Abstract
Automation has been an important part of biomedical research for decades, and the use of automated and robotic systems is now standard for such tasks as DNA sequencing, microfluidics, and high-throughput screening. Recently, Kodandaramaiah and colleagues (Nat Methods 9: 585–587, 2012) demonstrated, using anesthetized animals, the feasibility of automating blind patch-clamp recordings in vivo. Blind patch is a good target for automation because it is a complex yet highly stereotyped process that revolves around analysis of a single signal (electrode impedance) and movement along a single axis. Here, we introduce an automated system for blind patch-clamp recordings from awake, head-fixed mice running on a wheel. In its design, we were guided by 3 requirements: easy-to-use and easy-to-modify software; seamless integration of behavioral equipment; and efficient use of time. The resulting system employs equipment that is standard for patch recording rigs, moderately priced, or simple to make. It is written entirely in MATLAB, a programming environment that has an enormous user base in the neuroscience community and many available resources for analysis and instrument control. Using this system, we obtained 19 whole cell patch recordings from neurons in the prefrontal cortex of awake mice, aged 8–9 wk. Successful recordings had series resistances that averaged 52 ± 4 MΩ and required 5.7 ± 0.6 attempts to obtain. These numbers are comparable with those of experienced electrophysiologists working manually, and this system, written in a simple and familiar language, will be useful to many cellular electrophysiologists who wish to study awake behaving mice.
Keywords: automation, awake, behaving, mouse, patch clamp
the mouse has emerged as a leading model system for the study of the neural circuit basis of behavior (Dymecki and Kim 2007; Luo et al. 2008; O'Connor et al. 2009). Not only do mice have well-established advantages for cellular and molecular neurobiology, such as transgenic methods for identifying and manipulating specific cell populations, but also they have a rich behavioral repertoire that includes sensory, motor, and cognitive aspects (Guo et al. 2014). Moreover, recent experiments indicate that this behavioral repertoire is only partly diminished when mice are subjected to head fixation. Head-fixed mice have been trained to navigate mazes in virtual reality environments (Domnisoru et al. 2013; Harvey et al. 2009; Schmidt-Hieber and Häusser 2013); discriminate auditory, visual, tactile, and olfactory stimuli (Abraham et al. 2012; Andermann et al. 2010; Busse et al. 2011; Guo et al. 2014; Histed et al. 2012; Komiyama et al. 2010; Lee et al. 2012; Sanders and Kepecs 2012); and perform classic conditioning tasks (Chettih et al. 2011; Heiney et al. 2014). This is important because head fixation opens up the possibility of employing physiological and imaging methods that are extraordinarily difficult, although not impossible (Lee et al. 2014; Ziv et al. 2013), in freely moving animals.
Of special interest in this regard is whole cell patch-clamp electrophysiology (Harvey et al. 2009; Margrie et al. 2002; Petersen and Crochet 2013; Polack et al. 2013). Since its introduction more than 30 years ago, the patch-clamp has assumed a central place in cell-type-specific neurobiology: it allows for the measurement of suprathreshold and subthreshold synaptic- and ion-channel-mediated events with high signal quality and temporal fidelity; the manipulation of the subcellular processes that shape these events; and the physiological, morphological, and molecular identification of recorded cells. Most of the advances patch-clamp has made possible have come in reduced preparations (cell cultures and brain slices), in part because of the challenge of obtaining patch-clamp recordings in vivo, even when animals are anesthetized (Crochet 2011; DeWeese 2007; Häusser and Margrie 2014; Margrie et al. 2002). Patch-clamp in vivo is not conceptually difficult but requires an experimenter to monitor a living animal while executing a complex series of actions. The challenge is heightened when the animal is not merely living but awake and behaving and when the experimenter must also contend with behavioral equipment (e.g., cameras, locomotion and eye position monitors, virtual reality screens). These kinds of experiments place considerable and (for some) unreasonable demands on experimenter training, labor, expertise, and fortitude, and these demands in turn may limit the yield and quality of the resulting data.
Concerns of this sort are not limited to in vivo electrophysiology, and they have led to efforts to automate many types of biological experiments, moving burdens off of human experimenters and onto computers and other pieces of electronic equipment (Araci and Brisk 2014; Brunborg et al. 2014; Dunlop et al. 2008; King et al. 2009; Kodandaramaiah et al. 2013; Perin and Markram 2013; Slaughterbeck et al. 2007; Steinmeyer and Yanik 2012). In recent years, these efforts have been aided by advances in computer technology, the inclusion of computer control in standard scientific equipment, and the so-called “maker movement,” which aims to help amateurs and other nonspecialists interested in designing equipment to take advantage of modern technological advances such as those in embedded electronics (the Arduino microcontroller is a good example of this, https://www.arduino.cc/; Anderson 2014; Finley 2014; Morozov 2014; Teikari et al. 2012). In the case of patch-clamp in vivo, the benchmark effort has been that of Kodandaramaiah and colleagues (2012). These authors demonstrated the feasibility of automating blind patch-clamp recordings in anesthetized mice. Their system, written in the LabVIEW programming environment, produced patch recordings that matched those of experienced electrophysiologists. Subsequent work by these researchers (Holst et al. 2014; Kodandaramaiah et al. 2014) has pushed their project forward in several different directions, including automated pipette replacement and multiple simultaneous recordings.
Our own work can be viewed as both a complement and an alternative. It is complementary in that many of the advances (e.g., pipette replacement) that result from their project can be readily integrated into ours. It is alternative in that we have, in building an automated patch-clamp system for awake behaving mice, made different choices about hardware and software. One striking difference is that our system was written entirely in the MATLAB programming environment, which has a presence in the neuroscience community that is both broad and deep and which provides useful tools for analysis and instrument control (Wallisch et al. 2013). Another is that our system was constructed with awake, head-fixed mouse experiments in mind (Siegel et al. 2015). Our guiding ideal was a single, complete system that, with a minimum of human intervention, could obtain whole cell patch recordings from neurons in behaving mice while controlling and/or monitoring all relevant behavioral equipment and logging all useful data. All software and instructions for building and using equipment are included as supplemental material at the Journal of Neurophysiology Web site. In addition, software updates and a user discussion forum will be available (see endnote).
METHODS
An overview of the system is given schematically in Fig. 1. A mouse is head-fixed using a titanium head bar cemented to its skull; it is otherwise free to run on a running wheel. The equipment depicted around the mouse can be divided into equipment used to monitor behavior (cameras and rotation meter) and equipment used to obtain patch-clamp recordings (amplifier, pressure regulator, and micromanipulator). Every sensor and actuator is connected to a computer through a Universal Serial Bus (USB) port, a National Instruments data acquisition (NI DAQ) input-output device, or an Arduino microcontroller. The system requires MATLAB and its Data Acquisition Toolbox (The MathWorks). Other useful, although optional, toolboxes will be specified in what follows.
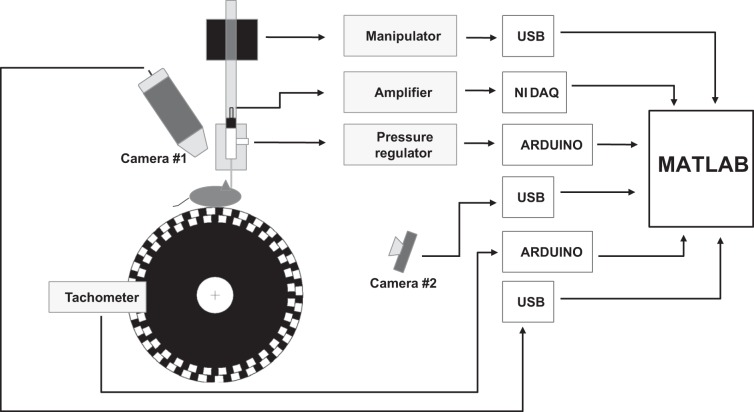
Fig. 1.
Schematic overview of the system. A head-fixed mouse runs on a wheel. The mouse's behavior is monitored by 2 cameras and a rotation meter (tachometer). Camera #1 has a high magnification (more than ×40) and is used to guide a patch electrode (inserted in a standard electrode holder) into a craniotomy; camera #2 is infrared-sensitive and is used to monitor the animal's behavior in the darkened Faraday cage. The rotation meter measures the speed and direction (clockwise or counterclockwise) of the animal's wheel running; toward this end, a quadrature pattern (black and white, as shown) is affixed to the side of the wheel. The patch-clamp equipment includes a custom pressure regulator, a commercial manipulator, and a custom data acquisition and analysis suite. Every sensor and actuator is connected to a computer through a Universal Serial Bus (USB) connection, a National Instruments data acquisition card (NI DAQ), or an Arduino microcontroller. The entire system is controlled by MATLAB code.
Many of the procedures and parameters used by the system can be changed without having to do any MATLAB programming. The ones that users are most likely to wish to change (e.g., the dorsal-ventral depth of the targeted brain area or the angle of electrode approach) can be specified using a graphical user interface (GUI). Others are contained in a single file saved to disk; users can change the numbers and specifications in this file to set, for example, the electrode pressure used to penetrate the dura or the step size used when hunting for a neuron. Choices made by users, whether through a GUI or through the file, are saved by the system and used in all subsequent experiments. The GUIs and the file, which is named “makefixedparametersfile,” are described in documents (“Program_structure” and “Installation”) included in the supplemental materials.
Animals and Surgical Procedures
All surgical and experimental procedures were approved by The University of Texas at Austin Institutional Animal Care and Use Committee and were in accordance with National Institutes of Health guidelines. Male C57BL/6 mice (6 wk old; The Jackson Laboratory) were housed in a room with a 12:12-h light-dark cycle for 1 wk before the first surgery and remained on this cycle throughout. All training and experiments were performed during the light period at approximately the same time of day (2-h range) for each mouse. Mice were allowed food and water ad libitum while in their home cages.
At 7 wk old, mice were surgically implanted with a custom-fabricated titanium head bar and a plastic or stainless steel recording chamber (3.1-mm inner diameter; Turnco Tool & Instrument; Fig. 2A). Animals were initially anesthetized with 3% isoflurane mixed with medical-grade oxygen and then maintained at 1–2% isoflurane throughout procedures. Each mouse was injected with 0.15 ml of Rimadyl [1 mg/ml subcutaneously (SQ)] and placed in a stereotaxic apparatus. The skull was prepared by scalping the crown and removing the fascia and scored with the tip of a scalpel blade. After the skull had been cleaned, dried, and leveled, the recording chamber was positioned so that its center was at a stereotaxic coordinate corresponding to area M2 (1.5 mm anterior to bregma, 1.0 mm left of the midline) and glued in place. A layer of low-viscosity cyanoacrylate was applied over the surface of the exposed skull. An initial layer of Metabond (Parkell) was applied over the cyanoacrylate, the titanium head bar was placed flush against the skull using a custom tool, and additional Metabond was used to cement the head bar to the skull and to encase the recording chamber.
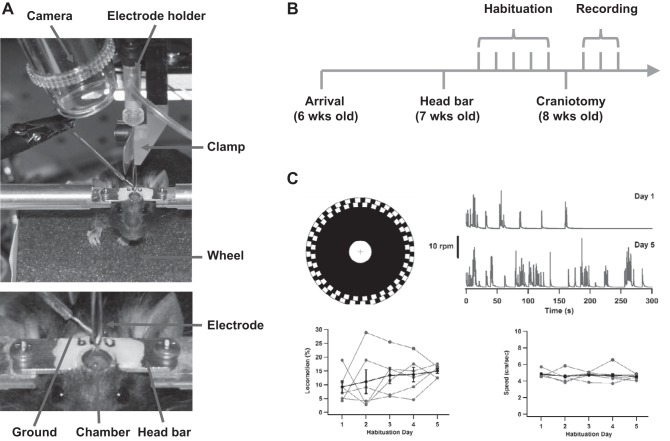
Fig. 2.
Surgery, habituation, and recording. A: the photographs show a head-fixed mouse atop the running wheel. Top: the patch electrode, clamped to a piece of hard plastic affixed to a rigid steel rod, enters the recording chamber vertically. The electrode is inserted in a standard electrode holder, which is present to provide a pressure port and a way of interfacing with the electrode wire. A high-magnification camera, with built-in light-emitting diodes (LEDs), is used to aid electrode placement and is seen at top left. A Teflon-coated silver wire, stripped and chlorided at the tip, served as a ground; it was passed through a glass pipette and held in saline near the craniotomy by the alligator clip visible at center left. Bottom: a closer view of the top of the mouse's head, where one can see the head bar, recording chamber, ground, and electrode. B: the timeline for surgery, habituation, and recording is illustrated schematically. Mice receive at least 5 days of habituation to the apparatus before the craniotomy surgery. Recordings begin the following day. C: locomotion on the running wheel is monitored using the black-and-white quadrature pattern (6-in. diameter) affixed to its side. Infrared light bounced off the pattern is used to detect black-white transitions (white is reflective, whereas black is not) and determine rotation speed. Differences between the 2 concentric rings, which are 90° out of phase, indicate whether the mouse is running forward (usually) or backward (occasionally). Even on the 1st day mice are introduced to the wheel, they do run and continue to do so throughout the habituation period. The bottom graphs show the percentage of time mice were active and the average running speed while active over the 5 days of habituation. Gray traces are the numbers for 6 individual animals, and black traces are means ± SE.
Mice were allowed 2 days of recovery before we began habituating them to the experimental apparatus (see next section). After at least 5 days of habituation (Fig. 2B), a second surgery was performed to make a craniotomy that would allow access for patch-clamp recordings. Mice were anesthetized with isoflurane as before, injected with 0.15 ml of Rimadyl (1 mg/ml SQ) and 0.03 ml of dexamethasone (2 mg/ml SQ), and placed again in the stereotaxic apparatus. The inside of the recording chamber was cleaned with 100% ethanol, and a small craniotomy (0.5- to 1.0-mm diameter) was made at the stereotaxic coordinate identified earlier. Care was taken to avoid damaging the dura, and bleeding was controlled with sterile saline and Gelfoam (Baxter). The recording chamber was filled with a thin layer of sterile saline and sealed with Kwik-Cast [World Precision Instruments (WPI)], and the animal was returned to its home cage. Electrophysiological recordings were usually performed the following day but occasionally as many as 3 days later (Fig. 2B).
Running Wheel
The running wheel (Fig. 2A) was adapted from a design by Medina and colleagues (Chettih et al. 2011; Heiney et al. 2014). It was cut from a 6-in. diameter foam roller to a 3.5-in. thickness. A stainless steel rod passed through the center of the wheel served as an axle. Both wheel and head-fixed mouse were held in place using mechanical components (Thorlabs) with some custom modifications. Arranged around the wheel assembly were mechanical stands (Sutter Instrument) that held micromanipulators and cameras. All of this equipment was fixed to a vibration isolation table and surrounded by a Faraday cage. Black photographic cloth covered the front of the cage, darkening its inside. An infrared-sensitive camera with built-in infrared light-emitting diodes (LEDs; Camera #2 of Fig. 1; Vimicro) controlled by the MATLAB program allowed an experimenter to monitor and record behavior inside the cage even when the cloth was down.
Habituation to this setup took place over at least 5 days. Mice were briefly (<15 s) anesthetized with isoflurane, placed on the wheel, and had their head bars secured to the wheel apparatus with screws. The animals woke up from the isoflurane in this situation. On the 1st day, mice were kept on the wheel for 15–20 min; over the course of the habituation period, this was extended to >1 h. On successive days, new elements of the experiment were introduced: the micromanipulators were turned on and moved; the LED lights of the high-magnification camera (Camera #1 of Fig. 1) were turned on and off at random intervals; and an experimenter reached into the cage to swap electrodes and lightly handle the mouse. For long training sessions (>30 min), the mouse was occasionally offered a drop of 5% sucrose in water.
Mouse movement on the running wheel was recorded using a black-and-white quadrature pattern glued to the side of the wheel (Fig. 2C). The pattern consisted of concentric black-and-white rings that were 90° out of phase with each other. Infrared light was reflected off each ring and recorded using an LED and a phototransistor (SparkFun), and black-white transitions for each ring were detected and used to calculate speed (rpm) and direction (clockwise or counterclockwise). This detection and calculation was done by an Arduino microcontroller at a rate of 20 Hz; the results were written to a text file on the hard disk along with a time stamp using the Processing language (https://www.processing.org/). The overarching automated patch program controlled the Processing program through a system command and read the data from the text file. A full description of the rotation meter, including a parts list and build instructions, is included in the supplemental material accompanying this article. The Processing program is also in the supplemental material in the running wheel subfolder. We found that mice took to the wheel readily. Even on the very 1st day, mice could run briskly (Fig. 2C). Over the course of 5 or more days of habituation, mice continued to run regularly. For six mice, we quantified locomotion during the habituation process by smoothing running speed with a sliding 1-s window and identifying active epochs as those for which wheel motion was >1 cm/s. The fraction of time mice were active varied considerably between mice and across days (Fig. 2C, bottom left), but their average speed when active was relatively uniform (Fig. 2C, bottom right).
Positioning Electrodes
Patch-clamp involves three basic elements: positioning the electrode, applying pressure to the back of the electrode, and analyzing the electrode signal (typically in terms of tip resistance). Our system includes separate components for all three, which we describe in turn in this section and the two that follow.
Patch electrodes were pulled on a Sutter P-97 electrode puller (Sutter Instrument) using fire-polished, filamented borosilicate glass pipettes (1.5-mm outer diameter, 0.86-mm inner diameter; Warner Instruments). Electrode resistances were 3–7 MΩ in saline with a taper such that their width was ∼100 μm ∼1 mm from the tip. Before use in vivo, programs for pulling electrodes were usually validated in vitro (see below). Electrodes were filled, using a MicroFil electrode filler (WPI), with an intracellular solution containing (in mM): 125 K-gluconate, 10 KCl, 4 NaCl, 10 HEPES, 4 Mg-ATP, 0.3 Tris-GTP, and 7 phosphocreatine (pH 7.4 at physiological temperatures). The recording chamber was filled with an extracellular solution containing (in mM): 135 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 5 HEPES (pH 7.4).
Patch electrodes were inserted into a standard electrode holder (Warner Instruments). In typical practice, such a holder would then be inserted into an amplifier headstage. Here, the conductive pin of the holder was connected via an intervening cable to a headstage that sat several inches away; the pressure port of the holder was connected to an automated pressure regulator (see next section). What held the electrode and its holder in place was that the electrode glass itself was clamped to a hard plastic piece affixed to a stainless steel rod (Fig. 2A). The rod was attached directly to a micromanipulator using a custom dovetail attachment. This design improves electrode mechanical stability and, with its slim profile, makes it easier to position other equipment close to the electrode. We found that inserting a cable between electrode holder and headstage did not increase noise levels.
The micromanipulators were widely used ones from Sutter Instrument (MP-285 and MPC-200) that allow movement along the three orthogonal axes with a precision of 64 nm. Both were controlled by the MATLAB program via serial commands issued through a USB port. In the case of the MPC-200, we made use of a MATLAB-compatible C++ library generously provided by J. D. Steinmeyer (Steinmeyer and Yanik 2012). Adapting the manipulator control procedures for micromanipulators from other manufacturers should not be difficult as long as those manipulators respond to serial commands.
For most of the patching process, controlling movement was straightforward as it only entailed moving the electrode by fixed amounts (e.g., 2-μm steps while searching for a neuron to patch). The one complicated part was lowering the electrode into the craniotomy (Fig. 3A). Usual (manual) practice for this is: to remove any covering saline, to get a clearer view of the cortical surface; to identify the position of the surface by looking through a stereomicroscope for a dimple as the electrode tip is pushed against it; to retract the electrode and cover the craniotomy again with saline; and finally to advance the electrode down past the identified position (DeWeese 2007). Rather than using this somewhat time-consuming procedure, which would have to be repeated for every electrode, we employed a high-magnification (up to ×90), long-range (∼5-cm) USB microscope (Dino-Lite) mounted to a Sutter micromanipulator. Images were acquired using the Image Acquisition Toolbox and analyzed using the Image Processing Toolbox (Fig. 3A).
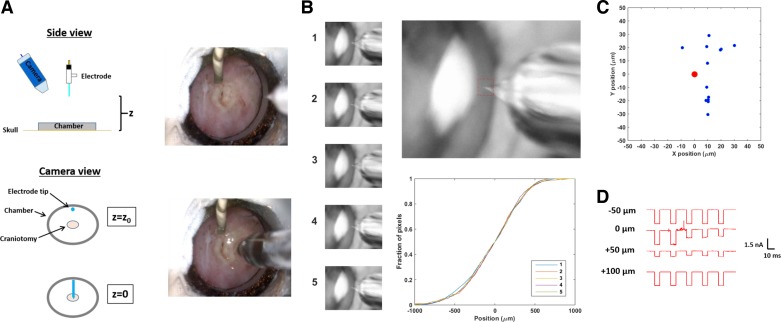
Fig. 3.
Positioning the electrode. A: on the left are schematics showing the relative positions of the patch electrode (in its holder), the high-magnification camera, the recording chamber, and the craniotomy. The electrode is centered above the craniotomy in the x–y plane and then lowered straight down along the z-axis. On the right are photographs, taken by the camera, showing an electrode above (top) and inside (bottom) the craniotomy. The stainless steel recording chamber has an inner diameter of 3.1 mm; the craniotomy (∼0.7-mm diameter) can be seen at its center. A Teflon-coated silver wire, stripped and chlorided at the tip, is dipped in the saline filling the chamber and serves as a ground. B: the vertical position of the electrode could be determined by the system using the camera alone. Before the pictures shown here were taken, the x–y coordinates of the craniotomy were determined manually by the user. A region of interest (ROI; the red dashed box seen in the top right panel) was then specified through a graphical user interface. Five electrodes were then moved automatically by the system, one after the other, to the same position in space, as seen in the 5 photographs at left. This automatic positioning was possible because, along the z-axis, electrode position could be determined by measuring the fraction of ROI pixels filled by the electrode as it was lowered through the ROI (bottom right); the shape of the resulting curve varied little from electrode to electrode. The fraction of pixels was determined by subtracting a ROI image without an electrode from each ROI image with one and then thresholding (MATLAB function “graythresh”). C: the custom-designed electrode holder enabled us to localize electrodes along the lateral (x–y) axes to within 30 μm. In this picture, 15 electrodes were lowered to a point in space, and their x–y positions, relative to a fixed target, were measured. The red dot indicates the 1st (target) location. Note that every subsequent electrode hit this location in the x–y plane to within 30 μm. This reproducibility was solely a consequence of the custom-designed electrode holder. D: the dura was left intact in these experiments to protect the cortex and to reduce pulsation. Under high positive pressure (≥4 psi), a patch electrode can penetrate the mouse dura (located at z = 0 μm) without clogging or breaking. The traces show the current deflections produced by −10-mV voltage steps as an electrode is lowered from 50 μm above to 100 μm below the dura. The transient noise and increased resistance associated with encountering the dura, as well as an accompanying baseline shift, can be used to locate its position along the z-axis.
Even though the magnification was not sufficient to resolve the very tip of the electrode, the properties of the camera were stable enough and the electrode shape stereotyped enough that, if the system could localize the visible part of an electrode to a landmark a known distance from the cortical surface (measured at the start of each session; Fig. 3B), it could bring the electrode tip down to the surface with good accuracy in the vertical (z-) direction (36 ± 8 μm, n = 18) and to within ±30 μm in the orthogonal (x- and y-) directions. That is, the automated system can localize the tip of an electrode to a position in space within a sphere of radius ∼40 μm.
The localization procedure is described in detail in the system documentation included in the supplemental material that accompanies this article. Here, we describe the steps concisely. The first electrode of a recording session on any given day is crucial: the system uses it to set the x–y position of all subsequent electrodes, identify a visual landmark against which to calibrate the z-position of other electrodes, and determine the vertical (z-axis) difference between landmark and cortical surface. Users place the first electrode in the holder and direct it under visual guidance (using the Dino-Lite camera) to a suitable part of the craniotomy (e.g., away from any visible blood vessels). The x–y position is then recorded. We have found empirically that clamping the electrode glass in our custom-designed holder, rather than relying on the rubber gaskets of conventional electrode holders, makes the x–y position reproducible to ≤30 μm (Fig. 3C); pictures of our holder are included in the supplemental material for those who wish to make their own. The user is then asked to use a simple GUI to identify a region of interest (ROI) in the visual field against which to reference subsequent electrodes. This landmark (a box of area ∼1 mm2) should be mostly flat and unlikely to change over the course of a recording session. Usually, we picked a piece of the (stainless steel) recording chamber for this purpose (Fig. 3B). As an electrode is lowered through the ROI, it fills the ROI pixels in a highly reproducible manner (Fig. 3B, graph at lower right), one that allows the system to determine the z-axis coordinate to ≤40 μm. Finally, the vertical distance between the cortical surface and the ROI can be established by lowering the electrode to the surface until a small resistance change is detected and then raising the electrode until it fills a fixed fraction of the ROI pixels (2 button presses in the main GUI are all that is required for this part). In total, these setup steps with the 1st electrode should take no more than 5 min. Every subsequent electrode in a recording session can then be placed automatically to a position in space to within ∼40 μm. It is worth noting explicitly that the positioning system does not depend on the precise position or angle of the camera, the pipette geometry, or where the craniotomy is located within the recording chamber; these properties must remain steady throughout a single recording session, but they can vary between sessions.
A second measure of surface position was given by a transient increase in electrode resistance and noise associated with penetrating the dura (Fig. 3D). In our experiments, the system used the resistance increase, which can be >100%, to identify the precise location of the dura for each electrode. In particular, the electrode position at which a >25% increase was first observed was taken to be the z-axis position of the dura. The system then used this number to adjust its estimate of the dorsal-ventral position of the electrode and to move the electrode to the desired target depth. If no resistance increase of >25% was observed, because, for example, the dura had been disrupted by previous electrode penetrations, then the system simply used the estimate of surface position obtained during the initial setup. Users can specify whether to employ this procedure, as well as the parameters (e.g., 25%), through the main parameters file. Note that the dura was left intact in all of our experiments. Resecting the mouse dura without damaging the underlying tissue or causing bleeding is very difficult to do, and we found, as others have, that it is not strictly necessary (Domnisoru et al. 2013; Harvey et al. 2009). Under high positive pressure (∼4 psi), a patch electrode inserted near vertically can pass through the mouse dura without clogging or breaking.
Regulating Pressure
Good control of pipette pressure is crucial for successful patch-clamping as it is used to keep electrode tips clean, form gigaohm seals, and rupture membranes. These pressures must range from a high of >4 psi (to enter the brain) down to a low of less than −2 psi (to establish whole cell mode) with the required temporal precision being <1 s. In manual patch-clamping, pressure is applied by syringe or mouth and held/released by closing/opening a stopcock. Commercial pressure regulators are available, but ones flexible and precise enough for this process tend to be very expensive.
For our system, we built an inexpensive pressure regulator around the Arduino microcontroller (a parts list is available online, as are build instructions). The basic design is shown in Fig. 4A. House air (+10 psi) and house vacuum (−10 psi) served as the positive and negative pressure sources, respectively. (A miniature vacuum/pressure pump, such as Cole-Parmer UX-79600-08, could be used instead if house pressure is unavailable.) The first solenoid valve (The Lee Company) determined whether positive or negative pressure would be applied. The proportional valve (Kelly Pneumatics) cut down the magnitude of the pressure at its output. The second solenoid valve determined whether the pipette would feel this pressure or atmospheric pressure. An electronic pressure monitor (Honeywell) measured the pressure output of the proportional valve and sent this number to the microcontroller (feedback), which compared it with the desired pressure specified by the MATLAB program (set) through a USB port, and calculated an error equal to their difference.
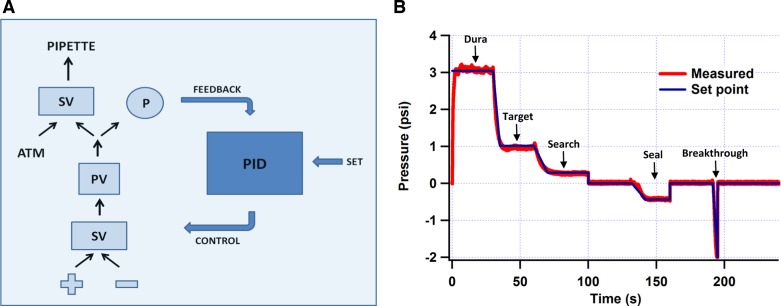
Fig. 4.
Low-cost pressure regulator. A: a schematic of the basic design. SV, solenoid valve; PV, proportional valve; P, pressure monitor; ATM, atmospheric pressure; PID, proportional-integral-derivative control algorithm; + and −, positive and negative pressure sources (±10 psi). The SVs determine which of 2 inputs is fed into an output. The PV regulates the pressure at its output. The PID algorithm, implemented on an Arduino microcontroller, is used to calculate the error between the desired pressure (set), given by the MATLAB program, and the actual pressure (feedback), measured by P, and then to use this error to control the other parts of the circuit (control). B: an example of the speed and accuracy of pressure control given by the regulator. Typical pressures for different steps in the in vivo patching process are labeled.
The microcontroller then used this error to determine an appropriate control signal to send back to the rest of the system so as to minimize the error. The control algorithm it employed to do this was proportional-integral-derivative (PID), which is generic and widely used (Åström and Murray 2008). PID has three parameters that can be adjusted to eliminate steady-state error, improve temporal response, and minimize “ringing” when the set value is changed suddenly. Although there are heuristic methods for tuning PID parameters, such as Ziegler-Nichols (Åström and Murray 2008), a certain amount of hand-tuning is useful here. With modest effort (2–4 h), we found a set of parameters that controlled pipette pressure well within the desired ranges (Fig. 4B) and that did not require retuning even after 6 mo of standard use.
In the supplemental material, we describe how to build and tune the pressure regulator we designed.
Analyzing Electrode Signals
Patch-clamp in vivo has generally been done blind (Margrie et al. 2002). Even when a two-photon microscope is available, neurons deeper than a few hundred micrometers from the cortical surface are not amenable to visualized patch. In vivo electrophysiologists therefore usually rely on the electrode signal itself to determine whether an electrode is clogged or broken, whether an electrode tip is close enough to a neuron to attempt patching, and whether a gigaohm seal or whole cell mode has been achieved. Our system uses electrode impedance to answer these questions.
Patching was done with the amplifier (MultiClamp 700A or 700B; Molecular Devices) in voltage-clamp mode. Brief (10–25 ms), small (−10 mV) voltage steps were applied to the electrode at all stages of the process. The resulting current deflections were used to estimate resistance. As we were only interested in (direct current) resistance, in making this estimate, 5–10 deflections were averaged (with outliers discarded), and electrode signals were band-pass filtered (1–300 Hz) using the Signal Processing Toolbox (The MathWorks). The filtering eliminated errors due to baseline drift and high-frequency noise. In some experiments, a hardware 60-Hz noise eliminator (HumBug) was also included, although this was not necessary. Of course, filtering distorted “real” aspects of the electrode signal along with drift and noise, such as biological rhythms and capacitive transients, giving the corners of current deflections a slightly rounded aspect. The only real concern in this regard is that filtering might affect pulsation in the electrode signal associated with mouse heartbeat (∼8 Hz) and visible when an electrode tip is very close to a neuron or other piece of membrane. This pulsation is often used by experienced electrophysiologists to determine whether an electrode has encountered a neuron (Margrie et al. 2002). The present version of our system does not make use of heartbeat pulsation.
Data acquisition and signal generation were done using a Peripheral Component Interconnect (PCI) card from National Instruments. The Data Acquisition Toolbox, which is a required component of our system, provides useful and simple commands for controlling and monitoring data acquisition devices from many manufacturers. Further details on this point are given in the supplemental documentation.
Patch-Clamp Process
As we noted above, there are three basic elements that make up the patch-clamp process (electrode movement, electrode pressure, and electrode signal). In results, we show a real example where the three elements are coordinated to obtain a whole cell patch recording. Here, we describe the basic ideas.
Some of the details were changed as we developed the system, and we will note those changes in this section, but the fundamental features remained constant. Moreover, in our most recent experiments (59 of the 67 electrode attempts described in results), a single set of parameters and procedures was employed. These are the numbers given and procedures described below. They are also the system defaults included in the supplemental material. Users have the choice to use the default settings or to change them through a GUI or the main parameters file (makefixedparametersfile).
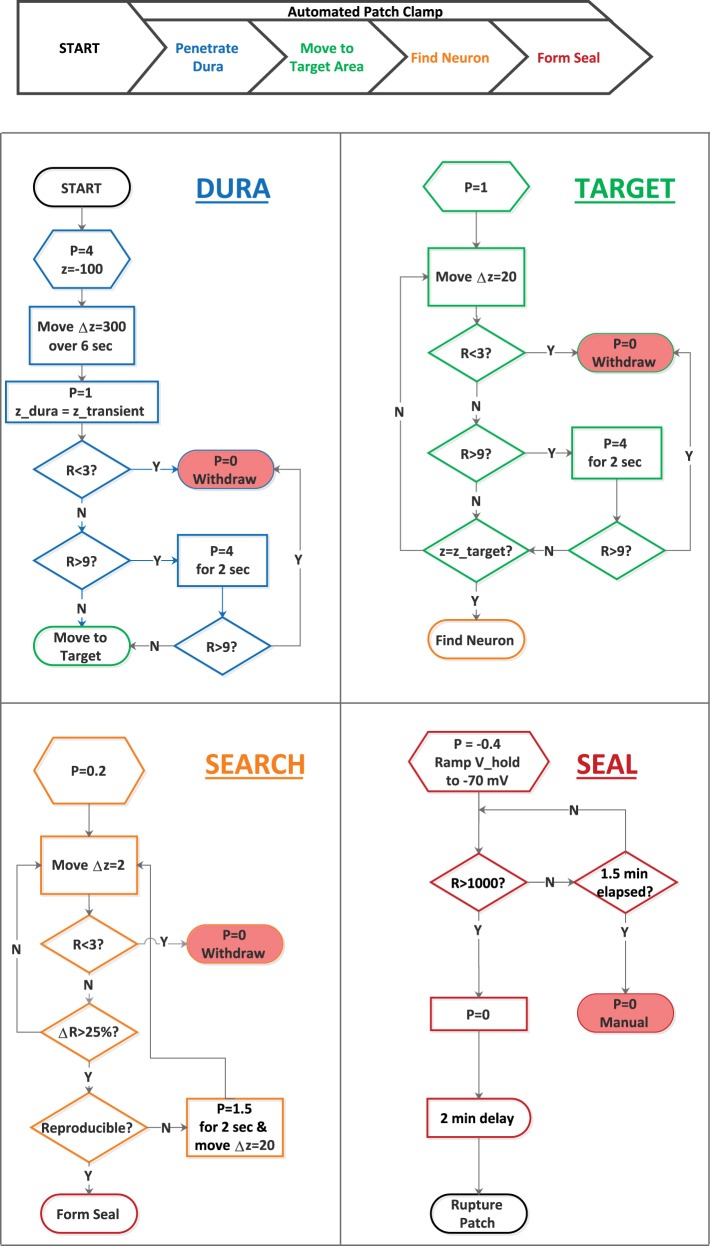
Whole cell patch-clamp in vivo has five phases: entering the brain (penetrating the dura), moving to the target region, searching for a neuron, sealing onto a neuron, and breaking through to whole cell mode. The first four of these are illustrated in flowchart form in Fig. 5. In the diagram, R stands for electrode resistance (in megohms), P for the pressure applied to the electrode (in psi), and z for the electrode position along the dorsal-ventral axis (in micrometers, with numbers becoming more positive as the electrode is lowered).
Fig. 5.
Flow control of the automated patching process. The flowchart diagrams illustrate the steps associated with penetrating the dura, moving to the target brain region, searching for a neuron, and forming a gigaohm seal. The symbols P, R, and Δz refer to the pressure applied to the electrode (in psi), the measured tip resistance (in megohms), and incremental movement down into the brain (in micrometers), respectively. The flowchart shapes have their standard meanings: oval, start or stop point in a process; rectangle, an operation; hexagram, a preparatory step; diamond, a decision point; rectangle with a curved right side, a fixed delay. Note that the patching process can be terminated by a sudden drop in resistance, indicative of a broken tip; a large increase in resistance (except when attempting to form a seal), indicative of a clogged tip; and formation of or failure to form a gigaohm seal. The position of the dura, z_dura, is identified by the position of the transient, z_transient, as the electrode crosses it. The position at which to begin searching for a neuron is designated z_target. Y, yes; N, no; V_hold, voltage-clamp holding potential.
Dura.
The first step is to move the electrode, under high positive pressure (4 psi), through the dura quickly (50 μm/s) and into the brain (200 μm). Once in the brain, P is reduced to 1 psi. (In early experiments, we instead set P to 50 mbar or 0.73 psi. As long as the pressure is sufficient to keep the electrode tip clean, its precise value should not matter.) If the electrode tip is broken (as indicated by a drop in R to <3 MΩ), the pressure applied to the electrode is immediately removed, to avoid spewing potassium-rich solution into the surrounding tissue, and the electrode is withdrawn. If the electrode tip is clogged (as indicated by an increase in R >9 MΩ), an attempt is made to flush it clean with the application of high positive pressure (4 psi for 2 s). If that attempt is unsuccessful, the pressure applied to the electrode is removed, and the electrode is withdrawn. While moving the electrode through the dura, the system attempts to identify the z-axis position of the dura by looking for a transient increase in R, as described in Positioning Electrodes.
Target.
The second step is to move the electrode under positive pressure (1 psi) to the targeted brain region at a moderate speed (20 μm/s). Again, the tip is constantly checked for breaking or clogging. If broken, the positive pressure applied to the electrode is removed, and the electrode is withdrawn. If clogged, an attempt is made to clear the electrode with application of high positive pressure (4 psi for 2 s). If this attempt is unsuccessful (R still >9 MΩ), the pressure application is terminated, and the electrode is withdrawn. If the attempt is successful, the system goes on to the next step.
Search.
The third step is for the system to hunt for a neuron to patch. In this step, P should be kept as low as possible. We have found through brain slice recordings (see results) that the dependence of R on proximity to a cell membrane is very sensitive to the value of P. For the blind patch technique, one wants the dependence to be as strong as possible. In most of our experiments, we used a pressure of 0.4 psi at this step, but, in recent experiments, we have found that 0.2 psi is sufficient to keep the electrode tip clean. The system default pressure at this step is now 0.2 psi. Having set the pressure, the system moves the electrode forward slowly (2 μm/s) while looking for a substantial (>25%) increase in R above baseline. If it finds one, it checks whether the increase is reproducible by moving the electrode back by 8 μm and then forward again 2–3 times. (In our initial attempts, we required that the resistance increase be reproduced 3 times, but we found with experience that reproducing the resistance increase twice was good enough; the number of reproductions is a parameter that a user of this system can select.) If the resistance increase cannot be reproduced, the system deems this detection a “false positive” and attempts to move beyond the obstruction by a brief (2 s) application of high (1.5 psi) pressure and a large (20 μm) z-axis movement. We have found that the occurrence of a false positive does not compromise the usefulness of an electrode. More than a quarter of electrodes that encountered a false positive were subsequently used to obtain a gigaohm seal, presumably because P was sufficient to keep the electrode tip clean.
Seal.
The fourth step is to form a seal (>1 GΩ) between the electrode and the cell membrane. Experienced electrophysiologists vary considerably in how they approach this step: some use mouth suction, others syringes; some apply a steady negative pressure, others pulse the negative pressure. In our first experiments, we used a protocol in which we ratcheted up negative pressure, increasing it from −0.3 to −0.6 to −0.9 psi and so on until a 1-GΩ seal was obtained or a time limit had been exceeded. However, in most of our experiments, we instead used the protocol illustrated at bottom right in Fig. 5. After neuron detection, P is set to −0.4 psi, and the voltage-clamp holding potential is ramped down from 0 to −70 mV over 10 s. The negative pressure is maintained until 90 s have elapsed or a >1-GΩ seal has formed, whichever comes first. The negative pressure is then removed, and the seal is allowed to improve for 1.5–2 min (the system default is 2 min). If the seal resistance at that point is >1 GΩ, the system moves on to the breakthrough step. If the seal resistance is <1 GΩ, the system default is to return to manual control, allowing the experimenter to decide whether to withdraw the electrode. However, users can specify, through the main parameters file, that the system should in this case remove any applied pressure and withdraw the electrode automatically in preparation for a new attempt.
Breakthrough.
The final step in the patch-clamp process is to rupture the membrane below the electrode tip to achieve whole cell mode. Our system applies negative pressure pulses (−2 psi, 1 s) until R drops below a threshold (300 MΩ) or four attempts have been made. Successful or unsuccessful, the system then restores manual control.
Our own criteria for a successful whole cell patch recording was a series resistance <75 MΩ and a resting membrane potential less than −50 mV.
Software and GUIs
The current version of the automated system was written using MATLAB 2014b on a computer running 64-bit Windows 7. We have tested elements of the system in 32- and 64-bit Windows XP, Vista, 7, and 8 and with MATLAB versions as early as 2012a and as advanced as 2015a (beta release). In writing the code, we attempted to minimize our use of specialized toolboxes, which add to expenses, and of older MATLAB features, which might limit the longevity of the system. Of special importance in this regard, we exclusively employed the “session-based” data acquisition interface, which was introduced in 2011. This interface incorporates ideas from modern object-oriented programming and is compatible with both 32- and 64-bit MATLAB, whereas the competing traditional, “legacy” data acquisition interface is limited to 32-bit MATLAB and will not be updated by The MathWorks after 2015 (http://www.mathworks.com/support/sysreq/roadmap.html). If a prospective user uses a MATLAB version older than 2011, it would not be difficult to transition back to the legacy interface (the original version of the present program used this interface), although we have not done so because the need for this seems unlikely.
The system as described here is designed to obtain whole cell patch recordings rather than acquire data from those recordings once obtained. However, indeed, we have available an entire MATLAB-based data acquisition suite suitable for current clamp, voltage-clamp, and field potential recordings from up to four electrodes and capable of basic analyses of the same. (See the documentation included in the supplemental material that accompanies the article.) This suite was developed some years ago (Desai and Walcott 2006) and has been tested by continuous use since then. As we noted in the Introduction, our guiding ideal was a single, complete system that could do the whole experiment from start to finish: move the electrode, obtain a whole cell patch recording, and record data in conjunction with behavior. Users may prefer their own software (pCLAMP, Ephus, AxoGraph, etc.) for data acquisition and analysis, and our system is flexible enough to allow them to make choices like this, but our purely MATLAB system will be available for online download.
Along with the basic code, we provide GUIs that allow users to manipulate parameters, choose processes, and generally run the system. (See the screenshots included in the supplemental material as well as the accompanying documentation that describes their use.) GUIs are available for both the patch-clamp process itself and the subsequent data acquisition and analysis process.
In Vitro Electrophysiology
The focus of this paper is on in vivo electrophysiology, but in developing the automated system we made extensive use of in vitro electrophysiology. We used whole cell patch recordings from neurons in brain slices to confirm that the patch electrodes produced by the puller program were suitable for cortical neurons and to validate the automated patch process.
The mice were C57BL/6 males (6–8 wk old; The Jackson Laboratory). Brain slices containing medial prefrontal cortex were prepared using standard procedures (Kalmbach et al. 2013). Shortly after receiving a lethal dose of ketamine/xylazine, mice were perfused transcardially with an ice-cold solution containing (in mM): 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 0.5 CaCl2, 7 MgCl2, 7 dextrose, 205 sucrose, 1.3 ascorbate, and 3 sodium pyruvate (bubbled with 95% O2-5% CO2 to maintain pH at ∼7.4). Brains were removed, and a vibratome was used to make 300-μm-thick coronal sections. Slices were cut in the same ice-cold saline used for perfusion and then were held for 30 min at 35°C in a chamber filled with artificial cerebrospinal fluid (aCSF) containing (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 2 CaCl2, 2 MgCl2, 25 dextrose, 1.3 ascorbate, and 3 sodium pyruvate (bubbled with 95% O2-5% CO2). Thereafter, they were maintained at room temperature in the same solution.
Over the next 5 h, recordings were made from layer 2/3 pyramidal neurons in area M2 in 32–35°C oxygenated aCSF (minus the ascorbate and sodium pyruvate) on an upright microscope with infrared, differential interference contrast (IR-DIC) optics. The patch-clamp equipment (amplifier, manipulator, and data acquisition board) were of the same type as used for the in vivo experiments, as were the pipettes and internal solution. The MATLAB code used to obtain recordings and electrophysiological data was identical in all important respects.
RESULTS
In the preceding section, we described a MATLAB-based system for obtaining whole cell patch recordings from awake mice running on a wheel. In this section, we describe how well the system has performed to date under real conditions. However, to get a sense of the landscape, we begin by describing how the automated system performed in vitro, specifically, in brain slices, which remain the gold standard for patch-clamp.
Automated Patching In Vitro
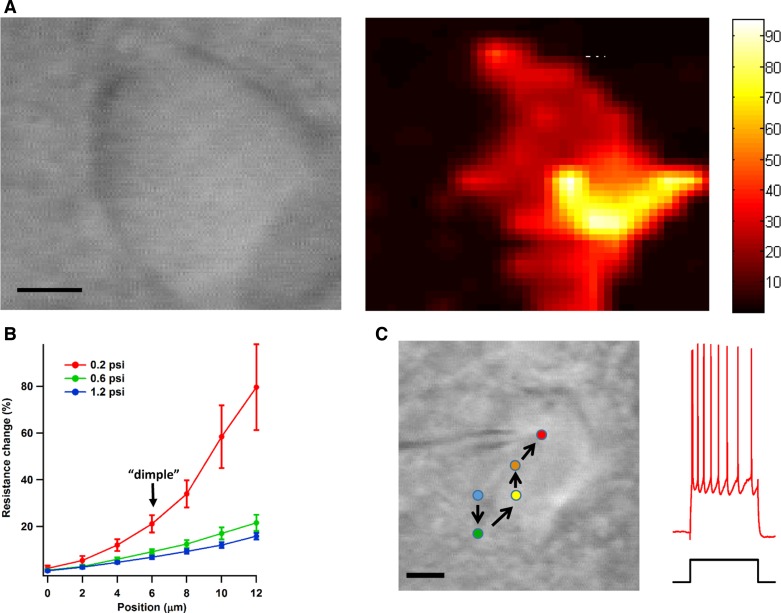
Standard descriptions of the blind patch-clamp process treat neurons as if they were point sources (DeWeese 2007; Margrie et al. 2002) rather than extended bodies with some spatial structure. We examined this issue systematically by probing neurons in brain slices with patch electrodes (n = 6). We identified the outlines of cell bodies under IR-DIC optics (Fig. 6A, left) and then had electrodes map them by moving in the x–y plane (2-μm increments) and, at each x–y point, pushing down by up to 15 μm while measuring tip resistance. We found that the resistance change induced by proximity to the neuronal membrane was sensitive to where on the membrane the electrode tip touched. At the periphery, even the maximal resistance change, when the tip was pressing down very strongly, was rarely >25%. However, near the center, at x–y positions experienced electrophysiologists would choose to place a patch electrode, resistance changes of near or over 100% were observable. The picture that emerged from these experiments (Fig. 6A, right) suggests an analogy with atomic force microscopy where distance-dependent forces allow a probe to map surface structure with good precision.
Fig. 6.
Validating the automated patch system in vitro. A: the photograph on the left shows the soma of a layer 2/3 neuron (scale bar: 5 μm). The color map on the right shows the maximum resistance change (in percent) elicited by moving a patch electrode down at different x–y positions. (Data points were linearly interpolated by 10× for better visualization.) Not only can one trace out the cell membrane in this way, but also one can identify the “center of mass” of the cell, made up of points that give maximal resistance changes and are the ones at which experienced electrophysiologists, working visually, would choose to place patch electrodes. B: the magnitude of the resistance change produced by encountering a neuron is sensitive to the applied pipette pressure. In 6 neurons, an electrode was positioned above the center of mass and lowered as its resistance was measured. The position 6 μm corresponds to the point where the pipette just dimples the cell membrane and at which most electrophysiologists would attempt to form a gigaohm seal; the position 12 μm corresponds to a point where the pipette deforms the membrane strongly and that most electrophysiologists would avoid. Note how much more sensitive resistance is to position when pipette pressure is small. C: an example of an automated recording in vitro. The pipette tip was initially positioned above a point at the edge of the cell body (scale bar: 4 μm). Probing the surface of the neuron by measuring resistance changes (as in A), the tip ended up at a position near the center of the neuron. There, the system successfully obtained a whole cell patch recording (right).
In these measurements, the pipette pressure was set at 0.2–0.4 psi, just enough to keep the tip clear of debris. To determine how much the blind patch process was affected by the absolute value of pipette pressure, we varied pressure between 0.2 and 1.2 psi as we moved the electrode tip down onto a neuron (n = 6) at an x–y point near its center, where the proverbial experienced electrophysiologist would have chosen to place it. We found that the resistance change was strongly sensitive to pressure (Fig. 6B). When pressures were ≥0.6 psi, resistances only crudely varied with the distance between the electrode tip and the cell membrane, presumably because the high positive pressure pushed away membrane and thereby cleared a path to ground. Only for small pressures (0.2 psi) was resistance change a good measure of distance, with a change of ∼20% being indicative of an electrode tip at just the right distance to merit patching.
To test the automated patch-clamp system, we had it attempt to patch layer 2/3 neurons using a pressure in the detection phase of 0.2 psi. The electrode tip was initially placed either directly above the center of the neuron (n = 3) or above a peripheral point (n = 2). In the former case, the system simply moved the electrode straight down until a 20% resistance change was detected. In the latter case, the system probed the membrane surface using resistance changes and thereby found the center for itself (Fig. 6C). The search algorithm used was a combination of a constrained gradient ascent (fixed number of possible angles and step sizes) and random exploration, with the system stopping at a point where a >50% increase could be measured. This method is reasonable because the center of a neuron, given typical cell body sizes, will be no more than 10 μm from its periphery and because it is not necessary to find the absolute peak of the resistance surface. All five in vitro attempts resulted in whole cell recordings.
Automated Patching In Vivo
We used the automated system to obtain 19 whole cell patch recordings from 16 awake mice, targeting neurons approximately 400–700 μm below the cortical surface. If an electrode failed to detect a neuron or obtain a gigaohm seal by the time it reached the end of that range, it was simply withdrawn, and an attempt with a new electrode was made. The other major reason for terminating an attempt early was clogging of the electrode tip. On average, 5.7 ± 0.6 attempts were required to obtain a single recording. In a few animals, 2 separate recordings were obtained. All recorded cells were confirmed to be neurons by the presence of action potentials. None were glial cells; we do not know why that was, but the propensity of blind patch to target neurons preferentially has also been observed by others (Kodandaramaiah et al. 2012; Margrie et al. 2002). In a few animals, we were unable to obtain any recordings, but in all of these cases our lack of success could be ascribed to visible damage to the cortex, especially bleeding, rather than to the automated system. Usually, if excessive blood was present in the craniotomy when the Kwik-Cast seal was removed or bleeding could not be controlled by saline or Gelfoam, we simply did not attempt to patch. Approximately one in four craniotomies was rejected for this reason. We did not attempt to establish formal criteria for distinguishing a suitable craniotomy from an unsuitable one because the difference was usually dramatic: in the latter case, blood filled the craniotomy to such an extent that no part of the cortical surface was clear. Protecting craniotomies for electrophysiological recording is a common issue, and there have been several recent efforts to design specialized recording chambers to address it (Adams et al. 2011; Goldey et al. 2014; Heiney et al. 2014).
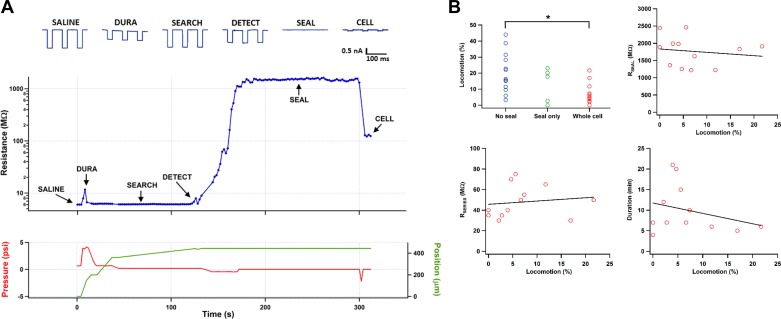
An example of automated patching is shown in Fig. 7A. The three basic elements of patch-clamp (movement, pressure, and resistance) are plotted together. This recording took 5 min to obtain, which was a typical time for successful recordings; the chief sources of variability were the time needed to detect a neuron and the time needed to form a gigaohm seal. In this example, penetrating the dura was marked by a striking increase (a doubling) in tip resistance, but in many cases a much smaller increase or none at all was observed, possibly because the dura had been disrupted during the craniotomy or by previous electrode penetrations. Rupturing the membrane to achieve whole cell mode was done in this case by the automated pressure regulator exclusively (−2 psi for 1 s), but in other cases we found it helpful to use manual suction to improve series (access) resistance; future versions of the system may include more reliable routines for rupturing membranes.
Fig. 7.
A: an example of the automated patch-clamp process. A patch electrode hovers in the saline above a craniotomy at time t = 0 s. Over the next 5 min, it penetrates the dura, moves to the target brain region, searches for a neuron, detects one, forms a gigaohm seal, and breaks through to whole cell mode. The traces at the top are the responses in voltage-clamp to −10-mV steps. The blue trace in the middle is the measured tip resistance. The red trace is the pressure applied to the back of the patch electrode. The green trace is the position of the electrode tip, where 0 μm is approximately the position of the dura and positive numbers indicate movement down into the brain. B: mouse locomotion affects sealing success but not recording quality. The sealing attempts of the system were divided into 3 categories: failed to obtain a gigaohm seal (No seal), obtained a gigaohm seal but failed to achieve whole cell mode (Seal only), and obtained a successful whole cell patch recording (Whole cell). In the top graph at left, average locomotion (percentage of time spent running) during the sealing attempt is shown for individual attempts in the 3 categories. Locomotion in the No seal condition was significantly larger than in the Whole cell condition (*P < 0.02, Wilcoxon rank sum test with Bonferroni correction). For attempts in which whole cell mode was achieved, locomotion during the sealing phase was not correlated with maximum seal resistance during patching (top right; r2 = 0.02), series resistance after whole cell breakthrough (bottom left; r2 = 0.02), or the duration of the subsequent recording (bottom right; r2 = 0.08).
The failure modes, when attempts did not result in whole cell patch recordings, varied in type, although the principal culprit was electrode clogging while entering the cortex or moving to the target area. We kept careful track of failure modes for the last 12 recordings. A total of 67 electrodes were used during these recording sessions: 25 clogged, 2 broke, 9 reached the end of the search range without detecting a neuron, 13 failed to form a gigaohm seal, 6 failed to break through into whole cell mode, and 12 resulted in successful recordings. Of the failure modes, the one that might most easily be remedied is clogging; one might apply a higher positive pressure to the electrode when entering the cortex and/or moving to the target area. Some researchers use pressures of >10 psi (Kodandaramaiah et al. 2012; Lee et al. 2014), although we did not because we were concerned about damaging the tissue with too high pressure.
A different possibility suggested by our data is to monitor locomotion and attempt to seal only when the mouse is not moving. We measured locomotion (percentage of time the mouse ran at speeds >1 cm/s) during the sealing phase for the 31 electrodes for which the system detected a neuron (Fig. 7B, top left). Locomotor activity was significantly higher for attempts in which the electrode did not form a gigaohm seal than for attempts in which the electrode achieved whole cell mode (20 ± 3 vs. 7 ± 2%, n = 13 and 12, respectively; P < 0.02 Bonferroni-corrected Wilcoxon rank sum). Importantly, this dependence on locomotion did not carry over once a seal had been formed: the maximum seal resistance before breakthrough (r2 = 0.02), the series resistance after breakthrough (r2 = 0.02), and the duration of the subsequent recording (r2 = 0.08) were not correlated with locomotion (Fig. 7B).
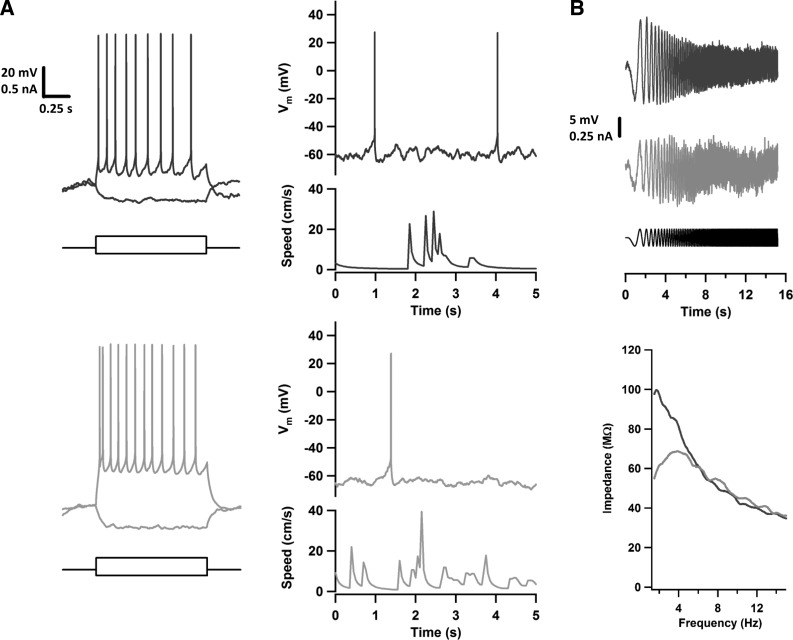
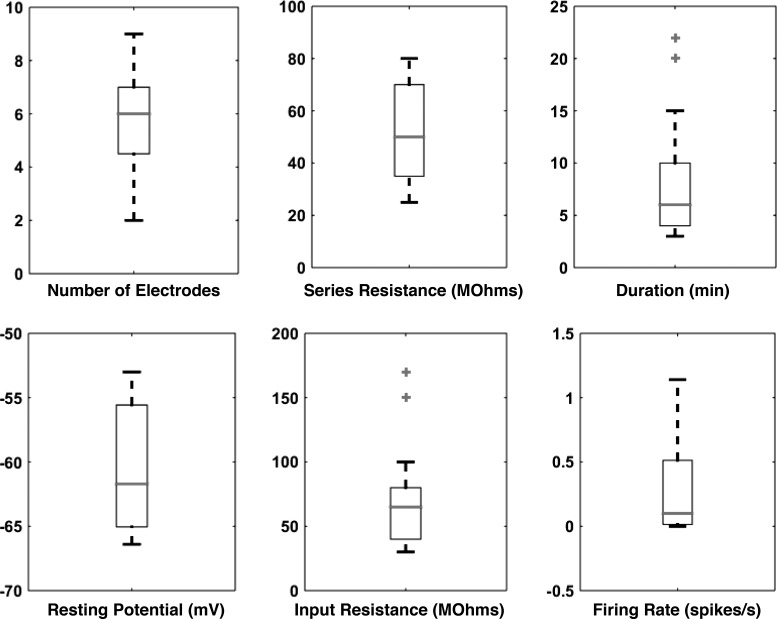
The recordings themselves were of good quality, comparable with those obtained by experienced electrophysiologists working manually, with series resistances that averaged 52 ± 4 MΩ. Four example recordings are shown in Fig. 8, and summary data are given in Fig. 9. Most neurons showed spontaneous spiking at a low (<1 Hz) but consistent rate, and recordings could be maintained even when animals ran on the wheel as fast as 30 cm/s (Fig. 8A, right). We were able to probe neurons not only with current steps (Fig. 8A, left), but also with more complex stimuli, such as a swept-sine (chirp) stimulus designed to characterize frequency preference (Fig. 8B). Recording durations averaged 8 ± 1 min. This is comparable with what has previously been obtained in some awake behaving mouse preparations (5–11 min; Domnisoru et al. 2013; Harvey et al. 2009; Schmidt-Hieber and Häusser 2013) but not in others (30 min; Polack et al. 2013; Zhou et al. 2014). We will return to this issue and what might be done about it in the discussion.
Fig. 8.
Four example recordings obtained in awake mice by the automated patch-clamp system. A: shown are the responses of 2 neurons to hyperpolarizing and depolarizing current steps and 5-s-long recordings of spontaneous activity in these same neurons as the mice ran at the speeds indicated. Note the recording stability despite animal locomotion. Vm, membrane potential. B: the frequency response of 2 neurons were probed by injecting a swept-sine current stimulus (black; 0–15 Hz over 15 s). The dark gray neuron responded as a low-pass filter and the lighter gray neuron as a band-pass (resonant) filter. This is seen more clearly by considering their impedance profiles (bottom), constructed by taking the ratio of the Fourier transform magnitudes of the responses and the stimulus.
Fig. 9.
Summary data. We obtained 19 whole cell patch recordings from neurons in awake mice using the automated system. The properties of the recordings (success rate, series resistance, and duration) and of the neurons (input resistance, Vm, and firing rate) are illustrated by the boxplots. Outliers are identified as data points >2.7 SD from the mean.
DISCUSSION
We have here introduced an automated system for obtaining whole cell patch-clamp recordings from awake mice running on a wheel. The system is written entirely in MATLAB and allows users to make use of the extensive collection of MATLAB toolboxes for signal and image processing, instrument control, and numerical analysis. Using this system, we obtained 19 recordings from prefrontal neurons in 8- to 9-wk-old mice (area M2, 400–700 μm below the cortical surface) with a success rate similar to that of experienced electrophysiologists.
Automated Patching
Automated patch-clamp systems are now standard for cellular electrophysiology experiments conducted on artificial cell lines, where maintaining a high throughput is essential, but have only recently been introduced for other preparations.
Working with acute brain slices, Perin and Markram (2013) developed a partially automated system that allows for multiple, simultaneous recordings from cortical neurons (up to 12 at a time). Their system moves electrodes into position near target neurons automatically but requires an experimenter to form gigaohm seals and rupture membranes manually. This is a reasonable strategy as patch recording in brain slices typically involves a fair amount of visual processing that would be difficult to capture in an algorithm. However, our pilot studies suggest that probing membranes using resistance measurements is a promising alternative that might make a fully automated slice system feasible.
Working with anesthetized mice, Kodandaramaiah and colleagues (2012) developed a blind patch-clamp system written in the LabVIEW programming environment that has received considerable, and well-deserved, attention. Their system achieved a success rate (percentage of attempts that resulted in recordings) of nearly 33%, which is considerably higher than our 17% rate. It seems reasonable to ascribe much of this disparity to the differences between an anesthetized, stationary mouse and an awake, locomoting one. Even so, it is intriguing to note that, in our experiments, an electrode that successfully reached the neuron hunting stage had a 30% (12 out of 40) chance of resulting in a whole cell recording, which is much closer to the 39% rate achieved by the Kodandaramaiah system under similar circumstances. That is, our electrodes were much more likely to clog before getting to the hunting stage than theirs were. This was probably because Kodandaramaiah and colleagues 1) used much a higher positive pressure (∼11 psi) to enter the cortex and move to the target area, and 2) removed the dura beforehand. Both of these measures could be incorporated into our protocols, although they have drawbacks: the possibility of damaging tissue and, in the case of dura removal, requiring that a mouse undergo a surgery the same day as the recording.
Hardware and Software
The system described here differs from conventional and other automated patch-clamp systems in ways that involve both hardware and software.
The two principal hardware advances are 1) our use of a USB camera and image-processing code to direct electrodes into craniotomies and 2) our design of an Arduino-based pressure regulator. The first hardware advance will likely be of interest even to electrophysiologists who do not wish to automate the patch-clamp process itself. At the price of a 5-min setup procedure, the system can thereafter direct the tip of a patch electrode to a point in space to within a sphere of radius ∼40 μm. The smallest craniotomies we used were ∼500 μm, but the system should certainly be useful, without modification or difficulty, even to researchers who use craniotomies of 100–200 μm. The system does require the purchase of a high-magnification, long-working-distance camera, but it obviates the need for a stereomicroscope, meaning that it does not add to fixed costs and may even reduce them. The second hardware advance, the pressure regulator, is not a conceptual or fundamental advance, as commercial regulators exist. Rather, in the spirit of the maker movement, it uses simple technology accessible to nonengineers to create a useful, low-cost device. Others have had similar ideas. Perin and Markram (2013) describe a clever system of pressure reservoirs switched by solenoid valves. However, our pressure regulator is distinguished by its flexibility: users can specify arbitrary pressure values with good temporal control through a GUI or by changing the numbers in a single file, and no daily setup procedure is required. (The documentation in the supplemental material describes how to build and tune the regulator and set pressure values.)
The principal software advance is that the system is written entirely in the scripted MATLAB language. For experimental biologists, MATLAB has many attractive features. 1) MATLAB is a procedural language written in text, consisting of a well-structured sequence of steps involving calls to statements, functions, and commands, which makes it similar to other procedural languages (e.g., C, IGOR Pro, and Pascal) that experimentalists are likely to have encountered. 2) MATLAB has support for object-oriented programming, which relates it to powerful and popular contemporary languages (e.g., C++, Java, and Python). 3) MATLAB has a large collection of toolboxes that provide simple means for users to control commercially available equipment (cameras, data acquisition devices, motors, signal generators, microcontrollers, and so forth) and to analyze signals and images using sophisticated algorithms. Finally, 4) MATLAB already has an enormous user base in the neuroscience community.
Success Rate and Duration
There are two areas in which the system might be improved: success rate and recording duration.
Our success rate was just over 17%. Published accounts indicate that blind patch experiments in vivo generally have success rates of 20–50% under good conditions (DeWeese 2007; Kodandaramaiah et al. 2012; Margrie et al. 2002). However, these numbers come from experiments performed on anesthetized animals, which suggests that our success rate with awake behaving animals is reasonable. Even so, there may be ways to improve it. One promising idea is to use heartbeat-associated pulsation of the current signal to determine when the electrode has encountered a neuron and seal formation should be attempted; it has been argued that the appearance of pulsation is a more reliable signature than resistance change (Margrie et al. 2002). A second idea is to monitor mouse locomotion during patching and pause the process if running speed exceeds some value, as brain movement associated with running might make seal formation more difficult (Fig. 7B). A third idea is, after encountering a neuron, to probe its cross-section by moving the electrode laterally while measuring resistance, as we did in the brain slice experiments, so as to identify a good position for patching.
Our durations averaged 8 min, with the longest being 22 min. This is long enough for many awake mouse experiments, as made clear by published studies that include data from recordings of as little as 5 min (Domnisoru et al. 2013; Harvey et al. 2009; Polack et al. 2013; Schmidt-Hieber and Häusser 2013). However, a short duration does constrain the kinds of experiments that can be done, and increasing recording duration is a chief aim of ongoing work. One simple improvement we have found is reducing the size of the craniotomy. Our earliest craniotomies were ∼1-mm diameter, whereas the later ones were ∼0.5 mm. Correspondingly, average recording duration increased over time from 5 min (1st 6 recordings) to 7 min (next 6) to 12 min (last 7). One thing that distinguishes studies in which durations of 30 min or more are observed is that patching in those studies is often done under visual guidance using 2-photon microscopy (Polack et al. 2013; Zhou et al. 2014). This suggests that optimal placement of the electrode against the neuron (near the center, with the tip just dimpling the surface) is important for recording stability. In the brain slice experiments, we found that the automated system could find this optimal position using resistance measurements alone. Doing so in vivo will be considerably more challenging because of increased movement, but that challenge will be borne by a computer algorithm rather than an experimenter. That fact, as well as anything, illustrates the potential power of automation in electrophysiology.
GRANTS
This study was supported by National Institutes of Health Grants MH-094839 and NS-084473 and the McKnight Foundation.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
N.S.D. and D.J. conception and design of research; N.S.D., J.J.S., W.T., and R.A.C. performed experiments; N.S.D. analyzed data; N.S.D., J.J.S., W.T., R.A.C., and D.J. interpreted results of experiments; N.S.D. prepared figures; N.S.D. drafted manuscript; N.S.D., J.J.S., W.T., R.A.C., and D.J. edited and revised manuscript; N.S.D., J.J.S., W.T., R.A.C., and D.J. approved final version of manuscript.
ENDNOTE
At the request of the author(s), readers are herein alerted to the fact that additional materials related to this manuscript may be found at the institutional Web site of one of the authors, which at the time of publication they indicate is: (https://clm.utexas.edu/robotpatch). These materials are not a part of this manuscript and have not undergone peer review by the American Physiological Society (APS). APS and the journal editors take no responsibility for these materials, for the Web site address, or for any links to or from it.
ACKNOWLEDGMENTS
We are grateful to members of the Johnston laboratory for useful discussions, Rick Gray for creating a Web site for this project, Betsy Walcott for reading the manuscript, David Tai for help with handling animals and preliminary investigations with the high-magnification camera, and Joe Steinmeyer for sharing his C++ library for controlling Sutter manipulators.
REFERENCES
- Abraham NM, Guerin D, Bhaukaurally K, Carleton A. Similar odor discrimination behavior in head-restrained and freely moving mice. PLoS One 7: e51789, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DL, Economides JR, Jocson CM, Parker JM, Horton JC. A watertight acrylic-free titanium recording chamber for electrophysiology in behaving monkeys. J Neurophysiol 106: 1581–1590, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andermann ML, Kerlin AM, Reid RC. Chronic cellular imaging of mouse visual cortex during operant behavior and passive viewing. Front Cell Neurosci 4: 3, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. Makers: The New Industrial Revolution. New York: Crown, 2014. [Google Scholar]
- Araci IE, Brisk P. Recent developments in microfluidic large scale integration. Curr Opin Biotechnol 25: 60–68, 2014. [DOI] [PubMed] [Google Scholar]
- Åström KJ, Murray RM. Feedback Systems: An Introduction for Scientists and Engineers. Princeton, NJ: Princeton Univ. Press, 2008. [Google Scholar]
- Brunborg G, Jackson P, Shaposhnikov S, Dahl H, Azqueta A, Collins AR, Gutzkow KB. High throughput sample processing and automated scoring. Front Genet 5: 373, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse L, Ayaz A, Dhruv NT, Katzner S, Saleem AB, Schölvinck ML, Zaharia AD, Carandini M. The detection of visual contrast in the behaving mouse. J Neurosci 31: 11351–11361, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettih SN, McDougle SD, Ruffolo LI, Medina JF. Adaptive timing of motor output in the mouse: the role of movement oscillations in eyelid conditioning. Front Integr Neurosci 5: 72, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crochet S. Intracellular whole-cell patch-clamp recordings of cortical neurons in awake head-restrained mice. Neuromethods 67: 219–235, 2011. [Google Scholar]
- Desai NS, Walcott EC. Synaptic bombardment modulates muscarinic effects in forelimb motor cortex. J Neurosci 26: 2215–2226, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWeese MR. Whole-cell recording in vivo. Curr Protoc Neurosci 6: 6–22., 2007. [DOI] [PubMed] [Google Scholar]
- Domnisoru C, Kinkhabwala AA, Tank DW. Membrane potential dynamics of grid cells. Nature 495: 199–204, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop J, Bowlby M, Peri R, Vasilyev D, Arias R. High-throughput electrophysiology: an emerging paradigm for ion-channel screening and physiology. Nat Rev Drug Discov 7: 358–368, 2008. [DOI] [PubMed] [Google Scholar]
- Dymecki SM, Kim JC. Molecular neuroanatomy's “three Gs”: a primer. Neuron 54: 17–34, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley K. Out in the open: the men supercharging neuroscience with open source hardware. Wired: March 10, 2014. [Google Scholar]
- Goldey GJ, Roumis DK, Glickfeld LL, Kerlin AM, Reid RC, Bonin V, Schafer DP, Andermann ML. Removable cranial windows for long-term imaging in awake mice. Nat Protoc 9: 2515–2538, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZV, Hires SA, Li N, O'Connor DH, Komiyama T, Ophir E, Huber D, Bonardi C, Morandell K, Gutnisky D, Peron S, Xu NL, Cox J, Svoboda K. Procedures for behavioral experiments in head-fixed mice. PLoS One 9: e88678, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CD, Collman F, Dombeck DA, Tank DW. Intracellular dynamics of hippocampal place cells during virtual navigation. Nature 46: 941–946, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusser M, Margrie TW. Two-photon targeted patching and electroporation in vivo. Cold Spring Harb Protoc 2014: 78–85, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiney SA, Wohl MP, Chettih SN, Ruffolo LI, Medina JF. Cerebellar-dependent expression of motor learning during eyeblink conditioning in head-fixed mice. J Neurosci 34: 14845–14853, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Histed MH, Carvalho LA, Maunsell JH. Psychophysical measurement of contrast sensitivity in the behaving mouse. J Neurophysiol 107: 758–765, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst G, Kodandaramaiah SB, Kolb I, Stoy W, Wickersham I, Singer A, Li L, Boyden ES, Zeng H, Forest CR. High-throughput fully-automated patch clamp robot for in-vivo electrophysiology and morphology. Society for Neuroscience, Washington, DC, November 18, 2014, no. 659.18/VV38. [Google Scholar]
- Kalmbach BE, Chitwood RA, Dembrow NC, Johnston D. Dendritic generation of mGluR-mediated slow afterdepolarization in layer 5 neurons of prefrontal cortex. J Neurosci 33: 13518–13532, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RD, Rowland J, Oliver SG, Young M, Aubrey W, Byrne E, Liakata M, Markham M, Pir P, Soldatova LN, Sparkes A, Whelan KE, Clare A. The automation of science. Science 324: 85–89, 2009. [DOI] [PubMed] [Google Scholar]
- Kodandaramaiah SB, Boyden ES, Forest CR. In vivo robotics: the automation of neuroscience and other intact-system biological fields. Ann NY Acad Sci 1305: 63–71, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodandaramaiah SB, Flores FJ, Franzesi GT, Singer AC, Holst G, Wickersham IR, Borgers C, Kopell NJ, Forest CR, Brown EN, Boyden ES. Automated multiple-cell patch clamp assessment of multineuron subthreshold dynamics in waking and anesthetized states. Society for Neuroscience, Washington, DC, November 18, 2014, no. 659.16/VV36. [Google Scholar]
- Kodandaramaiah SB, Franzesi GT, Chow BY, Boyden ES, Forest CR. Automated whole-cell patch-clamp electrophysiology of neurons in vivo. Nat Methods 9: 585–587, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama T, Sato TR, O'Connor DH, Zhang YX, Huber D, Hooks BM, Gabitto M, Svoboda K. Learning-related fine-scale specificity imaged in motor cortex circuits of behaving mice. Nature 464: 1182–1186, 2010. [DOI] [PubMed] [Google Scholar]
- Lee AK, Epsztein J, Brecht M. Whole-cell patch-clamp recordings in freely moving animals. Methods Mol Biol 1183: 263–276, 2014. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kwan AC, Zhang S, Phoumthipphavong V, Flannery JG, Masmanidis SC, Taniguchi H, Huang ZJ, Zhang F, Boyden ES, Deisseroth K, Dan Y. Activation of specific interneurons improves V1 feature selectivity and visual perception. Nature 488: 379–383, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron 37: 634–660, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margrie TW, Brecht M, Sakmann B. In vivo, low-resistance, whole-cell recordings from neurons in the anaesthetized and awake mammalian brain. Pflügers Arch 444: 491–498, 2002. [DOI] [PubMed] [Google Scholar]
- Morozov E. Making it. The New Yorker: January 13, 2014. [Google Scholar]
- O'Connor DH, Huber D, Svoboda K. Reverse engineering the mouse brain. Nature 461: 923–929, 2009. [DOI] [PubMed] [Google Scholar]
- Perin R, Markram H. A computer-assisted multi-electrode patch-clamp system. J Vis Exp 80: e50630, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CC, Crochet S. Synaptic computation and sensory processing in neocortical layer 2/3. Neuron 78: 28–48, 2013. [DOI] [PubMed] [Google Scholar]
- Polack PO, Friedman J, Golshani P. Cellular mechanisms of brain state-dependent gain modulation in visual cortex. Nat Neurosci 16: 1331–1339, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JI, Kepecs A. Choice ball: a response interface for two-choice psychometric discrimination in head-fixed mice. J Neurophysiol 108: 3416–3423, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Hieber C, Häusser M. Cellular mechanisms of spatial navigation in the medial entorhinal cortex. Nat Neurosci 16: 325–331, 2013. [DOI] [PubMed] [Google Scholar]
- Siegel JJ, Taylor W, Gray R, Kalmbach B, Zemelman BV, Desai NS, Johnston D, Chitwood RA. Trace eyeblink conditioning in mice is dependent upon the dorsal medial prefrontal cortex, cerebellum and amygdala: behavioral characterization and functional circuitry. Eneuro. First published July 2, 2015; doi: 10.1523/eneuro.0051-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughterbeck CR, Datta S, Smith S, Ritchie D, Wohnoutka P. High-throughput automated microscopy platform for the Allen brain atlas. J Lab Autom 12: 377–383, 2007. [Google Scholar]
- Steinmeyer JD, Yanik MF. High-throughput single-cell manipulation in brain tissue. PLoS One 7: e35603, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teikari P, Najjar RP, Malkki H, Knoblauch K, Dumortier D, Gronfier C, Cooper HM. An inexpensive Arduino-based LED stimulator system for vision research. J Neurosci Methods 211: 227–236, 2012. [DOI] [PubMed] [Google Scholar]
- Wallisch P, Lusignan ME, Benayoun MD, Baker TI, Dickey AS, Hatsopoulos NG. MATLAB for Neuroscientists. Waltham, MA: Academic Press, 2013. [Google Scholar]
- Zhou M, Liang F, Xiong XR, Li L, Li H, Xiao Z, Tao HW, Zhang LI. Scaling down of balanced excitation and inhibition by active behavioral states in auditory cortex. Nat Neurosci 17: 841–850, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci 16: 264–266, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]