Abstract
Understanding the structure of single neurons is critical for understanding how they function within neural circuits. BigNeuron is a new community effort that combines modern bioimaging informatics, recent leaps in labeling and microscopy, and the widely recognized need for openness and standardization to provide a community resource for automated reconstruction of dendritic and axonal morphology of single neurons.
Introduction
Although more than 100 years have passed since Santiago Ramón y Cajal was awarded the Nobel Prize for the neuron doctrine, we still lack an accepted catalog of neuron types and their names (DeFelipe et al, 2013). This remains an important challenge, as it is a major goal of neuroscience to understand the relationship between the algorithms implemented by the brain and the hardware used to implement them (Marr and Poggio, 1976; Koch 1999). The three-dimensional (3D) shape of a neuron – including its dendritic and axonal arbors – is central to determining its identity (phenotype), connectivity, synaptic integration, firing properties, and ultimately its role in the neural circuit. Characterizing and understanding the 3D morphology of individual neurons is fundamentally important for elucidating the breadth of neuronal diversity. Recent major neuroscience initiatives worldwide - such as the US BRAIN Initiative (http://braininitiative.nih.gov/) (Alivisatos et al., 2012), Europe’s Human Brain Project (http://www.humanbrainproject.eu/) (Kandel et al., 2013), and the Allen Cell Types Database (http://celltypes.brain-map.org/) are all based on the importance of understanding the diversity of cell types across multiple nervous systems as a step toward elucidating the relationship between the structure and function in the nervous system.
Quantifying the morphology of neurons and other tree-shaped biological structures (e.g. glial cells, brain vasculature, etc.) has been the focus of numerous studies over the past 30 years (Peng, et al, 2015; Gillette, et al, 2011). Yet the systematic characterization of even simple brain circuits at the level of their individual neurons is still limited by the lack of a robust system for fast and accurate reconstruction of neuronal branching arbors. Although tens of thousands of neurons have been digitized across multiple species, brain regions and laboratories worldwide, the variability introduced through different animal species, developmental stages and brain location as well as through distinct histological, imaging, and reconstruction protocols has made systematic analysis and comparison challenging.
The BigNeuron project (http://bigneuron.org/) is a community effort to define and advance the state-of-the-art of single neuron reconstruction, develop a toolkit of standardized reconstruction protocols, analyze neuron morphologies and establish a data resource for neuroscience. The project, announced on March 31, 2015, is sponsored by 14 neuroscience-related research organizations, and dozens of international research groups and individuals.
The initial goal of BigNeuron is to bench test a large set of open-source, automated neuron reconstruction algorithms, using community-contributed, openly-available 3D neuron image datasets that were acquired by a variety of light microscopy methods. Bench testing will be performed on a common open software platform running on supercomputers and the results will be compared and validated against manual segmentations using carefully defined consensus criteria from the computational and neuroscience communities. Ultimately, this will produce a large, community-generated database of single-neuron morphologies, open-source tools for neuroscience, and community-driven protocols intended to serve as the standard for digital reconstruction of single neurons.
Why BigNeuron is Needed
Since the birth of modern neuroscience, the prevailing approach for understanding neuronal morphology has been to spend many hours, days and weeks to manually delineate complicated neuronal shapes visualized using a variety of staining techniques. In the modern digital era, a typical workflow has three major steps. Neurons must first be labeled with a dye, antibody, or transgenic tracer to reveal neuronal structures. Next, one or more microscopy approaches are applied to digitally capture images. Lastly, neurons can be computationally traced or reconstructed, extracting their geometry from image pixel data (Meijering, 2010; Parekh and Ascoli, 2013). Substantial advances have been made in the last decade for both specimen preparation (e.g. genetic labeling (Cai, et al, 2013; Nern, et al, 2015), virus-based circuit tracing (Oh, et al, 2014), tissue clearing (Chung, et al, 2013), SCALE protocol (Hama et al, 2011) etc.) and advanced image capture (e.g. laser scanning microscopy, high-speed, high-resolution digital cameras, etc.). This has yielded hundreds of thousands of cell images, yet it is still unclear which approaches are most conducive to robust and accurate automated image processing.
The neuron reconstruction step has remained a key bottleneck in the workflow. Accurate neuronal reconstruction is still extremely resource intensive, relying on human labor for drawing, curating, and annotating neuron shape, even with the help of powerful computers. Before 2014, less than 10,000 dendritic reconstructions were available in the largest public neuronal morphology database, NeuroMorpho.Org (Halavi et al., 2012). Many of these digital morphologies, and the majority of neuronal structure data published to date, were collected using different manual tracing protocols and images of widely different quality, often resulting in incomplete and highly variable reconstructions. The lack of standardized imaging and reconstruction of neurons from different investigators greatly limits their future use in downstream analysis such as the computational reconstruction of neural circuitry. It is thus highly desirable to apply automation to both accelerate and standardize this process.
In the past years, a number of efforts have gone into improving the methods and algorithms for morphological reconstruction of neurons. Effectively, individual organizations – working chiefly with their own datasets – have taken steps to screen and document single neuron morphologies using automated neuron reconstruction. Several examples include Taiwan’s FlyCircuit.Org (Chiang, et al, 2011) and Janelia’s FlyLight project (Jenett, et al, 2012; Nern, et al, 2015), which focus on Drosophila CNS neurons, the BlueBrain project’s rat somatosensory neuron database (http://bluebrain.epfl.ch) and the Allen Institute’s Cell Types Database project (www.brain-map.org), which studies mouse and human neocortical neurons.
As a result of this heightened interest, many new algorithms for neuron reconstruction have emerged in the last five years (e.g. Wang, 2011; Peng, et al, 2011, Polavaram et al, 2014, Wan et al, 2015, Santamaría-Pang, 2015). Some have vastly enhanced the competitive performance of tracing accuracy compared to the independent, manually curated reconstructions in public databases (Xiao, et al, 2013; Chiang, et al, 2011).
Automated neuron reconstruction methods developed for different application scenarios typically have varying performance, especially when tried on neuron images of variable quality and different species. Because most of these methods have not been directly cross-tested thoroughly, it is unclear which methods are best matched with different imaging modalities or datasets. Furthermore, there are significant differences in coding platforms and languages, each requiring different image input paradigms and generating different output reconstruction formats. Thus it is hard to quantitatively compare these algorithms objectively. Many existing methods have been developed for very small images and their performance on much larger images is unknown (e.g. compare a single Drosophila interneuron to a human Betz cell).
Comparing different reconstruction algorithms to ground truth requires a substantial number of single-neuron datasets acquired under different conditions. Testing analysis methods thoroughly using the same core data will help characterize the landscape of analysis approaches to establish which are optimal for distinct image datasets corresponding to different species, brain regions, neuron types, and experimental conditions. Because community-contributed images used for BigNeuron were collected using a variety of paradigms, the analysis output will in turn provide guidelines for optimizing neuron labeling and sample preparation conditions, and matching them to the imaging methods and parameters that yield the most valuable information.
What BigNeuron will bring to the Community
BigNeuron is designed to benefit neuroscience in several practical ways:
First, at its completion, neurobiologists who are interested in understanding neuron morphology will be able to access a toolkit for analysis written by a broad range of experts. This will allow them to benefit from a community of ready-made collaborators, cutting down the costs and time to generate reconstruction data. Scientists who provide raw image data to the large-scale bench testing of BigNeuron will reduce the need to seek additional resources to quantify neuronal morphological data at small scale. Instead, they will have access to a worldwide neuron reconstruction method-developing community.
Second, BigNeuron will provide a common platform for neuron reconstruction method developers to compare and analyze algorithms. In a series of ongoing and planned hackathons worldwide, developers are learning (from each other) the relative pros and cons of various methods and how to leverage existing resources to refine or develop new algorithms. The BigNeuron platform will also entail real-world analysis of large data, thus serving as a practical guideline in determining the suitability of specific reconstruction methods for a variety of image datasets (as well as providing feedback regarding the utility of various sample preparation and imaging protocols). Bringing neuron reconstruction methods and results together will also encourage method developers to collaborate, share, and reuse each other’s software modules. To make these reconstruction methods and analysis approaches truly open, input and output data formats need to be standardized and implemented in a common computational infrastructure; hence the need for the community to come together and drive the BigNeuron project.
BigNeuron will also likely produce one of the largest community-derived phenotype databases for single neurons, cataloguing neuron shape and projection patterns from different species and different brain regions. Since all the neuron image data will be processed using the same protocols, it will be straightforward to compare the reconstructions. The rich dataset will not only attract more data analysts to examine neuron morphology, but also offer an opportunity to mine and query the patterns of neurons with distinct shapes. The database could ultimately be expanded to include functional data (e.g. physiology, gene expression) for reconstructed neurons.
Finally, the analysis from BigNeuron will provide an opportunity to improve the accuracy and efficiency in targeting, labeling and acquiring images of single neurons, using many of the powerful techniques recently developed for neuron specimen preparation and advanced imaging. As BigNeuron makes it easier to reconstruct neuron shapes, experimentalists can use the technical platform of BigNeuron in their experimental design, and will be able to refine their protocols and improve the raw image data quality at much lower cost.
Approach
The first (ongoing) year of BigNeuron is establishing the infrastructure, with a plan for initial data release in 2016. This first phase focuses on reconstructing sparsely labeled neurons in 3D neuron image-stacks. Test data include samples imaged using a variety of light microscopy modalities, especially laser-scanning microscopy (confocal and two-photon) and wide-field epifluorescence microscopy, as well as bright-field microscopy. The construction of a highly accessible database will enable analysis of neuronal morphology patterns across multiple species, with possible expansion to other related issues, such as resolving individual neurons within densely labeled samples, identifying connectivity, and analysis of multi-color images, time-lapse images, electron microscopic images, etc.
The initial BigNeuron operation will foster community involvement through several workshops for annotation and code development. First, BigNeuron is supporting three algorithm-porting hackathons to help international developers port their neuron reconstruction and analysis methods onto a common software platform. Second, we are organizing data collection days to meet with neurobiologists who will share raw image data. Third, a neuron annotation workshop has been organized to produce high-quality manual reconstructions that will serve as the “gold standard” for evaluating the performance of the neuron reconstruction algorithms. Finally, a data analysis hackathon will be held after a large number of reconstructions have been produced in the bench-testing phase.
It is important to attract and enable the contribution of a large variety of neuron image datasets for bench testing. Data collected to date include neuron image stacks from several species (e.g. fruitfly, dragonfly, silk moth, zebrafish, Xenopus, chick, mouse, rat, and human) and anatomical regions (e.g. cortical and subcortical areas, retina, and peripheral nervous system). These neurons have been labeled using different methods (e.g. transgenic fluorescent proteins introduced in a variety of ways as well as dyes introduced by intracellular injection) and will span a broad range of neuronal types (i.e. morphological and functional classes). Many of these neurons have been contributed from large-scale neuroinformatics projects, such as the Allen Institute’s Mouse and Human Cell Types projects, Taiwan’s FlyCircuit.Org (Chiang, et al, 2011), and Janelia’s FlyLight project (Nern, et al, 2015). In addition, a number of datasets are also being contributed directly by neuroscientists from a growing number of organizations. Several image datasets have already been reconstructed, manually curated, and/or proofread at the source laboratories. To ensure usability for bench testing, contributed image data will be pre-processed, including standardizing formats, adding essential metadata, and providing important information such as cell body position. Image data will then be archived for future bench testing.
Bench tests will employ an array of morphological metrics (e.g. individual neurite diameter, length, branching angles), summary statistics (average, s.d., minimum, maximum and total dendritic length), as well as histogram distributions or the dependency of one metric on another (e.g. Sholl analysis of number of branches versus distance from soma). These morphological metrics will be used to compare reconstructions from various automated reconstruction methods to gold-standard reconstructions obtained by expert manual reconstruction, with the most valuable metrics to be determined during the course of the project. These gold-standard reconstructions will also help characterize and quantify the types of errors in the automated neuron reconstructions.
The total number of potential bench tests equals the number of possible combinations of neuron images, reconstructions methods, and parameter configurations needed to obtain quality reconstructions. BigNeuron will bench test more than 20,000 single neuron image stacks, for about 20 reconstruction methods, each trying one to four parameter configurations to optimize the result. Therefore, over one million neuron reconstructions will be produced during bench testing. To assess the practical usability of an individual algorithm, the maximum running time of such an algorithm method will be constrained. Nevertheless, computational bench testing on this scale will still require several powerful supercomputers. BigNeuron has been granted access to supercomputing facilities run by the Oak Ridge National Laboratory, the Lawrence Berkeley National Laboratory, and the European Human Brain Project, along with other facilities. We expect that multiple bench testing on different supercomputers will ensure better reproducibility, foster interest and collaboration between participants, and reduce cost. These bench tests will be monitored to validate true performance of the various methods. The source code needed to generate the supercomputing job scripts will also be publicly shared, allowing anyone to run smaller or similar-scale bench tests using their own machines. Ultimately, BigNeuron’s platform will encourage development of accurate and computationally efficient algorithms.
Another important aspect of the project is the sharing of neuron reconstructions. We expect this bench test to generate a large number of reconstructions, roughly 20+ for each image dataset. The consensus reconstruction, as well as alternative ways to look at the population of reconstructions, such as the principal components, will be provided. All these data will be freely shared on a new public web-based database. The conditions associated with the generation of the data will also be documented and shared in the database. These metadata will also enable widespread use of the data, including mirroring by multiple sites such as NeuroMorpho.Org and other databases. The neuroscience community will be invited to analyze the reconstructions openly.
A common software platform is crucial to make BigNeuron a success. BigNeuron will employ the open source, cross-platform package Vaa3D (http://vaa3d.org) as the bench-test infrastructure. Vaa3D (Peng et al., 2010; Peng et al, 2014) is a visualization and analysis software suite created and maintained by Janelia Research Campus (HHMI) and the Allen Institute for Brain Science. This software performs 3D and higher dimensional dynamic reconstruction and rendering of very large image data sets and associated 3D surface objects, particularly those generated using a variety of modern microscopy methods. Vaa3D has a rich set of functions and plugins for neuron quantification and is compatible with well-established neuron analysis tools such as L-Measure (Scorcioni et al., 2008). Vaa3D is suitable for manual, semi-automatic, and completely automated digital tracing. This software has been used in several large neuroscience initiatives and a number of applications in other domains. For the BigNeuron project, 15 neuron-tracing algorithms have already been ported to Vaa3D as plugins. The software is also used for visualization, bench testing, and data analysis.
Given the increasing focus on Big Data and the grand challenges of understanding circuitry of the human brain in health and disease and the importance of the diverse population of neurons – arguably the brain’s fundamental units – the time is right for a project like BigNeuron to move the field toward a consensus on how to reconstruct and interpret neuronal morphology. Successful hackathons in Asia, Europe and in the US have already ported several new algorithms to the BigNeuron platform. A workshop in June 2015 will focus on getting expert consensus on gold-standard reconstructions and annotations. Bringing investigators together from the frontline of computational analysis of cellular neuroanatomy and morphology to establish open-source, automated neuron reconstruction algorithms using community-contributed, 3D neuron image datasets will enable the development of key benchmarks for future studies of neural circuitry, form and function in the coming years. BigNeuron offers an opportunity to take a much-needed step toward informing the evolution of accepted standards for reconstruction of highly complex neurons, and contributes critically to the mission of open and reproducible science that will be crucial to understand brains of all kinds – not least of all the brains working together to achieve its goals.
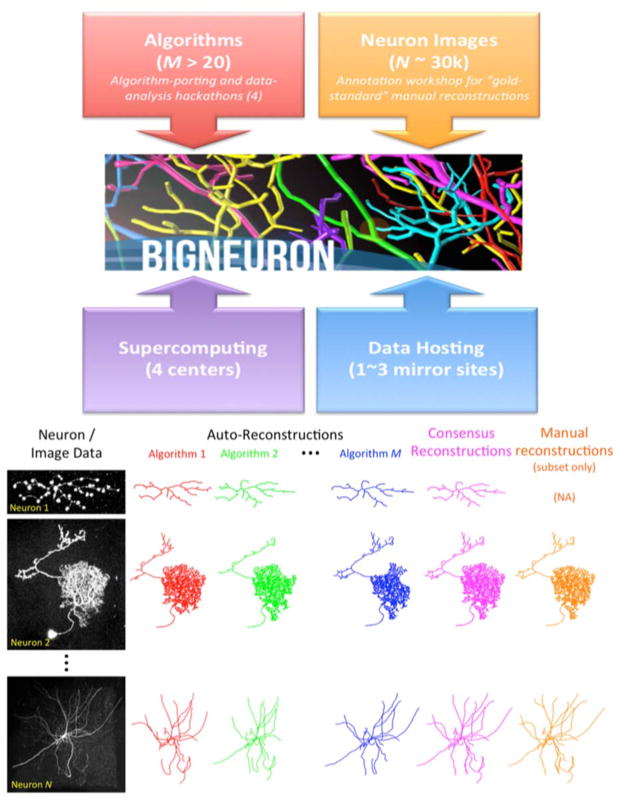
Figure 1. Bioimage Informatics Matchmaking.
In neuroscience labs worldwide, neuronal morphology image stacks are acquired through multiple microscopy modalities from a diverse array of experimental preparations. At the same time, a broad range of reconstruction algorithms are continuously developed using various programming languages, computational platforms, and custom test beds. Harnessing powerful supercomputers and the open source community, BigNeuron brings together tens of thousands of state-of-the-art 3D neuronal images with all major classes of automated tracing systems to generate an unprecedented number of digital morphological reconstructions. Expert cross-analysis of the initial bench testing dataset against manually curated gold standards will generate a unique toolbox of morphometric measurements aimed to stimulate further development.
References
- 1.DeFelipe J, López-Cruz PL, Benavides-Piccione R, Bielza C, Larrañaga P, Anderson S, … Ascoli GA. Nature Reviews Neuroscience. 2013;14(3):202–216. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marr D, Poggio T. AI Memo. Massachusetts Institute of Technology; 1976. Artificial Intelligence Laboratory. AIM–357. [Google Scholar]
- 3.Koch C. Biophysics of Computation: Information Processing in Single Neurons. Oxford University Press; New York: 1999. [Google Scholar]
- 4.Alivisatos AP, Chun M, Church GM, Greenspan RJ, Roukes ML, Yuste R. Neuron. 2012;74:970–974. doi: 10.1016/j.neuron.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kandel ER, Markram H, Matthews PM, Yuste R, Koch C. Nat Rev Neurosci. 2013;14:659–664. doi: 10.1038/nrn3578. [DOI] [PubMed] [Google Scholar]
- 6.Peng H, Meijering E, Ascoli GA. Neuroinformatics. 2015 doi: 10.1007/s12021-015-9270-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillette TA, Brown KM, Svoboda K, Liu Y, Ascoli GA. Neuroinformatics. 2011;9:303–304. doi: 10.1007/s12021-011-9117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meijering E. Cytometry A. 2010;77:693–704. doi: 10.1002/cyto.a.20895. [DOI] [PubMed] [Google Scholar]
- 9.Parekh R, Ascoli GA. Neuron. 2013;77:1017–1038. doi: 10.1016/j.neuron.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Nature methods. 2013;10(6):540–547. [PubMed] [Google Scholar]
- 11.Nern A, Pfeiffer BD, Rubin GM. Proceedings of the National Academy of Sciences. 2015:201506763. doi: 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, … Zeng H. Nature. 2014;508(7495):207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, … Deisseroth K. Nature. 2013;497(7449):332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Nat Neurosci. 2011;4(11):1481–8. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 15.Halavi M, Hamilton KA, Parekh R, Ascoli GA. Front Neurosci. 2012;6:49. doi: 10.3389/fnins.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, … Hwang JK. Current Biology. 2011;21(1):1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 17.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, … Zugates CT. Cell reports. 2012;2(4):991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Narayanaswamy A, Tsai CL, Roysam B. Neuroinformatics. 2011;9(2–3):193–217. doi: 10.1007/s12021-011-9110-5. [DOI] [PubMed] [Google Scholar]
- 19.Peng H, Long F, Myers G. Bioinformatics. 2011;27(13):i239–i247. doi: 10.1093/bioinformatics/btr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polavaram S, Gillette TA, Parekh R, Ascoli GA. Front Neuroanat. 2014;8 doi: 10.3389/fnana.2014.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan Y, Long F, Qu L, Xiao H, Hawrylycz M, Myers EW, Peng H. Neuroinformatics. 2015;2015 doi: 10.1007/s12021-015-9272-7. [DOI] [PubMed] [Google Scholar]
- 22.Santamaría-Pang A, Hernandez-Herrera P, Papadakis M, Saggau P, Kakadiaris IA. Neuroinformatics. 2015:1–24. doi: 10.1007/s12021-014-9253-2. [DOI] [PubMed] [Google Scholar]
- 23.Xiao H, Peng H. Bioinformatics. 2013;29(11):1448–1454. doi: 10.1093/bioinformatics/btt170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng H, Ruan Z, Long F, Simpson JH, Myers EW. Nat Biotechnol. 2010;28:348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng H, Tang J, Xiao H, Bria A, Zhou J, Butler V, … Long F. Nature Communications. 2014;5:4342. doi: 10.1038/ncomms5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scorcioni R, Polavaram S, Ascoli GA. Nat Protoc. 2008;3:866–876. doi: 10.1038/nprot.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]