Abstract
The vertebrate heart forms through successive phases of cardiomyocyte differentiation. Initially, cardiomyocytes derived from first heart field (FHF) progenitors assemble the linear heart tube. Thereafter, second heart field (SHF) progenitors differentiate into cardiomyocytes that are accreted to the poles of the heart tube over a well-defined developmental window. Although heart tube elongation deficiencies lead to life-threatening congenital heart defects, the variables controlling the initiation, rate and duration of myocardial accretion remain obscure. Here, we demonstrate that the AP-1 transcription factor, Fos-like antigen 2 (Fosl2), potentiates the rate of myocardial accretion from the zebrafish SHF. fosl2 mutants initiate accretion appropriately, but cardiomyocyte production is sluggish, resulting in a ventricular deficit coupled with an accumulation of SHF progenitors. Surprisingly, mutant embryos eventually correct the myocardial deficit by extending the accretion window. Overexpression of Fosl2 also compromises production of SHF-derived ventricular cardiomyocytes, a phenotype that is consistent with precocious depletion of the progenitor pool. Our data implicate Fosl2 in promoting the progenitor to cardiomyocyte transition and uncover the existence of regulatory mechanisms to ensure appropriate SHF-mediated cardiomyocyte contribution irrespective of embryonic stage.
KEY WORDS: AP-1, Fosl2, Cardiac development, Heart, Second heart field, Zebrafish
Summary: Early embryonic heart muscle differentiates in phases from two progenitor populations. AP-1 promotes the progenitor-to-cardiomyocyte transition specifically during the second phase.
INTRODUCTION
During vertebrate embryogenesis, cardiac muscle derives from progenitor cells in two successive waves (reviewed in Abu-Issa and Kirby, 2007; Vincent and Buckingham, 2010). The earliest cardiomyocytes differentiate bilaterally in lateral plate mesoderm (LPM) from first heart field (FHF) progenitors, migrate to the midline and assemble the linear heart tube. By contrast, second heart field (SHF) progenitors remain undifferentiated in LPM and come to reside in pharyngeal mesoderm where the poles of the linear heart tube attach to the embryo proper. Over a well-defined developmental window, SHF progenitors differentiate into nascent cardiomyocytes that are added progressively to the poles of the heart tube (Kelly et al., 2001; Mjaatvedt et al., 2001; Waldo et al., 2001). Termed myocardial accretion, this process significantly elongates the heart tube through the de novo production of new myocardial segments. At the arterial pole, SHF progenitors produce myocardium for the right ventricle (RV) and outflow tract (OFT) (Kelly et al., 2001; Rana et al., 2007; Verzi et al., 2005). They also produce a collar of smooth muscle at the base of the aortico-pulmonary trunk (Harmon and Nakano, 2013; Waldo et al., 2005). To balance cellular egress from the SHF resulting from cardiomyocyte differentiation, SHF cells proliferate to maintain the progenitor pool (Cai et al., 2003; Hutson et al., 2010; Tirosh-Finkel et al., 2010; van den Berg et al., 2009).
Genetic or environmental insults that compromise myocardial accretion at the arterial pole impede formation of the RV and OFT and lead to embryonic lethality (von Both et al., 2004; Cai et al., 2003; Prall et al., 2007). Milder defects leave the embryonic OFT shortened and susceptible to misalignment with the ventricles (Ward et al., 2005; Yelbuz et al., 2002). OFT misalignment can result in rightward shifting of the aorta at birth, a defining feature of the congenital heart defects tetralogy of Fallot and double outlet right ventricle (Nakajima, 2010). Therefore, subtle disturbances in SHF-mediated myocardial accretion are sufficient to cause serious congenital heart defects.
Successive phases of cardiomyocyte differentiation also generate the embryonic myocardium in zebrafish. Within the first 24 hours post-fertilization (hpf), FHF-derived cardiomyocytes form the linear heart tube comprising both atrial and ventricular cardiomyocytes (de Pater et al., 2009; Lazic and Scott, 2011; Yelon et al., 1999). Between 24 and 48 hpf, SHF progenitors differentiate into cardiomyocytes that are progressively accreted to the arterial pole of the heart tube (de Pater et al., 2009; Hami et al., 2011; Lazic and Scott, 2011; Zhou et al., 2011). SHF progenitors also proliferate during this developmental window (Nevis et al., 2013; Zeng and Yelon, 2014; Zhou et al., 2011). When accretion is complete, the FHF and SHF-derived cardiomyocytes inhabit roughly the proximal and distal halves of the ventricular myocardium, respectively. As in higher vertebrates, SHF progenitors in zebrafish also give rise to myocardium and smooth muscle in the OFT (Hami et al., 2011; Zeng and Yelon, 2014; Zhou et al., 2011). After SHF-mediated accretion is complete, the zebrafish ventricle grows through cardiomyocyte proliferation (Choi et al., 2013; Liu et al., 2010).
As a point of convergence for many signaling pathways, the dimeric transcription factor AP-1 has been implicated in myriad molecular, cellular, developmental and pathologic processes (reviewed in Eferl and Wagner, 2003). It comprises members of the c-Fos and c-Jun protein families and functions as a transcriptional activator or repressor depending on biological context (Suzuki et al., 1991). Prior work has implicated AP-1 in SHF-mediated OFT morphogenesis because Jun-null mice are born with rightward shifting of an unseptated OFT (Eferl et al., 1999). Conditional deletion of Jun within Isl1-expressing cells, including SHF progenitors, also causes rightward shifting of the aorta (Zhang et al., 2013b). Therefore, despite genetic evidence that AP-1 functions within SHF progenitors to ensure correct alignment of the embryonic OFT, the cellular basis of OFT misalignment in Jun-null mice remains incompletely understood. Moreover, because Fos-family member knockout mice have not been reported to exhibit defects in cardiac morphogenesis (Eferl and Wagner, 2003; Karreth et al., 2004), a potential role for Fos proteins in AP-1-dependent OFT alignment also remains unclear. Lastly, although an in vitro study implicated the Fos family member Fos-like (or related) antigen 2 (Fosl2 or Fra-2) and AP-1 complexes in the regulation of cardiomyocyte differentiation, this function has yet to be corroborated in vivo. To determine whether fosl2 regulates heart development in zebrafish, we created and characterized fosl2-null and fosl2-overexpressing zebrafish embryos. Our work implicates Fosl2-containing AP-1 complexes in potentiating the rate of cardiomyocyte differentiation specifically from SHF progenitors.
RESULTS
Isolation of fosl2-null alleles
Fosl2 is a highly conserved protein across vertebrate species (Fig. S1). To understand the function of Fosl2 during zebrafish development, we analyzed embryos injected with an anti-sense morpholino targeting a splice site in the fosl2 pre-mRNA. When compared with control animals, 72 hpf morphants displayed pericardial edema and smaller ventricles (Fig. 1A-D), suggesting preliminarily that Fosl2 supports ventricular morphogenesis and/or growth. However, because multiple studies have reported discrepancies between morphant and genetic loss-of-function phenotypes (den Broeder et al., 2009; Hinits et al., 2012; Kok et al., 2015; Rossi et al., 2015; Wright et al., 2004), we opted to isolate null alleles of fosl2 using TALEN-mediated genome editing before implicating Fosl2 in ventricular development.
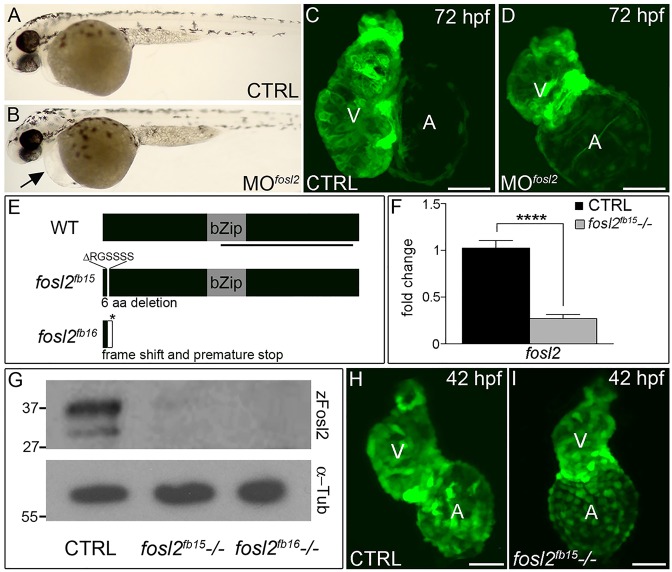
Fig. 1.
fosl2-null embryos exhibit defects in cardiogenesis. (A,B) Bright-field images of control (CTRL; A; n=44) and fosl2 morphant (MOfosl2; B; n=35) embryos at 72 hpf. Morphant embryos exhibit pericardial edema (black arrow). (C,D) Confocal images of GFP+ hearts in 72 hpf control (C; n=6) and fosl2 morphant (D; n=9) Tg(cmlc2:GFP) embryos. (E) Schematic diagrams of wild-type (WT) zebrafish Fosl2 with its basic leucine zipper domain (bZip) and the predicted protein products of two fosl2 mutant alleles, fosl2fb15 and fosl2fb16, generated through TALEN-mediated genome editing. The black line highlights the C-terminal region of the protein used to raise polyclonal antiserum. (F) Bar graph showing the relative levels of fosl2 mRNA in control and fosl2fb15−/− embryos at 48 hpf as measured by quantitative PCR (n=6 biological replicates per group). Error bars represent s.d. ****P<0.0001. (G) Western blots of 30 hpf whole-embryo lysates from control and mutant animals probed with Fosl2 (zFosl2) or α-tubulin (α-Tub) antiserum. (H,I) Confocal images of GFP+ hearts in 42 hpf control (H; n=10) and mutant (I; n=4) Tg(cmlc2:GFP) embryos. V, ventricle. A, atrium. Scale bars: 50 µm.
The first mutant allele we isolated, fosl2fb15, carries an 18 bp deletion that removes six amino acids near the N-terminus (Fig. 1E). Homozygous fosl2fb15 mutants express significantly lower levels of fosl2 transcript (Fig. 1F), demonstrating that the deletion undermines transcript production and/or stability. Consistent with this finding, fosl2fb15 mutant embryos have severely reduced Fosl2 protein levels as revealed through western blotting analysis (Fig. 1G) using an antibody we raised against the C-terminus of the zebrafish protein (Fig. 1E). The second mutant allele we isolated, fosl2fb16, contains a 16 bp deletion that shifts the open reading frame after codon 16 of 341 total. As a result, 16 additional divergent amino acids are encoded prior to a premature stop codon (Fig. 1E). As expected, we did not detect Fosl2 protein in fosl2fb16 mutant embryos (Fig. 1G) because the protein's epitopes are truncated by the deletion (Fig. 1E). Together, the severe reductions in mRNA and protein observed in fosl2fb15 homozygous embryos, the confirmed magnitude of the Fosl2fb16 truncation and the observation that both classes of mutants display indistinguishable phenotypes (see below) suggests strongly that both deletions create null alleles.
fosl2-null embryos exhibit small ventricles comprising fewer cardiomyocytes
When we analyzed cardiac chamber morphology, we learned that mutant ventricles appeared grossly smaller than those of siblings (Fig. 1H,I), a finding that is consistent with the morphant phenotype (Fig. 1C,D). Quantification of cardiomyocytes at 48 hpf revealed that both alleles reduced ventricular cell number by ∼18% (Fig. 2A-D) whereas gross atrial size (Fig. 1C,D) and cell number (Fig. 1A-D) were unaffected. Because ventricular and OFT morphogenesis are interrelated developmental processes (Hami et al., 2011; Zhou et al., 2011), we analyzed mutants for the production of Elastin 2 (Eln2) by OFT smooth muscle cells (Grimes et al., 2006; Miao et al., 2007). At 52 hpf, mutant OFTs were devoid of Eln2 when control animals were positive (Fig. 2E,F). Taken together, our data demonstrate that inactivation of zebrafish Fosl2 causes cardiomyocyte and smooth muscle marker deficiencies in the ventricle and OFT, respectively.
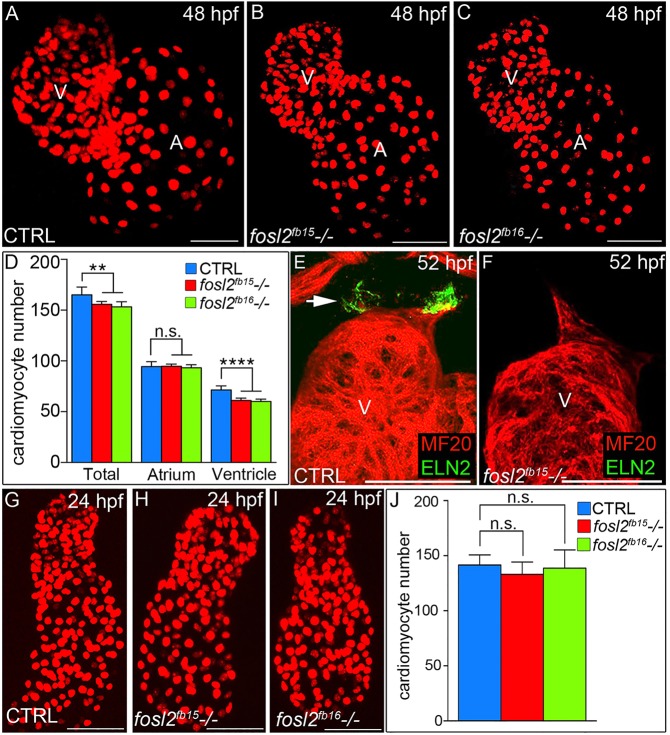
Fig. 2.
A ventricular cardiomyocyte deficit emerges in fosl2 mutant embryos after unperturbed linear heart tube morphogenesis. (A-C) Confocal images of fluorescent cardiomyocyte nuclei in hearts of 48 hpf control sibling (CTRL; A; n=11), fosl2fb15−/− (B; n=5) and fosl2fb16−/− (C; n=5) Tg(cmlc2:DsRed2-nuc) embryos. (D) Bar graph showing mean total, atrial and ventricular cardiomyocyte numbers in each experimental group. Error bars represent s.d. **P<0.01, ****P<0.0001, n.s., not significant. (E,F) Confocal images of ventricular and outflow tract (OFT) regions of 52 hpf control (E; n=10) and null mutant (F, n=4) embryos co-stained with antibodies recognizing striated muscle (MF20, red) or OFT smooth muscle (Elastin2, ELN2, green, arrow in E). (G-I) Confocal images of fluorescent cardiomyocyte nuclei in linear heart tubes of 24 hpf control (G; n=12), fosl2fb15−/− (H; n=5) and fosl2fb16−/− (I; n=5) Tg(cmlc2:DsRed2-nuc) embryos. (J) Bar graph showing mean cardiomyocyte numbers in each experimental group at 24 hpf. Error bars represent s.d. n.s., not significant. V, ventricle. A, atrium. Scale bars: 50 µm.
To narrow the developmental window during which the ventricular deficit emerges, we counted cardiomyocytes at 24 hpf when differentiation of the FHF-derived linear heart tube is largely complete (de Pater et al., 2009; Lazic and Scott, 2011). From this analysis, we learned that mutant heart tubes are indistinguishable from those in control animals (Fig. 2G-J). This observation supports three conclusions: (1) the ventricular deficiency in null animals becomes evident between 24 and 48 hpf; (2) inactivation of fosl2 does not negatively affect the rate of FHF progenitor differentiation; and (3) fosl2-null embryos do not suffer from generalized developmental delay, a finding confirmed independently by assaying the rate of hepatobiliary morphogenesis (Fig. S2).
Impaired cardiomyocyte differentiation from SHF progenitors in fosl2-null embryos
Next, we sought to decipher the cellular mechanism(s) underlying the ventricular deficiency that emerges in mutant animals between 24 and 48 hpf. In theory, a decrease in cardiomyocyte proliferation might explain fewer ventricular cells. However, previous work has demonstrated that cardiomyocyte proliferation is virtually non-existent in zebrafish embryos during this time frame (de Pater et al., 2009). We also ruled out increased cardiomyocyte apoptosis as a potential cause of the cellular deficiency (Fig. S3).
Because the ventricular cardiomyocyte deficit emerges during the SHF-mediated myocardial accretion window, we next assayed SHF function directly in fosl2 mutants. To that end, we used a cardiomyocyte photoconversion assay (de Pater et al., 2009; Lazic and Scott, 2011) to quantify FHF- and SHF-derived ventricular cardiomyocytes at 48 hpf. The assay employs the transgenic strain Tg(myl7:nlsKikGR), which continuously expresses the photoconvertible Kikume green-red (KikGR) protein in cardiomyocyte nuclei (Lazic and Scott, 2011). At 24 hpf, FHF-derived cardiomyocyte nuclei are labeled with photoconverted red protein by exposure to UV light. At 48 hpf, FHF-derived cardiomyocytes retain the red label while simultaneously expressing newly synthesized green KikGR. On the contrary, SHF-derived cardiomyocytes express green KikGR exclusively because they differentiate after photoconversion. Ultimately, counting the numbers of red and green cardiomyocyte nuclei in the proximal ventricle and green-only cardiomyocyte nuclei in the distal ventricle reveals the numbers of FHF- and SHF-derived cells, respectively.
Consistent with a previous report (Lazic and Scott, 2011), we found that ∼40% of cardiomyocytes in wild-type ventricles are derived from the SHF at 48 hpf (Fig. 3A-C,G). In fosl2 mutants, this percentage decreased to 28%, revealing an almost 30% reduction in SHF-derived cardiomyocytes (Fig. 3D-G). To evaluate the status of SHF progenitors themselves, we performed in situ hybridization for the SHF marker ltbp3 (Zhou et al., 2011). Mutant embryos harbored qualitative increases in ltbp3 transcripts (Fig. 3H,I) that we quantified as twofold higher with qPCR (Fig. 3J). Mutant embryos also expressed elevated levels of mef2cb (Fig. 3J), an additional marker of SHF progenitors (Lazic and Scott, 2011). We quantified SHF progenitor numbers by counting extra-cardiac nkx2.5:ZsYellow+ cell nuclei adjacent to the arterial pole (Paffett-Lugassy et al., 2013; Zhou et al., 2011) and learned that the elevation in SHF markers reflected a 38% increase in SHF progenitor cells (Fig. 3K-M). This accumulation cannot be explained by an increase in SHF progenitor proliferation (Fig. S3). Taken together, our data demonstrate that mutant embryos accumulate SHF progenitors at the expense of their differentiated progeny. These data support a model in which Fosl2 directly or indirectly facilitates the SHF progenitor to cardiomyocyte transition at the arterial pole.
Fig. 3.
Fosl2 potentiates the progenitor to cardiomyocyte transition during SHF-mediated ventricular growth. (A-G) Cardiomyocyte photoconversion assay. Control sibling (CTRL; A-C; n=23) and fosl2fb16−/− (D-F; n=6) Tg(myl7:nlsKikGR) embryos were photoconverted at 24 hpf and imaged by confocal microscopy at 48 hpf in the red (A,D) and green (B,E) channels. Merged images are shown in C,F. Dashed lines highlight boundaries between cardiomyocytes that differentiated before (bottom) or after (top) photoconversion. Arrowheads highlight SHF-derived green-only cardiomyocytes. (G) Bar graph showing the mean numbers of total ventricular cardiomyocytes, green and red positive cardiomyocytes, and green-only cardiomyocytes. Error bars represent s.d. ****P<0.0001, n.s., not significant. (H,I) Ventral images of 48 hpf control (H; n=16) and fosl2-null (I; n=5) embryos stained with a riboprobe for the SHF marker ltbp3. Arrowheads highlight extra-cardiac SHF progenitors. (J) Bar graph showing relative levels of the SHF markers ltbp3 (n=9 biological replicates per group) and mef2cb (n=3 biological replicates per group) in control and mutant embryos at 48 hpf as measured by quantitative PCR. Error bars represent s.d. ***P<0.001, *P<0.05. (K,L) Confocal images of the arterial poles in control (K; n=12) and fosl2 mutant (L; n=4) embryos carrying the Tg(nkx2.5:nZsYellow) and Tg(cmlc2:GFP) transgenes co-stained with antibodies recognizing ZsYellow (red) or GFP (green). (M) Bar graph showing the mean numbers of extra-cardiac SHF progenitor cells (red nuclei without yellow cytoplasm) in both experimental groups. Error bars represent s.d. ***P<0.001. (N-R) Double in situ hybridization analysis of fosl2 (blue) and cmlc2 (red) in whole mounted (N-Q) and sagittally crysectioned (R) 24 hpf (N,O) and 36 hpf (P-R) embryos. Anterior is to the left in N-P,R. Boxed region in N is magnified in O. (Q) Anterior and dorsal view of the embryo following removal of the head. Open arrowheads in O,Q,R highlight fosl2+ cells on either side of the arterial pole (AP; closed arrowheads and dashed line in O). More than 20 embryos per group were evaluated. VP, venous pole. Scale bars: 50 µm.
Consistent with a direct role, we learned that 24 hpf zebrafish embryos express fosl2 transcripts on both the ventricular and extra-cardiac sides of the arterial pole (Fig. 3N,O). At later time points, fosl2 expression is less evident in the heart proper but remains strongly expressed on the extra-cardiac side where SHF progenitors reside (Fig. 3Q,R, Fig. S4). fosl2 transcripts were also distributed broadly throughout the head but sparsely in the trunk and tail (Fig. 3N,P and data not shown). Transcripts encoding c-Jun, a potential AP-1 binding partner for Fosl2 (Eferl and Wagner, 2003) were also observed on both sides of the linear heart tube's arterial pole and later in the extra-cardiac region inhabited by SHF progenitors (Fig. S4). The similarities between fosl2 and jun expression are consistent with the possibility that Fosl2 pairs with c-Jun to form the AP-1 complex that potentiates the rate of cardiomyocyte differentiation from the zebrafish SHF.
fosl2-null embryos overcome sluggish accretion by extending the SHF differentiation window
To understand the natural course of the fosl2−/− cardiac phenotype, we evaluated mutant embryos at 72 hpf for ventricular cardiomyocyte numbers and OFT Eln2 expression. Unexpectedly, mutant embryos were indistinguishable from control siblings (Fig. 4A-E), demonstrating that phenotypic resolution had occurred between 48 and 72 hpf. Consistent with a full phenotypic recovery, mutant animals reach adulthood in the expected mendelian ratios and are fertile. Adult animals are also indistinguishable from siblings with regard to animal size, heart size, heart morphology and cardiac regenerative capacity (Fig. S5). By contrast, the ventricular cardiomyocyte deficiency observed in fosl2 morphants at 48 hpf persists at 72 hpf when severe OFT deficiencies also become evident (Fig. S6). The discordance between mutant and morphant phenotypes at 72 hpf suggests that morpholino toxicity and/or off-target effects contribute to the morphant phenotype or that genetic mutants are uniquely equipped to deploy compensatory mechanisms (Rossi et al., 2015). To test the former, we injected fosl2−/− embryos with the fosl2 morpholino and evaluated the compound mutant-morphant embryos for OFT deficiencies at 72 hpf. The OFT phenotype of compound mutant-morphants was indistinguishable from pure morphants (Fig. S6) demonstrating that the OFT deficiencies observed in morphants arise from morpholino toxicity and/or off-target effects independent of knocking down Fosl2 protein expression (Fig. S6).
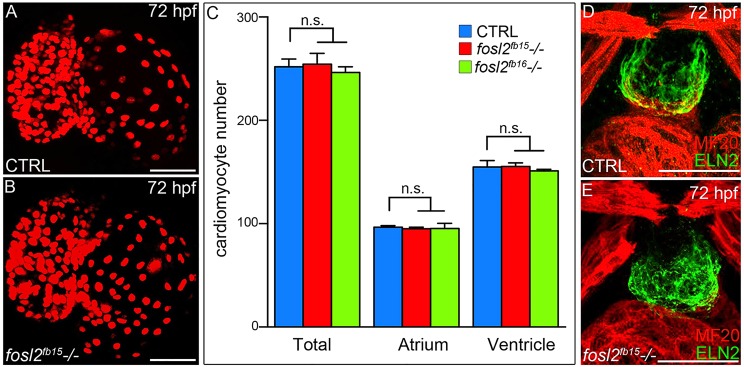
Fig. 4.
fosl2 mutants recover from their ventricular deficit. (A,B) Confocal images of hearts from 72 hpf control sibling (CTRL; A; n=12) and fosl2 (B; n=5) mutant Tg(cmlc2:DsRed2-nuc) embryos. (C) Bar graph showing the mean numbers of total, atrial and ventricular cardiomyocytes in control, fosl2fb15−/− and fosl2fb16−/− (n=3) embryos. Error bars represent s.d. n.s., not significant. (D,E) Confocal images of OFT regions in control (D; n=15) and fosl2 mutant (E; n=4) embryos co-stained with antibodies recognizing striated muscle (MF20, red) or OFT smooth muscle (Elastin2, ELN2, green). Scale bars: 50 µm.
Next, we sought to determine why fosl2 mutant embryos exhibit a deficit in SHF-derived ventricular cardiomyocytes at 48 hpf. First, we tested the hypothesis that mutant embryos exhibit a delay in initiating the accretion process. To that end, we photoconverted and analyzed control and mutant hearts immediately before (23 hpf) and shortly after (32 hpf) accretion commences, respectively. During this short time frame, control and mutant embryos produced equivalent numbers of SHF-derived cells (Fig. 5A-G), indicating that fosl2 mutants initiate accretion in a timely fashion. Taking into account that mutant animals produce fewer cardiomyocytes between 24 and 48 hpf (Fig. 2A-G), we deduced that the ventricular deficit at 48 hpf results from sluggish SHF differentiation between 32 and 48 hpf. To test this conclusion explicitly, we quantified the number of SHF-derived cardiomyocytes produced between 36 and 48 hpf and found that mutant animals accreted significantly fewer during this developmental window (Fig. S7). Taken together, these data demonstrate that the ventricular deficiency present at 48 hpf results from sluggish accretion between 36 and 48 hpf.
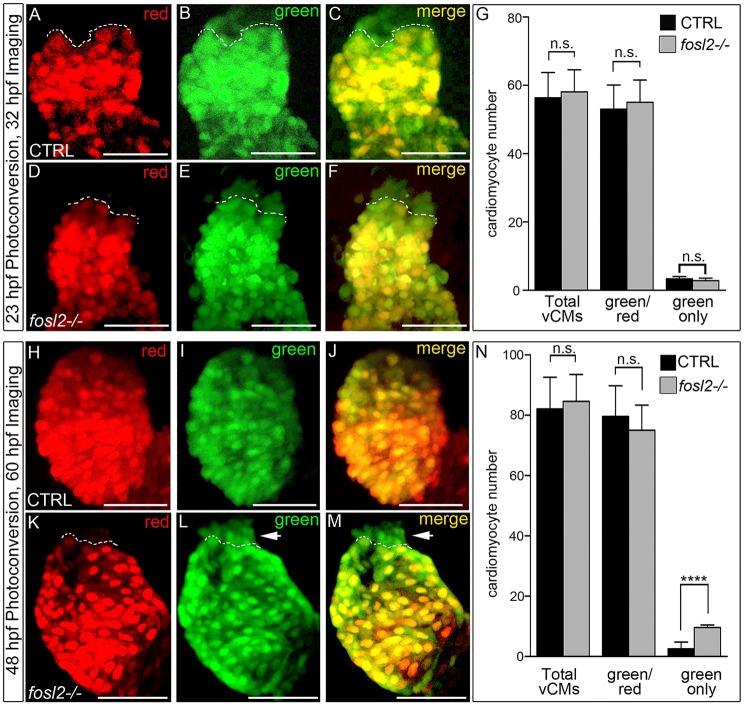
Fig. 5.
The ventricular deficit resolves in fosl2 mutants through extension of the SHF-mediated cardiomyocyte accretion window. Cardiomyocyte photoconversion assay. Control sibling (CTRL; A-C; n=11) and fosl2-null (D-F; n=7) Tg(myl7:nlsKiKGR) embryos were photoconverted at 23 hpf and imaged by confocal microscopy at 32 hpf in the red (A,D) and green (B,E) channels. Merged images are shown in C and F. Dashed lines highlight boundaries between cardiomyocytes that differentiated before (bottom) or after (top) photoconversion. (G) Bar graph showing the mean numbers of total ventricular cardiomyocytes, green and red positive cardiomyocytes, and green-only cardiomyocytes. Error bars represent s.d. n.s., not significant. Control (H-J; n=19) and fosl2 mutant (K-M; n=5) Tg(myl7:nlsKiKGR) embryos were photoconverted at 48 hpf and imaged by confocal microscopy at 60 hpf in the red (H,K) and green (I,J) channels. Merged images are shown in J,M. Dashed lines highlight boundaries between cardiomyocytes that differentiated before (bottom) or after (top) photoconversion. Arrowheads identify SHF-derived green-only cardiomyocytes that differentiated after 48 hpf specifically in fosl2-null animals. (N) Bar graph showing the mean numbers of total ventricular cardiomyocytes, green and red positive cardiomyocytes and green-only cardiomyocytes. Error bars represent s.d. n.s., not significant, ****P<0.0001. Scale bars: 50 µm.
Next, we ascertained whether the phenotypic recovery results from differentiation of the accumulated SHF progenitors (Fig. 3H-M) beyond 48 hpf, the developmental stage when accretion is largely complete in wild-type embryos. To that end, we photoconverted cardiomyocytes at 48 hpf and performed image analysis at 60 hpf. Whereas control embryos produced two SHF-derived cardiomyocytes on average during this 12 h window (Fig. 5H-J,N), null animals produced five times more (Fig. 3K-N). Because the number of cardiomyocytes accreted in mutant embryos after 48 hpf is roughly equivalent to the deficit observed (Fig. 2D), it is unlikely that reprogramming of atrial cardiomyocytes (Zhang et al., 2013a) contributes to the phenotypic recovery. Furthermore, an increase in ventricular cardiomyocyte proliferation between 48 and 72 hpf does not explain the phenotypic recovery (Fig. S8). Ultimately, these data demonstrate that mutant animals extend their SHF accretion window to offset the ventricular deficiency caused by sluggish differentiation.
Lastly, to determine whether another Fos family member might facilitate the phenotypic recovery by compensating for the loss of fosl2, we analyzed wild-type and fosl2−/− embryos for the expression of c-fos (fosab – ZFIN) at the arterial pole. In both wild-type and fosl2−/− embryos, c-fos expression was not observed in or near the heart despite the existence of strong staining in the brain (Fig. S9). These data suggest that upregulation of c-fos in the mutant does not account for the phenotypic recovery.
Overexpression of fosl2 perturbs SHF-mediated cardiomyocyte accretion
Lastly, we sought to characterize the phenotype of embryos overexpressing Fosl2. To that end, we injected one-cell stage zebrafish embryos with full-length fosl2 mRNA and quantified the numbers of atrial and ventricular cardiomyocytes at 48 hpf. Whereas atrial number was unaffected, ventricular cardiomyocytes were reduced by 31% (Fig. 6A-C). Using the cardiomyocyte photoconversion assay, we found that Fosl2 overexpression specifically reduced the number of SHF-derived ventricular cardiomyocytes by 86% (Fig. 6D-J). The cardiomyocyte deficit was accompanied by qualitative (Fig. 6K,L) and quantitative decreases in ltbp3 expression (>50%, Fig. 6M) and SHF progenitor cell number (>65%; Fig. 6N-P). The co-occurrence of reduced SHF progenitors and their myocardial progeny is consistent with a model wherein Fosl2 overexpression increases the propensity of SHF progenitors to differentiate, rather than proliferate, resulting in precocious depletion of the progenitor pool. This would significantly reduce the number of SHF progenitors capable of producing the full complement of ventricular cardiomyocytes. Although this model is consistent with the reported sufficiency of Fosl2 to increase the differentiation of osteoblasts (Bozec et al., 2010) and drive precocious differentiation of keratinocytes (Wurm et al., 2015), we cannot rule out other cellular mechanisms that might explain the overexpression phenotype.
Fig. 6.

Overexpression of Fosl2 compromises SHF-mediated ventricular growth. (A,B) Confocal images of hearts in 48 hpf control sibling (CTRL; A; n=11) and Fosl2-overexpressing (B; n=5) Tg(myl7:nlsDsRed-Express) embryos. (C) Bar graph showing the mean numbers of total, atrial and ventricular cardiomyocytes in both experimental groups. n.s., not significant, ***P<0.001. (D-I) Cardiomyocyte conversion assay. Control (n=7) and Fosl2-overexpressing (n=5) Tg(myl7:nlsKiKGR) embryos were photoconverted at 24 hpf and imaged by confocal microscopy at 48 hpf in the red (D,G) and green (E,H) channels. Merged images are shown in (F,I). Dashed lines highlight boundaries between cardiomyocytes that differentiated before (bottom) or after (top) photoconversion. Arrowheads identify SHF-derived green-only cardiomyocytes. (J) Bar graph showing the mean numbers of total ventricular cardiomyocytes, green and red positive cardiomyocytes, and green-only cardiomyocytes. Error bars represent s.d. ****P<0.0001, n.s., not significant. (K,L) Ventral images of control (n=30) and Fosl2 (n=35) overexpressing 48 hpf embryos stained with a riboprobe for the SHF marker ltbp3. (M) Bar graph showing the relative levels of ltbp3 mRNA at 48 hpf in both experimental groups as measured by quantitative PCR (n=3 biological replicates per group). **P<0.01. (N-O) Confocal images of the arterial poles in control (N; n=7) and Fosl2-overexpressing (N; n=6) embryos carrying the Tg(nkx2.5:nZsYellow) and Tg(cmlc2:AmCyan) transgenes co-stained with antibodies recognizing ZsYellow (red) or AmCyan (green). (P) Bar graph showing the mean numbers of extra cardiac SHF progenitor cells (red nuclei without yellow cytoplasm) in both experimental groups at 48 hpf. Error bars represent s.d. ****P<0.0001. Scale bars: 50 µm.
DISCUSSION
Our study implicates Fosl2, a component of the dimeric AP-1 transcription factor, in potentiating the rate of cardiomyocyte differentiation from SHF progenitors in zebrafish. In fosl2-null animals, the production of SHF-derived cardiomyocytes initiates appropriately, but sluggish differentiation results in a ventricular cardiomyocyte deficit and progenitor cell abundance. Null animals eventually resolve this phenotype by extending the accretion window until appropriate numbers of ventricular cells are achieved.
Our data provide in vivo relevance to a study demonstrating that Fosl2-containing AP-1 activity is required for cardiomyocyte differentiation from P19 embryonal carcinoma cells in vitro (Eriksson and Leppä, 2002). Furthermore, they are consistent with a report that conditional deletion of c-Jun in Isl1+ cells, including SHF progenitors, causes double outlet right ventricle (Zhang et al., 2013b), a manifestation of compromised SHF biology (Ward et al., 2005). Our data suggest that sluggish cardiomyocyte accretion from the mouse SHF might account for the rightward shifting aorta observed in Jun-null mice. Although overt congenital heart defects have not been reported in Fosl2-knockout mice (Karreth et al., 2004), genetic redundancy with other Fos family members might compensate completely or facilitate a phenotypic recovery from sluggish differentiation. If genetic redundancy exists in the mouse or zebrafish, then introducing additional Fos-family null alleles into the fosl2−/− animals would be predicted to reveal or exacerbate cardiac phenotypes, respectively. In that regard, our data suggest that Fosl2 mutations might cooperate with other genetic lesions that compromise SHF biology to cause congenital heart disease in the human population.
An important insight from our study is that the completion of myocardial accretion is not linked to developmental stage. At 48 hpf, when accretion has virtually ceased in WT embryos, fosl2−/− SHF progenitors continue producing cardiomyocytes to overcome the deficit. The cellular mechanism(s) ensuring that sufficient ventricular cardiomyocytes are produced, even in the face of sluggish differentiation, remain unknown. Perhaps SHF progenitors are capable of sensing ventricular size by monitoring contractility, shear stress and/or oxygenation. Under this scenario, if one or more of these variables were below threshold, then the SHF responds by continuing to produce cardiomyocytes until that threshold is crossed. Alternatively, perhaps SHF progenitors are pre-programmed with limited quantities of an intrinsic determinant that controls asymmetric cell divisions. If true, the appropriate number of cardiomyocyte progenitors would be produced independent of any delays to differentiation. Lastly, the mechanism(s) that extends the accretion window in fosl2−/− embryos also appears to delay the onset of smooth muscle production, perhaps through activation of a checkpoint.
The transcriptional targets of Fosl2 that positively regulate cardiomyocyte differentiation remain elusive. Although several transcriptional targets have been identified for Fosl2 (Bozec et al., 2010, 2008; Luther et al., 2014) and crucial regulators of SHF differentiation have been described (Rochais et al., 2009), we are not aware of any genes that fit into both categories. With regard to cellular differentiation, Fosl2 transcriptionally activates osteocalcin (Bozec et al., 2010) and members of the epidermal differentiation complex (Wurm et al., 2015) to positively regulate osteoclast and epidermal differentiation, respectively. However, because of the lineage restricted and non-cardiac nature of these particular targets, they are unlikely to mediate the role of Fosl2 in heart development. We attempted to identify transcriptional targets of Fosl2 using two unbiased approaches, ChIP sequencing and RNA sequencing of mutant embryos, but neither approach produced a convincing list of candidate targets, presumably because our antibody is not suitable for immunoprecipitation and the transcriptional changes underlying accumulation of SHF progenitors do not stand out in the context of whole embryos, respectively.
Together, the external development of zebrafish embryos combined with longitudinal assessments of cardiomyocyte production provide a highly sensitive method for uncovering SHF phenotypes that might otherwise go unnoticed in higher vertebrates. Despite the SHF phenotype, zebrafish fosl2 mutants appear grossly normal during embryogenesis. Therefore, specific assays for SHF function are required before ruling out SHF phenotypes in zebrafish mutants. The observation that Fosl2 overexpression might cause precocious differentiation of SHF progenitors in vivo provides rationale for testing the hypothesis that AP-1 activity would improve the efficiency and/or reduce the time required to differentiate SHF progenitors down the cardiomyocyte lineage in vitro for myocardial replacement therapies.
MATERIALS AND METHODS
Zebrafish strains
Zebrafish were grown and maintained according to animal protocols approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee. The following zebrafish strains were utilized: Tg(cmlc2:GFP)f1 (Burns et al., 2005), Tg(cmlc2:DsRed2-nuc)f2 (Mably et al., 2003), Tg(myl7:nlsKikGR)hsc6 (Lazic and Scott, 2011), Tg(nkx2.5nZsYellow)fb17 (Paffett-Lugassy et al., 2013), Tg(myl7:nlsDsRed-Express) hsc4 (Takeuchi et al., 2011), Tg(nkx2.5:ZsYellow)fb7 and Tg(cmlc2:CSY)fb2 (Zhou et al., 2011).
Morpholino injections
One-cell-stage wild-type Tg(cmlc2:GFP) or fosl2−/− embryos were injected with ∼1 nl of a morpholino (5′-GCGCTGAGACACATCTGTGCATACC-3′; 1-3 ng/nl; Gene Tools) targeting the first splice donor site in the fosl2 pre-mRNA. Sibling embryos were injected with a standard control oligo at the same dose (Gene Tools).
Isolation of fosl2-null alleles
DNA constructs encoding TALENs designed to cut between two binding sites (5′-TACGACACATCCTCCCGC-3′ and 5′-TGGTGTCCGGGTGCGCCG-3′) just downstream of the fosl2 ATG were obtained from the Genetic Perturbation Platform of the Broad Institute. Standard methods were utilized to generate and identify fosl2 alleles carrying insertions or deletions (Hwang et al., 2014). The fosl2fb15 allele harbors an 18 bp deletion (Δ5′-CTCCCGCGGCAGCAGCAG-3′) and the fosl2fb16 allele carries a 16 bp deletion (Δ5′-GCAGCAGCTCACCGGC-3′). Sibling embryos were used as controls in all experiments. Animals that are homozygous for either allele (fosl2fb15−/− and fosl2fb16−/−) are indistinguishable with respect to cardiomyocyte number at all stages analyzed. If not otherwise specified, fosl2fb15−/− embryos are shown.
Genotyping
Mutant genotypes were identified by capillary electrophoresis of carboxyfluorescein (6-FAM)-labeled amplicons generated with the following primers: forward, 5′-AAAAGGCAACATAAATTGGGAGTGC-3′ and reverse, 5′-GGCGCTGAGACACATCTGTGCATA-3′. We chose to attach a 6-FAM moiety on the forward primer. When compared with the wild-type amplicon (337 bp), mutant amplicons are smaller by 18 (fosl2fb15) or 16 (fosl2fb16) base pairs. For western blotting and qPCR analyses, embryonic tail tips were collected prior to lysing animals individually in wells of a 96-well plate. The lysates were frozen until genotyping was complete.
Quantitative polymerase chain reaction (qPCR)
The trunks and tails of embryos were excluded from the analysis as described (Zhou et al., 2011) to eliminate any potential influence of notochord (ltbp3) or skeletal muscle (mef2cb) expression. Animals were lysed individually in TRIzol Reagent (Life Technologies). After genotyping (see above), the lysates of at least three groups of 10 embryos from each cohort (control, fosl2−/− embryos or fosl2 mRNA-injected) were pooled prior to total RNA purification using the SV Total RNA Isolation System (Promega) and first strand synthesis using the Superscript III First-Strand Synthesis Kit (Life Technologies). Quantitative PCR analysis was performed using Fast SYBR Green Master Mix (Life Technologies) and an Applied Biosystems 7500 Real-Time PCR System (Life Technologies) according to the manufacturer's instructions. The 2−ΔΔCT method (Livak and Schmittgen, 2001) was used to measure differential expression levels after normalization to 18S ribosomal RNA (McCurley and Callard, 2008).
qPCR primer sequences used are as follows: ltbp3-F, 5′-CGCCCAAACAGGCTTGTAGTAGT-3′ and ltbp3-R, 5′-CACTCTTCGGTGAAAACGG-3′; mef2cb-F, 5′-CTCTCACTTATGTCAGGGTTCAAAT-3′ and mef2cb-R, 5′-GAGACTATCAGCCGGTGAGC-3′; fosl2-F1, 5′-AAGGAATCCCTTTGCTTGGA-3′ and fosl2-R1, 5′-GGTAACTGGAGGCGGGGATG-3′; 18S-F, 5′-TCGCTAGTTGGCATCGTTTATG-3′ and 18S-R, 5′-CGGAGGTTCGAAGACGATCA-3′.
Generation of custom polyclonal anti-serum recognizing zebrafish Fosl2
Using Gateway Cloning technology (Life Technologies), we generated a DNA construct for inducible expression of a 6×His-tagged fragment of Fosl2 (aa 151 to 317) in bacteria. The bacterially produced Fosl2 protein was affinity-purified using the HisPur Cobalt Purification Kit (Thermo Fisher Scientific), resolved by SDS-polyacrylamide gel electrophoresis and electroeluted from a Fosl2-containing gel slice prior to injection into rabbits (Rockland Immunochemicals). The IgG fraction was purified from rabbit serum internally designated ‘rabbit #1, bleed 2’ using the Protein A IgG Purification Kit (Thermo Fisher Scientific).
Western blotting
Animals were lysed individually in 4 µl lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl and 0.5% NP-40). After genotyping (see above), 10 wild-type or mutant whole embryo lysates were pooled and microfuged on high speed for 2 min at room temperature. The supernatant was diluted with 2× sample buffer and boiled for 5 min before being resolved by SDS-PAGE. The protein was transferred to PVDF membranes and blocked overnight in 5% milk. Fosl2 anti-serum and anti-α-tubulin antibody (Millipore, DM1A) were used at dilutions of 1:500 and 1:4000 in blocking solution, respectively. HRP-conjugated anti-rabbit (Cell Signaling, 7074) and anti-mouse (Cell Signaling, 7076) secondary antibodies were used at dilutions of 1:10,000. Blots were developed using Pierce ECL Western Blotting Substrate (Life Technologies).
Immunofluorescence, cell counting and photoconversion
Cardiomyocyte nuclei were counted in embryos carrying the myl7:nlsDsRed-Express or cmlc2:DsRed2-nuc transgene following immunostaining as described (Nevis et al., 2013; Zhou et al., 2011). Cardiomyocyte photoconversion and analysis were performed as described (Lazic and Scott, 2011; Paffett-Lugassy et al., 2013). MF20 and Eln2 immunostaining was performed as described (Zhou et al., 2011). SHF progenitor nuclei were visualized and counted in Tg(nkx2.5:nZsYellow), Tg(cmlc2:GFP) double transgenic animals after immunostaining as described (Nevis et al., 2013; Zhou et al., 2011). Embryos were imaged as described (Zhou et al., 2011).
In situ hybridization
A plasmid containing full-length zebrafish fosl2 (MGC:158347) was obtained from a commercial source (Open Biosystems). A cDNA encoding amino acids 31-339 of Fosl2 was PCR amplified and TOPO-cloned (Life Technologies) into pCRBTII to generate pCRBTII-fosl2partial. From this plasmid, an anti-sense riboprobe for detecting fosl2 was transcribed using T7 polymerase after linearization with BamHI. To generate a c-jun anti-sense riboprobe, a plasmid containing the zebrafish c-jun cDNA (MGC:77457) was obtained from a commercial source (Open Biosystems), linearized with EcoRI and transcribed with T7 polymerase. To generate a c-fos anti-sense riboprobe, a plasmid containing zebrafish c-fos (MGC:77885) was obtained from a commercial source (Open Biosystems). The plasmid was used to PCR amplify a 647 bp fragment of c-fos that was cloned into pCS2+. The resulting plasmid was linearized with BamHI and transcribed with T3 polymerase. Anti-sense riboprobes for detecting ltbp3 (Zhou et al., 2011), cmlc2 (Yelon et al., 1999) and nkx2.5 (Paffett-Lugassy et al., 2013) were produced as described. fosl2, ltbp3, nkx2.5, c-jun and c-fos probes were synthesized using the SP6/T7 DIG RNA labeling kit (Roche Diagnostics) substituting T3 polymerase as needed. cmlc2 probe was generated using the same kit but with Fluorescein RNA labeling mix (Roche Diagnostics, SP6/T7). Whole-mount in situ hybridizations were performed essentially as described (Thisse and Thisse, 2008). DIG and fluorescein-based in situ hybridizations were developed in NBT/BCIP (Roche Diagnostics) and INT/BCIP (Roche Diagnostics), respectively. Embryos were cryosectioned as described (Zhao et al., 2014). In photographs of stained embryos, the nkx2.5+ hepatobiliary system was circumscribed with the Lasso Tool in ImageJ (Schneider et al., 2012). The area of the circumscribed region was calculated using pixel number and size, the latter being determined empirically with a photograph of a stage micrometer taken under identical microscope settings.
Fosl2 overexpression
A full-length fosl2 cDNA was cloned into ClaI-digested pCS3+MT to generate pCS3+fosl2MT. One-cell-stage embryos were injected in the yolk with ∼1 nl (150 pg) of full-length fosl2 mRNA transcribed from pCS3+fosl2MT using the mMESSAGE mMACHINE SP6 Transcription Kit (Life Technologies) after linearization with HindIII.
Detection of apoptotic cell death
TUNEL (terminal deoxynucleotidyl transferase biotin-dUTP nick end-labeling) was performed on 36 hpf Tg(nkx2.5:ZsYellow) embryos as previously described (Espín et al., 2013).
Analysis of SHF cell proliferation
EdU labeling and staining was performed as previously described (Mahler et al., 2010; Nevis et al., 2013) using the Click-iT EdU imaging kit (Invitrogen). Briefly, 36 hpf Tg(nkx2.5:ZsYellow) embryos were incubated on ice for 30 min in 10 mM EdU, rinsed three times in E3 medium and chased until 48 hpf when embryos were processed for antibody staining with anti-rCFP and Click-iT Alexa Fluor 647 antibodies. Embryos were counterstained with DAPI and their distal ventricle and extra-cardiac ZsYellow+ regions were imaged by confocal microscopy. A SHF proliferation index (number of Edu+, ZsYellow+ double positive cells divided by the total number of DAPI+, ZsYellow+ cells) was calculated for each embryo and averaged.
Adult morphology and regeneration assessments
Adult hearts were dissected, fixed and processed as described (González-Rosa and Mercader, 2012). Apex amputations and Acid Fuchsin-Orange G (AFOG) staining were performed as described (Poss et al., 2002, Zhao et al., 2014).
Analysis of cardiomyocyte cell proliferation
BrdU labeling and staining was performed as previously described (Tkatchenko, 2006) with several modifications. Briefly, Tg(nkx2.5:nZsYellow) embryos were incubated in 5 mg/ml BrdU, 1% DMSO in E3 medium from 48 hpf to 72 hpf at 28°C, rinsed three times in E3 medium and fixed overnight in 4% PFA. Fixed embryos were rinsed in PBST, bleached in the dark for 20 min (using 0.8% KOH, 0.9% H2O2 and 1% Tween-20 in distilled water), permeabilized using 1% Triton-X100 in PBS for 2 h and equilibrated in DNase I buffer (40 mM Tris-HCl, pH 8.0, 10 mM MgSO4, 1 mM CaCl2) for 30 min at 37°C. Equilibrated embryos were treated with DNase I (1:50 in equilibration buffer) for 2 h at 37°C, rinsed three times in PBSTw (PBS+0.1% Tween20) and subjected to immunofluorescent staining with anti-rCFP and anti-BrdU antibodies. The ventricles of stained embryos were imaged and analyzed. A ventricular proliferation index (number of BrdU+, nZsYellow+ double positive cells divided by the total number of nZsYellow+ cells) was calculated for each embryo and averaged.
Protein sequence analyses
ClustalW2 (Larkin et al., 2007) was used to align Fosl2 proteins from several species. The bZIP domain was identified by the conserved domain feature (Marchler-Bauer et al., 2015) associated with the NCBI Standard Protein Blast.
Statistical analysis
Unpaired, two-tailed t-tests assuming equal standard deviations were used to calculate P-values in Prism6 software (GraphPad).
Acknowledgements
We thank Cong Zhu and John Doench of the Broad Institute for constructing TALENs targeting the zebrafish fosl2 locus. We thank Savo Lazic and Ian Scott (The Hospital for Sick Children, Toronto, Ontario, Canada) for providing the Tg(myl7:nlsKikGR) transgenic strain. Leila Jahangiri dedicates this work to her mother, Mary Teresa Jahangiri, who was her inspiration and passed away while this work was in progress.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
L.J. designed, performed and interpreted the majority of experiments. M.S., N.N., J.M.G.-R., A.B., K.N., N. P.-L., L.Z., M.A. and B.G.-A., designed, performed and interpreted experiments. C.E.B. and C.G.B. conceived the study, designed experiments, interpreted data and wrote the manuscript with input from all authors.
Funding
M.S. was supported by a National Science Foundation Graduate Research Fellowship. N.N., A.B. and M.A. were supported by the Cell and Molecular Training for Cardiovascular Biology [T32HL007208 (PI: Gerszten)] grant awarded to Massachusetts General Hospital. J.M.G.-R. was supported by an EMBO Long-Term Fellowship [ALTF 253-2014]. K.R.N. was supported by a National Institutes of Health National Research Service Award [1F32HL110627]. N. P.-L. was supported by the Harvard Stem Cell Institute Training Grant [5HL087735], a National Institutes of Health National Research Service Award [1F32HL112579] and an MGH Tosteson Award. L.Z. was supported by an AHA Post-doctoral Fellowship [14POST20380738]. B.G.-A. was supported by an American Heart Association (AHA) Post-doctoral Fellowship [10POST4170037] and an AHA Scientist Development Grant [14SDG19020018]. This work was supported by an R01 award to C.G.B. [5R01HL096816] from the National Institutes of Health. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.126136/-/DC1
References
- Abu-Issa R. and Kirby M. L. (2007). Heart field: from mesoderm to heart tube. Annu. Rev. Cell Dev. Biol. 23, 45-68. 10.1146/annurev.cellbio.23.090506.123331 [DOI] [PubMed] [Google Scholar]
- Bozec A., Bakiri L., Hoebertz A., Eferl R., Schilling A. F., Komnenovic V., Scheuch H., Priemel M., Stewart C. L., Amling M. et al. (2008). Osteoclast size is controlled by Fra-2 through LIF/LIF-receptor signalling and hypoxia. Nature 454, 221-225. 10.1038/nature07019 [DOI] [PubMed] [Google Scholar]
- Bozec A., Bakiri L., Jimenez M., Schinke T., Amling M. and Wagner E. F. (2010). Fra-2/AP-1 controls bone formation by regulating osteoblast differentiation and collagen production. J. Cell Biol. 190, 1093-1106. 10.1083/jcb.201002111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C. G., Milan D. J., Grande E. J., Rottbauer W., MacRae C. A. and Fishman M. C. (2005). High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 1, 263-264. 10.1038/nchembio732 [DOI] [PubMed] [Google Scholar]
- Cai C.-L., Liang X., Shi Y., Chu P.-H., Pfaff S. L., Chen J. and Evans S. (2003). Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877-889. 10.1016/S1534-5807(03)00363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.-Y., Gemberling M., Wang J., Holdway J. E., Shen M.-C., Karlstrom R. O. and Poss K. D. (2013). In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 140, 660-666. 10.1242/dev.088526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater E., Clijsters L., Marques S. R., Lin Y.-F., Garavito-Aguilar Z. V., Yelon D. and Bakkers J. (2009). Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 136, 1633-1641. 10.1242/dev.030924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Broeder M. J., van der Linde H., Brouwer J. R., Oostra B. A., Willemsen R. and Ketting R. F. (2009). Generation and characterization of FMR1 knockout zebrafish. PLoS ONE 4, e7910 10.1371/journal.pone.0007910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R. and Wagner E. F. (2003). AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859-868. 10.1038/nrc1209 [DOI] [PubMed] [Google Scholar]
- Eferl R., Sibilia M., Hilberg F., Fuchsbichler A., Kufferath I., Guertl B., Zenz R., Wagner E. F. and Zatloukal K. (1999). Functions of c-Jun in liver and heart development. J. Cell Biol. 145, 1049-1061. 10.1083/jcb.145.5.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M. and Leppä S. (2002). Mitogen-activated protein kinases and activator protein 1 are required for proliferation and cardiomyocyte differentiation of P19 embryonal carcinoma cells. J. Biol. Chem. 277, 15992-16001. 10.1074/jbc.M107340200 [DOI] [PubMed] [Google Scholar]
- Espín R., Roca F. J., Candel S., Sepulcre M. P., González-Rosa J. M., Alcaraz-Pérez F., Meseguer J., Cayuela M. L., Mercader N. and Mulero V. (2013). TNF receptors regulate vascular homeostasis in zebrafish through a caspase-8, caspase-2 and P53 apoptotic program that bypasses caspase-3. Dis. Model. Mech. 6, 383-396. 10.1242/dmm.010249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Rosa J. M. and Mercader N. (2012). Cryoinjury as a myocardial infarction model for the study of cardiac regeneration in the zebrafish. Nature Protocols 7, 782-788. 10.1038/nprot.2012.025 [DOI] [PubMed] [Google Scholar]
- Grimes A. C., Stadt H. A., Shepherd I. T. and Kirby M. L. (2006). Solving an enigma: arterial pole development in the zebrafish heart. Dev. Biol. 290, 265-276. 10.1016/j.ydbio.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Guner-Ataman B., Paffett-Lugassy N., Adams M. S., Nevis K. R., Jahangiri L., Obregon P., Kikuchi K., Poss K. D., Burns C. E. and Burns C. G. (2013). Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function. Development 140, 1353-1363. 10.1242/dev.088351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hami D., Grimes A. C., Tsai H.-J. and Kirby M. L. (2011). Zebrafish cardiac development requires a conserved secondary heart field. Development 138, 2389-2398. 10.1242/dev.061473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon A. W. and Nakano A. (2013). Nkx2-5 lineage tracing visualizes the distribution of second heart field-derived aortic smooth muscle. Genesis 51, 862-869. 10.1002/dvg.22721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y., Pan L., Walker C., Dowd J., Moens C. B. and Hughes S. M. (2012). Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 369, 199-210. 10.1016/j.ydbio.2012.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson M. R., Zeng X. L., Kim A. J., Antoon E., Harward S. and Kirby M. L. (2010). Arterial pole progenitors interpret opposing FGF/BMP signals to proliferate or differentiate. Development 137, 3001-3011. 10.1242/dev.051565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Peterson R. T. and Yeh J.-R. J. (2014). Methods for targeted mutagenesis in zebrafish using TALENs. Methods 69, 76-84. 10.1016/j.ymeth.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth F., Hoebertz A., Scheuch H., Eferl R. and Wagner E. F. (2004). The AP1 transcription factor Fra2 is required for efficient cartilage development. Development 131, 5717-5725. 10.1242/dev.01414 [DOI] [PubMed] [Google Scholar]
- Kelly R. G., Brown N. A. and Buckingham M. E. (2001). The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev. Cell 1, 435-440. 10.1016/S1534-5807(01)00040-5 [DOI] [PubMed] [Google Scholar]
- Kok F. O., Shin M., Ni C.-W., Gupta A., Grosse A. S., van Impel A., Kirchmaier B. C., Peterson-Maduro J., Kourkoulis G., Male I. et al. (2015). Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32, 97-108. 10.1016/j.devcel.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R. et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947-2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lazic S. and Scott I. C. (2011). Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev. Biol. 354, 123-133. 10.1016/j.ydbio.2011.03.028 [DOI] [PubMed] [Google Scholar]
- Liu J., Bressan M., Hassel D., Huisken J., Staudt D., Kikuchi K., Poss K. D., Mikawa T. and Stainier D. Y. R. (2010). A dual role for ErbB2 signaling in cardiac trabeculation. Development 137, 3867-3875. 10.1242/dev.053736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luther J., Ubieta K., Hannemann N., Jimenez M., Garcia M., Zech C., Schett G., Wagner E. F. and Bozec A. (2014). Fra-2/AP-1 controls adipocyte differentiation and survival by regulating PPARγ and hypoxia. Cell Death Differ. 21, 655-664. 10.1038/cdd.2013.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mably J. D., Burns C. G., Chen J.-N., Fishman M. C. and Mohideen M.-A. P. K. (2003). heart of glass regulates the concentric growth of the heart in zebrafish. Curr. Biol. 13, 2138-2147. 10.1016/j.cub.2003.11.055 [DOI] [PubMed] [Google Scholar]
- Mahler J., Filippi A. and Driever W. (2010). DeltaA/DeltaD regulate multiple and temporally distinct phases of notch signaling during dopaminergic neurogenesis in zebrafish. J. Neurosci. 30, 16621-16635. 10.1523/JNEUROSCI.4769-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I. et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222-D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurley A. T. and Callard G. V. (2008). Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 9, 102 10.1186/1471-2199-9-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M., Bruce A. E. E., Bhanji T., Davis E. C. and Keeley F. W. (2007). Differential expression of two tropoelastin genes in zebrafish. Matrix Biol. 26, 115-124. 10.1016/j.matbio.2006.09.011 [DOI] [PubMed] [Google Scholar]
- Mjaatvedt C. H., Nakaoka T., Moreno-Rodriguez R., Norris R. A., Kern M. J., Eisenberg C. A., Turner D. and Markwald R. R. (2001). The outflow tract of the heart is recruited from a novel heart-forming field. Dev. Biol. 238, 97-109. 10.1006/dbio.2001.0409 [DOI] [PubMed] [Google Scholar]
- Nakajima Y. (2010). Second lineage of heart forming region provides new understanding of conotruncal heart defects. Congenit. Anom. 50, 8-14. 10.1111/j.1741-4520.2009.00267.x [DOI] [PubMed] [Google Scholar]
- Nevis K., Obregon P., Walsh C., Guner-Ataman B., Burns C. G. and Burns C. E. (2013). Tbx1 is required for second heart field proliferation in zebrafish. Dev. Dyn. 242, 550-559. 10.1002/dvdy.23928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffett-Lugassy N., Singh R., Nevis K. R., Guner-Ataman B., O'Loughlin E., Jahangiri L., Harvey R. P., Burns C. G. and Burns C. E. (2013). Heart field origin of great vessel precursors relies on nkx2.5-mediated vasculogenesis. Nat. Cell Biol. 15, 1362-1369. 10.1038/ncb2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K. D., Wilson L. G. and Keating M. T. (2002). Heart regeneration in zebrafish. Science 298, 2188-2190. 10.1126/science.1077857 [DOI] [PubMed] [Google Scholar]
- Prall O. W. J., Menon M. K., Solloway M. J., Watanabe Y., Zaffran S., Bajolle F., Biben C., McBride J. J., Robertson B. R., Chaulet H. et al. (2007). An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 128, 947-959. 10.1016/j.cell.2007.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana M. S., Horsten N. C. A., Tesink-Taekema S., Lamers W. H., Moorman A. F. M. and van den Hoff M. J. B. (2007). Trabeculated right ventricular free wall in the chicken heart forms by ventricularization of the myocardium initially forming the outflow tract. Circ. Res. 100, 1000-1007. 10.1161/01.RES.0000262688.14288.b8 [DOI] [PubMed] [Google Scholar]
- Rochais F., Mesbah K. and Kelly R. G. (2009). Signaling pathways controlling second heart field development. Circ. Res. 104, 933-942. 10.1161/CIRCRESAHA.109.194464 [DOI] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M. and Stainier D. Y. R. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230-233. 10.1038/nature14580 [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. and Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Okuno H., Yoshida T., Endo T., Nishina H. and Iba H. (1991). Difference in transcriptional regulatory function between c-Fos and Fra-2. Nucleic Acids Res. 19, 5537-5542. 10.1093/nar/19.20.5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi J. K., Lou X., Alexander J. M., Sugizaki H., Delgado-Olguín P., Holloway A. K., Mori A. D., Wylie J. N., Munson C., Zhu Y. et al. (2011). Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat. Commun. 2, 187 10.1038/ncomms1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C. and Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59-69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Tirosh-Finkel L., Zeisel A., Brodt-Ivenshitz M., Shamai A., Yao Z., Seger R., Domany E. and Tzahor E. (2010). BMP-mediated inhibition of FGF signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development 137, 2989-3000. 10.1242/dev.051649 [DOI] [PubMed] [Google Scholar]
- Tkatchenko A. V. (2006). Whole-mount BrdU staining of proliferating cells by DNase treatment: application to postnatal mammalian retina. BioTechniques 40, 29-32. 10.2144/000112094 [DOI] [PubMed] [Google Scholar]
- van den Berg G., Abu-Issa R., de Boer B. A., Hutson M. R., de Boer P. A. J., Soufan A. T., Ruijter J. M., Kirby M. L., van den Hoff M. J. B. and Moorman A. F. M. (2009). A caudal proliferating growth center contributes to both poles of the forming heart tube. Circ. Res. 104, 179-188. 10.1161/CIRCRESAHA.108.185843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M. P., McCulley D. J., De Val S., Dodou E. and Black B. L. (2005). The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. 287, 134-145. 10.1016/j.ydbio.2005.08.041 [DOI] [PubMed] [Google Scholar]
- Vincent S. D. and Buckingham M. E. (2010). How to make a heart: the origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 90, 1-41. 10.1016/S0070-2153(10)90001-X [DOI] [PubMed] [Google Scholar]
- von Both I., Silvestri C., Erdemir T., Lickert H., Walls J. R., Henkelman R. M., Rossant J., Harvey R. P., Attisano L. and Wrana J. L. (2004). Foxh1 is essential for development of the anterior heart field. Dev. Cell 7, 331-345. 10.1016/j.devcel.2004.07.023 [DOI] [PubMed] [Google Scholar]
- Waldo K. L., Kumiski D. H., Wallis K. T., Stadt H. A., Hutson M. R., Platt D. H. and Kirby M. L. (2001). Conotruncal myocardium arises from a secondary heart field. Development 128, 3179-3188. [DOI] [PubMed] [Google Scholar]
- Waldo K. L., Hutson M. R., Ward C. C., Zdanowicz M., Stadt H. A., Kumiski D., Abu-Issa R. and Kirby M. L. (2005). Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev. Biol. 281, 78-90. 10.1016/j.ydbio.2005.02.012 [DOI] [PubMed] [Google Scholar]
- Ward C., Stadt H., Hutson M. and Kirby M. L. (2005). Ablation of the secondary heart field leads to tetralogy of Fallot and pulmonary atresia. Dev. Biol. 284, 72-83. 10.1016/j.ydbio.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Wright G. J., Leslie J. D., Ariza-McNaughton L. and Lewis J. (2004). Delta proteins and MAGI proteins: an interaction of Notch ligands with intracellular scaffolding molecules and its significance for zebrafish development. Development 131, 5659-5669. 10.1242/dev.01417 [DOI] [PubMed] [Google Scholar]
- Wurm S., Zhang J., Guinea-Viniegra J., García F., Muñoz J., Bakiri L., Ezhkova E. and Wagner E. F. (2015). Terminal epidermal differentiation is regulated by the interaction of Fra-2/AP-1 with Ezh2 and ERK1/2. Genes Dev. 29, 144-156. 10.1101/gad.249748.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelbuz T. M., Waldo K. L., Kumiski D. H., Stadt H. A., Wolfe R. R., Leatherbury L. and Kirby M. L. (2002). Shortened outflow tract leads to altered cardiac looping after neural crest ablation. Circulation 106, 504-510. 10.1161/01.CIR.0000023044.44974.8A [DOI] [PubMed] [Google Scholar]
- Yelon D., Horne S. A. and Stainier D. Y. R. (1999). Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 214, 23-37. 10.1006/dbio.1999.9406 [DOI] [PubMed] [Google Scholar]
- Zeng X.-X. I. and Yelon D. (2014). Cadm4 restricts the production of cardiac outflow tract progenitor cells. Cell Rep. 7, 951-960. 10.1016/j.celrep.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Han P., Yang H., Ouyang K., Lee D., Lin Y.-F., Ocorr K., Kang G., Chen J., Stainier D. Y. R. et al. (2013a). In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 498, 497-501. 10.1038/nature12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Liu J., Zhang J., Thekkethottiyil E. B., Macatee T. L., Ismat F. A., Wang F. and Stoller J. Z. (2013b). Jun is required in Isl1-expressing progenitor cells for cardiovascular development. PLoS ONE 8, e57032 10.1371/journal.pone.0057032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Borikova A. L., Ben-Yair R., Guner-Ataman B., MacRae C. A., Lee R. T., Burns C. G. and Burns C. E. (2014). Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 111, 1403-1408. 10.1073/pnas.1311705111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Cashman T. J., Nevis K. R., Obregon P., Carney S. A., Liu Y., Gu A., Mosimann C., Sondalle S., Peterson R. E. et al. (2011). Latent TGF-β binding protein 3 identifies a second heart field in zebrafish. Nature 474, 645-648. 10.1038/nature10094 [DOI] [PMC free article] [PubMed] [Google Scholar]