Abstract
Many studies on phage biology are based on isolation methods that may inadvertently select for narrow-host-range phages. Consequently, broad-host-range phages, whose ecological significance is largely unexplored, are consistently overlooked. To enhance research on such polyvalent phages, we developed two sequential multihost isolation methods and tested both culture-dependent and culture-independent phage libraries for broad infectivity. Lytic phages isolated from activated sludge were capable of interspecies or even interorder infectivity without a significant reduction in the efficiency of plating (0.45 to 1.15). Two polyvalent phages (PX1 of the Podoviridae family and PEf1 of the Siphoviridae family) were characterized in terms of adsorption rate (3.54 × 10−10 to 8.53 × 10−10 ml/min), latent time (40 to 55 min), and burst size (45 to 99 PFU/cell), using different hosts. These phages were enriched with a nonpathogenic host (Pseudomonas putida F1 or Escherichia coli K-12) and subsequently used to infect model problematic bacteria. By using a multiplicity of infection of 10 in bacterial challenge tests, >60% lethality was observed for Pseudomonas aeruginosa relative to uninfected controls. The corresponding lethality for Pseudomonas syringae was ∼50%. Overall, this work suggests that polyvalent phages may be readily isolated from the environment by using different sequential hosts, and this approach should facilitate the study of their ecological significance as well as enable novel applications.
INTRODUCTION
The total bacteriophage (phage) population on Earth is estimated at 1031 or more, making phages by far the most abundant biological entities on the planet (1, 2). As such, phages exert a significant influence over global biogeochemical cycles (3, 4) and are important drivers of bacterial diversity (5). Considering their ecological importance and value as a potential genetic resource, increasing our fundamental understanding of phage biology may facilitate the development of novel applications. However, research on phage diversity and ecology may be inadvertently limited by the use of biased isolation techniques that preferentially select for narrow-host-range phages (6, 7), while-broad host-range phages are consistently overlooked.
The classic approach to isolate and study phages is typically performed with high-density, nutrient-rich batch monocultures grown under planktonic conditions and most often results in the isolation of narrow-host-range phages (8–10).
Previous studies have demonstrated the existence of polyvalent phages, among which the temperate phages P1 and Mu are the most studied (11, 12). However, their broad-host-range properties were discovered accidently, and few studies have explored methods for isolating and enriching polyvalent phages. Jensen et al. developed a multiple-host enrichment method to identify phages capable of interclass infectivity (13), and Bielke et al. used a sequential isolation method to isolate phages with intergenus infectivity (14). However, these phage isolation methods have yielded variable results, with a later experiment showing no significant difference in phage host range (15). Furthermore, multiple-host enrichments tend to dramatically reduce phage infectivity, as measured by the efficiency of plating (EOP) on nonproduction hosts (13). Nevertheless, these studies demonstrate that polyvalent phages are more prevalent than previously appreciated, and changes in current isolation methods can shift the bias toward enrichment of polyvalent phages.
In order to facilitate research on the role of polyvalent phages in microbial ecology and evaluate their potential applications for microbial control or gene transfer, this study compared three different phage isolation and enrichment methods. These methods included a simultaneous multihost enrichment method and sequential multihost isolation approaches (linear and circular, with the latter involving a return to the original host). Polyvalent phages were enriched by a benign host (for safer production) and then tested for their intergenus infectivity and capacity to suppress the growth of model problematic bacteria. The utilities of phage libraries created by using either host enrichment or culture-independent concentration were also compared.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains in this study included Pseudomonas aeruginosa PAO1 (ATCC 15692), Pseudomonas aeruginosa HER1018 (ATCC BAA-47), Pseudomonas aeruginosa Migula (ATCC 700829), Pseudomonas sp. strain CF600 (16), Pseudomonas putida F1 (ATCC 700007), Pseudomonas syringae van Hall (ATCC 19310), Escherichia coli K-12 (ATCC 10798), and Escherichia coli C3000 (ATCC 15597). Culturing was performed by using tryptic soy broth (TSB) medium. The double-agar layer method was performed by using tryptone base layer agar (TBA) as a base agar and tryptone soft agar (TSA) as a soft agar (17). All bacteria were cultured at 37°C. Bacteriophages were stored at 4°C in SM buffer (50 mmol/liter Tris-HCl [pH 7.5], 0.1 mol/liter NaCl, 8 mmol/liter MgSO4, 0.01% gelatin) (18).
Preparation of phage stocks.
A sample of activated sludge was taken from the 69th Street Wastewater Treatment Plant in Houston, TX. A portion of this sample was initially mixed with an equal volume of TSB medium and incubated overnight at 37°C. Phages were detached from sediment particles by using the sodium pyrophosphate method (19). Centrifugation and filtration were used to remove particles larger than 0.2 μm, as previously described (18). The filtrate was treated as phage stock A. Another portion of the original sample was used to obtain phage stock B. Phages were similarly detached from sediment particles. Centrifugation and filtration were used to remove particles larger than 0.2 μm. The phage particles in the filtrate were further concentrated by polyethylene glycol 8000 (PEG 8000) precipitation (19) and resuspended in SM buffer to obtain the phage stock. The phage stocks were stored at 4°C and used within 1 week.
Bacteriophage isolation.
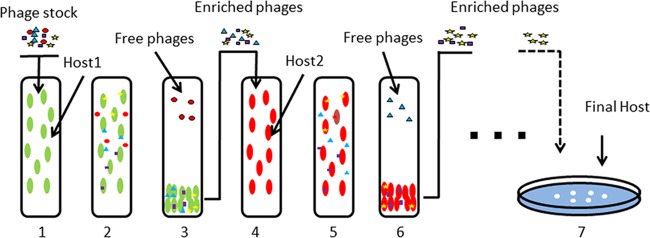
Two modified sequential multihost isolation methods were tested to isolate phages of interest. For sequential multihost isolation method A (Fig. 1), in step 1, the phage stock and host 1 were added to the upper layer of a double-layer agar plate, and the plate was incubated until plaques formed on the lawn of host 1. In step 2, all the plaques from step 1 were collected and cultured in batch with host 2 for 4 h. The phages and host 2 were subjected to double-layer plate assays, and the plates were incubated until plaques formed on the lawn of host 2. Similarly, in step 3, the plaques from step 2 were collected and cultured with host 3. The phages and host 3 were subjected to double-layer plate assays, and the plates were incubated until plaques formed on the lawn of host 3, and so on. For sequential multihost isolation method B (Fig. 2), the phage stock was added to host 1 at exponential phase to allow phages infecting host 1 to be adsorbed for 10 min. Free phages and adsorbed phages were then separated by centrifugation at 10,000 × g for 5 min. The supernatant was added to host 1 for another 10 min to allow the adsorption of phages with a low adsorption rate. Phages infecting host 1 were enriched with host 1 afterwards for 4 h. The enriched phages were added to host 2 at exponential phase, and the previous adsorption, separation, and enrichment procedures were repeated. The enriched phages were added to host 3 at exponential phase, and the same procedures were repeated again. Phages enriched by the last host were subjected to a double-layer plate assay, and the plates were incubated until plaques formed on the lawn of the last host.
FIG 1.
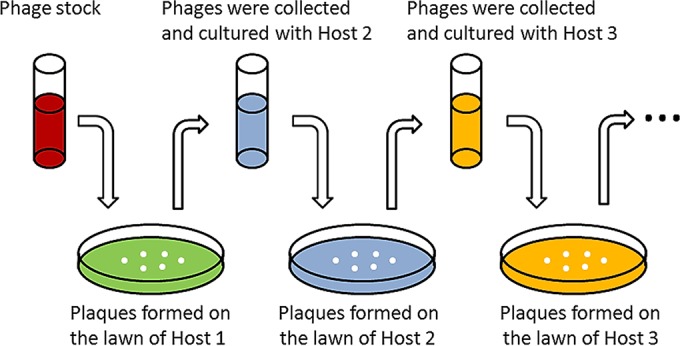
Sequential multihost isolation method A. In each step, one host was adopted to enrich and collect phages that can infect this host. The phage that can form plaques on the lawn of the last host is theoretically the one that can infect all the hosts.
FIG 2.
Sequential multihost isolation method B. (1) Phage stock is added to host 1 at exponential phase. (2) Phages infecting host 1 are adsorbed by host 1. (3) Free phages and adsorbed phages are separated by centrifugation. Phages infecting host 1 are enriched afterwards. (4 to 6) Enriched phages are added to host 2 at exponential phase, and the previous procedures are repeated with host 2. (7) Phages enriched by the last host are isolated by using the double-layer plate assay.
Bacteriophage purification.
A single phage plaque from the lawn of the last host was harvested and diluted in SM buffer. The phages were further purified three times by using standard procedures to ensure the removal of any contaminant phages (20). The phage titer was expressed as PFU per milliliter by using a double-layer plaque assay in triplicate. For morphological analysis, phage particles were further purified by ultracentrifugation using cesium chloride gradients (21) and then dialyzed in pure water to remove ions.
Transmission electron microscopy.
The purified and dialyzed phage (∼108 PFU/ml) was loaded onto carbon film copper grids and then negatively stained with 2% uranyl acetate (pH 4.5) (22). The excess stain was removed immediately, and the stained specimens were air dried for 30 min. The specimens were observed with a JEOL 2010 transmission electron microscope at 80 kV. Based on their morphology, phage identification and classification were conducted according to International Committee on Taxonomy of Viruses guidelines (23).
Bacteriophage host range and EOP.
The phage host range was initially determined by a spot test assay on the potential host lawn (24). In the spot test, 10 μl of the phage suspension (∼108 PFU/ml) was added to the potential host lawn and then incubated at 37°C overnight. The results were further confirmed by measuring the optical density at 600 nm (OD600) of the liquid medium in 96-well plates. Each well was inoculated with bacteria at the exponential phase to a final concentration of 106 CFU/ml. The initial multiplicity of infection (MOI) was 10, which is a common MOI used in bacterial challenge tests (25). The plate reader (Molecular Devices, MA) was set as follows: 600 nm, 37°C, 12-h measurement with an interval of 30 min, and shaking for 5 s before and after each measurement. The EOP was quantified by calculating the ratio of phage plaque titers obtained with alternative hosts to those obtained with the production host.
Bacteriophage adsorption rate constant, latent time, and burst size.
Adsorption rate constants for phages PX1 and PEf1 attaching to host cells were determined as proposed previously by Kropinski (26), based on the assumption that the adsorption of phage particles to bacterial cells followed first-order kinetics. One-step growth curve experiments were conducted to determine the latent time and burst size of these phages in multiple hosts, as previously described (27). All parameters were measured in TSB medium with shaking at 120 rpm at 30°C. Briefly, phages were added at an MOI of 0.01 to 1 ml of a mid-log-phase bacterial culture (diluted to an OD600 of 0.1) and allowed to adsorb for 5 min. Free phages were removed by centrifugation (7,000 × g for 2 min at 4°C), and cell pellets were resuspended with same volume of medium. Fifty microliters of the resuspended culture was transferred into 50 ml medium in a 100-ml flask. Samples were collected at an interval of 5 or 10 min and immediately subjected to plate assays for phage titration. Plaque counts were averaged and plotted to generate the one-step growth curve.
Bacteriophage host range stability and bacterial challenge tests.
To assess the stability of the host range, the purified phage stock (first-generation phage stock [P1]) was diluted until it formed several clear plaques on double-layer agar plates. A single plaque was selected and then cultured with P. putida F1 as the host in TSB medium at 37°C for 12 h (Fig. 3). The enriched phage in tube 2 was considered the second generation. Similarly, we enriched phages with the same hosts for the third and fourth generations. Spot tests were used to test the host range. The OD600 (measured during bacterial batch growth in liquid culture) was used to assess the inactivation of multiple hosts by first- and fourth-generation phages. To avoid the interference caused by cell debris, the viable bacterial density was measured when the uninfected (control) bacteria reached stationary phase by using plate assays and expressed as CFU. Specifically, The culture in the well was collected and rinsed (centrifugation at 4°C and resuspension) three times with 200 μl of phosphate-buffered saline (PBS) at 0°C, serial dilution was performed by using PBS at 0°C, and the culture was plated at 37°C overnight. Lethality was calculated as (CFUc − CFUp)/CFUc × 100%, where CFUc is the CFU from the control culture without phage and CFUp is the CFU from the culture amended with a phage (P1 or P4). To assess whether differences in bacterial growth were statistically significant, Microsoft Excel was used for Student's t test at the 99% confidence level.
FIG 3.
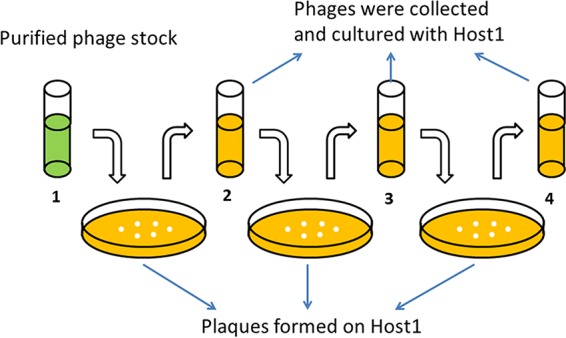
Host range stability test. In each step, one single plaque that formed on the lawn of host 1 was collected and then enriched in liquid culture with host 1 as the host overnight. Phages of all generations were subjected to a spot test to determine their host range.
RESULTS
Simultaneous multihost enrichment methods are not sufficient to isolate broadly polyvalent phages.
Initially, the isolation of polyvalent phages was attempted by using a previously reported multiple-host enrichment method (13). In this method, phage enrichment is first conducted by using environmental water samples amended with growth medium containing multiple hosts, with bacteriophages subsequently being detected by using plaque assays. This method was duplicated with the exception of using activated sludge as the phage source. Using this method, we isolated multiple phages capable of infecting different strains of either E. coli or Pseudomonas aeruginosa but no phages capable of infecting more than one of the four Pseudomonas species tested (Table 1).
TABLE 1.
Influence of phage stock and isolation method on phage host range
| Phage stock | Isolation methoda | Phage(s) | Host range |
|---|---|---|---|
| A | Multiple-host enrichment | Group 1 | E. coli K-12, E. coli C3000 |
| Group 2 | P. aeruginosa PAO1, P. aeruginosa HER1018, P. aeruginosa ATCC 700829 | ||
| B | Multiple-host enrichment | PPJ1 | P. putida F1, Pseudomonas sp. CF600 |
| A | Method A | PX1, PX2, PX3, PX4 | P. aeruginosa PAO1, P. aeruginosa HER1018, P. putida F1, Pseudomonas sp. CF600, P. syringae van Hall |
| B | Method A | PEa1, PEa2, PEa3 | E. coli K-12, E. coli C3000, P. aeruginosa PAO1, P. aeruginosa HER1018 |
| A | Method B | PPJ2 | P. aeruginosa PAO1, P. aeruginosa HER1018, P. putida F1, Pseudomonas sp. CF600, P. syringae van Hall |
| PEf3 | E. coli K-12, E. coli C3000, P. aeruginosa PAO1, P. aeruginosa HER1018, P. putida F1 | ||
| B | Method B | PEf1, PEf2 | E. coli K-12, E. coli C3000, P. aeruginosa PAO1, P. aeruginosa HER1018, P. putida F1 |
Method A is sequential multihost isolation method A, and method B is sequential multihost isolation method B.
This method was then modified for use of a purified and concentrated phage stock from the same sludge sample (phage stock B). This resulted in the isolation of phage PPJ1, which is capable of infecting both P. putida F1 as well as Pseudomonas sp. CF600. However, further phage characterization yielded an EOP of 3.08 × 10−4 for Pseudomonas sp. CF600 relative to P. putida F1 (Table 2). No other phages that were capable of interspecies infectivity were isolated under these particular conditions and using these particular hosts.
TABLE 2.
EOP of selected phages on their hosts
| Phage | Production host | Indicator host | Titer (PFU/ml)a | EOP |
|---|---|---|---|---|
| PPJ1 | P. putida F1 | P. putida F1 | 1.05 × 108 | 1.00 |
| Pseudomonas sp. CF600 | 3.40 × 103 | 3.08 × 10−4 | ||
| PX1 | P. putida F1 | P. putida F1 | 1.08 × 108 | 1.00 |
| P. aeruginosa PAO1 | 8.69 × 107 | 0.80 | ||
| Pseudomonas sp. CF600 | 9.06 × 107 | 0.84 | ||
| P. syringae van Hall | 1.24 × 108 | 1.15 | ||
| PPJ2 | P. putida F1 | P. putida F1 | 1.15 × 108 | 1.00 |
| P. aeruginosa PAO1 | 5.25 × 107 | 0.45 | ||
| Pseudomonas sp. CF600 | 7.05 × 107 | 0.61 | ||
| P. syringae van Hall | 7.50 × 108 | 0.65 | ||
| PEa1 | E. coli K-12 | E. coli K-12 | 1.04 × 108 | 1.00 |
| E. coli C3000 | 8.53 × 107 | 0.82 | ||
| P. aeruginosa PAO1 | 6.40 × 107 | 0.61 | ||
| PEf1 | E. coli K-12 | E. coli K-12 | 1.01 × 108 | 1.00 |
| E. coli C3000 | 1.07 × 108 | 1.06 | ||
| P. aeruginosa PAO1 | 6.05 × 107 | 0.60 | ||
| P. putida F1 | 7.50 × 107 | 0.75 |
Data shown are the means of results from triplicate independent experiments.
Sequential multihost isolation methods can successfully isolate polyvalent phages.
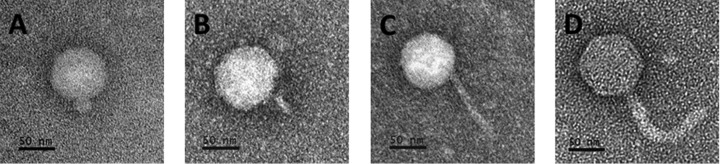
Both sequential multihost isolation methods enabled the isolation of phages capable of infecting all four species of Pseudomonas tested from phage stock A (Table 1). Phages PX1, PX2, PX3, and PX4 were isolated with method A using the same hosts (P. aeruginosa PAO1, Pseudomonas sp. CF600, P. putida F1, and P. syringae van Hall) but with different infection sequences (see Table S2 in the supplemental material). Phage PPJ2 was isolated with method B according to the sequence P. aeruginosa PAO1 > Pseudomonas sp. CF600 > P. putida F1 > P. syringae van Hall > P. putida F1 > Pseudomonas sp. CF600 > P. aeruginosa PAO1. Both method A and method B allowed the isolation of phages capable of infecting both E. coli and Pseudomonas from phage stock B. Phages PEa1, PEa2, and PEa3 were isolated with method A according to the sequence E. coli K-12 > P. aeruginosa PAO1 > P. putida F1 or P. aeruginosa PAO1 > E. coli K-12 > P. putida F1. However, the plaques collected from the second host could not infect P. putida F1 in step 3. In other words, PEa1, PEa2, and PEa3 could infect only E. coli K-12 and P. aeruginosa PAO1. Phages PEf1, PEf2, and PEf3 were isolated with method B according to the sequence E. coli K-12 > P. aeruginosa PAO1 > P. putida F1 > P. aeruginosa PAO1 > E. coli K-12. PX1, PPJ2, PEa1, and PEf1 showed almost no differences in EOP on the hosts used during their isolation (Table 2). Figure 4 shows electron microscopic images of these four phages. Based on their morphology, PX1 and PPJ2 belong to the Podoviridae family, while PEa1 and PEf1 belong to the Siphoviridae family.
FIG 4.
Electron microscopic images of isolated polyvalent phages. Panels depict phages PX1 (Podoviridae) (A), PPJ2 (Podoviridae) (B), PEa1 (Siphoviridae) (C), and PEf1 (Siphoviridae) (D). Phages were negatively stained with 2% uranyl acetate. Bar, 50 nm.
Bacteriophage growth parameters.
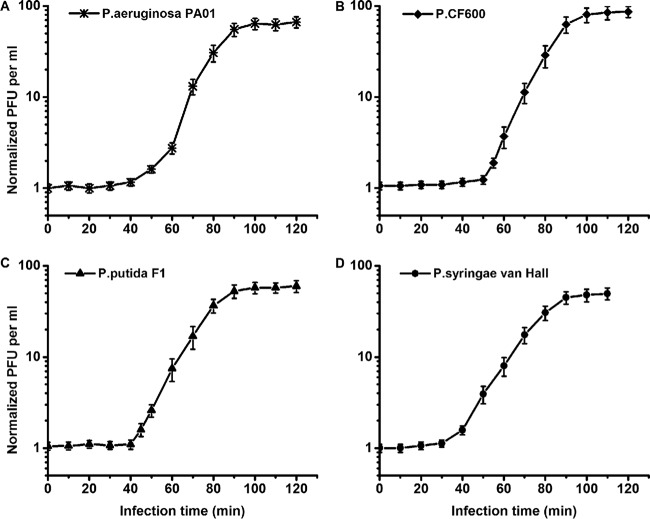
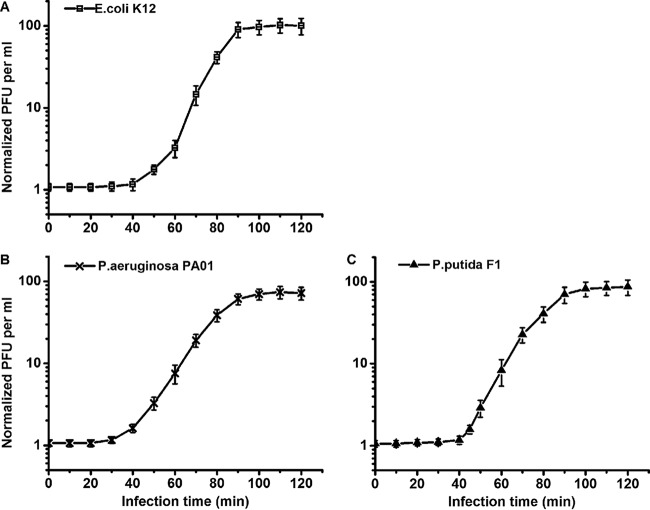
Both phages PX1 and PEf1 had variable (host-dependent) adsorption rate constants, latent times, and burst sizes (Tables 3 and 4). The adsorption rate constant of PX1 was (3.54 to 5.88) × 10−10 ml/min, and the adsorption rate constant of PEf1 was (5.96 to 8.53) × 10−10 ml/min. These values are lower than those of prototypical narrow-host-range phage T4 (2.4 × 10−9 ml/min), T7 (2.0 × 10−9 to 4.0 × 10−9 ml/min), or λ (1.3 × 10−9 to 9.9 × 10−9 ml/min) (7, 10, 28). Figures 5 and 6 show the one-step growth curves of PX1 and PEf1, respectively, with different hosts. The phage latent time ranged from 40 to 55 min, and the burst size ranged from 45 to 99 PFU/cell. For each phage, the latent time and burst size in different hosts were positively correlated (R2 = 0.906 for PX1; R2 = 0.948 for PEf1), which is consistent with previously observed results (8). The adsorption rate constants were negatively correlated with latent time (R2 = 0.805 for PX1; R2 = 0.920 for PEf1).
TABLE 3.
Growth parameters of phage PX1 and corresponding host lethalitya
| Host | Adsorption rateb (10−10 ml/min) | Latent time (min) | Burst size (PFU/cell) | Lethality (%)c |
|---|---|---|---|---|
| P. aeruginosa PAO1 | 3.63 | 50 | 60 | 62 |
| Pseudomonas sp. CF600 | 3.54 | 55 | 84 | 57 |
| P. putida F1 | 5.88 | 45 | 54 | 52 |
| P. syringae van Hall | 5.83 | 40 | 45 | 50 |
Data shown are the means of results from triplicate independent experiments.
Adsorption rates were measured with 10 mM Ca2+. Calculations are shown in Fig. S2 and Table S1 in the supplemental material.
The initial MOI was 10 (measured after 12 h of treatment).
TABLE 4.
Growth parameters of phage PEf1 and corresponding host lethalitya
| Host | Adsorption rateb (10−10 ml/min) | Latent time (min) | Burst size (PFU/cell) | Lethality (%)c |
|---|---|---|---|---|
| P. aeruginosa PAO1 | 8.53 | 40 | 72 | 65 |
| P. putida F1 | 6.59 | 45 | 80 | 57 |
| E. coli K-12 | 5.96 | 50 | 99 | 90 |
Data shown are the means of results from triplicate independent experiments.
The adsorption rate was measured with 10 mM Ca2+. Calculations are shown in Fig. S3 and Table S1 in the supplemental material.
The initial MOI was 10 (measured after 12 h of treatment).
FIG 5.
One-step growth curves of phage PX1 in multiple hosts. (A) P. aeruginosa PAO1; (B) Pseudomonas sp. CF600; (C) P. putida F1; (D) P. syringae van Hall. Error bars indicate standard deviations of the means of data from triplicate independent experiments.
FIG 6.
One-step growth curves of phage PEf1 in different hosts. (A) E. coli K-12; (B) P. aeruginosa PAO1; (C) P. putida F1. Error bars indicate standard deviations of the means of data from triplicate independent experiments.
Polyvalent phages isolated by sequential multihost isolation have stable host ranges and display significant host growth inhibition.
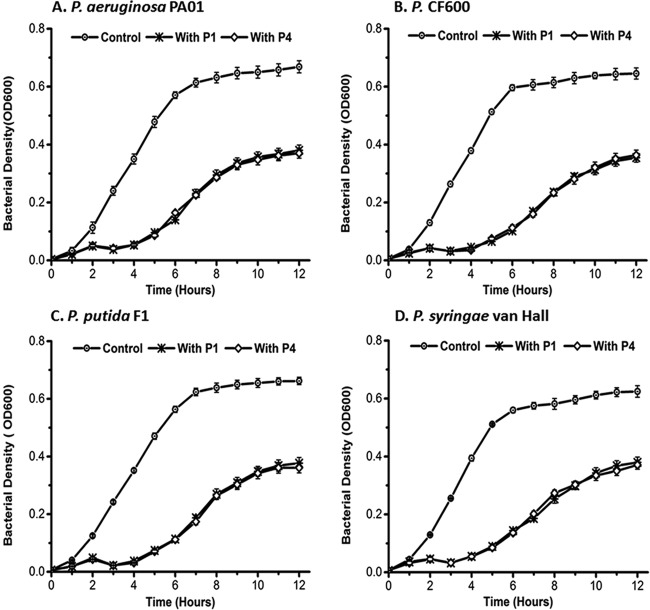
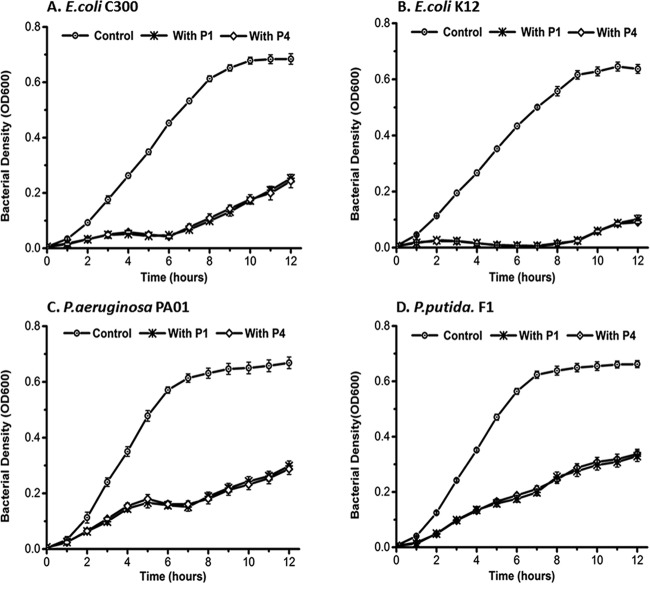
Spot tests revealed that phage PX1, isolated with phage stock A, formed clear lytic zones on P. aeruginosa PAO1, Pseudomonas sp. CF600, P. putida F1, and P. syringae van Hall but had no effect on E. coli. Phage PEf1 formed clear lytic zones on P. aeruginosa PAO1, P. putida F1, E. coli K-12, and E. coli C3000. Halos were formed around the lytic zone of E. coli K-12 and E. coli C3000. However, PEf1 showed no infectivity toward Pseudomonas sp. CF600 or P. syringae van Hall. Phages PX1 and PEf1 were enriched by P. putida F1 individually, and the following generations were subjected to spot tests. The spot test results for subsequent generations corresponded to those of the first generation (see Fig. S1 in the supplemental material). This indicates that PX1 (isolated with method A) and PEf1 (isolated with method B) retained their broad host range when enriched with a single host.
Liquid culture experiments were used for bacterial challenge tests and to confirm phage host range. The first generation (P1) and fourth generation (P4) of PX1 and PEf1 were tested for their impact on growth at an MOI of 10. Host growth in the test groups (either P1 or P4) was significantly lower than that in the control group (P < 0.001). There was little difference in host growth inhibition between P1 and P4 (Fig. 7 and 8). The use of either P1 or P4 of PX1 similarly delayed the onset of exponential growth for each host by ∼3 h and reduced the maximum viable bacterial density (CFU reaching the stationary phase) by >50% except for P. syringae van Hall (Fig. 7 and Table 3). PEf1 significantly decreased both the growth rate and final bacterial concentration for all hosts (Fig. 8 and Table 4). The viable bacterial counts of E. coli K-12 decreased by 90% and those of P. aeruginosa PAO1 decreased by 65% over the course of the experiment. Under the favorable growth conditions tested (e.g., substrate abundance and no competitive exclusion pressure), the isolated polyvalent phages alone could not prevent the emergence of phage-resistant bacteria in monocultures.
FIG 7.
Host range tests of polyvalent phage PX1 using batch growth experiments. The initial MOI was 10. Error bars indicate standard deviations of the means of data from triplicate independent experiments.
FIG 8.
Host range tests of polyvalent phage PEf1 using batch growth experiments. The initial MOI was 10. Error bars indicate standard deviations of the means of data from triplicate independent experiments.
DISCUSSION
This study contributes to the study of viral ecology and the advancement of biological control of bacteria by developing methods to isolate and enrich broad-host-range bacteriophages (i.e., polyvalent phages). We show that the use of different sequential hosts selects for polyvalent phages and is apparently biased against narrow-host-range phages, likely due to continual dilution after exposure to the initial host. Spatial separation of the target hosts, which differentiates our approach from previous attempts to isolate broad-host-range phages (13, 14), is critical for the selection of polyvalence. Our data also demonstrate that these polyvalent phages can be enriched by benign hosts and potentially be used for microbial control of pathogenic bacteria (Fig. 5 to 8).
In order to isolate polyvalent phages, we first attempted to replicate previously reported methods that utilized the simultaneous addition of multiple hosts for enrichment prior to isolation by plaque assays. However, this method was not sufficient to isolate phages with broad infectivity. The EOP of the isolated polyvalent phage (PPJ1) was greatly reduced on the second host. We hypothesized that the separation of hosts would help prevent such large decreases in EOP and devised two sequential multihost isolation methods that utilize an optimized incubation time followed by isolation via plaque assays.
Sequential multihost isolation method A eliminates the initial host enrichment step to maintain phage library diversity. The isolation of phages by sequential plaque assays ensures that specialist phages are eliminated and that the isolated generalist phages have an EOP comparable to that of the previous host. Sequential multihost isolation method B was designed to separate specialist phages from generalists by using multiple adsorption and centrifugation steps. In each step, the phages are exposed to a single host, and the adsorbed phages are separated by centrifugation, along with the host. These adsorbed phages are then enriched by this host and introduced to a new host. We postulate that specialist phages that associate with the initial host are separated during centrifugation of subsequent hosts, which increases the relative abundance of polyvalent phages.
It is unclear why the sequence of host exposure was a critical factor in determining phage EOP among its different hosts. Specifically, polyvalent phages isolated by using a circular host sequence (with the initial host and final host being the same; isolation occurs from this strain) did not suffer significant losses in EOP. In contrast, polyvalent phages isolated with a linear host sequence (with no duplicate hosts; isolation occurs from the last strain) exhibited dramatically lower cross-infectivity, with an EOP of at least 10−4 or lower on other bacteria. Regardless of the underlying mechanisms responsible for this interesting phenomenon, this observation suggests the importance of circular host isolation approaches when using pooled phage libraries, to enhance both the recovery and infectivity of polyvalent phages.
To assess the effect of the phage stock preparation on the isolation of polyvalent phages, two different phage stocks were compared. Phage stock A was prepared by using an enrichment step intended to increase the phage concentration, which was considered culture dependent. Phage stock B was only filtered and concentrated, which represents a culture-independent method. We were not able to isolate phages capable of infecting both E. coli and Pseudomonas with method A from phage stock A but succeeded at this using stock B. It is generally recognized that during the enrichment process, the bacterial community structure changes, likely becoming much less diverse, and the viral community structure evolves accordingly (29). The optimal foraging theory posits that host discrimination (narrowing of host range) is beneficial under conditions of high phage growth rates and host abundance (7). For phages with a very narrow host range living under planktonic conditions, it is important to bind with the greatest affinity when coming into contact with a potential host and to be able to replicate quickly. Importantly, manipulation of phage adsorption rate constants through tail fiber mutagenesis also affects optimal lysis time; phages with higher adsorption constants have lower optimal lysis times and vice versa (10). Lower lysis times are directly related to a smaller burst size as well (8). However, high adsorption rates may be detrimental to phage fitness in biofilm-like environments due to lower levels of phage emigration after lysis (30). An implication of these results is that phages with high adsorption constants for a particular host will generally outcompete those with low absorption constants under planktonic conditions and with high host densities. While each individual phage will have fewer progeny, the shorter lysis time will allow the progeny to take advantage of the high host abundance and propagate more quickly. Consequently, the bias caused by the enrichment processes should be considered when isolating polyvalent phages, although it may be unavoidable at times.
Interestingly, the infectivity of phages PEa1, PEa2, and PEa3 (isolated by using method A) and of phages PEf1, PEf2, and PEf3 (isolated by using method B) was limited to the species used during their isolation. Accordingly, we cannot guarantee the isolation of extremely polyvalent phages capable of infecting a large number of genera without verification using a larger host reference library. Nevertheless, our results suggest the possibility that this approach may enable the selection of polyvalent phages with relatively specific host ranges, which may be beneficial for phage production in benign hosts or for minimizing off-target kills in microbial control applications.
Overall, we developed two novel methods for the rapid isolation and safer enrichment of broadly polyvalent phages and demonstrated that the isolation of phages capable of interorder infectivity can be easily achieved with the use of different sequential hosts. This corroborates the emerging perception that polyvalent phages are more widespread than previously perceived (32) and may lead to an increased understanding of phage host range and ecology. This work may also incentivize research to harness the extremely broad targeting capability of polyvalent phages for microbial control (31) or gene delivery in uncharacterized environments, which may greatly expand and enhance many biomedical and environmental engineering applications.
Supplementary Material
ACKNOWLEDGMENT
We thank Jing Wang for her help on phage characterization in this research.
Funding Statement
This work was supported by an NSF PIRE grant (R76200) and the China Scholarship Council. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02382-15.
REFERENCES
- 1.Bergh O, Borsheim KY, Bratbak G, Heldal M. 1989. High abundance of viruses found in aquatic environments. Nature 340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 2.Chibani-Chennoufi S, Bruttin A, Dillmann M-L, Brüssow H. 2004. Phage-host interaction: an ecological perspective. J Bacteriol 186:3677–3686. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Wilhelm SW, Suttle CA. 1999. Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49:781–788. doi: 10.2307/1313569. [DOI] [Google Scholar]
- 5.Weinbauer MG, Rassoulzadegan F. 2004. Are viruses driving microbial diversification and diversity? Environ Microbiol 6:1–11. [DOI] [PubMed] [Google Scholar]
- 6.Guyader S, Burch CL. 2008. Optimal foraging predicts the ecology but not the evolution of host specialization in bacteriophages. PLoS One 3:e1946. doi: 10.1371/journal.pone.0001946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heineman RH, Springman R, Bull JJ. 2008. Optimal foraging by bacteriophages through host avoidance. Am Nat 171:E149–E157. doi: 10.1086/528962. [DOI] [PubMed] [Google Scholar]
- 8.Bull JJ. 2006. Optimality models of phage life history and parallels in disease evolution. J Theor Biol 241:928–938. doi: 10.1016/j.jtbi.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 9.Duffy S, Turner PE, Burch CL. 2006. Pleiotropic costs of niche expansion in the RNA bacteriophage phi 6. Genetics 172:751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao Y, Wang IN. 2008. Bacteriophage adsorption rate and optimal lysis time. Genetics 180:471–482. doi: 10.1534/genetics.108.090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott JR. 1968. Genetic studies on bacteriophage P1. Virology 36:564–574. doi: 10.1016/0042-6822(68)90188-8. [DOI] [PubMed] [Google Scholar]
- 12.Mizuuchi K, Craigie R. 1986. Mechanism of bacteriophage Mu transposition. Annu Rev Genet 20:385–429. doi: 10.1146/annurev.ge.20.120186.002125. [DOI] [PubMed] [Google Scholar]
- 13.Jensen EC, Schrader HS, Rieland B, Thompson TL, Lee KW, Nickerson KW, Kokjohn TA. 1998. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl Environ Microbiol 64:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bielke L, Higgins S, Donoghue A, Donoghue D, Hargis BM. 2007. Salmonella host range of bacteriophages that infect multiple genera. Poult Sci 86:2536–2540. doi: 10.3382/ps.2007-00250. [DOI] [PubMed] [Google Scholar]
- 15.Knezevic P, Kostanjsek R, Obreht D, Petrovic O. 2009. Isolation of Pseudomonas aeruginosa specific phages with broad activity spectra. Curr Microbiol 59:173–180. doi: 10.1007/s00284-009-9417-8. [DOI] [PubMed] [Google Scholar]
- 16.Powlowski J, Shingler V. 1994. Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation 5:219–236. doi: 10.1007/BF00696461. [DOI] [PubMed] [Google Scholar]
- 17.Adams MH. 1959. Bacteriophages, p 624 Interscience Publishers, New York, NY. [Google Scholar]
- 18.Park M, Lee J-H, Shin H, Kim M, Choi J, Kang D-H, Heu S, Ryu S. 2012. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl Environ Microbiol 78:58–69. doi: 10.1128/AEM.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danovaro R, Middelboe M. 2010. Separation of free virus particles from sediments in aquatic systems, p 74–81. In Wilhelm SW, Weinbauer MG, Suttle CA (ed), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX. [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed, vol 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 21.Lawrence JE, Steward GF. 2010. Purification of viruses by centrifugation, p 166–181. In Wilhelm SW, Weinbauer MG, Suttle CA (ed), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX. [Google Scholar]
- 22.Ackermann H-W, Heldal M. 2010. Basic electron microscopy of aquatic viruses, p 182–192. In Wilhelm SW, Weinbauer MG, Suttle CA (ed), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX. [Google Scholar]
- 23.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed). 2012. Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on the Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 24.Kutter E. 2009. Phage host range and efficiency of plating, p 141–149. In Clokie MRJ, Kropinski AM (ed), Bacteriophages, vol 1 Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 25.Abedon ST. 2009. Kinetics of phage-mediated biocontrol of bacteria. Foodborne Pathog Dis 6:807–815. doi: 10.1089/fpd.2008.0242. [DOI] [PubMed] [Google Scholar]
- 26.Kropinski A. 2009. Measurement of the rate of attachment of bacteriophage to cells, p 151–155. In Clokie MRJ, Kropinski AM (ed), Bacteriophages. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 27.Hyman P, Abedon ST. 2009. Practical methods for determining phage growth parameters, p 175–202. In Clokie MRJ, Kropinski AM (ed), Bacteriophages. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 28.Kasman LM, Kasman A, Westwater C, Dolan J, Schmidt MG, Norris JS. 2002. Overcoming the phage replication threshold: a mathematical model with implications for phage therapy. J Virol 76:5557–5564. doi: 10.1128/JVI.76.11.5557-5564.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gill JJ, Hyman P. 2010. Phage choice, isolation, and preparation for phage therapy. Curr Pharm Biotechnol 11:2–14. doi: 10.2174/138920110790725311. [DOI] [PubMed] [Google Scholar]
- 30.Gallet R, Shao Y, Wang IN. 2009. High adsorption rate is detrimental to bacteriophage fitness in a biofilm-like environment. BMC Evol Biol 9:241. doi: 10.1186/1471-2148-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharjee AS, Choi J, Motlagh AM, Mukherji ST, Goel R. 2015. Bacteriophage therapy for membrane biofouling in membrane bioreactors and antibiotic-resistant bacterial biofilms. Biotechnol Bioeng 112:1644–1654. doi: 10.1002/bit.25574. [DOI] [PubMed] [Google Scholar]
- 32.Khan MA, Satoh H, Katayama H, Kurisu F, Mino T. 2002. Bacteriophages isolated from activated sludge processes and their polyvalency. Water Res 36:3364–3370. doi: 10.1016/S0043-1354(02)00029-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.