Abstract
AIM: To assess the associations of human leukocyte antigen (HLA) class II DQB1*0301 and/or DRB1*1101 allele with spontaneous hepatitis C virus (HCV) clearance by meta-analysis of individual dataset from all studies published till date.
METHODS: To clarify the impact of HLA class II polymorphisms on viral clearance, we performed a meta-analysis of the published data from 11 studies comparing the frequencies of DQB1*0301 and DRB1*1101 alleles in individuals with spontaneous resolution to those with persistent infection. As we identified the heterogeneity between studies, summary statistical data were calculated based on a random-effect model.
RESULTS: Meta-analyses yielded summary estimates-odds ratio (OR) of 2.36 [95%CI (1.62, 3.43), P<0.00001] and 2.02 [95%CI (1.56, 2.62), P<0.00001] for the effects of DQB1*0301 and DRB1*1101 alleles on spontaneous clearance of HCV, respectively.
CONCLUSION: These results support the hypothesis that specific HLA class II alleles might influence the susceptibility or resistance to persistent HCV infection. Both DQB1*0301 and DRB1*1101 are protective alleles and present HCV epitopes more effectively to CD4+T lymphocytes than others, and subjects with these two alleles are at a lower risk of developing chronic HCV infection. Large, multi-ethnic confirmatory and well-designed studies are needed to determine the host genetic determinants of HCV infection.
Keywords: Human leukocyte antigen, Genetic polymorphism, DQB1*0301, DRB1*1101, Hepatitis C virus, Spontaneous clearance, Meta-analysis
INTRODUCTION
HCV infection presents with diverse clinical manifestations. Approximately 20% of infected patients successfully eliminate the virus, whereas the majority of patients develop chronic infection with a wide spectrum of liver lesions, ranging from minimal inflammation to cirrhosis and hepatocelluar carcinoma (HCC)[1]. The mechanisms underlying the spontaneous viral clearance or development of chronic HCV infection have not yet been identified[2]. Apart from viral characteristics (viral genotype, quasi-species distribution and viral load), it is generally accepted that cellular immune responses play an important role in viral clearance and disease resolution[3]. Clearance of the virus during acute infection has been shown to be associated with strong and sustained HCV-specific CD4+ and CD8+ T-cell responses[4-8]. Though CD8+ T cells are primary effector cells directly eliminating HCV-infected cells, CD4+ T cells play a central regulatory role, which modulates the CD8+ T-cell responses and is crucial for the production of neutralizing antibodies. A number of recent studies have suggested that early sustaining vigorous and multi-specific CD4+ T-cell responses might be a critical determinant for the primary HCV infection[9,10]. The importance to CD4+ T-cell response is the presentation of HCV antigens in the context of HLA class II molecules. Therefore, the differences in HLA class II alleles may strongly influence the clinical course of hepatitis C.
HLA genes located on chromosome 6 encode peptides involved in host immune response. Moreover, the HLA loci display an unprecedented degree of diversity in human genome, presumably an evolutionary adoption to immune pressure from various infectious agents. Polymorphisms in HLA class II molecules can give rise to amino acid substitutions with different peptide-binding characteristics and determine antigenic specificities, the strength of immune response to HCV[11,12]. Consequently, distinct HLA class II alleles may be expected to exert an impact on the development of host immune responses against HCV infection. However, few studies have demonstrated consistency in different populations. In several Irish studies performed in women exposed to HCV genotype 1b from a single inoculum, DRB1*0101 and DQB1*0501 alleles are found to be associated with viral elimination[13-17]. One German study suggests that HLA-DR15 (B1*15011) might constitute an important genetic factor for the clearance of HCV[18]. Thio et al[19] reported that HLA-A*1101, HLA-B*57, and HLA-Cw*0102 contribute to the spontaneous resolution of primary HCV infection. It was reported that the DQA1*03 and DQB1*0302 alleles promote viral clearance and confer protection against chronic HCV infection in Caucasians[20]. A number of studies have been conducted in different ethnic populations to investigate the associations of DQB1*0301 or DRB1*1101 allele with spontaneous HCV clearance[13,21-30]. However, the results from these molecular epidemiological studies are confusing rather than being conclusive. Single study might be underpowered to detect the association between DQB1*0301 and/or DRB1*1101 allele and HCV infection because of the limited sample size. The aim of this study was to assess the associations of HLA class II DQB1*0301 and/or DRB1*1101 allele(s) with spontaneous HCV clearance by meta-analysis of individual dataset from all studies published till date.
MATERIALS AND METHODS
Identification and eligibility of studies
PubMed was used to search for relevant reports published between 1997 and 2004 (using the key words: human leukocyte antigen, genetic polymorphism, hepatitis C virus, spontaneous clearance). The inclusion criteria for our analysis were as follows: studies reporting odds ratio (OR) or risk ratio (RR) calculated by comparing those with self-limiting infection to those with chronic infection as a measure of association; study designs such as cohort study, population-based case control study, hospital-based case-control study; studies using HLA class II molecular genotyping technique; studies including DQB1*0301 and DRB1*1101 alleles. A total of 11 candidate papers were identified.
Data extraction
Data were collected on the DQB1*0301 and DRB1*1101 allele frequencies, authors, journals, years of publication, country of origin, demographics, selection, definition of spontaneous viral clearance (anti-HCV positive and HCV-RNA negative at least for 6 mo), chronically persistent infection (anti-HCV positive and HCV-RNA positive for more than 1 year) and racial descents (categorized as Asian, European, and North American descents). Furthermore, we examined whether matching was used and HLA class II molecular-typing assay was validated.
Statistical analysis
The OR of sustained HCV clearance associated with HLA class II polymorphisms (DQB1*0301 and DRB1*1101 alleles) was estimated for each study. The primary analysis was to estimate the allele frequencies between the groups with viral elimination and chronic infection. For each genetic group, we estimated the heterogeneity between the studies[31]. Data were combined using both fixed effect (Mantel-Haenszel) and random effect (DerSimonian and Laird) models[32]. In the absence of heterogeneity, the two methods provided identical results. Random effects were more appropriate, when heterogeneity was present. Analyses were performed by the software of SAS 8.0 (SAS Institute, Cary, NC, USA) and Review Manager 4.2 (Cochrance Collaboration, Oxford, UK). All P values were two-sided.
RESULTS
Characteristics of the studies are shown in Table 1. Multiple studies were conducted in the European population (n = 9). The number of individuals with spontaneous resolution and persistent infection in DRB1*1101 and DQB1*0301 studies was 706 and 1 524, 714 and 1 497, respectively. All the subjects of these studies were selected based on a histological diagnosis of strict criteria. With two exceptions[24,25] where the mean age of two groups differed and one where the age status was not stated[26], all other studies were matched for age. Most studies also stated the use of matching on sex status except for two[27,30]. Compared to other studies, only one was homogenous in terms of gender, source of HCV infection, HCV-serotype and ethnicity[13]. This subject population was derived from a cohort of females inoculated with anti-D immunoglobulin in 1977 that was contaminated with HCV from a single source.
Table 1.
Characteristics of studies included in the meta-analysis
| First author (year) [reference] | Country (ethnicity) | Spontaneous resolution | Persistent infection | Matching | Anti-HCV tests | HLA typing |
| Alric (1997) | France (European) | 25, M/F: 9/16 | 103, M/F: 58/45 | Sex, age, source of HCV infection, HCV-serotype | 2G EIA and RIBA | PCR-SSOP |
| [21] | Age: 40.6±15.7 yr | Age: 45.4±12.4 yr | ||||
| Cramp (1998) | UK (European) | 49, M/F: 30/19 | 55, M/F: 31/24 | Sex, age, source of HCV infection and duration | 2G line immunoassay | PCR-SSOP |
| [22] | Duration: 15.5 (3-42) yr | Duration: 14.2 (2-40) yr | ||||
| Minton (1998) | UK (European) | 35, M/F: 19/16 | 138, M/F: 87/51 | Sex, age, source of HCV infection | 2G ELISA and RIBA | PCR-SSOP |
| [23] | Age: 37.9±10.8 yr | Age: 37.2±10.1 yr | ||||
| Mangia (1999) | Italy (European) | 35 | 149 | Sex, HCV-serotype, not age, | RIBA and 3G EIA | PCR-SSP |
| [24] | not duration | |||||
| Thursz (1999) | European | 85, M/F: 37/48 | 170, M/F: 74/96 | Sex, center, not age | ELISA and RIBA | PCR-SSP |
| [25] | Age: 45±14 yr | Age: 50±16 yr | ||||
| Vejbaesya (2000) | Thailand (Asian) | 43 Blood donor | 57 | Sex | 2G EIA and RIBA | PCR-SSOP |
| [26] | M/F: 25/18 | M/F: 31/18 | ||||
| Alric (2000) | France (European) | 63, M/F: 21/42 | 282, M/F: 150/132 | Age, source of HCV infection and duration, not sex | 2G EIA and RIBA | PCR-SSOP |
| [27] | Age: 42.1±15.4 yr | Age: 46±12.3 yr | ||||
| Fanning (2000) | Irish (European) | 85 Female | 72 Female | From single source | RIBA | Reverse line probe hybridization |
| [13] | ||||||
| Thio (2001) | North America | 200, M/F: 166/34 | 374, M/F: 310/64 | Age, sex, race | 2G EIA and RIBA | PCR-SSP PCR-SSCP |
| [28] | Age: 25.7 yr | Age: 27.8 yr | ||||
| Azocar (2003) | Hispanic (European) | 40,M/F: 33/7 | 72, M/F: 54/18 | Age, sex | EIA and RIBA | PCR-SSOP PCR-SSP |
| [29] | Age: 37.9 yr | Age: 39.2 yr | ||||
| Spada (2004) | Italy (European) | 10, M/F: 5/5 | 24,M/F:22/2 | Not sex, age, source of HCV infection, HCV-serotype | 3G ELISA and RIBA | PCR-SSP |
| [30] | Age: 40.5 (20-61) yr | Age: 29 (20-56) yr |
EIA: enzyme-immunoassay; RIBA: recombinant immunoblot assay; ELISA: enzyme-linked-immunosorbent assay; 2G: second generation; 3G: third generation; SSOP: sequence-specific oligonucleotide probes; SSP: sequence specific primers; SSCP: single stranded conformational polymorphisms.
With regard to the anti-HCV antibody testing, most studies used a combination of two tests, while only one used a single test[13]. Five studies used low resolution molecular typing (PCR-SSOP) for HLA, while others used high resolution molecular typing (PCR-SSP) for HLA[33]. Low resolution molecular typing methods for HLA could not identify the specific alleles. Accurate methods for HLA class II typing should involve the combination of PCR-SSOP, PCR-SSP, and PCR-SSCP[12].
Table 2 shows the OR on HCV spontaneous clearance associated with DRB1*1101 allele. Overall, individuals with DRB1*1101 allele had a significantly reduced risk for chronic hepatitis C compared to individuals without it [OR = 2.02, 95%CI (1.56, 2.62) (P<0.00001)]. Similarly, subjects with DQB1*0301 allele appeared to be more likely to clear the virus, while those lacking it were more prone to develop chronic infections [OR = 2.36, 95%CI (1.62, 3.43), P<0.00001] (Table 3). These analyses were based on the data from 11 studies regardless of ethnics.
Table 2.
Effect of DRB1*1101 allele on self-limiting HCV infection
| Study | Spontaneous resolution n/N | Persistent infection n/N | OR (random) 95%CI |
| Alric | 10/25 | 10/102 | 6.13 (2.18, 17.22) |
| Minton | 11/35 | 11/135 | 5.17 (2.01, 13.27) |
| Cramp | 9/49 | 6/55 | 1.84 (0.60, 5.60) |
| Mangia | 7/35 | 24/149 | 1.30 (0.51, 3.32) |
| Thursz 1 | 26/85 | 29/170 | 2.14 (1.16, 3.94) |
| Thursz 2 | 14/57 | 15/152 | 2.97 (1.33, 6.65) |
| Fanning | 4/68 | 4/64 | 0.94 (0.22, 3.92) |
| Alric | 20/59 | 25/170 | 2.97 (1.50, 5.91) |
| Vejbaesya | 2/43 | 3/57 | 0.88 (0.14, 5.50) |
| Thio | 15/200 | 24/374 | 1.18 (0.61, 2.31) |
| Azocar | 4/40 | 11/72 | 0.62 (0.18, 2.08) |
| Spada | 2/10 | 2/24 | 2.75 (0.33, 22.92) |
| Total (95%CI) | 124/706 | 164/1 524 | 2.02 (1.56, 2.62) |
Test for heterogeneity: χ2 = 19.38, df = 11 (P<0.05), I2 = 43.2%. Test for overall effect: Z = 5.30 (P<0.00001).
Table 3.
Effect of DQB1*0301 allele on self-limiting HCV infection
| Study | Spontaneous resolution n/N | Persistent infection n/N | OR (random) 95%CI |
| Alric | 21/25 | 28/91 | 11.81 (3.71, 37.61) |
| Minton | 18/35 | 33/135 | 3.27 (1.51, 7.07) |
| Cramp | 26/49 | 10/55 | 5.09 (2.10, 12.33) |
| Mangia | 17/33 | 42/143 | 2.56 (1.18, 5.53) |
| Thursz 1 | 39/85 | 47/170 | 2.22 (1.29, 3.82) |
| Thursz 2 | 25/57 | 37/152 | 2.43 (1.28, 4.61) |
| Fanning | 25/78 | 18/67 | 1.28 (0.63, 2.64) |
| Alric | 38/59 | 45/157 | 4.50 (2.39, 8.50) |
| Vejbaesya | 24/43 | 18/57 | 2.74 (1.20, 6.22) |
| Thio | 49/200 | 71/374 | 1.38 (0.92, 2.09) |
| Azocar | 6/40 | 13/72 | 0.80 (0.28, 2.30) |
| Spada | 4/10 | 15/24 | 0.40 (0.09, 1.81) |
| Total (95%CI) | 292/714 | 377/1 497 | 2.36 (1.62, 3.43) |
Test for heterogeneity: χ2 = 33.33, df = 11 (P = 0.0005), I2 = 67.0%. Test for overall effect: Z = 4.48 (P<0.00001).
We performed tests for homogeneity with respect to the studies of DRB1*1101 and DQB1*0301 alleles and viral clearance and obtained statistically significant results (DRB1*1101: χ2 = 19.38, P<0.05; DQB1*0301: χ2 = 33.33, P = 0.0005). Therefore, summary statistics were calculated based on a random-effect model.
Figure 1 shows the funnel plot analysis to detect publication bias of each study for DRB1*1101 and DQB1*0301, respectively. The shape of the funnel plot seemed to be asymmetrical, suggesting that publication bias might affect the findings of our meta-analysis. Furthermore, an Egger's test was used to provide statistical evidence for funnel plot symmetry[45]. In the linear regression analysis, the intercept value provided a measure of asymmetry, the larger the deviation the more pronounced the asymmetry. The intercept value in our study was 2.33 for DQB1*0301 and 2.03 for DRB1*1101, respectively, both of them were significantly deviated from zero (P<0.0001 for DQB1*0301 and P<0.0022 for DRB1*1101).
Figure 1.
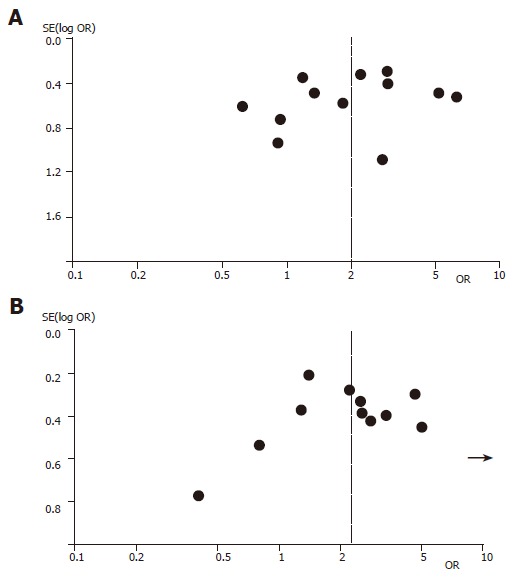
Funnel plot analysis of publication bias for DRB1*1101 allele (A) and DQB1*0301 allele (B). Each data point represents a separate study for the indicated association.
DISCUSSION
DQB1*0301 allele is reproducibly involved in spontaneous viral clearance of HCV in different populations[21-26,28,34-36]. Studies conducted in patients from France[21], Britain[22], Italy[24,35] with different genetic backgrounds have identified significant genetic associations with different DQB1*0301-bearing haplotypes, suggesting that DQB1*0301 might be one of the most prominent factors in HCV clearance[37,38].
Another allele correlated with self-limiting HCV is DRB1*1101[21,23,25,27,34,39]. But disagreement exists over the allele. In Italy[24,35] BRB1*1104 correlates with HCV clearance, while in Japan[40] DR13 is linked to self-limiting infection.
It is difficult to determine whether HLA-DRB1*1101 or DQB1*0301 allele is the more relevant factor as HLA-DRB1*11 is associated with DQB1*0301. Some studies found that DRB1*1101 rather than DQB1*0301 is closely associated with viral elimination[23,30,41,42]. While some findings suggest that DQB1*0301 is dominant in determining the outcome of HCV infection[21,22,27,43,44]. In our meta-analysis, the OR for the two alleles was virtually identical, but which one is responsible for the viral elimination is not clear.
The outcome of HCV infection varies substantially among the individuals. Though the exact mechanisms involved in viral clearance have not been fully elucidated, there is evidence that cellular immune response, especially T-helper lymphocyte responses to HCV, plays an important role in the control of persistent HCV infection. DQB1*0301, DRB1*1101 alleles are closely associated with self-limiting HCV infection. Our meta-analysis demonstrated that subjects with HLA-DRB1*1101 allele reduced 102% risk of developing chronic HCV infection and those carrying HLA-DQB1*0301 allele reduced 136% risk of persistent infection. HLA-DQB1*0301 and DRB1*1101 alleles might act as the most prominent factors favoring the elimination of HCV and conferring protection against chronic evolution.
There are two possible explanations for these observations. (1) Because HCV is not cytopathic for infected cells, the immune response may play an important role in HCV infection. Binding to HCV peptides of HLA molecules is the first step for the initiation of this immune response. Due to the extensive polymorphisms of HLA, the strength of the response may vary among the individuals. It is assumed that DQB1*0301 and DRB1*1101 alleles may present HCV epitopes more effectively to CD4+ Th cells than others, resulting in vigorous proliferative response to HCV and probably disease recovery[37,43,46-47]. CD4+ Th responses have been characterized in patients with self-limiting infection and most immunodominant epitopes are present in the context of DQB1*0301 and DRB1*1101 molecules[48]. Therefore, individuals with these two alleles may present peptides derived from HCV in a more efficient manner than those without, thus clearing the infection spontaneously. (2) Another explanation for the association between the two alleles and viral elimination may be due to the other protective HLA loci[22,36]. Interpreting results of genetic studies can be complicated by linkage disequilibrium of the implicated gene linked to the true gene of interest[12]. Other candidate susceptible genes located in the HLA class II region also require antigen presentation. An excess of DQB1*0301 and DRB1*1101-restricted peptide-specific responses in individuals, who clear the virus, needs to be shown in order to prove conclusively whether the mechanism of the gene effect is direct or indirect.
While there are some consistent observations on DQB1*0301 and DRB1*1101 with self-limiting infection, many results are not uniform. These inconsistencies may be due to ethnic differences, patient selection, sample size, HCV-serotype, and HLA typing technique. Firstly, a primary cause for the difference in results by different authors may be related to the great variability of the frequency of HLA alleles in different populations. It is quite possible that one ethnic group may have the preferential use of different allele(s) in viral eradication compared to other ethnic groups. This DRB1*1101 association with viral clearance was not observed in the Irish anti-D population, which might be explained by the lower frequency of DRB1*1101 in Irish[16] (6.4%) than in French[21] control populations (25.2%). The association of DRB1*0101 and DQB1*0501 alleles with viral clearance has been documented in Caucasian Americans, whereas in African–American patients viral clearance is mainly associated with DQB1*0301[28]. Another study showed that there are marked differences in the frequency of viral clearance in Caucasians and African-Americans[49]. Racial heterogeneity has to be taken into account in the future studies[50,51]. Secondly, many studies were conducted on relatively smaller samples. Insufficient number of individuals might decrease the power to detect a difference in the distribution of DQB1*0301 and DRB1*1101 alleles between patients with resolving infection and those with chronic evolution, though a true difference exists[52]. Lack of an association may not mean that associations do not exist. Thirdly, a number of HLA studies concerning the association between HLA class II and outcome of infection have compared allele frequency in HCV patients to that in healthy controls[20,53,54]. Studies including both HCV RNA positive and negative patients have failed to discover particular genetic associations[20]. Fourthly, effects of interactions in other environmental/behavioral and/or viral factors may be inevitable. There is a clear correlation between the HLA haplotype of individuals and outcome of HCV infection[55]. A complex interplay between various genes is likely to modulate the anti-HCV response rather than a single allele. Moreover, several variables including young age at infection and female sex have been suggested to determine HCV clearance[17,30,56]. Another possible reason for the discrepancy between these studies lies in the viral genotype.
In conclusion, large and well-designed studies should be carried out to investigate the host genetic determinants for HCV infection. Finding an association between specific alleles and favorable clinical outcomes in HCV patients might open new avenues to explore and understand the pathogenesis of HCV.
Footnotes
Supported by the National Natural Science Foundation of China, No. 30200232
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
References
- 1.Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36:S35–S46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 2.Sun J, Li K, Shata MT, Chan TS. The immunologic basis for hepatitis C infection. Curr Opin Gastroenterol. 2004;20:598–602. doi: 10.1097/00001574-200411000-00016. [DOI] [PubMed] [Google Scholar]
- 3.Neumann-Haefelin C, Blum HE, Chisari FV, Thimme R. T cell response in hepatitis C virus infection. J Clin Virol. 2005;32:75–85. doi: 10.1016/j.jcv.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Grüner NH, Gerlach TJ, Jung MC, Diepolder HM, Schirren CA, Schraut WW, Hoffmann R, Zachoval R, Santantonio T, Cucchiarini M, et al. Association of hepatitis C virus-specific CD8+ T cells with viral clearance in acute hepatitis C. J Infect Dis. 2000;181:1528–1536. doi: 10.1086/315450. [DOI] [PubMed] [Google Scholar]
- 5.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, Robbins G, Phillips R, Klenerman P, Walker BD. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thimme R, Bukh J, Spangenberg HC, Wieland S, Pemberton J, Steiger C, Govindarajan S, Purcell RH, Chisari FV. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci USA. 2002;99:15661–15668. doi: 10.1073/pnas.202608299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper S, Erickson AL, Adams EJ, Kansopon J, Weiner AJ, Chien DY, Houghton M, Parham P, Walker CM. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 9.Eckels DD, Wang H, Bian TH, Tabatabai N, Gill JC. Immunobiology of hepatitis C virus (HCV) infection: the role of CD4 T cells in HCV infection. Immunol Rev. 2000;174:90–97. doi: 10.1034/j.1600-0528.2002.017403.x. [DOI] [PubMed] [Google Scholar]
- 10.Wertheimer AM, Miner C, Lewinsohn DM, Sasaki AW, Kaufman E, Rosen HR. Novel CD4+ and CD8+ T-cell determinants within the NS3 protein in subjects with spontaneously resolved HCV infection. Hepatology. 2003;37:577–589. doi: 10.1053/jhep.2003.50115. [DOI] [PubMed] [Google Scholar]
- 11.de Andrade DR, de Andrade DR. The influence of the human genome on chronic viral hepatitis outcome. Rev Inst Med Trop Sao Paulo. 2004;46:119–126. [PubMed] [Google Scholar]
- 12.Thio CL, Thomas DL, Carrington M. Chronic viral hepatitis and the human genome. Hepatology. 2000;31:819–827. doi: 10.1053/he.2000.4316. [DOI] [PubMed] [Google Scholar]
- 13.Fanning LJ, Levis J, Kenny-Walsh E, Wynne F, Whelton M, Shanahan F. Viral clearance in hepatitis C (1b) infection: relationship with human leukocyte antigen class II in a homogeneous population. Hepatology. 2000;31:1334–1337. doi: 10.1053/jhep.2000.7437. [DOI] [PubMed] [Google Scholar]
- 14.McKiernan SM, Hagan R, Curry M, McDonald GS, Kelly A, Nolan N, Walsh A, Hegarty J, Lawlor E, Kelleher D. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 15.McKiernan SM, Hagan R, Curry M, McDonald GS, Nolan N, Crowley J, Hegarty J, Lawlor E, Kelleher D. The MHC is a major determinant of viral status, but not fibrotic stage, in individuals infected with hepatitis C. Gastroenterology. 2000;118:1124–1130. doi: 10.1016/s0016-5085(00)70365-9. [DOI] [PubMed] [Google Scholar]
- 16.Barrett S, Ryan E, Crowe J. Association of the HLA-DRB1*01 allele with spontaneous viral clearance in an Irish cohort infected with hepatitis C virus via contaminated anti-D immunoglobulin. J Hepatol. 1999;30:979–983. doi: 10.1016/s0168-8278(99)80249-9. [DOI] [PubMed] [Google Scholar]
- 17.Barrett S, Goh J, Coughlan B, Ryan E, Stewart S, Cockram A, O'Keane JC, Crowe J. The natural course of hepatitis C virus infection after 22 years in a unique homogenous cohort: spontaneous viral clearance and chronic HCV infection. Gut. 2001;49:423–430. doi: 10.1136/gut.49.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lechmann M, Schneider EM, Giers G, Kaiser R, Dumoulin FL, Sauerbruch T, Spengler U. Increased frequency of the HLA-DR15 (B1*15011) allele in German patients with self-limited hepatitis C virus infection. Eur J Clin Invest. 1999;29:337–343. doi: 10.1046/j.1365-2362.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- 19.Thio CL, Gao X, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, O'Brien SJ, Karacki P, Astemborski J, Carrington M, et al. HLA-Cw*04 and hepatitis C virus persistence. J Virol. 2002;76:4792–4797. doi: 10.1128/JVI.76.10.4792-4797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tibbs C, Donaldson P, Underhill J, Thomson L, Manabe K, Williams R. Evidence that the HLA DQA1*03 allele confers protection from chronic HCV-infection in Northern European Caucasoids. Hepatology. 1996;24:1342–1345. doi: 10.1053/jhep.1996.v24.pm0008938158. [DOI] [PubMed] [Google Scholar]
- 21.Alric L, Fort M, Izopet J, Vinel JP, Charlet JP, Selves J, Puel J, Pascal JP, Duffaut M, Abbal M. Genes of the major histocompatibility complex class II influence the outcome of hepatitis C virus infection. Gastroenterology. 1997;113:1675–1681. doi: 10.1053/gast.1997.v113.pm9352872. [DOI] [PubMed] [Google Scholar]
- 22.Cramp ME, Carucci P, Underhill J, Naoumov NV, Williams R, Donaldson PT. Association between HLA class II genotype and spontaneous clearance of hepatitis C viraemia. J Hepatol. 1998;29:207–213. doi: 10.1016/s0168-8278(98)80005-6. [DOI] [PubMed] [Google Scholar]
- 23.Minton EJ, Smillie D, Neal KR, Irving WL, Underwood JC, James V. Association between MHC class II alleles and clearance of circulating hepatitis C virus. Members of the Trent Hepatitis C Virus Study Group. J Infect Dis. 1998;178:39–44. doi: 10.1086/515599. [DOI] [PubMed] [Google Scholar]
- 24.Mangia A, Gentile R, Cascavilla I, Margaglione M, Villani MR, Stella F, Modola G, Agostiano V, Gaudiano C, Andriulli A. HLA class II favors clearance of HCV infection and progression of the chronic liver damage. J Hepatol. 1999;30:984–989. doi: 10.1016/s0168-8278(99)80250-5. [DOI] [PubMed] [Google Scholar]
- 25.Thursz M, Yallop R, Goldin R, Trepo C, Thomas HC. Influence of MHC class II genotype on outcome of infection with hepatitis C virus. The HENCORE group. Hepatitis C European Network for Cooperative Research. Lancet. 1999;354:2119–2124 DOI : 10.1016/S0140-6736(99)91443-5. doi: 10.1016/s0140-6736(99)91443-5. [DOI] [PubMed] [Google Scholar]
- 26.Vejbaesya S, Songsivilai S, Tanwandee T, Rachaibun S, Chantangpol R, Dharakul T. HLA association with hepatitis C virus infection. Hum Immunol. 2000;61:348–353. doi: 10.1016/s0198-8859(99)00131-7. [DOI] [PubMed] [Google Scholar]
- 27.Alric L, Fort M, Izopet J, Vinel JP, Bureau C, Sandre K, Charlet JP, Beraud M, Abbal M, Duffaut M. Study of host- and virus-related factors associated with spontaneous hepatitis C virus clearance. Tissue Antigens. 2000;56:154–158. doi: 10.1034/j.1399-0039.2000.560207.x. [DOI] [PubMed] [Google Scholar]
- 28.Thio CL, Thomas DL, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, O'Brien SJ, Karacki P, Marti D, Astemborski J, et al. Racial differences in HLA class II associations with hepatitis C virus outcomes. J Infect Dis. 2001;184:16–21. doi: 10.1086/321005. [DOI] [PubMed] [Google Scholar]
- 29.Azocar J, Clavijo OP, Yunis EJ. MHC class II genes in HCV viral clearance of hepatitis C infected Hispanic patients. Hum Immunol. 2003;64:99–102. doi: 10.1016/s0198-8859(02)00722-x. [DOI] [PubMed] [Google Scholar]
- 30.Spada E, Mele A, Berton A, Ruggeri L, Ferrigno L, Garbuglia AR, Perrone MP, Girelli G, Del Porto P, Piccolella E, et al. Multispecific T cell response and negative HCV RNA tests during acute HCV infection are early prognostic factors of spontaneous clearance. Gut. 2004;53:1673–1681. doi: 10.1136/gut.2003.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 32.Petitti D. B. Meta-analysis, decision analysis, and cost-effectiveness analysis. New York: Oxford University Press; 1994. [Google Scholar]
- 33.Welsh K, Bunce M. Molecular typing for the MHC with PCR-SSP. Rev Immunogenet. 1999;1:157–176. [PubMed] [Google Scholar]
- 34.Yenigün A, Durupinar B. Decreased frequency of the HLA-DRB1*11 allele in patients with chronic hepatitis C virus infection. J Virol. 2002;76:1787–1789. doi: 10.1128/JVI.76.4.1787-1789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zavaglia C, Martinetti M, Silini E, Bottelli R, Daielli C, Asti M, Airoldi A, Salvaneschi L, Mondelli MU, Ideo G. Association between HLA class II alleles and protection from or susceptibility to chronic hepatitis C. J Hepatol. 1998;28:1–7. doi: 10.1016/s0168-8278(98)80195-5. [DOI] [PubMed] [Google Scholar]
- 36.Wawrzynowicz-Syczewska M, Underhill JA, Clare MA, Boron-Kaczmarska A, McFarlane IG, Donaldson PT. HLA class II genotypes associated with chronic hepatitis C virus infection and response to alpha-interferon treatment in Poland. Liver. 2000;20:234–239. doi: 10.1034/j.1600-0676.2000.020003234.x. [DOI] [PubMed] [Google Scholar]
- 37.Donaldson PT. The interrelationship between hepatitis C virus and HLA. Eur J Clin Invest. 1999;29:280–283. doi: 10.1046/j.1365-2362.1999.00465.x. [DOI] [PubMed] [Google Scholar]
- 38.Thursz M. MHC and the viral hepatitides. QJM. 2001;94:287–291. doi: 10.1093/qjmed/94.6.287. [DOI] [PubMed] [Google Scholar]
- 39.Tillmann HL, Chen DF, Trautwein C, Kliem V, Grundey A, Berning-Haag A, Böker K, Kubicka S, Pastucha L, Stangel W, et al. Low frequency of HLA-DRB1*11 in hepatitis C virus induced end stage liver disease. Gut. 2001;48:714–718. doi: 10.1136/gut.48.5.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuzushita N, Hayashi N, Moribe T, Katayama K, Kanto T, Nakatani S, Kaneshige T, Tatsumi T, Ito A, Mochizuki K, et al. Influence of HLA haplotypes on the clinical courses of individuals infected with hepatitis C virus. Hepatology. 1998;27:240–244. doi: 10.1002/hep.510270136. [DOI] [PubMed] [Google Scholar]
- 41.Amoroso A, Berrino M, Canale L, Cornaglia M, Guarrera S, Mazzola G, Savoldi S, Scolari F, Sällberg M, Clementi M, et al. Are HLA class II and immunoglobulin constant region genes involved in the pathogenesis of mixed cryoglobulinemia type II after hepatitis C virus infection? J Hepatol. 1998;29:36–44. doi: 10.1016/s0168-8278(98)80176-1. [DOI] [PubMed] [Google Scholar]
- 42.Harcourt GC, Lucas M, Sheridan I, Barnes E, Phillips R, Klenerman P. Longitudinal mapping of protective CD4+ T cell responses against HCV: analysis of fluctuating dominant and subdominant HLA-DR11 restricted epitopes. J Viral Hepat. 2004;11:324–331. doi: 10.1111/j.1365-2893.2004.00516.x. [DOI] [PubMed] [Google Scholar]
- 43.Harcourt G, Hellier S, Bunce M, Satsangi J, Collier J, Chapman R, Phillips R, Klenerman P. Effect of HLA class II genotype on T helper lymphocyte responses and viral control in hepatitis C virus infection. J Viral Hepat. 2001;8:174–179. doi: 10.1046/j.1365-2893.2001.00289.x. [DOI] [PubMed] [Google Scholar]
- 44.De Re V, Caggiari L, Talamini R, Crovatto M, De Vita S, Mazzaro C, Cannizzaro R, Dolcetti R, Boiocchi M. Hepatitis C virus-related hepatocellular carcinoma and B-cell lymphoma patients show a different profile of major histocompatibility complex class II alleles. Hum Immunol. 2004;65:1397–1404. doi: 10.1016/j.humimm.2004.08.183. [DOI] [PubMed] [Google Scholar]
- 45.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hüe S, Cacoub P, Renou C, Halfon P, Thibault V, Charlotte F, Picon M, Rifflet H, Piette JC, Pol S, et al. Human leukocyte antigen class II alleles may contribute to the severity of hepatitis C virus-related liver disease. J Infect Dis. 2002;186:106–109. doi: 10.1086/341086. [DOI] [PubMed] [Google Scholar]
- 47.Hamed NA, Hano AF, Raouf HA, Gamal M, Eissa M. Relationship between HLA-DRB1*0101, DRB1*0301 alleles and interleukin-12 in haemophilic patients and hepatitis C virus positive hepatocellular carcinoma patients. Egypt J Immunol. 2003;10:17–26. [PubMed] [Google Scholar]
- 48.Lamonaca V, Missale G, Urbani S, Pilli M, Boni C, Mori C, Sette A, Massari M, Southwood S, Bertoni R, et al. Conserved hepatitis C virus sequences are highly immunogenic for CD4(+) T cells: implications for vaccine development. Hepatology. 1999;30:1088–1098. doi: 10.1002/hep.510300435. [DOI] [PubMed] [Google Scholar]
- 49.Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, Nolt K, Nelson KE, Strathdee SA, Johnson L, et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–456. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- 50.Sarmiento OL, Ford CL, Newbern EC, Miller WC, Poole C, Kaufman JS. The importance of assessing effect modification when asserting racial differences in associations between human leukocyte antigen class II alleles and hepatitis C virus outcomes. J Infect Dis. 2002;185:266–268. doi: 10.1086/338198. [DOI] [PubMed] [Google Scholar]
- 51.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothman KJ, Greenland S. Modern Epidemiology. Lippincott- Raven Publishers: Philadelphia; 1998. [Google Scholar]
- 53.Higashi Y, Kamikawaji N, Suko H, Ando M. Analysis of HLA alleles in Japanese patients with cirrhosis due to chronic hepatitis C. J Gastroenterol Hepatol. 1996;11:241–246. doi: 10.1111/j.1440-1746.1996.tb00069.x. [DOI] [PubMed] [Google Scholar]
- 54.Höhler T, Gerken G, Notghi A, Knolle P, Lubjuhn R, Taheri H, Schneider PM, Meyer zum Büschenfelde KH, Rittner C. MHC class II genes influence the susceptibility to chronic active hepatitis C. J Hepatol. 1997;27:259–264. doi: 10.1016/s0168-8278(97)80169-9. [DOI] [PubMed] [Google Scholar]
- 55.McKiernan S, Kelleher D. Immunogenetics of hepatitis C virus. J Viral Hepat. 2000;7 Suppl 1:13–14. doi: 10.1046/j.1365-2893.2000.00022.x. [DOI] [PubMed] [Google Scholar]
- 56.Renou C, Halfon P, Pol S, Cacoub P, Jouve E, Bronowicki JP, Arpurt JP, Rifflet H, Picon M, Causse X, et al. Histological features and HLA class II alleles in hepatitis C virus chronically infected patients with persistently normal alanine aminotransferase levels. Gut. 2002;51:585–590. doi: 10.1136/gut.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]


