131038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn TW, Maaser C, Eckmann L, Heidemann J, Lugering A, Newberry R, Domschke W, Herbst H, Kucharzik T. The lymphotoxin-beta receptor is critical for control of murine Citrobacter rodentium-induced colitis. Gastroenterology. 2004;127:1463–1473. doi: 10.1053/j.gastro.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- Tumanov AV, Koroleva EP, Guo X, Wang Y, Kruglov A, Nedospasov S, Fu YX. Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay V, Poroyko V, Kim TJ, Devkota S, Fu S, Liu D, Tumanov AV, Koroleva EP, Deng L, Nagler C, et al. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nat Immunol. 2012;13:947–953. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallance BA, Deng W, Jacobson K, Finlay BB. Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium. Infect Immun. 2003;71:3443–3453. doi: 10.1128/IAI.71.6.3443-3453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waaij D, Berghuis-de Vries JM, Lekkerkerk L-v. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond) 1971;69:405–411. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, Flach M, Bengsch B, Thimme R, Holscher C, et al. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Koroleva EP, Kruglov AA, Kuprash DV, Nedospasov SA, Fu YX, Tumanov AV. Lymphotoxin beta receptor signaling in intestinal epithelial cells orchestrates innate immune responses against mucosal bacterial infection. Immunity. 2010;32:403–413. doi: 10.1016/j.immuni.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang F, Fuss IJ, Yang Z, Strober W. Transcription of RORgammat in developing Th17 cells is regulated by E-proteins. Mucosal Immunol. 2014;7:521–532. doi: 10.1038/mi.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]