Abstract
In recent years, remarkable progress has been made in the field of virus environmental ecology. In marine ecosystems, for example, viruses are now thought to play pivotal roles in the biogeochemical cycling of nutrients and to be mediators of microbial evolution through horizontal gene transfer. The diversity and ecology of viruses in soils are poorly understood, but evidence supports the view that the diversity and ecology of viruses in soils differ substantially from those in aquatic systems. Desert biomes cover ∼33% of global land masses, and yet the diversity and roles of viruses in these dominant ecosystems remain poorly understood. There is evidence that hot hyperarid desert soils are characterized by high levels of bacterial lysogens and low extracellular virus counts. In contrast, cold desert soils contain high extracellular virus titers. We suggest that the prevalence of microbial biofilms in hyperarid soils, combined with extreme thermal regimens, exerts strong selection pressures on both temperate and virulent viruses. Many desert soil virus sequences show low values of identity to virus genomes in public databases, suggesting the existence of distinct and as-yet-uncharacterized soil phylogenetic lineages (e.g., cyanophages). We strongly advocate for amplification-free metavirome analyses while encouraging the classical isolation of phages from dominant and culturable microbial isolates in order to populate sequence databases. This review provides an overview of recent advances in the study of viruses in hyperarid soils and of the factors that contribute to viral abundance and diversity in hot and cold deserts and offers technical recommendations for future studies.
INTRODUCTION
Over recent decades, the critical roles that viruses play in the environment have become increasingly recognized by the research community (1). It has been estimated by direct counts of extracellular (“free-floating”) virus-like particles (VLPs) that the global “virosphere” may contain up to 1031 viral particles (2), suggesting that viruses may be the most abundant biological entities on the planet and, potentially, the greatest reservoir of genetic diversity (3–5). Appreciation of the ecological importance of viruses on a global scale has predominantly emerged from studies of marine and freshwater microbial communities (6–12), where viruses have been linked to core processes such as biogeochemical nutrient cycling (6, 7, 10), microbial population control through viral lysis (7, 8), and microbial evolution via horizontal gene transfer (11). Research on the virus ecology of soil environments has progressed more slowly and has received proportionally less attention (12–14). However, enumeration of virus particles by electron microscopy (EM) performed on several soil types (15–17) has shown high viral abundance values ranging from 1.5 ×108 to 6.4 ×108 per gram (dry weight) of soils. Soil ecosystems are subject to unique abiotic ecological pressures, in part due to their wide compositional spectrum and spatial heterogeneity in terms of physicochemical properties (18, 19). Environmental stresses are even greater in extremely arid soil systems, where soil organisms and communities may be simultaneously exposed to pulsed-water events and to the effects of desiccation-, solute-, and UV-B radiation-induced oxidative stresses (20, 21). Deserts represent the single largest terrestrial ecosystem type on Earth, covering ∼33.6% of the global land mass, excluding Antarctica (22), and are classified in terms of their aridity index, a ratio between precipitation (P) and potential of evapotranspiration (PET) (23). This results in four desert categories: dry-semiarid (0.5 < P/PET < 0.65), semiarid (0.2 < P/PET < 0.5), arid (0.05 < P/PET < 0.2), and hyperarid (P/PET < 0.05). Hyperarid deserts generally receive annual precipitation of ≤70 mm and are often associated with intrinsic characteristics such as high (∼7 to 9) pH, high salinity levels, high surface radiation fluxes, long periods of desiccation, and low water activity (24). Desert soil microbial ecology research has primarily focused on bacterial communities, which have been shown to be largely responsible for primary production and provision of key ecosystem services (25–28). Soil virus populations and functions are seldom taken into consideration, thereby omitting a crucial variable within ecological models designed to predict microbial population dynamics. As a result, the ecological roles, survival mechanisms (against biotic and abiotic factors), and spatial and temporal changes in viral community structures (virus biogeography) and viral phylogenetic diversity are still poorly understood in desert soils. Within the field of soil virus ecology (13), several desert soil ecosystems have been recently investigated (Table 1). With the advances in next-generation-sequencing (NGS) technologies, culture-independent methods have become the standard for determinations of viral diversity (41). However, the rapidly growing volume of viral environmental sequence data has revealed that most sequences (∼70%) have no homologs in public databases, and such sequences are typically labeled “viral dark matter” (42, 43). Here, we discuss the current understanding of hot and cold desert soil virus diversity and function, propose alternative technical approaches to virus concentration methods, and identify key areas of future research.
TABLE 1.
Medium-to-high-throughput soil-based studies pertaining to viral ecology since 2005
| Authors, yr of publication (reference no.) | Soil type(s) | Location(s) of sample collection | Method(s) useda |
|---|---|---|---|
| Prigent et al., 2005 (29) | Hot desert surface sand | Sahara Desert in Morocco and Tunisia | EM, PFGE, lytic induction |
| Williamson et al., 2005 (16) | Agricultural, forest | Delaware, USA | Epifluorescence microscopy, EM |
| Fierer et al., 2007 (30) | Hot arid desert, tallgrass prairie, tropical rainforest | USA, Peru | Sanger sequencing of random viral metagenomics clones |
| Williamson et al., 2007 (17) | Loamy and sandy soils, agricultural, forested wetlands | Antarctica (Tom and Obelisk Pond); USA (Delaware) | Induction assays, epifluorescence counting |
| Prestel et al., 2008 (31) | Surface sand | Namib Desert | EM, PFGE, Sanger sequencing of cloned DNA fragments (LASL) |
| Swanson et al., 2009 (32) | Dystric-fluvic Cambisol soil | Dundee, Scotland | EM, epifluorescence counting |
| Meiring et al., 2012 (33) | Soil under hypoliths | Miers Valley, Antarctica | Lytic induction, EM, phage isolation from culture |
| Pearce et al., 2012 (34) | Surface soil | Alexander Island, Antarctica | Shotgun metagenome pyrosequencing |
| Swanson et al., 2012 (35) | Surface soil (Antarctica) | Antarctica | EM, lytic induction, phage isolation |
| Prestel et al., 2013 (36) | Dune surface sand | Mojave Desert, USA | EM, random amplification for viral DNA (Sanger) |
| Srinivasiah et al., 2013 (37) | Surface soil (Antarctica); silt loamy soil (USA) | Antarctica (Tom and Obelisk pond); Delaware, USA | RAPD viral community fingerprinting |
| Adriaenssens et al., 2015 (38) | Soil-associated rocks (hypoliths) | Namib Desert | Shotgun viral metagenome sequencing (Illumina) |
| Zablocki et al., 2014 (39) | Antarctic surface soil and hypoliths | Miers Valley, Antarctica | Shotgun viral metagenome sequencing (Illumina) |
| Srinivasiah et al., 2015 (40) | Silt loamy soil | Delaware, USA | Microcosms, RAPD viral community fingerprinting, epifluorescence counting |
EM, electron microscopy; PFGE, pulse field gel electrophoresis; LASL, linker amplified shotgun library; RAPD, random amplified polymorphic DNA.
DIVERSITY AND ABUNDANCE OF VIRUSES IN DESERT SOILS
Hot deserts.
Viral community analyses have been conducted on surface soil samples from three hot hyperarid deserts: the Sahara (29), Namib (31, 38), and Mojave (30, 36). In each of those studies, difficulties in detecting extracellular VLPs by electron microscopy (EM) or pulse field gel electrophoresis (PFGE) profiling were reported, suggesting a very low viral abundance within these soils. However, the inclusion of a lytic induction step (prophage excision stimulated by the addition of mitomycin C [44]) in the soil extraction protocol substantially increased the recovery of virus particles (29, 31). For Sahara Desert surface sand samples, induced phage genomes were estimated to range in size from 45 to 270 kb. EM of the induced phage fraction showed a majority of tailed virus morphotypes belonging to the Myoviridae family, some of which showed peculiar ribbon-like structures located at the tail tip of the virions (45). In the Namib Desert soil samples, 20 distinct morphotypes were identified, all members of the Myoviridae and Siphoviridae families with no apparent Podoviridae-like virions (31). PFGE profiles from Namib soils indicated an average genome size of 55 to 65 kb, with several genomes of up to 350 kb in size (31). EM visualization of Mojave Desert sand samples showed 11 distinct tailed morphotypes, belonging to all three families of the Caudovirales (36). Sanger sequencing of randomly selected cloned phage fragments from the Mojave Desert soil virus communities showed that 36% of the sequenced clones had no homologs in public sequence databases (36). Within the identified virus sequences, the majority were homologous to bacteriophages infecting common soil bacteria such as members of the Proteobacteria, including Bacillus and Rhizobium. From the same samples, 38 bacterial isolates were grown in pure culture and 84% were shown to harbor at least one SOS-inducible phage. A similar study on loamy sand from a different area of the Mojave Desert showed that a large majority of randomly selected metaviral clone sequences had no database homologs (30). Of those clones with significant sequence identity (tBLASTx search using an E-value cutoff value of 10−3), phages associated with Actinoplanes, Mycobacterium, Myxococcus, and Streptomyces were the most common. Other virus signals detected included archaeal (Haloarcula phage) and herpes-like viruses. Using a similar methodology, 50% of the viral sequences from three Namib Desert surface sand samples had no homologs in public sequence databases, with most positive hits showing homology to Siphoviridae phages linked to Gram-positive bacteria (31). Most recently, a shotgun NGS approach was used to investigate the metavirome of Namib Desert hypoliths (38), cyanobacterium-dominated microbial niche communities on the ventral surfaces of translucent rocks (46). The most abundant sequences belonged to Geobacillus- and Bacillus-infecting phages, while cyanophage markers were unexpectedly found only in low numbers. The distinct phylogenetic clustering of assembled phoH genes (a cyanophage marker [47]) suggested that desert soil cyanophages were related only distantly to their well-studied marine counterparts (38) and that the dominance of marine cyanophage sequences in sequence databases might account for the low cyanophage hit rate of homologous sequences in the Namib Desert hypolithon metavirome. This observation has wider implications for studies of soil metaviromics, where an underestimation of cyanophage abundance and diversity may skew estimates of the functional importance (and population dynamics) of soil cyanobacteria, arguably the most important taxonomic group in desert soil microbial communities (27, 28).
Cold deserts.
Studies of viral communities in cold hyperarid desert soils have been almost exclusively conducted in the major ice-free regions of Antarctica (e.g., the East Antarctic McMurdo Dry Valleys). Direct viral counts by epifluorescence microscopy (17) showed high VLP densities, in the range of 2.3 × 108 to 6.4 × 108 extracellular VLPs per gram of dry soil. The prevalence of bacterial lysogens within these soils was between 4.6% and 21.1%, a much lower occurrence level than has been estimated for bacteria in hot desert soils (84% [30]). Using epifluorescence direct counts of extractable bacteria and extracellular virus particles, virus-to-bacteria ratios (VBR) ranging from 170 to 8,200 were calculated, the highest recorded for any soil ecosystem (17). Antarctic soil bacterial isolates have yielded several unique virus genomic structures. The distinct temperate siphoviruses (SpaA1 and BceA1) isolated from Staphylococcus pasteuri and Bacillus cereus both contained almost-complete additional phage genomes (MZTP02) (35). This “Russian doll” gene arrangement had not been previously described for soil bacteriophages and has led to speculation that it may represent a “fast-track” route for virus evolution and horizontal gene transfer, with a possible role in host range expansion. Pyrosequencing of Antarctic soil metagenomic DNA has identified a wide diversity of bacteria, archaea, microeukaryotes, and viruses (34). From the total sequence data set, 494 phage-related hits (0.18% of the total number of sequences) were identified. Top BLAST hits in public databases were related to phages known to infect Mycobacteria, Burkholderia, Bordetella, Pseudomonas, Enterobacteria, Flavobacterium, Myxococcus, Synechococcus, Prochlorococcus, and Sinorhizobium. However, viral DNA was not specifically enriched in this study, and this may have resulted in an underestimation of viral diversity. The spatial composition and dynamics of viral communities along an Antarctic soil transect have been recently reported (37). Using random amplified polymorphic DNA (RAPD-PCR) assays, viral community fingerprints were used to assess short-term changes in the composition of viral communities. To maximize the number of virus sequences amplified, RAPD-PCR primer design was based on the identification of recurring dodecamer sequences (G+C content, ≥70%) within 22 selected viral metagenomes. Qualitative comparisons of the Antarctic fingerprint patterns demonstrated that heterogeneous soil conditions and associated environmental factors (e.g., carbon levels, moisture content, pH, and light exposure frequency) impacted the composition of viral assemblages across geographic distances of as little as 20 meters. The RAPD-PCR fingerprint data also suggested that virus assemblages were not present as inactive, inert particles but were dynamically involved in infection of coexisting microbial hosts. Furthermore, the authors suggested that environmental pressures (e.g., low moisture levels) known to influence bacterial community structures in the Antarctic desert (17) were shown to have a similarly influential role in virus community dynamics. Abundance estimates (17) suggest that Antarctic desert soils contain a substantially higher proportion of free extracellular VLPs than hot hyperarid desert soils, where a lysogenic lifestyle appears to be prevalent (29, 31, 36). A sequence-based metagenomic comparison of viral assemblages (single- and double-stranded DNA viruses only) in surface soils and hypolithic communities in the Antarctic McMurdo Dry Valleys (39) demonstrated that bacteriophages constituted the majority of the identified viruses, representing all Caudovirales families. Mycobacterium phage sequences were the most highly represented in the viral fraction (34). No archaeal virus sequences were recorded, in line with previous observations that archaea are either absent in this environment or present in very low numbers (26, 48). Within the hypolith metavirome data set, the fraction of cyanophage sequences was underrepresented, with low sequence similarities to known cyanophages. Dry Valley surface soils also contained a number of other virus signatures, including phycodnaviruses, mimiviruses, and virophage capsid protein genes (39), many of which are most commonly identified in aquatic systems.
FACTORS SHAPING VIRAL COMMUNITY STRUCTURES IN DESERT SOILS
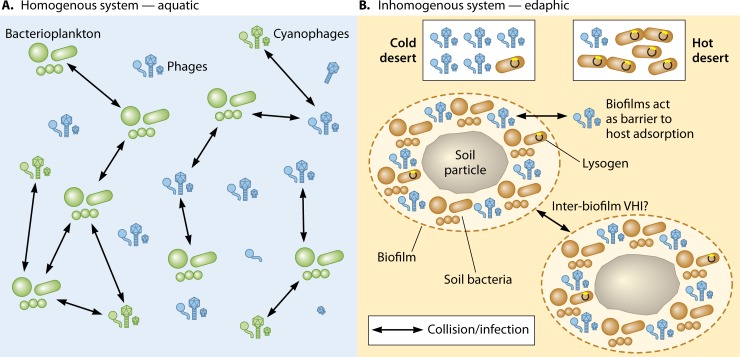
Soil virus populations display dynamics different from those in marine and freshwater systems (37) (Fig. 1). In marine systems, two major factors influence viral abundance: the biological productivity of the system and microbial diversity and abundance (3, 5). Viral abundance has been shown to increase as bacterial productivity in a system increases (49). Cooccurring virus host communities also influence viral abundance, as in the case of microbial bloom events, which increase the number of lytic infections, thereby releasing additional phage particles (50, 51). Marine environment-associated abiotic parameters such as temperature, salinity, and pH are stable on relatively large spatial scales (13) and do not appear to significantly affect viral abundance. Soil systems, particularly desert soils, are inhomogeneous, in that soil particles are semidiscrete. Extended periods of desiccation and oligotrophy are typical characteristics of hyperarid desert soils. Under these environmental constraints, microbial populations often form discrete biofilms, where cells embedded in extracellular polymeric substance (EPS) matrices are adsorbed to particle surfaces (52–54). The EPS matrix serves a protective role, sequesters nutrients, and provides a defense barrier against virulent phages (55). Temperate phages in their prophage state have been shown to stabilize biofilms, whereas a switch to the lytic cycle aids in biofilm dispersal. Within the biofilm, temperate phages have been shown to contribute to the life cycle of biofilm by aiding in biofilm dispersal (55). While this has been described for Pseudomonas aeruginosa biofilms (56), we argue that a similar mechanism may be present in hyperarid desert soil biofilms. Such a mechanism would drive the positive selection of temperate phages in this ecosystem and negatively influence the presence of virulent phages. This suggestion is consistent with the observation that extracellular VLPs are readily extracted only after induction of prophages in hot desert soils (29, 31, 36, 57). In Antarctic hyperarid desert soils, where biofilm communities also frequently occur (58), high VLP counts have been recorded. We suggest that the effects of temperature may explain the apparent differences between hot and cold desert soil systems. Temperature has been shown to be one of the major factors controlling viral survival rates in soils (59, 60), with lower temperatures enabling higher survival rates, extended latent periods, and reduced burst sizes. Warmer temperatures have been associated with reduced virus proliferation and higher inactivation rates (61, 62). Thus, in Antarctic soils, colder temperatures may allow the preservation of extracellular VLPs, making them more abundant and detectable (63). In contrast, the high-temperature settings (e.g., maxima of ≥50°C in the Namib Desert [48]) of hot deserts may increase the rate of degradation of extracellular virus particles.
FIG 1.
Virus community dynamics in aquatic (A) and soil (B) ecosystems. Marine and freshwater systems can be regarded as homogenous systems, where the distributions of virus particles (e.g., phages) and host organisms (e.g., bacterioplankton) are relatively even. (A) Such a continuous medium allows rapid phage-host dispersion and increases the rates of phage-host collisions, leading to high infection rates. (B) In contrast, hyperarid soil microbial communities exist as discrete systems, embedded in protective biofilms. The level of virus-host interactions (VHI) within and between individual biofilm communities remains an open issue, but diffusion rates are expected to be low on both small and large spatial scales.
Viral operational taxonomic unit (OTU) abundance estimates from low-throughput Sanger sequencing of metaviromes have provided some insights into the factors that shape the diversity of viral communities in desert soils (30). Comparisons of viral community compositions across three contrasting soil ecosystems (prairie, desert, and rainforest) have demonstrated that microbial communities were both locally and globally diverse. Comparative phylogenetic analyses showed little taxonomic overlap of soils sampled from the three different habitats, as well as low values of identity to annotated sequences in public databases. However, the factors that may be responsible for the observed niche specialization are as yet unknown.
Similar habitat-specific viral community compositions that have been determined through the use of hierarchical clustering of metaviromes, based on dinucleotide frequencies, have been reported (64). This method is especially useful for gaining ecological insights from metagenomic data sets containing a majority of unaffiliated reads that have been provided to public databases. Dinucleotide frequencies within metaviromes have shown distinct virus community clustering within single habitat types such as desert soils. Although reported from a single study which analyzed two sets of pooled samples, cluster analysis of hypolith and open-soil metaviromes from Antarctic and Namib Desert soil samples has shown that the two hypolith metaviromes clustered at a single node whereas, in contrast, the two open-soil metaviromes displayed identical patterns (65). Despite great geographic distances or differing environmental conditions, similar habitat types harbored more closely related viral communities. The most obvious common factor in the two contrasting deserts is very limited water availability, which may be a key driver of community speciation and recruitment in these soils.
TECHNICAL RECOMMENDATIONS AND FUTURE RESEARCH
Research on desert soil viruses is technically challenging, partly due to the physical properties of soil. Desert soils frequently produce suboptimal (≤10 ng/μl) viral DNA yields (66), forcing the inclusion of a random PCR amplification step for NGS library construction. The use of whole-genome amplification (WGA) by multiple-displacement amplification (MDA) or random-priming, sequence-independent, single-primer amplification (RP-SISPA) (67) almost certainly results in biased amplification of certain virus groups (68–71) and prevents the accurate determination of viral abundances and diversity. While viral amplification is widely accepted as a necessity in metaviromic studies (72), we argue that amplification of virus metagenomic DNA should be avoided where possible. It would be preferable to focus efforts on improving virus concentration methods in order to reach the minimum concentrations required for sequencing. Sequential rounds of centrifugation and the pooling of samples should increase the number of viruses recovered. Methodological improvements in virus concentration would also allow more-precise virus counts using microscopy (73). Thus, the development of more-efficient and -effective metaviromic DNA extraction technologies, so as to obviate the use of WGA, would represent a substantial advance in the field. Efforts to achieve this goal are further facilitated by recent technical improvements in sequencing chemistries where, for example, Illumina paired-end sequencing library construction kits (ThruPLEX; Rubicon Genomics) have reduced the minimum genomic DNA requirement to around 50 pg.
Sequence-based identification of viral communities, using either multiple gene markers (74) or full-virome sequencing (75), is becoming more routine. In marine virus ecology, the use of conserved viral marker genes such as DNA polymerases (76), ribonucleotide reductases (77), and T4-related structural proteins (78, 79) has provided detailed data on viral biodiversity, on intra- and interviral evolutionary relationships, and on oceanic viral turnover rates. The use of these methods to study virus diversity and biogeography in desert soils is relatively new and most commonly involves the sequencing of whole metaviromes (37–39). However, metaviromic approaches generally result in a large number of unknown sequences (43). In addition, we warn that the taxonomic affiliation of single genes and/or virus genome fragments (using BLAST analysis of public databases) in metavirome data sets may not be evidence for the presence of these viruses in the sample (80), and results should be carefully inspected by additional read mapping to a reference genome. While a metaviromics approach provides the opportunity for virus discovery, it may also be valuable to use, in parallel, a high-throughput sequencing approach focusing on conserved signature genes. Such a combinatorial strategy could provide both informative data on viral richness and insights into the functional roles of viruses in soil ecosystems.
A common feature of many desert ecosystems is the occurrence of hypolithic niches (81). These rock-associated cryptic microbial communities are usually dominated by photosynthetic cyanobacteria but contain a wide diversity of members of the phyla Actinobacteria, Acidobacteria, Bacteroidetes, and Proteobacteria (26, 82–84). Cyanobacteria are of particular importance, due to their key roles in primary productivity and nitrogen input in depauperate ecosystems (27, 28). To date, no fully characterized desert soil-associated cyanophage isolates have been reported. Preliminary metagenomic data on Antarctic and Namib hypoliths (38, 39) have shown evidence of novel soil cyanophage lineages, the sequences of which have low identities to characterized marine cyanophage genomes. As cyanobacteria are readily amenable to culturing (85), this provides opportunities for the isolation of their phages and access to full-length soil cyanophage genomes. Such data would support downstream applications such as primer design for targeted amplification of related taxa and monitoring of these assemblages within desert soil ecosystems.
CONCLUSION
Research on phage ecosystem ecology in hyperarid desert soils has demonstrated that desert soil viruses are numerous and diverse and carry novel genes whose functions are yet to be determined. In order to understand how viruses contribute to desert soil ecosystem functioning, critical research questions, addressing both micro- and macroscale issues, must be addressed. The microscale complexity of the soil matrix drives the distribution, maintenance, metabolic state, and biodiversity of microbial communities (86). Consequently, investigating the dynamics of virus-host interactions at the microscale level will contribute significantly to our understanding of the factors which determine virus distribution and diversity of bacteriophages in soil systems. In addition, the effects of extreme physicochemical conditions (e.g., intense UV radiation and temperature) on the preservation of virion particles and the kinetics of virus decay in hyperarid deserts remain unexplored. At the macroscale level (i.e., the ecosystem level), the contributions of viruses to ecosystem services such as nutrient cycling and energy flow and the sequestration of nutrients remain open issues. It would also be highly informative to understand the kinetics and scales of virus transport processes within and between hyperarid ecosystems, potentially important factors in understanding phage phylogeography.
ACKNOWLEDGMENTS
This work was funded by the South African National Research Foundation and the University of Pretoria Genomics Research Institute. E.M.A. is funded by the Claude Leon Foundation Postdoctoral Fellowship program.
Biographies

Olivier Zablocki received his B.Sc. and M.Sc. in Microbiology from the University of Pretoria, South Africa. While pursuing his M.Sc., he trained as a plant virologist and participated in the implementation of NGS for plant disease diagnosis for the South African citrus industry, with a focus on Citrus tristeza virus. In 2013, he started a Ph.D. program under the supervision of Don Cowan, the current director of the Centre for Microbial Ecology and Genomics at the University of Pretoria. For his thesis, he used both metaviromics and soil physicochemical analyses to assess virus community structure and dynamics in hyperarid desert soil ecosystems. He has recently joined the Institute for Microbial Biotechnology and Metagenomics at the University of the Western Cape (South Africa) to continue research in virus ecology using “omics” strategies.

Evelien Adriaenssens received her B.Sc. and M.Sc. from the faculty of Bioscience Engineering of the University of Leuven (KU Leuven), Belgium. During her Ph.D. research, she worked on the isolation and characterization of bacteriophages of the potato pathogen Dickeya solani for applications in plant protection. This was a collaborative effort between the Laboratory of Gene Technology of Rob Lavigne (KU Leuven), the Plant Production Laboratory of Maurice De Proft (KU Leuven), and the Unit Plant-Crop Protection of the Institute for Agricultural and Fisheries Research with Martine Maes. After obtaining the degree in 2012, she joined the Centre of Microbial Ecology and Genomics at the University of Pretoria, South Africa, in 2013 to work as a Postdoctoral Fellow heading the viral metagenomics project, investigating viral communities in hot and cold deserts.

Don Cowan was educated in New Zealand at the University of Waikato and completed a period of postdoctoral study there before moving to University College London (United Kingdom) as a Lecturer in 1985. After 16 years in London, he accepted the position as Professor of Microbiology in the Department of Biotechnology at the University of the Western Cape, Cape Town, where he was a Senior Professor and Director of the Institute for Microbial Biotechnology and Metagenomics. In May 2012 he moved to the University of Pretoria as director of both the Genomics Research Institute and the Centre for Microbial Ecology and Genomics. His research activities encompass a wide range of projects in the field of ecogenomics: the use of genomic and metagenomic methods to understand the diversity and function of microorganisms in different environments.
REFERENCES
- 1.Rohwer F, Prangishvili D, Lindell D. 2009. Roles of viruses in the environment. Environ Microbiol 11:2771–2774. doi: 10.1111/j.1462-2920.2009.02101.x. [DOI] [PubMed] [Google Scholar]
- 2.Suttle CA. 2005. Viruses in the sea. Nature 437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 3.Weinbauer MG, Rassoulzadegan F. 2004. Are viruses driving microbial diversification and diversity? Environ Microbiol 6:1–11. [DOI] [PubMed] [Google Scholar]
- 4.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 5.Dennehy JJ. 2014. What ecologists can tell virologists. Annu Rev Microbiol 68:17–135. doi: 10.1146/annurev-micro-091313-103436. [DOI] [PubMed] [Google Scholar]
- 6.Fuhrman JA. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 7.Wommack KE, Colwell RR. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64:69–114. doi: 10.1128/MMBR.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bettarel Y, Sime-Ngando T, Amblard C, Dolan J. 2004. Viral activity in two contrasting lake ecosystems. Appl Environ Microbiol 70:2941–2951. doi: 10.1128/AEM.70.5.2941-2951.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winget DM, Williamson KE, Helton RR, Wommack KE. 2005. Tangential flow diafiltration: an improved technique for estimation of virioplankton production. Aquat Microb Ecol 41:221–232. doi: 10.3354/ame041221. [DOI] [Google Scholar]
- 10.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasić L, Thingstad TF, Rohwer F, Mira A. 2009. Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 12.Breitbart M. 2012. Marine viruses: truth or dare. Ann Rev Mar Sci 4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- 13.Kimura M, Jia ZJ, Nakayama N, Asakawa S. 2008. Ecology of viruses in soils: past, present and future perspectives. Soil Sci Plant Nutr 54:1–32. doi: 10.1111/j.1747-0765.2007.00197.x. [DOI] [Google Scholar]
- 14.Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. 2008. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol 159:349–357. doi: 10.1016/j.resmic.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Ashelford KE, Day MJ, Fry JC. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl Environ Microbiol 69:285–289. doi: 10.1128/AEM.69.1.285-289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson KE, Radosevich M, Wommack KE. 2005. Abundance and diversity of viruses in six Delaware soils. Appl Environ Microbiol 71:3119–3125. doi: 10.1128/AEM.71.6.3119-3125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williamson KE, Radosevich M, Smith DW, Wommack KE. 2007. Incidence of lysogeny within temperate and extreme soil environments. Environ Microbiol 9:2563–2574. doi: 10.1111/j.1462-2920.2007.01374.x. [DOI] [PubMed] [Google Scholar]
- 18.Schlesinger WH, Reynolds JF, Cunningham GL, Huenneke LF, Jarrell WM, Virginia RA, Whitford WG. 1990. Biological feedbacks in global desertification. Science 247:1043–1048. doi: 10.1126/science.247.4946.1043. [DOI] [PubMed] [Google Scholar]
- 19.Palmer TM. 2003. Spatial habitat heterogeneity influences competition and coexistence in an African acacia ant guild. Ecology 84:2843–2855. doi: 10.1890/02-0528. [DOI] [Google Scholar]
- 20.Austin AT, Yahdjian L, Stark JM, Belnap J, Porporato A, Norton U, Ravetta DA, Schaeffer SM. 2004. Water pulses and biogeochemical cycles in arid and semiarid ecosystems. Oecologia 141:221–235. doi: 10.1007/s00442-004-1519-1. [DOI] [PubMed] [Google Scholar]
- 21.Chen LZ, Wang GH, Hong S, Liu A, Li C, Liu YD. 2009. UV-B-induced oxidative damage and protective role of exopolysaccharides in desert cyanobacterium Microcoleus vaginatus. J Integr Plant Biol 51:194–200. doi: 10.1111/j.1744-7909.2008.00784.x. [DOI] [PubMed] [Google Scholar]
- 22.Meigs P. 1952. Arid and semiarid climatic types of the world, p 135–138. In Proceedings of the VIII General Assembly and XVII International Congress. International Geographical Union, Washington, DC. [Google Scholar]
- 23.United Nations Environmental Programme (UNEP). 2013. Global environment outlook 2000. Routledge, London, United Kingdom. [Google Scholar]
- 24.Shmida A, Wilson M. 1985. Biological determinants of species diversity. J Biogeogr 12:1–20. doi: 10.2307/2845026. [DOI] [Google Scholar]
- 25.Harel Y, Ohad I, Kaplan A. 2004. Activation of photosynthesis and resistance to photoinhibition in cyanobacteria within biological desert crust. Plant Physiol 136:3070–3079. doi: 10.1104/pp.104.047712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pointing SB, Chan Y, Lacap DC, Lau MCY, Jurgens JA, Farrell RL. 2009. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci U S A 106:19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tracy CR, Streten-Joyce C, Dalton R, Nussear KE, Gibb KS, Christian KA. 2010. Microclimate and limits to photosynthesis in a diverse community of hypolithic cyanobacteria in northern Australia. Environ Microbiol 12:592–607. doi: 10.1111/j.1462-2920.2009.02098.x. [DOI] [PubMed] [Google Scholar]
- 28.Cowan DA, Pointing SB, Stevens MI, Craig Cary S, Stomeo F, Tuffin IM. 2011. Distribution and abiotic influences on hypolithic microbial communities in an Antarctic dry valley. Polar Biol 34:307–311. doi: 10.1007/s00300-010-0872-2. [DOI] [Google Scholar]
- 29.Prigent M, Leroy M, Confalonieri F, Dutertre M, DuBow MS. 2005. A diversity of bacteriophage forms and genomes can be isolated from the surface sands of the Sahara Desert. Extremophiles 9:289–296. doi: 10.1007/s00792-005-0444-5. [DOI] [PubMed] [Google Scholar]
- 30.Fierer N, Breitbart M, Nulton J, Salamon P, Lozupone C, Jones R, Robeson M, Edwards RA, Felts B, Rayhawk S, Knight R, Rohwer F, Jackson RB. 2007. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol 73:7059–7066. doi: 10.1128/AEM.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prestel E, Salamitou S, Dubow MS. 2008. An examination of the bacteriophages and bacteria of the Namib Desert. J Microbiol 46:364–372. doi: 10.1007/s12275-008-0007-4. [DOI] [PubMed] [Google Scholar]
- 32.Swanson MM, Fraser G, Daniell TJ, Torrance L, Gregory PJ, Taliansky M. 2009. Viruses in soils: morphological diversity and abundance in the rhizosphere. Ann Appl Biol 155:51–60. doi: 10.1111/j.1744-7348.2009.00319.x. [DOI] [Google Scholar]
- 33.Meiring TL, Marla Tuffin I, Cary C, Cowan DA. 2012. Genome sequence of temperate bacteriophage Psymv2 from Antarctic Dry Valley soil isolate Psychrobacter sp. MV2. Extremophiles 16:715–726. doi: 10.1007/s00792-012-0467-7. [DOI] [PubMed] [Google Scholar]
- 34.Pearce DA, Newsham KK, Thorne MAS, Calvo-Bado L, Krsek M, Laskaris P, Hodson A, Wellington EM. 2012. Metagenomic analysis of a southern maritime Antarctic soil. Front Microbiol 3:403. doi: 10.3389/fmicb.2012.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson MM, Reavy B, Makarova KS, Cock PJ, Hopkins DW, Torrance L, Koonin EV, Taliansky M. 2012. Novel bacteriophages containing a genome of another bacteriophage within their genomes. PLoS One 7:e40683. doi: 10.1371/journal.pone.0040683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prestel E, Regeard C, Salamitou S, Neveu J, Dubow MS. 2013. The bacteria and bacteriophages from a Mesquite Flats site of the Death Valley desert. Antonie Van Leeuwenhoek 103:1329–1341. doi: 10.1007/s10482-013-9914-4. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasiah S, Lovett J, Polson S, Bhavsar J, Ghosh D, Roy K, Fuhrmann JJ, Radosevich M, Wommack KE. 2013. Direct assessment of viral diversity in soils by random PCR amplification of polymorphic DNA. Appl Environ Microbiol 79:5450–5457. doi: 10.1128/AEM.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adriaenssens EM, Van Zyl L, De Maayer P, Rubagotti E, Rybicki E, Tuffin M, Cowan DA. 2015. Metagenomic analysis of the viral community in Namib Desert hypoliths. Environ Microbiol 17:480–495. doi: 10.1111/1462-2920.12528. [DOI] [PubMed] [Google Scholar]
- 39.Zablocki O, van Zyl L, Adriaenssens EM, Rubagotti E, Tuffin M, Cary SC. 2014. High-level diversity of tailed phages, eukaryote-associated viruses and virophage-like elements in the metaviromes of Antarctic soils. Appl Environ Microbiol 80:6888–6897. doi: 10.1128/AEM.01525-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivasiah S, Lovett J, Ghosh D, Roy K, Fuhrmann JJ, Radosevich M, Wommack KE. 2015. Dynamics of autochthonous soil viral communities parallels dynamics of host communities under nutrient stimulation. FEMS Microbiol Ecol 91:fiv063. doi: 10.1093/femsec/fiv063. [DOI] [PubMed] [Google Scholar]
- 41.Rosario K, Breitbart M. 2011. Exploring the viral world through metagenomics. Curr Opin Virol 1:289–297. doi: 10.1016/j.coviro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Youle M, Haynes M, Rohwer F. 2012. Scratching the surface of biology's dark matter, p 61–81. In Viruses: essential agents of life. Springer, New York, NY. [Google Scholar]
- 43.Hatfull GF. 27 May 2015. Dark matter of the biosphere: the amazing world of bacteriophage diversity. J Virol doi: 10.1128/JVI.01340-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ackermann H-W, DuBow MS. 1987. Viruses of prokaryotes. CRC Press, Boca Raton, FL. [Google Scholar]
- 45.Prestel E, Regeard C, Andrews J, Oger P, DuBow MS. 2012. A novel bacteriophage morphotype with a ribbon-like structure at the tail extremity. Res J Microbiol. [Google Scholar]
- 46.Makhalanyane TP, Valverde A, Lacap DC, Pointing SB, Tuffin MI, Cowan DA. 2013. Evidence of species recruitment and development of hot desert hypolithic communities. Environ Microbiol Rep 5:219–224. doi: 10.1111/1758-2229.12003. [DOI] [PubMed] [Google Scholar]
- 47.Goldsmith DB, Crosti G, Dwivedi B, McDaniel LD, Varsani A, Suttle CA, Weinbauer MG, Sandaa RA, Breitbart M. 2011. Development of phoH as a novel signature gene for assessing marine phage diversity. Appl Environ Microbiol 77:7730–7739. doi: 10.1128/AEM.05531-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makhalanyane TP, Valverde A, Birkeland N-K, Cary SC, Tuffin IM, Cowan DA. 2013. Evidence for successional development in Antarctic hypolithic bacterial communities. ISME J 7:2080–2090. doi: 10.1038/ismej.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maranger R, Bird DF. 1995. Viral abundance in aquatic systems: a comparison between marine and fresh waters. Mar Ecol Prog Ser 121:217–226. doi: 10.3354/meps121217. [DOI] [Google Scholar]
- 50.Ortmann AC, Lawrence JE, Suttle CA. 2002. Lysogeny and lyric viral production during a bloom of the cyanobacterium Synechococcus spp. Microb Ecol 43:225–231. doi: 10.1007/s00248-001-1058-9. [DOI] [PubMed] [Google Scholar]
- 51.Bratbak G, Heldal M, Norland S, Thingstad T. 1990. Viruses as partners in spring bloom microbial trophodynamics. Appl Environ Microbiol 56:1400–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davey ME, O'Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donlan RM. 2002. Biofilms: Microbial life on surfaces. Emerg Infect Dis 8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gorbushina AA. 2007. Life on the rocks. Environ Microbiol 9:1613–1631. doi: 10.1111/j.1462-2920.2007.01301.x. [DOI] [PubMed] [Google Scholar]
- 55.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. 2012. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10:39–50. [DOI] [PubMed] [Google Scholar]
- 56.Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, Hauser A, McDougald D, Webb JS, Kjelleberg S. 2009. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J 3:271–282. doi: 10.1038/ismej.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thurber RV. 2009. Current insights into phage biodiversity and biogeography. Curr Opin Microbiol 12:582–587. doi: 10.1016/j.mib.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 58.Aguilera A, Souza-Egipsy V, Amils R. 2012. Photosynthesis in extreme environments. Artif Photosynth InTech Eur Rijeka 271–288. [Google Scholar]
- 59.Hurst CJ, Gerba CP, Cech I. 1980. Effects of environmental variables and soil characteristics on virus survival in soil. Appl Environ Microbiol 40:1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nasser AM, Oman SD. 1999. Quantitative assessment of the inactivation of pathogenic and indicator viruses in natural water sources. Water Res 33:1748–1752. doi: 10.1016/S0043-1354(98)00380-7. [DOI] [Google Scholar]
- 61.Straub TM, Pepper IL, Gerba CP. 1992. Persistence of viruses in desert soils amended with anaerobically digested sewage sludge. Appl Environ Microbiol 58:636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leonardopoulos J, Papaconstantinou A, Georgakopoulou-Papandreou E. 1996. The meaning of soil characteristics and temperature for the survival of bacteriophages in naturally contaminated soil samples. Acta Microbiol Hell 41:309–316. [Google Scholar]
- 63.Williamson KE. 2011. Soil phage ecology: abundance, distribution, and interactions with bacterial hosts, p 113–136. In Biocommunication in soil microorganisms. Springer, New York, NY. [Google Scholar]
- 64.Willner D, Thurber RV, Rohwer F. 2009. Metagenomic signatures of 86 microbial and viral metagenomes. Environ Microbiol 11:1752–1766. doi: 10.1111/j.1462-2920.2009.01901.x. [DOI] [PubMed] [Google Scholar]
- 65.Zablocki O, van Zyl L, Adriaenssens EM, Rubagotti E, Tuffin M, Cary SC, Cowan D. 2014. Niche-dependent genetic diversity in Antarctic metaviromes. Bacteriophage 4:e980125. doi: 10.4161/21597081.2014.980125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim KH, Bae JW. 2011. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl Environ Microbiol 77:7663–7668. doi: 10.1128/AEM.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weynberg KD, Wood-Charlson EM, Suttle CA, van Oppen MJH. 2014. Generating viral metagenomes from the coral holobiont. Front Microbiol 5:206. doi: 10.3389/fmicb.2014.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim K-H, Chang H-W, Nam Y-D, Roh SW, Kim M-S, Sung Y, Jeon CO, Oh H-M, Bae J-W. 2008. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl Environ Microbiol 74:5975–5985. doi: 10.1128/AEM.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Polson SW, Wilhelm SW, Wommack KE. 2011. Unraveling the viral tapestry (from inside the capsid out). ISME J 5:165. doi: 10.1038/ismej.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karlsson OE, Belák S, Granberg F. 2013. The effect of preprocessing by sequence-independent, single-primer amplification (SISPA) on metagenomic detection of viruses. Biosecur Bioterror 11(Suppl1:S227–S234. doi: 10.1089/bsp.2013.0008. [DOI] [PubMed] [Google Scholar]
- 71.Rosseel T, Van Borm S, Vandenbussche F, Hoffmann B, van den Berg T, Beer M, Höper D. 2013. The origin of biased sequence depth in sequence-independent nucleic acid amplification and optimization for efficient massive parallel sequencing. PLoS One 8:e76144. doi: 10.1371/journal.pone.0076144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delwart EL. 2007. Viral metagenomics. Rev Med Virol 17:115–131. doi: 10.1002/rmv.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williamson KE, Wommack KE, Radosevich M. 2003. Sampling natural viral communities from soil for culture-independent analyses. Appl Environ Microbiol 69:6628–6633. doi: 10.1128/AEM.69.11.6628-6633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adriaenssens EM, Cowan DA. 2014. Using signature genes as tools to assess environmental viral ecology and diversity. Appl Environ Microbiol 80:4470–4480. doi: 10.1128/AEM.00878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sullivan MB. 2015. Viromes, not gene markers, for studying double-stranded DNA virus communities. J Virol 89:2459–2461. doi: 10.1128/JVI.03289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen F, Suttle CA, Short SM. 1996. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl Environ Microbiol 62:2869–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sakowski EG, Munsell EV, Hyatt M, Kress W, Williamson SJ, Nasko DJ, Polson SW, Wommack KE. 2014. Ribonucleotide reductases reveal novel viral diversity and predict biological and ecological features of unknown marine viruses. Proc Natl Acad Sci U S A 111:15786–15791. doi: 10.1073/pnas.1401322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hambly E, Tétart F, Desplats C, Wilson WH, Krisch HM, Mann NH. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc Natl Acad Sci U S A 98:11411–11416. doi: 10.1073/pnas.191174498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Short CM, Suttle CA. 2005. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl Environ Microbiol 71:480–486. doi: 10.1128/AEM.71.1.480-486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Emerson JB, Thomas BC, Andrade K, Allen EE, Heidelberg KB, Banfielda JF. 2012. Dynamic viral populations in hypersaline systems as revealed by metagenomic assembly. Appl Environ Microbiol 78:6309–6320. doi: 10.1128/AEM.01212-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas DSG. 2011. Arid environments: their nature and extent, p 1–16. In Thomas DSG. (ed), Arid zone geomorphology: process, form and change in drylands, 3rd ed John Wiley & Sons, Ltd., Chichester, United Kingdom. doi: 10.1002/9780470710777.ch1. [DOI] [Google Scholar]
- 82.Schlesinger WH, Pippen JS, Wallenstein MD, Hofmockel KS, Klepeis DM, Mahall BE. 2003. Community composition and photosynthesis by photoautotrophs under quartz pebbles, southern Mojave Desert. Ecology 84:3222–3231. doi: 10.1890/02-0549. [DOI] [Google Scholar]
- 83.Warren-Rhodes KA, Rhodes KL, Pointing SB, Ewing SA, Lacap DC, Gómez-Silva B, Amundson R, Friedmann EI, McKay CP. 2006. Hypolithic cyanobacteria, dry limit of photosynthesis, and microbial ecology in the hyperarid Atacama Desert. Microb Ecol 52:389–398. doi: 10.1007/s00248-006-9055-7. [DOI] [PubMed] [Google Scholar]
- 84.Warren-Rhodes KA, Rhodes KL, Boyle LN, Pointing SB, Chen Y, Liu S, Zhuo P, McKay CP. 2007. Cyanobacterial ecology across environmental gradients and spatial scales in China's hot and cold deserts. FEMS Microbiol Ecol 61:470–482. doi: 10.1111/j.1574-6941.2007.00351.x. [DOI] [PubMed] [Google Scholar]
- 85.Garcia-Pichel F, López-Cortés A, Nübel U. 2001. Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado Plateau. Appl Environ Microbiol 67:1902–1910. doi: 10.1128/AEM.67.4.1902-1910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vos M, Wolf AB, Jennings SJ, Kowalchuk GA. 2013. Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol Rev 37:936–954. doi: 10.1111/1574-6976.12023. [DOI] [PubMed] [Google Scholar]