Abstract
A 2-year-old female donkey (Equus asinus) was euthanized in the Pathology Department of Firat University, Elazig, Turkey. Necropsy disclosed the presence of 7 hydatid cysts distributed throughout the lung parenchyma. One of those cysts represented the parasite material of the present study and was molecularly identified through sequencing of a fragment of cytochrome c oxidase subunit 1 (CO1) and nicotinamide adenine dinucleotide dehydrogenase subunit 1 (NADH1) gene, as Echinococcus equinus. The generated CO1 sequence supports the presence of the dominant haplotype as has been described in Europe and Africa. The NADH1 sequence was found similar to sequences reported in equids in Egypt and the United Kingdom. The molecular identification of E. equinus in a donkey is being reported for the first time in Turkey.
Keywords: Echinococcus equinus, donkey, molecular identification, Turkey
The parasite Echinococcus equinus, formerly known as the horse strain (G4) of Echinococcus granulosus, has been firmly established as an independent species according to the second taxonomic revision [1]. There is no evidence of significant intraspecific variation in E. equinus [2]. The presence of the dominant haplotype EQUK01 in various hosts and regions of the world along with 2 other haplotypes was indicated with the use of the mitochondrial cytochrome oxidase c subunit 1 (CO1) gene [3]. A significant amount of CO1 and nicotinamide adenine dinucleotide dehydrogenase subunit 1 (NADH1) sequence variation among E. granulosus genotypes has been found by the pioneers Bowles, Blair, and McManus [4,5] indicating the high discriminatory power of those 2 genes and consequently their importance as markers of species and strain identities.
E. equinus infections have been identified in horses in Italy [6], Spain [7], and recently in Germany [8] and the United Kingdom (UK) [3]. Since Kumaratilake et al. [9] based upon morphology described the occurrence of the equine strain in a naturally infected zebra in Southwestern Africa, data concerning similar infections in intermediate hosts other than horses are becoming increasingly common. Cases of infection in donkeys have been described in Egypt [10] and Tunisia [11], while Simsek and Cevik [12] reported its occurrence in a mule in Turkey. E. equinus was recently molecularly characterized for the first time in a zebra (Equus burchellii) and a lemur (Varecia rubrai) in the UK [13]. Although human infection with E. equinus has never been recorded, the finding of a primate (lemur) naturally infected with the parasite, raises the possibility of zoonotic susceptibility of humans to this species [13]. Interestingly, eggs of E. equinus were recently found in feces of lions (Panthera leo) and jackals (Canis mesomelas) in Namibia. Α genuine sylvatic life cycle is strongly indicated given the absence of domestic animals in the territory of those in the wild [14].
Here we present the genotypic characterization of the etiological agent of a hydatid cyst that was incidentally found in a naturally infected donkey in Turkey.
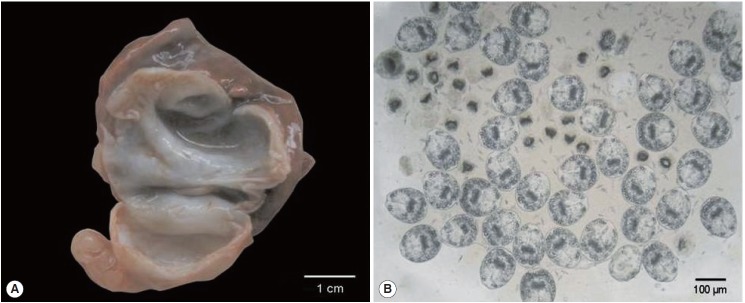
A 2-year-old female donkey that was born and raised in Turkey was euthanized with intravenous administration of pentobarbital as a part of the course of veterinary medical education of students in Firat University, Elazig, Turkey. The animal underwent a complete postmortem examination. Minor lesions of degenerative character were detected in the renal parenchyma and the stomach epithelium of the carcass along with the presence of 7 fluid-filled hydatid cysts distributed throughout the lung parenchyma. A 3-cm in diameter hydatid cyst was isolated from the left cranial lobe of the lung and represented the parasite material of the present study (Fig. 1A). Following the removal of the cyst, the hydatid fluid was aspirated and centrifuged. Microscopical examination of wet unstained preparations revealed the presence of abundant protoscolices (Fig. 1B).
Fig. 1.
Gross appearance of the Echinococcus equinus hydatid cyst and wet unstained preparation of the hydatid sand. (A) The cut surface of the hydatid cyst found in the animal’s lung. The whitish germinal layer represented the source of the template DNA. (B) Abundant invaginated E. equinus protoscoleces in wet unstained preparation.
Total genomic DNA was isolated from the germinal layer of the hydatid cyst with the use of a commercially available DNA extraction kit (MBI Fermentas, Vilnius, Lithuania) according to the manufacturer’s instructions. Three sets of primers previously described were used to amplify fragments of 3 genetic mitochondrial loci under the published PCR conditions. The primer pair Eg.ss1.f (5´ GTATTTTGTAAAGTTGTTCTA 3´) and Eg.ss1. rev. (5´ CTAAATCACATCATCTTACAAT 3´) [15] which amplifies a fragment (254 bp) of the mitochondrial 12S rRNA gene, selectively for the G1-G3 genotypes of E. granulosus, was firstly used in order to evaluate the presence of one of the aforementioned genotypes. Thermocycling consisted of a hold of 3 min at 95˚C and 40 cycles (30 sec/94˚C, 1 min/57˚C, 40 sec/72˚C) followed by a final extension step (5 min/72˚C). A fragment (446 bp) of the CO1 gene was amplified with the use of the JB3 (5´ TTTTTTGGGCATCCTGAGGTTTAT 3´), and JB4.5 (5´ TAAAGAAAGAACATAATGAAAATG 3´) primer set [4] under the following conditions: Pre-denaturating step at 95˚C for 5 min and 35 cycles (50 sec/94˚C, 50 sec/45˚C, 50 sec/72˚C) followed by a final extension step (10 min/72˚C). A third fragment of 520 bp belonging to the NADH1 gene was amplified with the use of the primer set JB11 (5´ AGATTCGTAAGGGGCCTAATA 3´) and JB12 (5´ ACCACTAACTAATTCACTTTC 3´) [5] under the following conditions: Hold (5 min/95˚C) and 35 cycles (30 sec/95˚C, 30 sec/52˚C, 30 sec/72˚C) followed by a final extension step (5 min/72˚C). All 3 PCR reactions were carried out in a final volume of 50 μl containing 5 μl 10× PCR buffer, 2.5 mM MgCl2, 250 μM of each dNTPs, 20 pmol of each primer, 1.25 U TaqDNA polymerase (MBI Fermentas), and 5 μl of template genomic DNA.
Amplicons were electrophoretically separated on an ethidium bromide stained 1.4% agarose gel and visualized under UV. The 12S rRNA PCR assay did not amplify the 254 bp expected fragment, thus the presence of G1-G3 genotypes was immediately excluded. The CO1 and NADH1 PCR assays yielded a 446 and a 520 bp band, respectively, each one typical of other E. granulosus strains as well.
The CO1 and NADH1 bands were excised from the gel and purified with the use of QIA quick Gel Extraction Kit (Qiagen, Hilden, Germany). The sequences were automatically obtained by ABI PRISM Sequence Detection System, and their identity was determined by an NCBI BLAST [16] search. Chromatograms’ quality was evaluated, the edges were trimmed, and the ambiguities were corrected in FinchTV 1.4.0 (Geospiza Inc., Seattle Washington, USA) (http://www.geospiza.com).
Two unquestionable sequences of 378 bp (CO1 fragment) and 390 bp (NADH1 fragment) were aligned with the use of Clustal Omega algorithm at www.ebi.ac.uk/Tools/msa/clustalo/ with sequences of E. equinus previously reported. The CO1 sequence showed 100% similarity with the common haplotype EQUK01 (KP101614) described from horses, dogs, zebra, and lemur in the UK as well as from donkeys in Tunisia (KM014645) and a mule in Turkey (KC953029). The NADH1 sequence was found to be 100% identical to the E. equinus sequences reported from donkeys in Egypt (JN191322) and horses in Germany (GQ420652) and the UK (AJ508084).
Both sequences were deposited in GenBank under the accession nos. KM525658 (CO1) and KP725298 (NADH1). Phylogenetic trees were inferred by the neighbor-joining algorithm [17] with the Kimura-80 parameters using the CLC Sequence viewer 7 software [18]. Bootstrap analysis of 1,000 replicates determined the robustness of the trees. CO1 and NADH1 sequences of Taenia saginata (GenBank accession nos. JQ756979 and AM503345) and Taenia multiceps (GenBank accession nos. JQ710587 and KF233942) were used as the outgroups (Fig. 2).
Fig. 2.
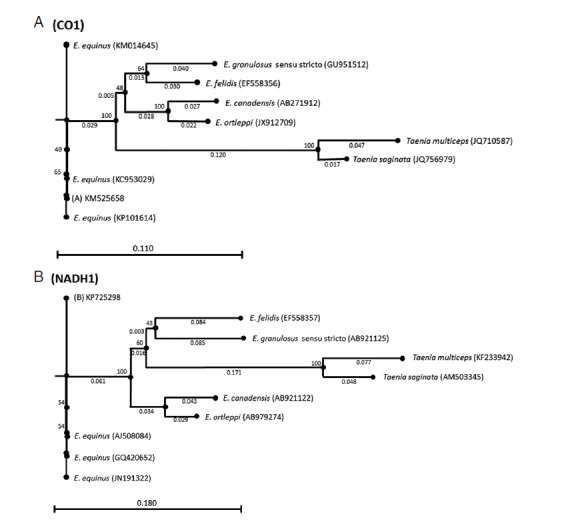
Genetic relationships inferred by the neighbor-joining algorithm among the CO1 and NADH1 Echinococcus equinus sequences from the donkey in Turkey and the reference sequences of Echinococcus species. Taenia saginata and Taenia multiceps sequences were used as outgroups.
Recent taxonomic classification based on genetic information from both mitochondrial and nuclear DNA genes suggests the presence of 5 valid species within the E. granulosus species complex: E. granulosus sensu stricto (G1-G3 genotypes), E. equinus (G4 genotype), E. ortleppi (G5 genotype), E. canadensis (G6-G10 genotypes), and E. felidis [1].
E. equinus appears exclusively to use equines as intermediate hosts [19]. Donkeys, which are used in Turkey for work, milk, and in some areas as guards to protect flocks from predators, get infected by intaking parasite eggs while grazing in contaminated fields. Improper disposal of infected donkey carcasses in the environment, instead of burying or burning them, is an essential factor for the maintenance of the parasite life cycle. Stray dogs and wild carnivores that feed on an infected carcass actively complete the cycle.
There are few reports concerning the occurrence of E. equinus in Asia. Two cases of dogs harboring this species have been reported in Jordan [21] and Kyrgyzstan [20], while Simsek and Cevik [12] reported its presence in a mule in Turkey. The finding of a fertile E. equinus hydatid cyst in a donkey as well indicates that the species is most likely well established in the country. The donkey case, although an isolated case, could be viewed as an important potential risk factor for the dissemination of this parasite species. It is necessary that all equine carcasses be necropsied and strain identification be performed. It is suggested that dogs, as well as wild carnivores, in Turkey be screened so as the prevalence be estimated.
The presence of a mitochondrial CO1 gene dominant haplotype is confirmed. Contrary to the genotype G1 which seems to be a continuously evolving strain presented with a great variety of CO1 haplotypes worldwide, the haplotypic variation of E. equinus appears to be restricted.
This is the first time that the molecular identification of E. equinus in a donkey has been reported in Turkey and in Asia.
Acknowledgments
This study was supported by a grant from Firat University Research Foundation (project no: VF.14.05) to S. Simsek and H. Eroksuz.
Footnotes
We have no conflict of interest related to this work.
REFERENCES
- 1.Nakao M, Yanagida T, Konyaev S, Lavikainen A, Odnokurtsev VA, Zaikov VA, Ito A. Mitochondrial phylogeny of the genus Echinococcus (Cestoda: Taeniidae) with emphasis on relationships among Echinococcus canadensis genotypes. Parasitology. 2013;140:1625–1636. doi: 10.1017/S0031182013000565. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins DJ, Romig T, Thompson RCA. Emergence/re-emergence of Echinococcus spp.-a global update. Int J Parasitol. 2005;35:1205–1219. doi: 10.1016/j.ijpara.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 3.Boufana B, Lett WS, Lahmar S, Buishi I, Bodell AJ, Varcasia A, Casulli A, Beeching NJ, Campbell F, Terlizzo M, McManus DP, Craig PS. Echinococcus equinus and Echinococcus granulosus sensu stricto from the United Kingdom: genetic diversity and haplotypic variation. Int J Parasitol. 2014;45:161–166. doi: 10.1016/j.ijpara.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. 1992;54:165–174. doi: 10.1016/0166-6851(92)90109-w. [DOI] [PubMed] [Google Scholar]
- 5.Bowles J, McManus DP. NADH dehydrogenase 1 sequences compared for species and strains of the genus Echinococcus. Int J Parasitol. 1993;23:969–972. doi: 10.1016/0020-7519(93)90065-7. [DOI] [PubMed] [Google Scholar]
- 6.Varcasia A, Garippa G, Pipia AP, Scala A, Brianti E, Giannetto S, Battelli G, Poglayen G, Micagni G. Cystic echinococcosis in equids in Italy. Parasitol Res. 2008;102:815–818. doi: 10.1007/s00436-007-0862-7. [DOI] [PubMed] [Google Scholar]
- 7.González LM, Mwambete KD, Montero E, Rosenzvit MC, McManus DP, Gárate T, Bandera CC. Further molecular discrimination of Spanish strains of Echinococcus granulosus. Exp Parasitol. 2002;102:46–56. doi: 10.1016/s0014-4894(02)00146-7. [DOI] [PubMed] [Google Scholar]
- 8.Blutke A, Hamel D, Hüttner M, Gehlen H, Romig T, Pfister K, Hermanns W. Cystic echinococcosis due to Echinococcus equinus in a horse from Southern Germany. J Vet Diagn Invest. 2010;22:458–462. doi: 10.1177/104063871002200323. [DOI] [PubMed] [Google Scholar]
- 9.Kumaratilake LM, Thompson RCA, Eckert J. Echinococcus granulosus of equine origin from different countries possess uniform morphological characteristics. Int J Parasitol. 1986;16:529–540. doi: 10.1016/0020-7519(86)90089-5. [DOI] [PubMed] [Google Scholar]
- 10.Aboelhadid SM, El-Dakhly KM, Yanai T, Fukushi H, Hassanin KM. Molecular characterization of Echinococcus granulosus in Egyptian donkeys. Vet Parasitol. 2013;193:292–296. doi: 10.1016/j.vetpar.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Boufana B, Lahmar S, Rebai W, Ben Safta Z, Jebabli L, Ammar A, Kachti M, Aouadi S, Craig PS. Genetic variability and haplotypes of Echinococcus isolates from Tunisia. Trans R Soc Trop Med Hyg. 2014;108:706–714. doi: 10.1093/trstmh/tru138. [DOI] [PubMed] [Google Scholar]
- 12.Simsek S, Cevik A. First detection and molecular characterization of Echinococcus equinus in a mule in Turkey. Acta Parasitologica. 2014;59:773–777. doi: 10.2478/s11686-014-0308-1. [DOI] [PubMed] [Google Scholar]
- 13.Boufana B, Stidworthy MF, Bell S, Chantrey J, Masters N, Unwin S, Wood R, Lawrence RP, Potter A, McGarry J, Redrobe S, Killick R, Foster AP, Mitchell S, Greenwood AG, Sako Y, Nakao M, Ito A, Wyatt K, Lord B, Craig PS. Echinococcus and Taenia spp. from captive mammals in the United Kingdom. Vet Parasitol. 2012;190:95–103. doi: 10.1016/j.vetpar.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 14.Wassermann M, Aschenborn O, Aschenborn J, Mackenstedt U, Romig T. A sylvatic lifecycle of Echinococcus equinus in the Etosha National Park, Namibia. Int J Parasitol: Parasites and Wildlife. 2014;doi:10. doi: 10.1016/j.ijppaw.2014.12.002. 1016/j.ijppaw.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinkel A, Njoroge EM, Zimmermann A, Wälz M, Zeyhle E, Elmahdi IE, Mackenstedt U, Romig T. A PCR system for detection of species and genotypes of the Echinococcus granulosus complex, with reference to the epidemiological situation in eastern Africa. Int J Parasitol. 2004;34:645–653. doi: 10.1016/j.ijpara.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSL-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Knudsen B, Knudsen T, Flensborg M, Sandmann H, Heltzen M, Andersen A, Dickenson M, Bardram J, Steffensen PJ, Mønsted S, Lauritzen T, Forsberg R, Thanbichler A, Bendtsen JD, Gorlitz L, Rasmussen J, Tordrup D, Vaerum M, Ravn MN, Hachenberg C, Fisker E, Dekker P, Hachenberg C, Schultz J, Hein AMK, Sinding JB. CLC Main Workbench. Version 5.5. Aarhus, Denmark: CLC Bio; 2007. [Google Scholar]
- 19.Romig T, Dinkel A, Mackenstedt U. The present situation of echinococcosis in Europe. Parasitol Int. 2006;55(suppl):S187–S191. doi: 10.1016/j.parint.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 20.Ziadinov I, Mathis A, Trachsel D, Rysmukhambetova A, Abdyjaparov TA, Kuttubaev OT, Deplazes P, Torgerson PR. Canine echinococcosis in Kyrgyzstan: using prevalence data adjusted for measurement error to develop transmission dynamics models. Int J Parasitol. 2008;38:1179–1190. doi: 10.1016/j.ijpara.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Qaoud KM, Abdel-Hafez SK, Craig PS. Canine echinococcosis in northern Jordan: increased prevalence and dominance of sheep/dog strain. Parasitol Res. 2003;90:187–191. doi: 10.1007/s00436-002-0793-2. [DOI] [PubMed] [Google Scholar]