Abstract
We used whole-genome sequencing to determine evolutionary relationships among 20 outbreak-associated clinical isolates of Listeria monocytogenes serotypes 1/2a and 1/2b. Isolates from 6 of 11 outbreaks fell outside the clonal groups or “epidemic clones” that have been previously associated with outbreaks, suggesting that epidemic potential may be widespread in L. monocytogenes and is not limited to the recognized epidemic clones. Pairwise comparisons between epidemiologically related isolates within clonal complexes showed that genome-level variation differed by 2 orders of magnitude between different comparisons, and the distribution of point mutations (core versus accessory genome) also varied. In addition, genetic divergence between one closely related pair of isolates from a single outbreak was driven primarily by changes in phage regions. The evolutionary analysis showed that the changes could be attributed to horizontal gene transfer; members of the diverse bacterial community found in the production facility could have served as the source of novel genetic material at some point in the production chain. The results raise the question of how to best utilize information contained within the accessory genome in outbreak investigations. The full magnitude and complexity of genetic changes revealed by genome sequencing could not be discerned from traditional subtyping methods, and the results demonstrate the challenges of interpreting genetic variation among isolates recovered from a single outbreak. Epidemiological information remains critical for proper interpretation of nucleotide and structural diversity among isolates recovered during outbreaks and will remain so until we understand more about how various population histories influence genetic variation.
INTRODUCTION
Listeria monocytogenes is a bacterial pathogen that is almost exclusively transmitted by food. Invasive listeriosis typically presents as sepsis or meningoencephalitis in older adults, those with certain chronic illnesses, and people undergoing immunosuppression. Infections during pregnancy can cause fever and other nonspecific symptoms in the mother with severe outcomes such as fetal loss, premature labor, and neonatal illness and death. Although listeriosis is relatively rare (∼1,600 cases occur annually in the United States), approximately 20% of cases are fatal and outbreaks are not uncommon. There are 13 known serotypes of L. monocytogenes, though the majority of human illnesses are caused by serotypes 1/2a, 1/2b, and 4b (1, 2). Molecular subtyping methods have differentiated L. monocytogenes isolates into 4 distinct genetic lineages, with isolates of serotypes 4b and 1/2b typically belonging to lineage I (LI) and isolates of serotype 1/2a typically belonging to lineage II (LII) (3). Strains of lineages III and IV rarely cause listeriosis in humans. Historically, isolates of serotype 4b have caused the greatest proportion of listeriosis outbreaks and the largest number of cases per outbreak (2). In 2011, however, serotypes 1/2a and 1/2b were implicated in the largest listeriosis outbreak in U.S. history. Whole cantaloupes from a single farm were identified as the source, highlighting the potential for L. monocytogenes transmission via fresh produce (4). Ultimately, five different pulsed-field gel electrophoresis (PFGE) patterns associated with outbreak-related illness were identified (4).
Although standardized PFGE is currently the established subtyping method for detecting clusters of disease and confirming the source of listeriosis outbreaks, the method cannot be used to infer evolutionary relationships. Establishing the evolutionary relatedness among subtypes of L. monocytogenes allows us to identify groups of strains that account for a larger proportion of sporadic listeriosis cases and are more often associated with outbreaks. A strength of nucleic acid sequencing-based subtyping is that it enables categorization of strains into related subgroups that may share important genetic characteristics because of common ancestry. Multilocus sequence typing (MLST) (5) and multivirulence-locus sequence typing (MvLST) (6) are amenable to evolutionary analysis, and these approaches have been used to categorize isolates into higher-level groups: a retrospective study of L. monocytogenes clinical isolates in Canada used MLST and MvLST to demonstrate that isolates with similar PFGE patterns recovered over 2 decades all belonged to the same clonal group (7). A recent comparison of the MLST and MvLST typing schemes (8) demonstrated correspondence in both phylogenetic clustering and discriminatory power between clonal complexes (CCs) (as determined by MLST) and epidemic clones (ECs) (as determined by MvLST) with prevalent, globally disseminated CCs in LI and LII encompassing ECs. An EC has been defined as a group of isolates that are genetically related, some of which have been implicated in temporally and geographically unrelated outbreaks. An implicit assumption in the definition of ECs is that not all L. monocytogenes isolates are equivalent with respect to their potential to cause outbreaks. In contrast, the CC is solely framed in evolutionary biology terms as a group of isolates that descended from a common ancestor and accumulated differences mainly through mutations; no involvement in listeriosis epidemics is implied from the designation CC.
A previous study analyzed concatenated virulence and housekeeping gene sequences to show that the isolates from the U.S. cantaloupe-associated outbreak fell into three groups: one isolate was not related to other outbreak strains, but the other isolates fell into two separate groups that contained strains from previous outbreaks, and thus, each of these two groups were designated novel epidemic clones, including the first described for serotype 1/2b (9). We expanded upon these findings by using whole-genome sequences to reconstruct the evolutionary relationships among L. monocytogenes strains of serotypes 1/2a and 1/2b that were implicated in outbreaks over the last 2 decades, including the outbreak associated with cantaloupe. We first present a high-level overview by placing the strains into a phylogenetic (clonal) framework to begin to understand how outbreak-associated strains are distributed within the diversity of L. monocytogenes. We then examine in more detail three pairs of closely related isolates from three CCs to understand more about genome-level variation within different outbreaks. Lastly, we delve into a more detailed analysis of a single pair of epidemiologically related isolates to illustrate the complexity of genetic changes that can be present among isolates in a single outbreak.
MATERIALS AND METHODS
Strain selection, characterization, and phylogenetic analysis.
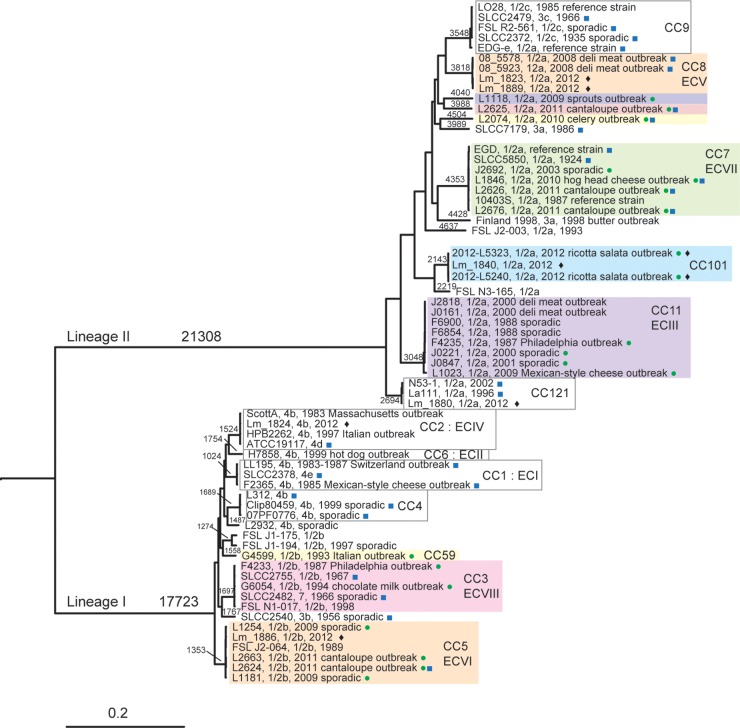
We selected a total of 20 isolates of serotypes 1/2a (13 isolates) and 1/2b (7 isolates) from our collection of clinical isolates, including representative isolates from U.S. outbreaks and sporadic isolates that were similar to these outbreak strains based on previously generated PFGE and multilocus genotyping (MLGT) data (10) (see Table S1 in the supplemental material). Genomic DNA was extracted using the ArchivePure DNA cell/tissue kit (5 PRIME, Inc., Gaithersburg, MD). Whole-genome sequences were determined on a GAIIx using standard Illumina chemistry to generate 76-bp reads, which were assembled using Velvet version 1.0.2.8 (11) and VelvetOptimiser-2.2.4. In addition, we included 49 publicly available genomes in the analyses, for a total of 69 sequences. This data set comprises 52 genomes for isolates of serotypes 1/2a and 1/2b and phylogenetically related serotypes (1/2c and 3). These 52 genomes include 20 outbreak-associated isolates representing 11 outbreaks and isolates from clinical, food, and environmental sources, for example, one isolate from an implicated production facility (ricotta salata) that matched a human case (12). We also included 17 additional genome sequences from isolates of serotypes 4b, 4d, and 4e to root the phylogenetic tree. Gene sequences corresponding to the 7 loci for the Institut Pasteur L. monocytogenes MLST scheme (5) were compiled from the genome sequences. MLST sequence types (STs) were determined based on comparisons with the Institut Pasteur MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Lmono.html). We assigned strains to CCs based on STs as presented in the work of Ragon et al. (5) and in the Pasteur database. We used the previously defined method for classification of CCs, which is that CCs are groups of allelic profiles sharing 6 out of 7 alleles with at least one other member of the group (5). As the older genome sequences do not have reads available and cannot be used with high-quality, single nucleotide polymorphism (hqSNP) analysis, single nucleotide polymorphisms (SNPs) were called using kSNP v2.0 (13) with a k-mer size of 19; the resulting SNP matrix was used as the input for a maximum likelihood phylogenetic analysis using RAxML v7.4.2 (14). Values on the branches in Fig. 1 indicate SNP differences for the main lineage and clonal complexes as inferred by kSNP, which included only SNPs that were present in 50% or more of the isolates. Based on previous comparisons (H. C. den Bakker, unpublished data), these SNP differences are comparable to SNP counts obtained from the core genome. Additionally, due to its core algorithm, kSNP will not identify clustered SNPs, as opposed to our hqSNP method, and there were some hot-spot regions which kSNP undercalled SNPs, e.g., the clustered SNPs found in Fig. 3. Thus, SNP counts from the kSNP method are considerably lower than pairwise hqSNP counts, which would include clustered SNPs and more SNPs in the accessory genome.
FIG 1.
Listeria monocytogenes phylogeny based on single variable nucleotides. Variable nucleotides were found using kSNP v2.0, and RAxML v7.4.2 was used to infer the phylogeny. Interior branches that define lineages I and II are labeled alongside the respective branches. Clonal complex (CC) and epidemic clone (EC) clades grouped together and are labeled next to a vertical bar to indicate all CC or EC members. All lineage, CC, and EC clades are supported with 100% of the RAxML bootstrap repetitions. The scale bar at the bottom represents the point variations per variable site. Values along the branches represent the numbers of variable nucleotides along that branch. Colored boxes represent groups that contain one or more 1/2a or 1/2b outbreak-associated isolates. Green circles indicate isolates that were newly sequenced for this study, and blue squares indicate isolates for which a completed genome is available. Diamonds are used to indicate isolates that were used in more detailed downstream analyses.
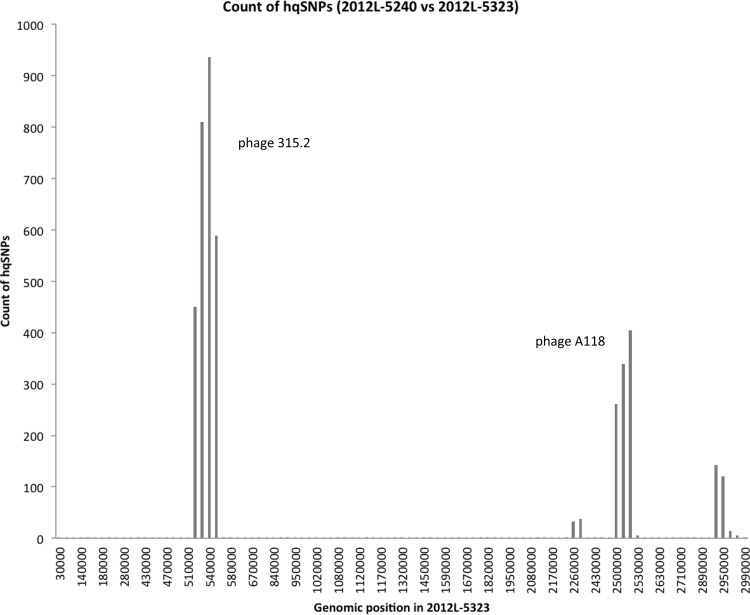
FIG 3.
hqSNP counts between CC101 clinical isolates 2012L-5240 and 2012L-5323 plotted against the position of a pseudochromosome for 2012L-5323. High-quality SNPs were determined using the raw reads of 2012L-5240 and mapping them against the assembly of 2012L-5323. The graph was produced by counting the number of hqSNPs per 10,000 positions in the assembly. Two phages as inferred by PHAST were each located in a region with high hqSNP counts; the two phages are identified near the corresponding hqSNP peak.
Comparison of epidemiologically linked isolate pairs.
We performed more detailed comparisons on pairs of genome sequences from epidemiologically linked isolate pairs within CCs. We used Lyve-SET v0.7 (https://github.com/lskatz/lyve-SET) to identify hqSNPs (15). SnpEff (16) was used to annotate SNPs as falling into protein-encoding (coding DNA sequence [CDS]) or non-protein-encoding regions, with the former also designated synonymous or nonsynonymous SNPs. The CDS SNPs were also annotated as core genome versus accessory genome by searching against a list of core genes obtained from a Markov clustering (MCL) method implemented in ITEP (Integrated Toolkit for Exploration of microbial Pan-genomes) (17). In short, a gene presence-absence matrix was constructed for finished genomes used in this study, using an MCL inflation value of 2.0 and a BLAST maxbit score of 0.4. One pairwise comparison with high SNP counts was further examined by ordering contigs of one genome against a reference genome using the Mauve Contig Mover (18) and concatenating sorted contigs to create a pseudochromosome. Reads from the second closely related genome were then mapped against the pseudochromosome to examine the distribution of pairwise SNP differences. The reciprocal mapping of reads between the two closely related strains was also performed. PHAST (19) was used to identify phages within the two genomes, and phage sequences were extracted using a custom script, extractSequence.pl (https://github.com/lskatz/lskScripts), and the exact coordinates given by PHAST. Phage phylogenies were inferred using RAxML and visualized with MEGA5 (20).
Nucleotide sequence accession numbers.
Newly determined sequence data were deposited in GenBank under accession numbers CP007689, CP007687, CP007688, CP007684, CP007685, CP007686, JMUA00000000, JPTW00000000, JNGR00000000, JNFI00000000, JNHA00000000, JPTX00000000, JNGJ00000000, JNGK00000000, JNGL00000000, JNGM00000000, JNGN00000000, JNGO00000000, JNGY00000000, and JNGQ00000000.
RESULTS AND DISCUSSION
Phylogenetic relationships of outbreak-associated strains show that multiple outbreak strains are unrelated to clonal complexes or epidemic clones.
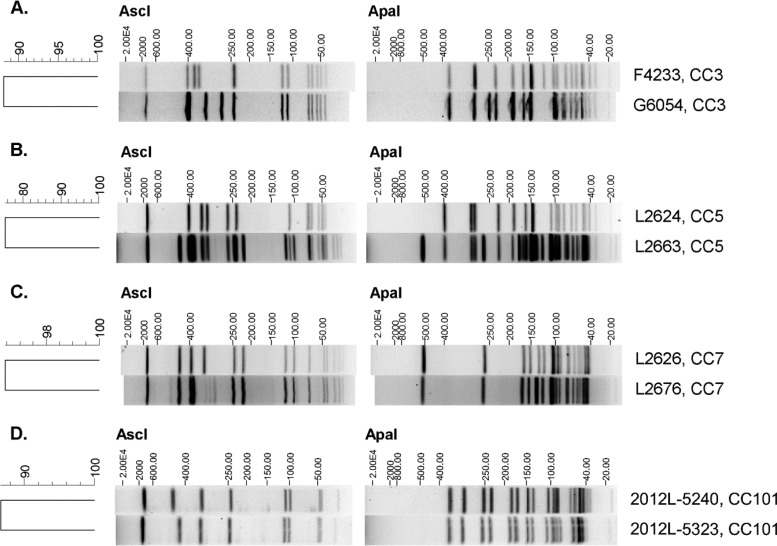
We used SNP data from whole-genome sequencing (WGS) to characterize the relationships among the 1/2a and 1/2b outbreak isolates and place them in the larger context of L. monocytogenes genetic diversity. A total of 20 outbreak-associated isolates of serotypes 1/2a and 1/2b were included in the data set, and these isolates belonged to 10 distinct phylogenetic groups. The majority of isolates (11, or 55%) were categorized into four groups corresponding to common CCs that represent the four recognized epidemic clones for serotypes 1/2 a (III, V, VII) and 1/2b (VI) (Fig. 1). The remaining 9 isolates represented six branches in the phylogenetic tree; one branch included two 1/2b isolates (F4233 and G6054) that represented two historically unrelated outbreaks (Pennsylvania and chocolate milk outbreaks, respectively) (Table 1). The genome sequences of the two isolates differed by 181 hqSNPs. Representative isolates from the different groups of serotype 1/2b differed by approximately 7,000 to 8,500 SNPs; thus, F4233 and G6054 are closely related to one another relative to the diversity within serotype 1/2b. The genetic relationship among strains F4233 and G6054 cannot be inferred by PFGE; however, there were only a few band differences comparing the AscI and ApaI profiles of these strains (Fig. 2A).The two isolates represent CC3, which ranks among the four most common L. monocytogenes clones across the globe (21). By using a subtyping technique that can be used for evolutionary inference, we were able to demonstrate that CC3 may be a previously unrecognized EC (VIII), the second to be described for serotype 1/2b. However, the chocolate milk outbreak was gastroenteritis and not invasive listeriosis; whether the designation of epidemic clone should be applied in this case could be debated. It is unclear why a large, globally disseminated clone has not been implicated in numerous outbreaks; as application of WGS increases, CC3 may be recovered from multiple listeriosis outbreaks and thus designated unambiguously as an epidemic clone.
TABLE 1.
Isolate information and molecular characterization of the L. monocytogenes genomes used in this study
| Isolate | Type | U.S. state or country | Outbreak associateda | Yr | Source | Pasteur ST | CCb | EC | Reference(s)c | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| F2365 | 4b | California | Yes, Mexican-style cheese | 1985 | Cheese | 1 | 1 | I | 31, 32 | NC_002973.6 |
| SLCC 2378 | 4e | NAe | No | NA | Poultry | 73 | 1 | I | 33 | FR733644 |
| LL195 | 4b | Switzerland | Yes, Vacherin Mont d'Or cheese | 1983 | Human | 1 | 1 | I | 34 | HF558398.1 |
| Lm_1824 | 4b | Italy | Nod | 2012 | Cheese processing facility | 2 | 2 | IV | 25 | AZIV00000000 |
| ATCC 19117 | 4d | NA | No | NA | Sheep | 2 | 2 | IV | 33 | FR733643 |
| HPB2262 | 4b | Italy | Yes, corn (gastroenteritis) | 1997 | Human | 2 | 2 | IV | 35, 36 | AATL00000000 |
| ScottA | 4b | Massachusetts | Yes, milk | 1983 | Human | 290 | 2 | IV | 37, 38 | CM001159.1 |
| F4233 | 1/2b | Pennsylvania | Yes, unknown | 1987 | Human | 3 | 3 | VIII | 39 | JMUA00000000 |
| FSL N1-017 | 1/2b | NA | No | 1998 | Food | 3 | 3 | VIII | 36 | AARP00000000 |
| G6054 | 1/2b | Illinois | Yes, chocolate milk (gastroenteritis) | 1994 | Human | 3 | 3 | VIII | 40 | JPTW00000000 |
| SLCC 2482 | 7 | NA | No | 1966 | Human | 3 | 3 | VIII | 33 | FR720325 |
| SLCC 2755 | 1/2b | NA | No | 1967 | Chinchilla | 66 | 3 | VIII | 33 | FR733646 |
| 07PF0776 | 4b | USA | No | NA | Human | 4 | 4 | 41, 42 | NC_017728.1 | |
| Clip80459 | 4b | NA | No | 1999 | Human | 4 | 4 | NC_012488.1 | ||
| L312 | 4b | NA | Unknown | NA | Cheese | 4 | 4 | 30 | FR733642 | |
| FSL J2-064 | 1/2b | USA | No | 1989 | Human | 5 | 5 | VI | AARO00000000.2 | |
| L1181 | 1/2b | USA | No | 2009 | Human | 5 | 5 | VI | JNGR00000000 | |
| L1254 | 1/2b | USA | No | 2009 | Human | 5 | 5 | VI | JNFI00000000 | |
| L2624 | 1/2b | Multistate, USA | Yes, cantaloupe | 2011 | Human | 5 | 5 | VI | 4 | CP007686 |
| L2663 | 1/2b | Multistate, USA | Yes, cantaloupe | 2011 | Human | 5 | 5 | VI | 4 | JNHA00000000 |
| Lm_1886 | 1/2b | Italy | Nod | 2012 | Cheese processing facility | 5 | 5 | VI | 25 | AZIX00000000 |
| H7858 | 4b | Multistate, USA | Yes, frankfurter | 1998 | Frankfurter | 6 | 6 | II | 32 | AADR00000000 |
| G4599 | 1/2b | Italy | Yes, unknown (gastroenteritis) | 1993 | Human | 59 | 59 | 43 | JPTX00000000 | |
| FSL J1-175 | 1/2b | USA | No | NA | Water | 87 | 36 | AARK00000000 | ||
| FSL J1-194 | 1/2b | USA | No | 1997 | Human | 88 | 36 | AARJ00000000 | ||
| SLCC 2540 | 3b | USA | No | 1956 | Human | 617 | 33 | FR733645 | ||
| 10403S | 1/2a | USA | No | 1987 | Reference strain | 85 | 7 | VII | 36 | AARZ00000000 |
| J2692 | 1/2a | USA | No | 2003 | Human | 7 | 7 | VII | JNGJ00000000 | |
| L1846 | 1/2a | Louisiana | Yes, hog head cheese | 2010 | Human | 7 | 7 | VII | 44 | CP007688 |
| L2626 | 1/2a | Multistate, USA | Yes, cantaloupe | 2011 | Human | 561 | 7 | VII | 4 | CP007684 |
| L2676 | 1/2a | Multistate, USA | Yes, cantaloupe | 2011 | Human | 7 | 7 | VII | 4 | CP007685 |
| SLCC 5850 | 1/2a | United Kingdom | No | 1924 | Rabbit | 12 | 7 | VII | 33 | FR733647 |
| EGD | 1/2a | NA | No | NA | Reference strain | 12 | 7 | VII | 45 | NC_022568.1 |
| 08_5578 | 1/2a | Canada | Yes, deli meat | 2008 | Human | 292 | 8 | V | 26 | NC_013766.1 |
| 08_5923 | 1/2a | Canada | Yes, deli meat | 2008 | Human | 120 | 8 | V | 26 | NC_013768.1 |
| Lm_1823 | 1/2a | Italy | Nod | 2012 | Cheese processing facility | 8 | 8 | V | 25 | AZIU00000000 |
| Lm_1889 | 1/2a | Italy | Nod | 2012 | Cheese processing facility | 8 | 8 | V | 25 | AZIY00000000 |
| EGDe | 1/2a | NA | No | NA | Reference strain | 35 | 9 | 46 | NC_003210.1 | |
| FSL R2-561 | 1/2c | USA | No | NA | Human | 9 | 9 | 36 | AARS00000000 | |
| LO28 | 1/2c | NA | No | 1985 | Reference strain | 210 | 9 | 36 | AARY00000000 | |
| SLCC 2372 | 1/2c | United Kingdom | No | 1935 | Human | 122 | 9 | 33 | FR733648 | |
| SLCC 2479 | 3c | NA | No | 1966 | NA | 9 | 9 | 33 | FR733649 | |
| F4235 | 1/2a | Pennsylvania | Yes, unknown | 1987 | Human | 11 | 11 | III | 39 | JNGK00000000 |
| F6854 | 1/2a | Oklahoma | No | 1988 | Human | 11 | 11 | III | 32 | AADQ00000000 |
| F6900 | 1/2a | USA | No | 1988 | Human | 86 | 11 | III | 27 | AARU00000000.2 |
| J0161 | 1/2a | Multistate, USA | Yes, turkey deli meat | 2000 | Human | 11 | 11 | III | 27, 47 | AARW00000000 |
| J0221 | 1/2a | USA | No | 2000 | Human | 11 | 11 | III | JNGL00000000 | |
| J0847 | 1/2a | USA | No | 2001 | Human | 11 | 11 | III | JNGM00000000 | |
| J2818 | 1/2a | Multistate, USA | Yes, turkey deli meat | 2000 | Turkey deli meat | 86 | 11 | III | 27, 47 | AARX00000000 |
| L1023 | 1/2a | Multistate, USA | Yes, Mexican-style cheese | 2009 | Human | 11 | 11 | III | 48 | JNGN00000000 |
| 2012-L5240 | 1/2a | Multistate, USA | Yes, ricotta salata | 2012 | Human | 101 | 101 | 12 | JNGO00000000 | |
| 2012L-5323 | 1/2a | Multistate, USA | Yes, ricotta salata | 2012 | Human | 101 | 101 | 12 | JNGY00000000 | |
| Lm_1840 | 1/2a | Italy | Nod | 2012 | Cheese processing facility | 101 | 101 | 25 | AZIW00000000 | |
| Finland1988 | 3a | Finland | Yes, butter | 1988 | Unknown | 155 | 36 | AART00000000 | ||
| FSL J2-003 | 1/2a | USA | No | 1993 | Human | 89 | AARM00000000 | |||
| FSL N3-165 | 1/2a | USA | No | NA | Soil | 90 | AARQ00000000 | |||
| L1118 | 1/2a | Multistate, USA | Yes, sprouts | 2009 | Human | 573 | 2 | JNGQ00000000 | ||
| L2074 | 1/2a | Texas | Yes, celery | 2010 | Human | 378 | 49 | CP007689 | ||
| L2625 | 1/2a | Multistate, USA | Yes, cantaloupe | 2011 | Human | 29 | 4 | CP007687 | ||
| SLCC 7179 | 3a | Austria | No | 1986 | Cheese | 91 | 33 | FR733650 | ||
| N53-1 | 1/2a | Denmark | No | 2002 | Salmon | 121 | 121 | 50 | HE999705.1 | |
| La111 | 1/2a | Denmark | No | 1996 | Salmon processing facility | 121 | 121 | 50 | HE999704.1 | |
| Lm_1880 | 1/2a | Italy | Nod | 2012 | Cheese processing facility | 121 | 121 | 25 | AZIZ00000000 | |
| FSL J2-071 | 4c | USA | No | 1994 | Animal | 131 | 71 | 36 | AARN00000000 | |
| SLCC 2376 | 4c | NA | No | NA | Poultry | 71 | 71 | 33 | FR733651 | |
| HCC23 | 4a | USA | No | NA | Catfish | 201 | 51 | NC_011660.1 | ||
| L99 | 4a | Netherlands | No | 1950 | Cheese | 201 | 52 | FM211688 | ||
| M7 | 4a | China | No | NA | Milk | 201 | 53 | CP002816.1 | ||
| FSL J1-208 | 4a | Georgia | No | 1998 | Animal | 569 | 54 | AARL00000000 |
Outbreaks designated according to food source, if known. All outbreaks were invasive listeriosis unless otherwise noted.
Clonal complex numbers assigned based on Pasteur MLST ST as well as data presented by Ragon et al. (5).
References are those describing the genome sequence of a given isolate as well as any publication describing the outbreak with which a given isolate was associated.
Isolates from ricotta salata processing facility; not classified as outbreak related because no matching human cases were identified.
NA, not available.
FIG 2.
AscI and ApaI PFGE profiles of Listeria monocytogenes isolates representing clonal complexes. (A) CC3 (Pennsylvania and chocolate milk outbreaks); (B) CC5 (cantaloupe outbreak); (C) CC7 (cantaloupe outbreak); (D) CC101 (ricotta salata outbreak). Strain identification and CC are given to the right of each profile. The AscI and ApaI patterns were compared in BioNumerics v.6.6.10 using the unweighted pair group method with arithmetic mean; the dendrogram shown to the left of each pair of profiles was produced using the Dice similarity coefficient, with the tolerance and optimization set at 1.5%.
Four of the remaining branches were represented by single outbreak isolates (L1118, L2074, L2625, and G4599) that were not closely related to any of the other isolates (Fig. 1). Lastly, two isolates (2012L-5240 and 2012L-5323) with similar PFGE subtypes from the U.S. 2012 ricotta salata outbreak (12) were determined to be related to each other but were not closely related to other L. monocytogenes phylogenetic groups. The isolates from this outbreak form a distinct genetic group (Fig. 1) and were classified as members of CC101.
Magnitude and pattern of genetic divergence varies widely between the epidemiologically linked isolate pairs.
WGS is being adopted by public health agencies for many functions, including routine surveillance (22) and outbreak detection and investigation (23, 24). Interpreting genetic variation will present new challenges to such investigations. To gain insight into the genome diversity within L. monocytogenes subgroups as defined by more traditional typing methods (i.e., MLST and PFGE), we examined the hqSNP differences between three pairs of epidemiologically linked isolates within CCs that had distinct PFGE subtypes (Table 2; Fig. 2B to D). We examined two isolate pairs from the 2011 cantaloupe outbreak that belong to two different CCs; the pair of isolates in CC5 (L2624 and L2663) had only 7 hqSNPs between them, which were all located in the accessory genome. In contrast, the pair of cantaloupe outbreak isolates from CC7 (L2626 and L2676) had 169 hqSNPs, with the majority of SNPs located in the core genome. The differences as assessed by PFGE were not concordant with WGS, as the CC5 isolates showed 16 discernible band differences (∼75% similarity), whereas the CC7 isolates had only 1 discernible difference (∼96% similarity). The pair of clinical isolates from the 2012 ricotta salata outbreak (2012L-5240 and 2012L-5323; CC101) had the greatest number of hqSNPs between them, with a total of 4,244 SNPs, 3,782 of which were located in the accessory genome. The reciprocal reference comparison identified a similar number of SNPs (4,562); for comparative purposes, we report the more conservative estimate in Table 2. The two isolates were nearly identical by PFGE (Fig. 2D).
TABLE 2.
Variation in hqSNPs within outbreak strains of L. monocytogenes from the same clonal complex
| Outbreak isolates | Clonal complex | No. of total hqSNPsa | No. of hqSNPs in protein coding genesb | No. per core/ no. per accessoryc |
No. for S/ no. for Nd |
|---|---|---|---|---|---|
| L2624 vs L2663 | 5 | 7 | 7 | 0/7 | 2/5 |
| L2626 vs L2676 | 7 | 207 | 169 | 139/30 | 108/61 |
| 2012L-5240 vs 2012L-5323 | 101 | 4,244 | 3,868 | 80/3,788 | 2,775/1,093 |
Total number of high-quality SNPs determined as described by Katz et al. (15).
Number of hqSNPs located within protein coding genes.
Number of hqSNPs in protein coding genes that are either the core genome or the accessory genome.
Number of hqSNPs in protein coding genes that lead to a synonymous (S) or nonsynonymous (N) change.
On the surface, the three isolate pairs appeared to have similar levels of genetic difference based on MLST (i.e., 0 or 1 differences in allelic profiles). However, overall genetic differences between the pairs as assessed by WGS varied by more than 2 orders of magnitude across the three comparisons. In addition, the patterns of mutation and the processes contributing to the divergence of the pairs of isolates also differed. The two isolates we examined in CC7 appeared to have diverged predominantly by point mutations within the core genome. In contrast, the CC101 pair has undergone substantial divergence within the accessory genome, with the clusters of SNPs likely introduced by horizontal gene transfer (HGT) from other Listeria strains. Because we compared only two isolates from each CC, it is unlikely that the results represent differences between the CCs; instead, they illustrate the various magnitudes of genetic difference that can occur among members of a clonal complex within single outbreaks. We next explored in more detail the genetic differences between the two CC101 isolates using publicly available draft genomes of L. monocytogenes strains from the implicated ricotta salata cheese processing facility (25).
HGT in phage regions contributes substantially to genetic divergence between clonally related strains within a single outbreak.
The cheese processing facility where the ricotta salata originated was shown to be contaminated with genetically diverse subtypes of L. monocytogenes (25): two isolates from CC8 were recovered and sequenced, as were one isolate each from CC121, CC5, CC2, and CC101 (Fig. 1). Such diversity in the microbial community raises the possibility of recombination between Listeria strains in the environment. We tested the hypothesis that HGT among other L. monocytogenes isolates present in the processing environment contributed to the diversification of CC101 isolates as follows. We created a histogram of hqSNP differences for the two clinical isolates in CC101 and then overlaid the distribution of point mutations onto a genome map (Fig. 3). In the histogram, we found two large peaks in hqSNP differences (Fig. 3); each peak corresponded to regions with sequences that were homologous to phages. We found specific phage-like sequences from the two regions that were present in the majority of CCs from the cheese facility and so examined specific elements within these two regions in more detail. The first phage-like element fell within the first large peak of SNP counts, and it was identified by BLAST as being chimeric: it showed similarity to a phage sequence found in Streptococcus pyogenes (six genes with similarity to phage 315.2; GenBank accession no. NC_004585) and also showed similarity to two phages previously found in Listeria (six genes in each of phages A006 [NC_009815] and LP-101 [NC_024387]). We refer to this genetic element here as phage 315.2′ for simplicity and hypothesize that this is a novel phage in Listeria with regions that share homology with phages in Streptococcus. The second phage-like element was present in the second large peak of SNP counts and was identified by BLAST as being most similar to a phage sequence found in L. monocytogenes (phage A118) (although this phage was also chimeric).
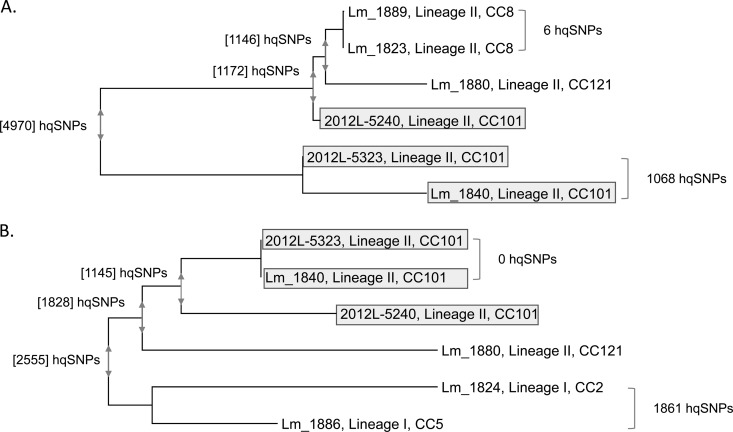
The phage sequences for each of the two genome regions were extracted when present, and phylogenies were generated for each phage segment (Fig. 4). The phylogeny for phage 315.2 differed from the L. monocytogenes WGS phylogeny, as it showed that one of the CC101 clinical isolates, 2012L-5240, was more closely related to the phage sequences from CC8 processing facility isolates Lm_1823 and Lm_1889 (Fig. 4A). The phylogeny for phage 315.2 thus bolstered the notion that CC101 isolates have diverged through recombination with other L. monocytogenes strains present in the environment. It is possible that recombination occurred between CC8 strains and CC101 strains; alternatively, both strains could have received genetic material from an additional unsampled strain in the environment. The other two CC101 isolates also showed extensive diversity in the same region and differed by 1,068 SNPs. The phylogeny for the second phage (homologous to L. monocytogenes phage A118) (Fig. 4B) showed that it was also divergent between the CC101 isolates and thus accounted for some SNP diversity between the outbreak isolates. The divergence in the second phage sequence between CC101 isolates also was likely a result of recombination.
FIG 4.
RAxML phylogenies constructed from phage sequences from the L. monocytogenes ricotta salata outbreak clinical isolates and isolates from the implicated cheese processing facility. (A) Phylogeny constructed from the phage sequence with similarity to S. pyogenes phage 315.2 (associated with the first large peak in SNP counts in Fig. 3); (B) phylogeny constructed from the second phage sequence, homologous to L. monocytogenes phage A118 (associated with the second large peak count in Fig. 3). Numbers beside brackets to the right of the trees indicate hqSNP differences between the bracketed isolates. Numbers within brackets at the nodes of the tree indicate the maximum pairwise hqSNP difference within the group of isolates encompassed by the vertical arrows at the node. The lineage and clonal complex are provided for each isolate; those in CC101 are highlighted in gray. Within CC101, 2012L-5240 and 2012L-5323 are human clinical isolates, and Lm_1840 is an isolate from the cheese processing facility. Note that the mapping of reads to the phage regions was performed at a lower mismatch threshold than mapping to the whole genome, which resulted in higher SNP counts.
Previous studies have shown epidemiologically linked isolates to be divergent with respect to acquisition of bacteriophage, including the CC7 isolates studied in this investigation, where the comK prophage was present in L2676 but not in L2626 (9). Gilmour et al. also found two isolates from a single foodborne outbreak that differed in the presence of a prophage which they designated ϕLMC1 (26). A different scenario was described by Orsi et al., who compared the genomes of isolates from the same processing environment that were recovered 12 years apart. While only 11 SNPs were found in the core genome among four isolates, the comK phage differed by 1,274 SNPs between the pair of isolates from 1988 and the pair from 2000 (27). The results presented here for the CC101 isolates show even greater divergence in phage regions within the context of a single outbreak, between concurrently recovered strains.
For the three pairs of clinical isolates examined in this study (L2624 and L2663, L2626 and L2676, and 2012L-5240 and 2012L-5323), interviews indicated that the patients consumed the implicated product, cantaloupe or ricotta salata. The pairwise comparisons demonstrate the complexity of interpreting genetic variation even within the context of an outbreak investigation with an identified source. In one example, the isolates were nearly identical, as would typically be expected for isolates from a point source outbreak. However, this will be true only if the bacterial population has undergone a genetic bottleneck (i.e., the inoculum contains only a single genotype) and the time frame is too short for substantial genetic diversity to accumulate. More complex bacterial population histories and more diverse bacterial communities in the production chain will influence genetic diversity and thus complicate interpretation of molecular subtyping results. Genetic elements such as phages that have been horizontally transferred can be ignored in analyses to establish the strain phylogeny; however, such hypervariable regions could yield epidemiologically relevant information, particularly when examined in the context of an evolutionary framework established using core genes.
Wang et al. recently presented detailed characterization of accessory genes that lent additional support to linking food and clinical isolates in an outbreak setting because those strains all shared novel elements in hypervariable regions (28). For the ricotta salata outbreak, we show a contrasting case where HGT has resulted in divergence between clonally related isolates from a single outbreak that apparently evolved within the context of a rich microbial background. Although the timing of genetic exchange in CC101 is not known (i.e., either prior to or following introduction into the cheese processing facility), these data raise the issue that horizontal gene transfer could be common among L. monocytogenes strains when multiple strain types are present in the production chain, leading to extensive diversification of genetically related isolates, even within the context of a single outbreak. The ricotta salata outbreak investigation was conducted with PFGE, and the full scope of genetic variation among the CC101 isolates was unknown. In retrospect, had the CC101 subtypes been fully characterized, the diversity in phage regions could have been seen a strong indication that other strains were present at some point in the processing facility or product; as it was, only CC101 isolates were linked in the outbreak, and it is unknown whether other CCs were present as minority members of the population in the product sold in the United States and whether the scope of the outbreak was accurately tallied. How the accessory genome can best inform outbreak investigations is a topic that warrants further investigation.
Conclusions.
In this study, we constructed an evolutionary framework to better understand how outbreak-associated isolates of L. monocytogenes serotypes 1/2a and 1/2b are related to each other and to L. monocytogenes clones shown previously to be prevalent and globally disseminated (21). Most outbreak-related strains in our study were associated with large CCs, an insight that could not be gained from PFGE subtyping. Our study also revealed that genetic divergence between epidemiologically related isolates within the same CC varied greatly as assessed by WGS, and we demonstrated that substantial divergence between clonally related isolates from a single outbreak coincided with a diverse microbial community in the processing environment. The complexity of genome-level variation is such that epidemiological and product-related information will continue to be critical for outbreak investigations. In addition, the genetic diversity we found among closely related isolates from a single outbreak highlights the need to more fully characterize outbreaks by sequencing multiple isolates from food products and production environments.
The variation in prevalence and distribution of CCs implies that there are genetic differences among strains that lead some groups to realize higher levels of transmission (5, 29, 30). How this relates to epidemic potential is not entirely clear, however, as L. monocytogenes clones not associated with large CCs or ECs were also implicated in outbreaks in this study. It is possible that some strains in the study were associated with an outbreak merely because of chance events that enhanced their transmission (i.e., any LI or LII strain could cause an epidemic given the right circumstances). Alternatively, some genetic change could have occurred—such as a change in gene content or a nucleotide substitution that led to altered gene function or expression—and these L. monocytogenes strains thus represent emerging epidemic clones. The lack of sequence data from closely related sporadic isolates does not allow us to examine the alternative hypothesis in the current study. This limitation highlights the need to conduct routine surveillance with molecular techniques that are amenable to evolutionary analysis—such as WGS and MLST—so that we can more fully understand the population context from which listeriosis epidemics arise. Understanding how both sporadic and outbreak isolates are distributed into clonal groups provides a framework for further investigation into the biological characteristics of more successful groups, with the goal of identifying the genetic basis for the traits that enable higher rates of transmission and virulence. The insights provided by examining diversity in an evolutionary framework may ultimately lead us to better control methods for L. monocytogenes.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Gerner-Smidt and three anonymous reviewers for insightful comments.
Material support was provided by the National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or other government agencies.
Funding Statement
T.M.B. and H.C.D.B. thank M. Wiedmann for support from USDA NIFA Special Research Grants 2008-34459-19043 and 2009-34459-19750.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02440-15.
REFERENCES
- 1.Silk BJ, Date KA, Jackson KA, Pouillot R, Holt KG, Graves LM, Ong KL, Hurd S, Meyer R, Marcus R, Shiferaw B, Norton DM, Medus C, Zansky SM, Cronquist AB, Henao OL, Jones TF, Vugia DJ, Farley MM, Mahon BE. 2012. Invasive listeriosis in the Foodborne Diseases Active Surveillance Network (FoodNet), 2004–2009: further targeted prevention needed for higher-risk groups. Clin Infect Dis 54(Suppl 5):S396–S404. [DOI] [PubMed] [Google Scholar]
- 2.Cartwright EJ, Jackson KA, Johnson SD, Graves LM, Silk BJ, Mahon BE. 2013. Listeriosis outbreaks and associated food vehicles, United States, 1998–2008. Emerg Infect Dis 19:1–9; quiz, 184. doi: 10.3201/eid1901.120393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward TJ, Gorski L, Borucki MK, Mandrell RE, Hutchins J, Pupedis K. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J Bacteriol 186:4994–5002. doi: 10.1128/JB.186.15.4994-5002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCollum JT, Cronquist AB, Silk BJ, Jackson KA, O'Connor KA, Cosgrove S, Gossack JP, Parachini SS, Jain NS, Ettestad P, Ibraheem M, Cantu V, Joshi M, DuVernoy T, Fogg NW Jr, Gorny JR, Mogen KM, Spires C, Teitell P, Joseph LA, Tarr CL, Imanishi M, Neil KP, Tauxe RV, Mahon BE. 2013. Multistate outbreak of listeriosis associated with cantaloupe. N Engl J Med 369:944–953. doi: 10.1056/NEJMoa1215837. [DOI] [PubMed] [Google Scholar]
- 5.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, Jayarao BM, Knabel SJ. 2004. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl Environ Microbiol 70:913–920. doi: 10.1128/AEM.70.2.913-920.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knabel SJ, Reimer A, Verghese B, Lok M, Ziegler J, Farber J, Pagotto F, Graham M, Nadon CA, Canadian Public Health Laboratory N, Gilmour MW. 2012. Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J Clin Microbiol 50:1748–1751. doi: 10.1128/JCM.06185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantinelli T, Chenal-Francisque V, Diancourt L, Frezal L, Leclercq A, Wirth T, Lecuit M, Brisse S. 2013. “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J Clin Microbiol 51:3770–3779. doi: 10.1128/JCM.01874-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lomonaco S, Verghese B, Gerner-Smidt P, Tarr C, Gladney L, Joseph L, Katz L, Turnsek M, Frace M, Chen Y, Brown E, Meinersmann R, Berrang M, Knabel S. 2013. Novel epidemic clones of Listeria monocytogenes, United States, 2011. Emerg Infect Dis 19:147–150. doi: 10.3201/eid1901.121167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward TJ, Ducey TF, Usgaard T, Dunn KA, Bielawski JP. 2008. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl Environ Microbiol 74:7629–7642. doi: 10.1128/AEM.01127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zerbino DR. 2010. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics Chapter 11:Unit 11.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heiman KE, Garalde VB, Gronostaj M, Jackson KA, Beam S, Joseph L, Saupe A, Ricotta E, Waechter H, Wellman A, Adams-Cameron M, Ray G, Fields A, Chen Y, Datta A, Burall L, Sabol A, Kucerova Z, Trees E, Metz M, Leblanc P, Lance S, Griffin PM, Tauxe RV, Silk BJ. 30 June 2015. Multistate outbreak of listeriosis caused by imported cheese and evidence of cross-contamination of other cheeses, USA, 2012. Epidemiol Infect doi: 10.1017/S095026881500117X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 15.Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Guo Y, Wang S, Paxinos EE, Orata F, Gladney LM, Stroika S, Folster JP, Rowe L, Freeman MM, Knox N, Frace M, Boncy J, Graham M, Hammer BK, Boucher Y, Bashir A, Hanage WP, Van Domselaar G, Tarr CL. 2013. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. mBio 4(4):e00398-13. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benedict MN, Henriksen JR, Metcalf WW, Whitaker RJ, Price ND. 2014. ITEP: an integrated toolkit for exploration of microbial pan-genomes. BMC Genomics 15:8. doi: 10.1186/1471-2164-15-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. 2009. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chenal-Francisque V, Lopez J, Cantinelli T, Caro V, Tran C, Leclercq A, Lecuit M, Brisse S. 2011. Worldwide distribution of major clones of Listeria monocytogenes. Emerg Infect Dis 17:1110–1112. doi: 10.3201/eid/1706.101778,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.den Bakker HC, Allard MW, Bopp D, Brown EW, Fontana J, Iqbal Z, Kinney A, Limberger R, Musser KA, Shudt M, Strain E, Wiedmann M, Wolfgang WJ. 2014. Rapid whole-genome sequencing for surveillance of Salmonella enterica serovar Enteritidis. Emerg Infect Dis 20:1306–1314. doi: 10.3201/eid2008.131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson BR, Salter M, Tarr C, Conrad A, Harvey E, Steinbock L, Saupe A, Sorenson A, Katz L, Stroika S, Jackson KA, Carleton H, Kucerova Z, Melka D, Strain E, Parish M, Mody RK, Centers for Disease Control and Prevention . 2015. Notes from the field: listeriosis associated with stone fruit—United States, 2014. MMWR Morb Mortal Wkly Rep 64:282–283. [PMC free article] [PubMed] [Google Scholar]
- 24.CDC. 2015. Multistate outbreak of listeriosis linked to Blue Bell Creameries products (final update). Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/listeria/outbreaks/ice-cream-03-15/index.html Accessed 13 October 2015. [Google Scholar]
- 25.Chiara M, D'Erchia AM, Manzari C, Minotto A, Montagna C, Addante N, Santagada G, Latorre L, Pesole G, Horner DS, Parisi A. 2014. Draft genome sequences of six Listeria monocytogenes strains isolated from dairy products from a processing plant in southern Italy. Genome Announc 2(2):e00282-14. doi: 10.1128/genomeA.00282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. doi: 10.1186/1471-2164-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, Galagan JE, Birren BW, Ivy RA, Sun Q, Graves LM, Swaminathan B, Wiedmann M. 2008. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9:539. doi: 10.1186/1471-2164-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Holmes N, Martinez E, Howard P, Hill-Cawthorne G, Sintchenko V. 26 August 2015. It is not all about SNPs: comparison of mobile genetic elements and deletions in Listeria monocytogenes genomes links cases of hospital-acquired listeriosis to the environmental source. J Clin Microbiol doi: 10.1128/JCM.00202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.den Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, Degoricija L, Barker M, Petrauskene O, Furtado MR, Wiedmann M. 2010. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 11:688. doi: 10.1186/1471-2164-11-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, Goesmann A, Barbuddhe S, Hain T, Chakraborty T. 2013. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14:47. doi: 10.1186/1471-2164-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CDC. 1985. Listeriosis outbreak associated with Mexican-style cheese—California. MMWR Morb Mortal Wkly Rep 34:357–359. [PubMed] [Google Scholar]
- 32.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT, Peterson J, White O, Nelson WC, Nierman W, Beanan MJ, Brinkac LM, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Haft DH, Selengut J, Van Aken S, Khouri H, Fedorova N, Forberger H, Tran B, Kathariou S, Wonderling LD, Uhlich GA, Bayles DO, Luchansky JB, Fraser CM. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res 32:2386–2395. doi: 10.1093/nar/gkh562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haase JK, Murphy RA, Choudhury KR, Achtman M. 2011. Revival of Seeliger's historical ‘Special Listeria Culture Collection.’ Environ Microbiol 13:3163–3171. doi: 10.1111/j.1462-2920.2011.02610.x. [DOI] [PubMed] [Google Scholar]
- 34.Weinmaier T, Riesing M, Rattei T, Bille J, Arguedas-Villa C, Stephan R, Tasara T. 2013. Complete genome sequence of Listeria monocytogenes LL195, a serotype 4b strain from the 1983–1987 listeriosis epidemic in Switzerland. Genome Announc 1(1):e00152-12. doi: 10.1128/genomeA.00152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aureli P, Fiorucci GC, Caroli D, Marchiaro G, Novara O, Leone L, Salmaso S. 2000. An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria monocytogenes. N Engl J Med 342:1236–1241. doi: 10.1056/NEJM200004273421702. [DOI] [PubMed] [Google Scholar]
- 36.den Bakker HC, Desjardins CA, Griggs AD, Peters JE, Zeng Q, Young SK, Kodira CD, Yandava C, Hepburn TA, Haas BJ, Birren BW, Wiedmann M. 2013. Evolutionary dynamics of the accessory genome of Listeria monocytogenes. PLoS One 8:e67511. doi: 10.1371/journal.pone.0067511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fleming DW, Cochi SL, MacDonald KL, Brondum J, Hayes PS, Plikaytis BD, Holmes MB, Audurier A, Broome CV, Reingold AL. 1985. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N Engl J Med 312:404–407. doi: 10.1056/NEJM198502143120704. [DOI] [PubMed] [Google Scholar]
- 38.Briers Y, Klumpp J, Schuppler M, Loessner MJ. 2011. Genome sequence of Listeria monocytogenes Scott A, a clinical isolate from a food-borne listeriosis outbreak. J Bacteriol 193:4284–4285. doi: 10.1128/JB.05328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz B, Hexter D, Broome CV, Hightower AW, Hirschhorn RB, Porter JD, Hayes PS, Bibb WF, Lorber B, Faris DG. 1989. Investigation of an outbreak of listeriosis: new hypotheses for the etiology of epidemic Listeria monocytogenes infections. J Infect Dis 159:680–685. doi: 10.1093/infdis/159.4.680. [DOI] [PubMed] [Google Scholar]
- 40.Dalton CB, Austin CC, Sobel J, Hayes PS, Bibb WF, Graves LM, Swaminathan B, Proctor ME, Griffin PM. 1997. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N Engl J Med 336:100–105. doi: 10.1056/NEJM199701093360204. [DOI] [PubMed] [Google Scholar]
- 41.Alonzo F III, Bobo LD, Skiest DJ, Freitag NE. 2011. Evidence for subpopulations of Listeria monocytogenes with enhanced invasion of cardiac cells. J Med Microbiol 60:423–434. doi: 10.1099/jmm.0.027185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMullen PD, Gillaspy AF, Gipson J, Bobo LD, Skiest DJ, Freitag NE. 2012. Genome sequence of Listeria monocytogenes 07PF0776, a cardiotropic serovar 4b strain. J Bacteriol 194:3552. doi: 10.1128/JB.00616-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franciosa G, Pourshaban M, Gianfranceschi M, Aureli P. 1998. Genetic typing of human and food isolates of Listeria monocytogenes from episodes of listeriosis. Eur J Epidemiol 14:205–210. doi: 10.1023/A:1007448210169. [DOI] [PubMed] [Google Scholar]
- 44.CDC. 2011. Outbreak of invasive listeriosis associated with the consumption of hog head cheese—Louisiana, 2010. MMWR Morb Mortal Wkly Rep 60:401–405. [PubMed] [Google Scholar]
- 45.Bécavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, Wu Z, Kuhbacher A, Brisse S, Pucciarelli MG, Garcia-del Portillo F, Hain T, Portnoy DA, Chakraborty T, Lecuit M, Pizarro-Cerda J, Moszer I, Bierne H, Cossart P. 2014. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. mBio 5:e00969-14. doi: 10.1128/mBio.00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couve E, de Daruvar A, Dehoux P, Domann E, Dominguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, Portillo FG, Garrido P, Gautier L, Goebel W, Gomez-Lopez N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Perez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, Tierrez A, Vázquez-Boland J-A, Voss H, Wehland J, Cossart P. 2001. Comparative genomics of Listeria species. Science 294:849–852. [DOI] [PubMed] [Google Scholar]
- 47.Olsen SJ, Patrick M, Hunter SB, Reddy V, Kornstein L, MacKenzie WR, Lane K, Bidol S, Stoltman GA, Frye DM, Lee I, Hurd S, Jones TF, LaPorte TN, Dewitt W, Graves L, Wiedmann M, Schoonmaker-Bopp DJ, Huang AJ, Vincent C, Bugenhagen A, Corby J, Carloni ER, Holcomb ME, Woron RF, Zansky SM, Dowdle G, Smith F, Ahrabi-Fard S, Ong AR, Tucker N, Hynes NA, Mead P. 2005. Multistate outbreak of Listeria monocytogenes infection linked to delicatessen turkey meat. Clin Infect Dis 40:962–967. doi: 10.1086/428575. [DOI] [PubMed] [Google Scholar]
- 48.Jackson KA, Biggerstaff M, Tobin-D'Angelo M, Sweat D, Klos R, Nosari J, Garrison O, Boothe E, Saathoff-Huber L, Hainstock L, Fagan RP. 2011. Multistate outbreak of Listeria monocytogenes associated with Mexican-style cheese made from pasteurized milk among pregnant, Hispanic women. J Food Prot 74:949–953. doi: 10.4315/0362-028X.JFP-10-536. [DOI] [PubMed] [Google Scholar]
- 49.Gaul LK, Farag NH, Shim T, Kingsley MA, Silk BJ, Hyytia-Trees E. 2012. Hospital-acquired listeriosis outbreak caused by contaminated diced celery—Texas, 2010. Clin Infect Dis doi: 10.1093/cid/cis817. [DOI] [PubMed] [Google Scholar]
- 50.Holch A, Webb K, Lukjancenko O, Ussery D, Rosenthal BM, Gram L. 2013. Genome sequencing identifies two nearly unchanged strains of persistent Listeria monocytogenes isolated at two different fish processing plants sampled 6 years apart. Appl Environ Microbiol 79:2944–2951. doi: 10.1128/AEM.03715-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steele CL, Donaldson JR, Paul D, Banes MM, Arick T, Bridges SM, Lawrence ML. 2011. Genome sequence of lineage III Listeria monocytogenes strain HCC23. J Bacteriol 193:3679–3680. doi: 10.1128/JB.05236-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hain T, Ghai R, Billion A, Kuenne CT, Steinweg C, Izar B, Mohamed W, Mraheil MA, Domann E, Schaffrath S, Karst U, Goesmann A, Oehm S, Puhler A, Merkl R, Vorwerk S, Glaser P, Garrido P, Rusniok C, Buchrieser C, Goebel W, Chakraborty T. 2012. Comparative genomics and transcriptomics of lineages I, II, and III strains of Listeria monocytogenes. BMC Genomics 13:144. doi: 10.1186/1471-2164-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J, Xia Y, Cheng C, Fang C, Shan Y, Jin G, Fang W. 2011. Genome sequence of the nonpathogenic Listeria monocytogenes serovar 4a strain M7. J Bacteriol 193:5019–5020. doi: 10.1128/JB.05501-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.den Bakker HC, Bowen BM, Rodriguez-Rivera LD, Wiedmann M. 2012. FSL J1-208, a virulent uncommon phylogenetic lineage IV Listeria monocytogenes strain with a small chromosome size and a putative virulence plasmid carrying internalin-like genes. Appl Environ Microbiol 78:1876–1889. doi: 10.1128/AEM.06969-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.