Abstract
Cardiogenesis involves the coordinated regulation of multiple biological processes by a finite set of transcription factors (TFs). Here, we show that the Forkhead TFs Checkpoint suppressor homologue (CHES-1-like) and Jumeau (Jumu), which govern cardiac progenitor cell divisions by regulating Polo kinase activity, play an additional, mutually redundant role in specifying the cardiac mesoderm (CM) as eliminating the functions of both Forkhead genes in the same Drosophila embryo results in defective hearts with missing hemisegments. This process is mediated by the Forkhead TFs regulating the fibroblast growth factor receptor Heartless (Htl) and the Wnt receptor Frizzled (Fz): CHES-1-like and jumu exhibit synergistic genetic interactions with htl and fz in CM specification, thereby implying that they function through the same genetic pathways, and transcriptionally activate the expression of both receptor-encoding genes. Furthermore, ectopic overexpression of either htl or fz in the mesoderm partially rescues the defective CM specification phenotype in embryos lacking both Forkhead genes. Together, these data emphasize the functional redundancy that leads to robustness in the cardiac progenitor specification process, and illustrate the pleiotropic functions of Forkhead TFs in different aspects of cardiogenesis.
KEY WORDS: Forkhead domain transcription factors, Signaling pathway receptors, Fibroblast growth factor receptor, Wnt signaling pathway receptors, Gene regulation, Cardiac progenitor specification, Cardiac mesoderm specification, Heart development, Cardiogenesis, Organogenesis
Summary: Checkpoint suppressor homologue and Jumeau, which are known to govern cardiac progenitor cell divisions, play additional, mutually redundant roles in specifying cardiac mesoderm in Drosophila.
INTRODUCTION
The rhythmically contracting heart of Drosophila exhibits remarkable similarities to the vertebrate heart at the primitive linear tube stage of development in terms of structure, morphogenetic origins and regulatory mechanisms (Bodmer and Frasch, 2010; Cripps and Olson, 2002; Olson, 2006). In both Drosophila and vertebrates, the heart tube originates from two bilaterally symmetrical rows of mesodermal cells that have migrated most distally from the point of invagination during gastrulation. This migration ensures that these cells end up in stereotyped locations where, in response to appropriate position-specific inductive signals such as bone morphogenetic proteins, Wnt proteins and fibroblast growth factors, they initiate gene expression programs involving numerous conserved transcription factors (e.g. GATA, FOG, Forkhead domain, NK homeodomain, LIM homeodomain and T-box proteins) and become determined as the cardiac mesoderm (CM), i.e. the cardiac progenitors that are the precursors of the embryonic heart. Subsequent refinement and modulation of these gene expression programs bring about the division and differentiation of these CM cells into distinct cardiac subtypes, such as the inner tube of Myocyte enhancer factor 2 (Mef2)-expressing contractile cardial cells (CCs) and the external sheath of Zn finger homeodomain 1 (zfh1)-expressing nephrocytic pericardial cells (PCs) in Drosophila.
Cardiogenesis thus requires the integration of multiple signaling pathways and transcription factor-mediated gene expression programs to orchestrate diverse developmental processes. This raises two intriguing questions: how are the numerous complex processes involved in cardiogenesis orchestrated by a finite set of regulators, and how is the requisite coordination between these distinct regulatory mechanisms achieved?
One family of transcription factors (TFs) that has been implicated in cardiogenesis in both vertebrates and Drosophila is the Forkhead (Fkh/Fox) domain family of proteins. At least four Fkh TFs are known to be required for proper cardiac development in mammals, and mutations in three Fkh genes have been linked to human congenital heart defects (Evans-Anderson et al., 2008; Hu et al., 2004; Korver et al., 1998; Roessler et al., 2008; Wang et al., 2004; Yu et al., 2010). We have previously also shown a cardiogenic role for two Drosophila Fkh genes, jumeau (jumu) and Checkpoint suppressor homologue (CHES-1-like). Both genes are initially maternally expressed, with jumu and CHES-1-like showing subsequent zygotic expression in the cells fated to become the CM from embryonic Stages 11 to 13, and from Stages 11 to 12, respectively. Each of these two Fkh genes determines cardiac cell subtypes, numbers and positions by regulating a Polo kinase-dependent pathway to mediate three distinct categories of cardiac progenitor cell divisions (Ahmad et al., 2014, 2012). In addition, our prior findings revealed that Fkh TF binding sites are significantly enriched in combination with those of other known cardiogenic TFs in the enhancers of genes expressed in the heart, and that overexpression of Jumu in the mesoderm resulted in elevated expression levels of many known cardiac genes (Ahmad et al., 2014, 2012; Zhu et al., 2012). Collectively, these results suggested that these Fkh TFs mediate additional cardiogenic processes beyond solely cardiac progenitor cell divisions by regulating many downstream target genes.
Here, we show that the Fkh genes jumu and CHES-1-like also play a significant role in specifying the CM, and that this process is achieved by the Fkh TFs transcriptionally regulating Heartless (Htl), which is a fibroblast growth factor receptor (FGFR), and Frizzled (Fz), which is a receptor of the Wingless/Wnt signaling pathway.
RESULTS
Loss of function of both CHES-1-like and jumu results in embryos missing entire rows of heart cells in random hemisegments
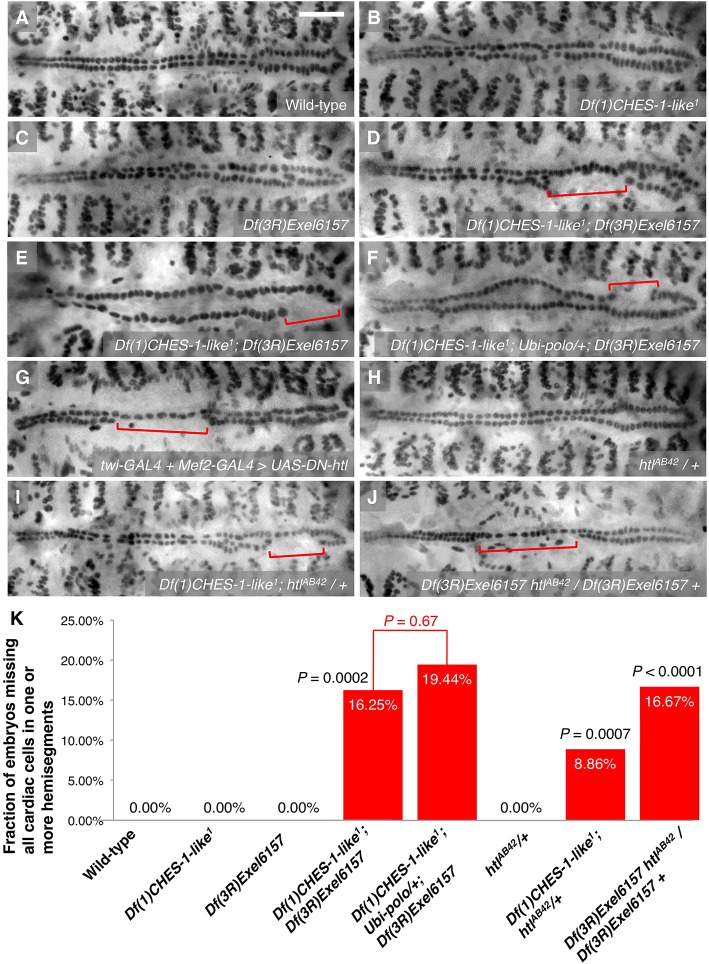
Our previous study showed that embryos homozygous for either the jumu null deficiency Df(3R)Exel6157 or the CHES-1-like null mutation Df(1)CHES-1-like1 exhibit hemisegments with localized increases or decreases in CC number, occasional enlarged CC nuclei, or mispositioned CCs as a consequence of defective cardiac progenitor cell divisions when compared with wild-type embryos (Fig. 1A-C) (Ahmad et al., 2012). However, a significantly large fraction (P=0.0002) of embryos lacking both jumu and CHES-1-like functions exhibits a more severe phenotype that never occurs in embryos missing just one of these two Fkh genes: 16.25% of embryos that were doubly homozygous for both the jumu null deficiency and the CHES-1-like null mutation exhibit one or more hemisegments missing entire rows of cardial cells (Fig. 1D,E,K; Table 1). Pericardial cells were also absent in the hemisegments missing CCs (Fig. S1). The location of the hemisegments lacking all heart cells was random: no significant difference (P=0.83) was detected in the frequency of this ‘missing cardiac hemisegments’ (MCH) phenotype between the anterior aorta and the wider posterior section of the heart (Table S1).
Fig. 1.
MCH phenotypes associated with CHES-1-like, jumu and htl. (A-J) Mef2 antibody staining of CCs, illustrating the presence or absence of the MCH phenotype (square brackets) in representative Stage 16 embryos that are (A) wild type, (B) homozygous for the CHES-1-like null mutation, (C) homozygous for the jumu null deficiency, (D,E) doubly homozygous for both the CHES-1-like mutation and the jumu deficiency, (F) doubly homozygous for both the CHES-1-like mutation and the jumu deficiency, but ubiquitously expressing polo, (G) expressing a dominant-negative version of the Htl FGFR pan-mesodermally, (H) heterozygous for a htl null mutation, (I) homozygous for the CHES-1-like null mutation and heterozygous for the htl null mutation, or (J) homozygous for the jumu null deficiency and heterozygous for the htl mutation. Scale bar: 50 μm. (K) Quantification and significance of the MCH phenotypes. From left to right, the P-values indicate the significances of the difference in phenotype between embryos lacking both Fkh genes and embryos lacking either the CHES-1-like or jumu gene; the difference in phenotype between embryos missing both Fkh genes and embryos missing both Fkh genes while ubiquitously expressing polo; the difference between the phenotype of embryos both heterozygous for the htl null mutation and homozygous for the CHES-1-like null mutation and the additive effects of the phenotypes in htl heterozygotes and the CHES-1-like homozygotes; and the difference between the phenotype of embryos both heterozygous for the htl null mutation and homozygous for the jumu null deficiency and the additive effects of the phenotypes in htl heterozygotes and the jumu homozygotes.
Table 1.
Quantification of the missing cardiac hemisegments (MCH) phenotypes associated with different genotypes
Similar MCH phenotypes were also observed when both jumu and CHES-1-like functions were simultaneously knocked down by CM precursor-targeted RNA interference directed by the tinD-GAL4 and Hand-GAL4 drivers (Fig. S2), indicating that the requirement of both these Fkh genes for proper heart development is autonomous to the cells fated to become cardiac progenitors.
This MCH phenotype could not be a consequence of the cardiac progenitor cell division defects caused by mutations in jumu or CHES-1-like for at least two reasons. First, because four of the six CCs in each hemisegment, the Tinman-expressing CCs (Tin-CCs), arise by two symmetric cell divisions from two cardiac progenitor cells (Han and Bodmer, 2003; Ward and Skeath, 2000), the aforementioned cell division defects would have left at least these two progenitor cells intact in the CC row, rather than resulting in all six CCs being lost. Second, we had previously shown that the cardiac progenitor cell division defects caused by mutations in jumu or CHES-1-like could be partially rescued by ubiquitously expressing their downstream gene polo, with many of the rescued hearts looking the same as wild type (Ahmad et al., 2012). However, no significant rescue (P=0.67) of this MCH phenotype was detected when polo was ubiquitously expressed in embryos lacking both jumu and CHES-1-like functions: 19.44% of these embryos exhibited the phenotype (Fig. 1F,K; Table 1).
Collectively, these data suggest that the MCH phenotype is likely to be a consequence of a defect at an earlier step in cardiogenesis: either a failure of the mesodermal cells to migrate far enough dorsally to reach the locations where they could receive the position-specific signals to become the CM, or a defect in the signal transduction mechanisms activated by these position-specific cues.
The MCH phenotype of embryos lacking both CHES-1-like and jumu functions is not caused by defects in mesoderm migration
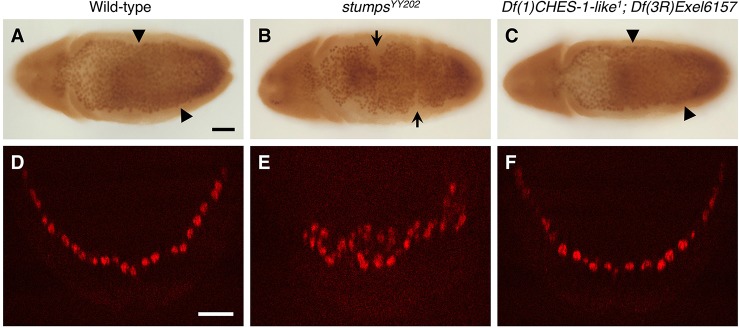
To determine whether the MCH phenotype in the double homozygotes reflects defects in mesoderm migration, we examined and compared wild-type embryos, embryos homozygous for a mutation of stumps, a gene essential for proper mesoderm migration (Imam et al., 1999; Michelson et al., 1998a; Vincent et al., 1998), and embryos that were doubly homozygous for both the CHES-1-like and jumu null mutations.
Ventral views of the migrating mesoderm in wild-type embryos reveal a smooth dorsolateral margin at Stage 9 (Fig. 2A). Transverse views show the mesoderm migrating smoothly as a monolayer with the dorsalmost mesodermal cells reaching locations by Stage 10 (Fig. 2D) where they can receive position-specific signals to become specified as cardiac progenitors. By contrast, the mesoderm of the stumps mutants display a ragged margin characteristic of defects in migration at Stage 9 (Fig. 2B), with transverse section views at Stage 10 showing mesodermal cells aggregated ventrally and not reaching the dorsalmost positions reached by cells in wild-type embryos (Fig. 2E).
Fig. 2.
The MCH phenotype in embryos lacking both CHES-1-like and jumu functions is not caused by defects in mesoderm migration. (A-C) Mef2 antibody staining of the mesoderm, illustrating the presence or absence of mesoderm migration defects in ventral views of representative Stage 9 embryos that are (A) wild type, (B) homozygous for the stumpsYY202 mutation and (C) lack both Fkh genes. Note that the dorsolateral margin of the mesoderm in the stumps mutant (B) is ragged and exhibits discontinuities (arrows) characteristic of migration defects. By contrast, the migrating mesoderm of both wild-type embryos (A) and embryos lacking both CHES-1-like and jumu (C) exhibit smooth dorsolateral margins (arrowheads). Scale bar: 50 μm. (D-F) Mef2 antibody staining (red) of the migrating mesoderm in representative transverse views of the ventral halves of Stage 10 embryos that are (D) wild type, (E) lack stumps function, and (F) lack both Fkh genes. Note that the mesoderm migrates dorsolaterally as a monolayer in both the wild-type embryo and the CHES-1-like; jumu double homozygote, but remains aggregated ventrally and unable to reach the dorsalmost positions in the stumps mutant. Scale bar: 25 μm.
None of the 110 Stage 9 doubly homozygous CHES-1-like; jumu embryos we examined displayed the ragged mesodermal margin characteristic of migration defects (Fig. 2C). Furthermore, examination of 20 Stage 10 transverse views of these double homozygotes showed that mesodermal cells always migrated as a monolayer and were always able to reach the dorsalmost positions necessary for receiving instructive cardiogenic signals (Fig. 2F). Collectively, these results indicate that migration defects are not responsible for this MCH phenotype.
Loss of the late function of the FGFR Heartless phenocopies the MCH phenotype of embryos lacking both CHES-1-like and jumu functions
As the MCH phenotype was not due to defects in mesoderm migration, we considered the alternative possibility that this phenotype reflected defects in signaling mechanisms involved in cardiac progenitor specification. A previously known example of such CM specification defects involves disruption of signal transduction through the FGFR Heartless (Htl) (Michelson et al., 1998b). Although htl initially plays a role in mesoderm migration, it also has a second, later function in specifying the CM. When this late function of htl is disrupted by expressing a dominant-negative version of the FGFR throughout the entire mesoderm such that migration is not affected, MCH phenotypes identical to those in the CHES-1-like; jumu double mutants are observed (Fig. 1G).
Synergistic genetic interactions between the Fkh genes and htl
The observation that disrupting late htl function phenocopies the MCH phenotype of CHES-1-like; jumu double mutants raised the possibility that htl and the Fkh genes could be acting together to specify the CM. In such a case, strong genetic interactions might occur between either of the Fkh genes and htl. To examine this possibility, we quantified and compared the MCH phenotypes in embryos heterozygous for a htl null mutation, in embryos homozygous for a CHES-1-like null mutation, in embryos homozyogous for a jumu null deficiency, in embryos both heterozygous for the htl mutation and homozygous for the CHES-1-like mutation, and in embryos heterozygous for the htl allele and homozygous for the jumu deficiency (Fig. 1K; Table 1). Whereas the MCH phenotype was never detected in the htl heterozygotes (Fig. 1H) or in the CHES-1-like homozygotes (Fig. 1B), embryos that were both heterozygous for the htl allele and homozygous for the CHES-1-like allele (Fig. 1I) exhibited this phenotype with a frequency that was significantly greater (P=0.0007) than the additive effects of the phenotypes in htl heterozygotes and the CHES-1-like homozygotes (Fig. 1K). Similarly, the frequency of MCH phenotypes in embryos both heterozygous for the htl mutation and homozygous for the jumu deficiency (Fig. 1J) was significantly greater (P<0.0001) than the additive effects of the htl heterozygotes and jumu homozygotes (Fig. 1C,H,K). Collectively, these results demonstrate synergistic genetic interactions between CHES-1-like and htl, and between jumu and htl, and are consistent with all three genes acting in the same CM specification pathway.
CHES-1-like and jumu transcriptionally activate htl expression in the precursors of the cardiac mesoderm
Although the synergistic genetic interactions between CHES-1-like, jumu and htl suggests that they work together in the same cardiac progenitor specification pathway, they do not indicate whether the Fkh TFs act either exclusively or redundantly upstream of htl to regulate its expression, or whether signaling through Htl instead mediates the expression of the Fkh genes. To discriminate between these possibilities, we examined the expression of htl in embryos that were wild type, embryos that were mutant for one or the other of the two Fkh genes, and embryos that lacked both Fkh genes.
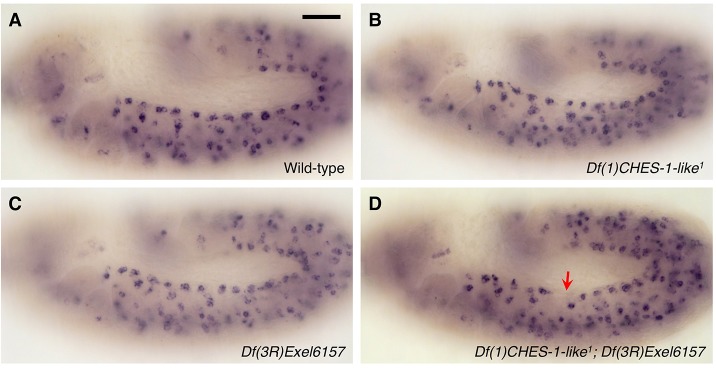
Whole embryo in situ hybridizations showed that htl transcripts are expressed in an evenly distributed pattern in every hemisegment in wild-type embryos when the CM is specified at Stage 11 (Fig. 3A). Although there was no significant change in htl expression in embryos homozygous for mutations in either CHES-1-like or jumu alone (Fig. 3B,C), hemisegments lacking htl expression in cells that would normally have become the CM were frequently observed in embryos lacking both CHES-1-like and jumu functions (Fig. 3D). These results are consistent with both Fkh genes acting in a mutually redundant manner with either one being sufficient to activate htl expression.
Fig. 3.
CHES-1-like and jumu activate htl transcription in the cardiac mesoderm. (A-D) htl mRNA in Stage 11 wild-type embryos (A), embryos lacking CHES-1-like function (B), embryos lacking jumu function (C), and embryos lacking both CHES-1-like and jumu functions (D) detected by in situ hybridization. The arrow in D indicates the absence of htl expression in cells that would normally have become the CM. Scale bar: 50 μm.
CHES-1-like and jumu transcriptionally activate htl expression by binding to the cardiac enhancer ChIPCRM2610
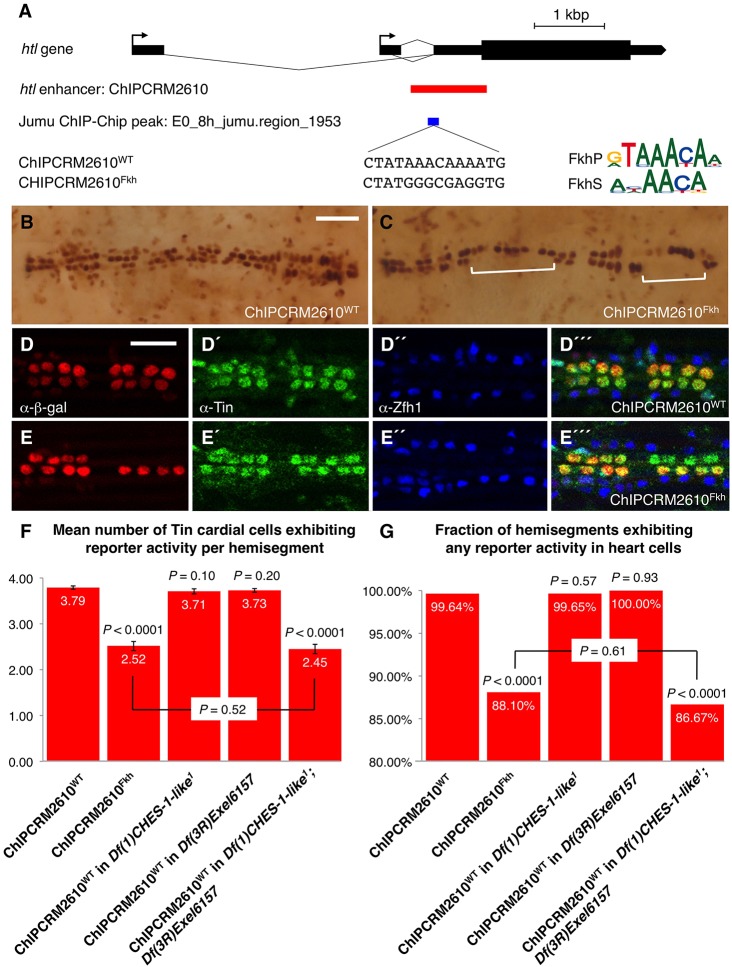
We next attempted to ascertain whether this Fkh-mediated transcriptional regulation was achieved by direct binding of the CHES-1-like and Jumu TFs to an htl enhancer. A particularly promising candidate for the htl enhancer through which this Fkh-mediated activation might be brought about is ChIPCRM2610, which drives reporter expression in the CM (Gisselbrecht et al., 2013). Chromatin immunoprecipitation data from the modENCODE project (Nègre et al., 2011) indicated that Jumu protein binds in vivo to ChIPCRM2610 at the CM specification stage (Fig. 4A). Utilizing known binding specificities of mouse Fkh TFs (Badis et al., 2009; Robasky and Bulyk, 2011) that we had previously used successfully to analyze Fkh function in Drosophila enhancers (Zhu et al., 2012), we identified a single putative Fkh binding site in ChIPCRM2610 the sequence of which matched those of both the canonical primary Fkh binding motif (FkhP) and a secondary alternative Fkh binding motif (FkhS), and the location of which corresponded with the Jumu chromatin immunoprecipitation data (Fig. 4A).
Fig. 4.
CHES-1-like and Jumu are activators of a htl cardiac enhancer. (A) Relative positions of the htl gene, the htl enhancer ChIPCRM2610, the Jumu ChIP peak, and the Fkh TF binding sequence. Logo representations of the PWMs of Fkh primary (FkhP) and secondary (FkhS) TF binding motifs obtained from protein-binding microarrays are shown, as are the Fkh binding sequences in the wild-type (ChIPCRM2610WT) and the mutated (ChIPCRM2610Fkh) enhancer. (B,C) Representative Stage 16 hearts showing β-galactosidase reporter activity driven by the wild-type (B) and mutant (C) enhancers. Square brackets indicate entire hemisegments lacking reporter activity in the case of the mutated enhancer (C). Scale bar: 50 μm. (D-E‴) Enhancer activity at single-cell resolution. The posterior-most four CCs in a hemisegment are marked by Tin expression (green), and the PCs are marked by Zfh1 expression (blue). (D-D‴) Reporter activity (red) driven by the wild-type enhancer is located primarily in the Tin-expressing CCs. (E-E‴) Mutating the Fkh binding site results in significant reduction of reporter activity. Scale bar: 25 μm. (F,G) Quantification and significance of the effects of mutating the Fkh binding site in the ChIPCRM2610 htl enhancer, and the effects of loss of function of CHES-1-like, jumu, or both CHES-1-like and jumu on the wild-type ChIPCRM2610 htl enhancer activity. The P-value over each column indicates the significance of the difference in reporter activity between the relevant enhancer genotype combination and the wild-type ChIPCRM2610WT enhancer in wild-type embryos. Error bars in F indicate 95% confidence intervals.
To test whether the Fkh TFs regulate ChIPCRM2610 through this single putative Fkh binding site, we used relevant protein binding microarray data to mutate the wild-type Fkh binding site to a sequence that, based on prior findings (Robasky and Bulyk, 2011; Zhu et al., 2012), results in significant loss of Fkh TF binding (Fig. 4A). The wild-type and mutant versions of the ChIPCRM2610 htl enhancer were cloned into β-galactosidase reporter vectors and independently inserted into the same location on the second chromosome. Thus, any differences in reporter expression between the wild-type and mutant htl enhancers can be attributed solely to the Fkh binding site mutation and not to local positional effects.
As the Drosophila heart at embryonic Stage 16 consists of multiple cells arranged in a metamerically repeated and stereotyped pattern, the perdurance of the expression patterns of the wild-type and mutant ChIPCRM2610 enhancer in each repeated hemisegment at this stage provides a simple means of scoring enhancer activity. The wild-type enhancer (ChIPCRM2610WT) drives reporter expression in virtually every cardiac hemisegment (99.64% of all examined hemisegments) primarily in the four Tin-CCs (Fig. 4B,D-D‴,F,G; Table 2; Fig. S3A). By contrast, mutating the Fkh binding site (resulting in the ChIPCRM2610Fkh enhancer) causes significant reduction (P<0.0001) of enhancer-driven reporter expression, with many hemisegments (11.90%) showing no cardiac reporter expression at all, and an overall decrease in the mean number of Tin-CCs per hemisegment in which the reporter is expressed from 3.79 to 2.52 (Fig. 4C,E-E‴,F,G; Table 2; Fig. S3B).
Table 2.
Quantification of the effects of mutating the Fkh binding site in the ChIPCRM2610 htl enhancer, and the effects of loss of function of CHES-1-like, jumu, or both CHES-1-like and jumu on the activity of the wild-type ChIPCRM2610 htl enhancer
We next assessed whether a similar phenotype could be obtained by eliminating the function of one or both of the Fkh TFs in trans. Embryos lacking either CHES-1-like or jumu exhibited reporter expression driven by the wild-type ChIPCRM2610WT enhancer that was not significantly different (P=0.10 and P=0.20, respectively, for mean numbers of Tin-CCs per hemisegment exhibiting reporter expression; P=0.57 and P=0.93, respectively, for the fractions of all hemisegments exhibiting any reporter expression) from that in wild-type embryos (Fig. 4F,G; Fig. S3C-D‴; Table 2).
By contrast, embryos that lacked both Fkh TFs demonstrated a significant reduction (P<0.0001) in the number of cells with visible reporter expression, with expression being limited to a mean of 2.45 Tin-CCs per hemisegment, and 13.33% of the hemisegments examined showing no cardiac expression (Fig. 4F,G; Fig. S3E-E‴; Table 2). Of note, there was no significant difference in reporter expression (P=0.52 for mean numbers of Tin-CCs exhibiting reporter expression; P=0.61 for fractions of all hemisegments exhibiting any reporter expression) between these double homozygotes bearing the ChIPCRM2610WT enhancer and otherwise wild-type embryos bearing the ChIPCRM2610Fkh enhancer (Fig. 4F,G). These observations indicate that abolition of Fkh TF binding by either eliminating both CHES-1-like and Jumu TFs, or by mutating the binding site on the enhancer results in similar levels of reduction in enhancer activity.
Collectively, the convergence of results between these cis and trans experiments demonstrate (1) that the CHES-1-like and Jumu TFs activate htl expression in the precursors of the CM by directly binding to the ChIPCRM2610 enhancer, and (2) that either of these two Fkh TFs is sufficient by itself to activate expression in wild-type numbers of cells.
Mesoderm-targeted ectopic expression of Htl partially rescues the MCH phenotype in embryos lacking both CHES-1-like and jumu functions
The results from our genetic interaction experiments together with our cis and trans analysis of htl expression suggest that CHES-1-like and jumu transcriptionally activate htl expression to bring about proper CM specification. To test this hypothesis, full length Htl (Michelson et al., 1998b) was expressed pan-mesodermally under the control of the twi-GAL4 driver. If htl acts downstream of the Fkh TFs in this CM specification pathway, then ectopic pan-mesodermal expression of the FGFR should at least be able to partially rescue the MCH phenotype of the CHES-1-like; jumu double mutants. We found that expression of the FGFR throughout the mesoderm does indeed significantly reduce the severity of the MCH phenotype in embryos lacking both CHES-1-like and jumu functions (Fig. 5; Table 1), indicating a downstream requirement of htl for correct CM specification mediated by the Fkh genes.
Fig. 5.
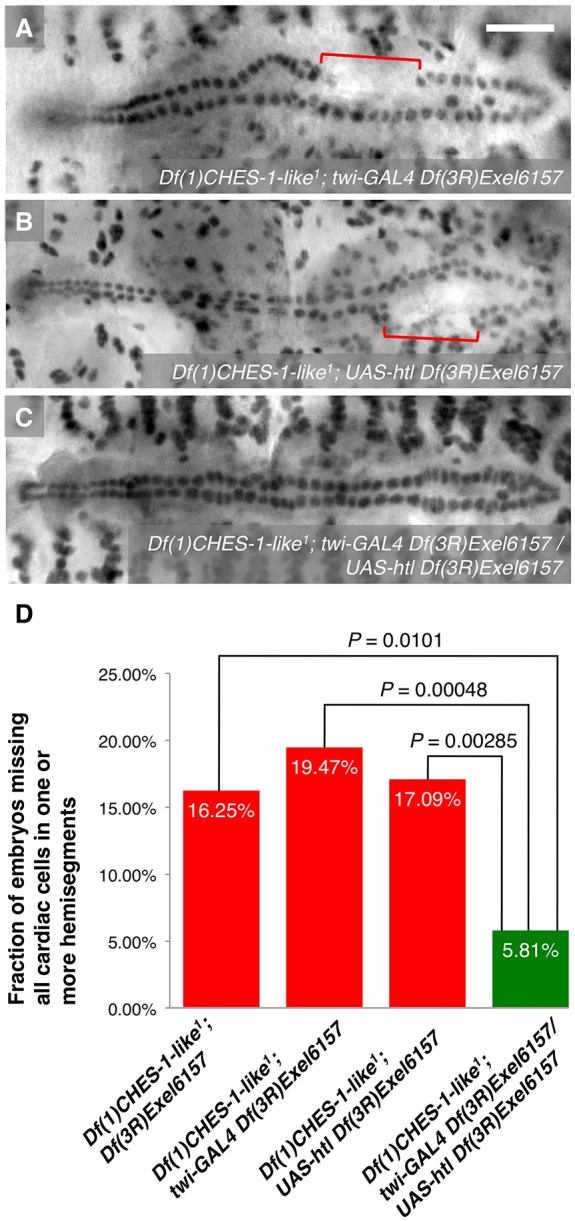
Mesoderm-targeted ectopic expression of Htl partially rescues the MCH phenotype in embryos lacking both CHES-1-like and jumu functions. (A-C) Mef2 antibody staining of CCs, illustrating the presence or absence of the MCH phenotype (square brackets), in representative Stage 16 embryos lacking both functional CHES-1-like and jumu genes but possessing (A) twi-GAL4, (B) UAS-htl or (C) both twi-GAL4 and UAS-htl transgenes, and therefore expressing Htl constitutively throughout the entire mesoderm. Scale bar: 50 μm. (D) Quantification and significance of the MCH phenotypes.
Microarray-based genome-wide RNA expression profiling indicates that CHES-1-like and jumu differentially regulate the expression of many known cardiac genes
Our previous findings had shown that Fkh TF binding sites are significantly enriched in combination with those of other known cardiogenic TFs in the enhancers of genes expressed in the CM or heart (Ahmad et al., 2014; Zhu et al., 2012), suggesting that these Fkh TFs regulate numerous downstream cardiac genes. To test this hypothesis, we used flow cytometry and Affymetrix microarrays to examine the effects of homozygous null mutations for either CHES-1-like or jumu on gene expression in mesodermal cells from Stage 11 embryos.
Consistent with the observation that CHES-1-like and jumu both activate cardiac genes, such as htl, and repress others, such as Nidogen (Zhu et al., 2012), our genome-wide expression profiling identified 42 and 56 known heart genes that were downregulated and upregulated, respectively, in the mesoderm of CHES-1-like null mutants, and 38 and 39 known cardiac genes that were downregulated and upregulated, respectively, in jumu null mutants (Table S2A-D). These numbers represent statistically significant over-representations (P=1.907×10−11 for CHES-1-like; P=6.987×10−9 for jumu) of known cardiac genes among those for which expression is differentially regulated by the Fkh genes, thus confirming that the Fkh TFs do indeed regulate many heart genes.
Of note, significant reduction in the expression levels of htl is not detected in the microarray data for either CHES-1-like or jumu null mutants alone (Table S2A,C). The most conservative explanation for this observation is that it reflects the functional redundancy already demonstrated between CHES-1-like and Jumu in regulating htl expression: either of these two Fkh TFs is sufficient by itself to activate wild-type levels of htl expression. Consistent with this explanation, microarray-based genome-wide expression profiling data from mesodermal cells ectopically overexpressing jumu (Ahmad et al., 2012) showed elevated expression levels of htl (log2 fold change=0.990804; P=8.97×10−5) compared with wild type (Table S2F).
If jumu is indeed sufficient to activate htl, then ectopic overexpression of Jumu in the mesoderm would be expected to partially resemble the effects of ectopic overexpression of activated Htl. This result is indeed observed in our expression profiling data: there is significant overlap (odds ratio=4.566; P<8.8×10−16) between genes with significantly elevated expression levels as a consequence of ectopic mesodermal overexpression of the constitutively activated form of Htl (Ahmad et al., 2012; Estrada et al., 2006) and genes that are upregulated when jumu is ectopically overexpressed in the mesoderm (Table S2F,G).
Expression profiling indicates that CHES-1-like and jumu also activate mesodermal expression of the Wingless/Wnt receptor-encoding gene frizzled
CM specification in both Drosophila and vertebrates also involves other signaling mechanisms besides FGF/FGFR, such as the Wingless/Wnt, Bone morphogenetic protein and Notch pathways (Bodmer and Frasch, 2010; Vincent and Buckingham, 2010). This raises the question of whether components of these other signaling pathways involved in cardiac progenitor specification are also regulated by the Fkh TFs. To address this, we again examined the expression profiling data for CHES-1-like and jumu null mutants.
We found that frizzled (fz), which encodes a receptor of the Wnt signaling protein Wingless (Wg), had its expression levels significantly reduced in the mesoderm of embryos lacking either CHES-1-like (log2 fold change=−1.257736; P=3.16×10−7) or jumu function (log2 fold change=−1.2118035; P=2.59×10−7) compared with wild type (Table S2A,C). These results indicate that both jumu and CHES-1-like activate fz expression in the mesoderm.
As Fz is one of the receptors of Wg, reductions in the expression levels of fz in the mesoderm would be expected to partially resemble the effects of inhibiting or reducing Wg signaling, such as with null mutations in wg. Expression profiling data on flow cytometry-purified mesodermal cells show that there is indeed significant overlap between genes downregulated in the mesoderm of embryos homozygous for a null mutation in wg (Ahmad et al., 2012; Estrada et al., 2006) and genes downregulated in either CHES-1-like (odds ratio=2.131; P<8.8×10−16) or jumu (odds ratio=1.786; P<8.8×10−16) null mutants, thus providing further evidence for the Fkh TF-mediated activation of fz (Table S2A,C,E).
Synergistic genetic interactions between the Fkh genes and fz
The largely ubiquitous presence of the fz transcript at Stages 10-11 due to maternal expression (Park et al., 1994) made it impossible for us to independently verify its Fkh-mediated activation in the subset of mesodermal cells normally destined to become cardiac progenitors by performing RNA in situ hybridization in appropriate wild-type and Fkh mutant backgrounds. The absence of known cardiac-specific enhancers for fz also precluded us from carrying out cis and trans analyses as in the case of htl. However, we hypothesized that if one or both of the Fkh genes were indeed regulating fz expression in the CM specification process, then we might expect to see synergistic, i.e. more than merely additive, genetic interactions between the Fkh TFs and the Wg receptor.
Previous work had already identified a role for fz in CM specification: embryos lacking both Fz and another Wg receptor, Frizzled 2 (Fz2), fail to develop cardiac progenitors (Bhanot et al., 1999; Chen and Struhl, 1999). Given this redundancy between the two Wg receptors and the maternal expression of fz, when we examined Stage 16 embryos homozygous for fzR52, a strong hypomorphic allele of fz resulting in a truncated protein (Jones et al., 1996), we found that only one out of 117 homozygotes (0.85%) exhibited the MCH phenotype (Fig. 6A,D; Table 1).
Fig. 6.
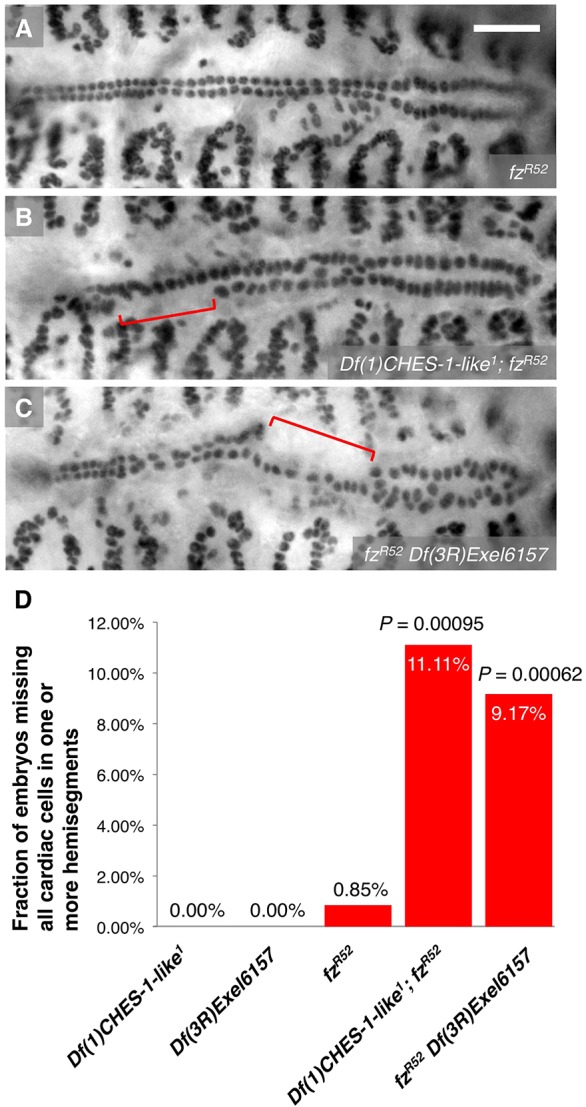
Synergistic genetic interactions between CHES-1-like and fz, and between jumu and fz. (A-C) Mef2 antibody staining of CCs, illustrating the presence or absence of the MCH phenotype (square brackets), in representative Stage 16 embryos that are (A) homozygous for fzR52, a strong hypomorphic mutation of fz, (B) doubly homozygous for both the fz mutation and the CHES-1-like null mutation, and (C) doubly homozygous for both the fz mutation and the jumu null deficiency. Whereas the MCH phenotype is rarely detected in the fzR52 homozygotes, embryos doubly homozygous for the fz mutation and one of the Fkh gene mutations exhibit significant instances of the MCH phenotype. Scale bar: 50 μm. (D) Quantification and significance of the MCH phenotypes. From left to right, the P-values indicate the difference between the phenotype of embryos doubly homozygous for the fz and CHES-1-like mutations and the additive effects of the phenotypes in fz homozygotes and CHES-1-like homozygotes; and the difference between the phenotype of embryos doubly homozygous for the fz mutation and the jumu null deficiency and the additive effects of the phenotypes in fz homozygotes and jumu homozygotes.
Embryos individually homozygous for either the CHES-1-like null mutation (Fig. 1B) or the jumu null deficiency (Fig. 1C) do not exhibit the MCH phenotype (Fig. 6D; Table 1). However, embryos that were doubly homozygous for mutations in both CHES-1-like and fz exhibited the MCH phenotype with a frequency (11.11%) significantly greater (P=0.00095) than the additive effects of the phenotypes in CHES-1-like homozygotes and the fz homozygotes (Fig. 6B,D; Table 1). Similarly, the frequency of MCH phenotypes (9.17%) in embryos doubly homozygous for both the fz hypomorphic mutation and the jumu null deficiency was significantly greater (P=0.00062) than the additive effects of the single fz and jumu homozygotes (Fig. 6C,D; Table 1).
Collectively, these results demonstrate synergistic genetic interactions between CHES-1-like and fz, and between jumu and fz. In light of our expression profiling results showing regulation of fz in the mesoderm by both CHES-1-like and jumu, the most conservative explanation for these synergistic genetic interactions is that both Fkh genes activate fz expression to bring about the proper specification of cardiac progenitors. Although these data cannot completely rule out the possibility that the Fkh genes and fz specify cardiac CM by independent parallel pathways, we note that in the latter scenario the genetic interactions between the Fkh genes and fz would be far more likely to be additive, rather than synergistic.
Mesoderm-targeted ectopic expression of Fz partially rescues the MCH phenotype in embryos lacking both CHES-1-like and jumu functions
To determine conclusively whether CHES-1-like and jumu also activate fz expression to bring about the proper CM specification, full length Fz was expressed pan-mesodermally under the control of the twi-GAL4 driver. If fz acts downstream of the Fkh TFs in this CM specification pathway, then ectopic pan-mesodermal expression of Fz should be able to partially rescue the MCH phenotype of the CHES-1-like; jumu double mutants in a manner similar to that of ectopic pan-mesodermal expression of Htl. We found that expression of Fz throughout the mesoderm significantly reduces the severity of the MCH phenotype in embryos lacking both CHES-1-like and jumu functions (Fig. 7; Table 1), thereby confirming that the Fkh genes do indeed activate fz expression to bring about the proper specification of cardiac progenitors.
Fig. 7.
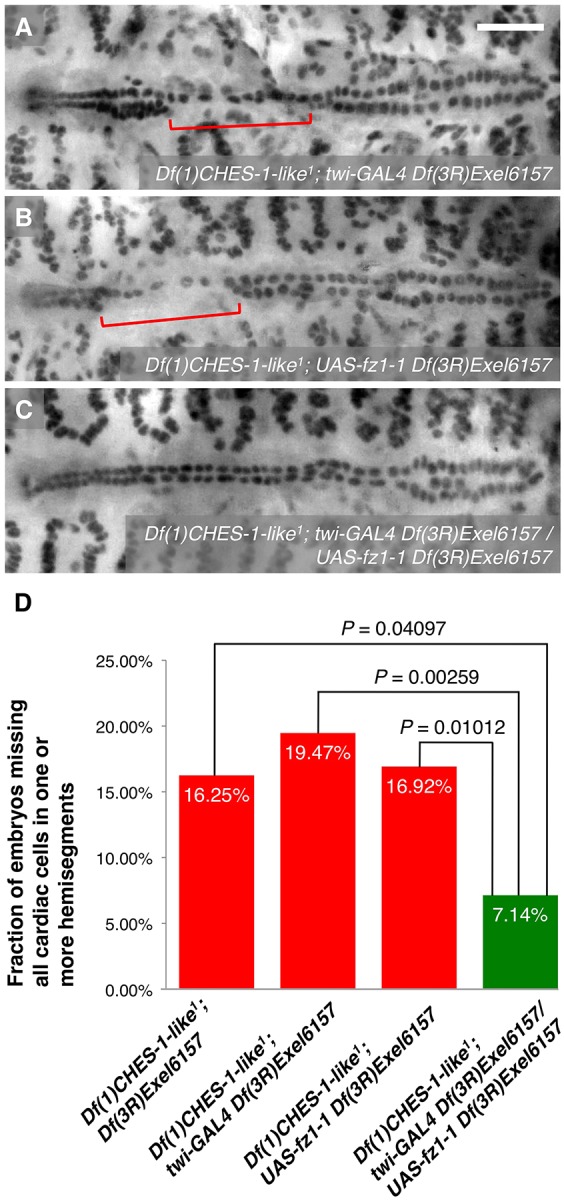
Mesoderm-targeted ectopic expression of Fz partially rescues the MCH phenotype in embryos lacking both CHES-1-like and jumu functions. (A-C) Mef2 antibody staining of CCs, illustrating the presence or absence of the MCH phenotype (square brackets), in representative Stage 16 embryos lacking both functional CHES-1-like and jumu genes but possessing (A) twi-GAL4, (B) UAS-fz1-1 or (C) both twi-GAL4 and UAS-fz1-1 transgenes, and therefore expressing Fz constitutively throughout the entire mesoderm. Scale bar: 50 μm. (D) Quantification and significance of the MCH phenotypes.
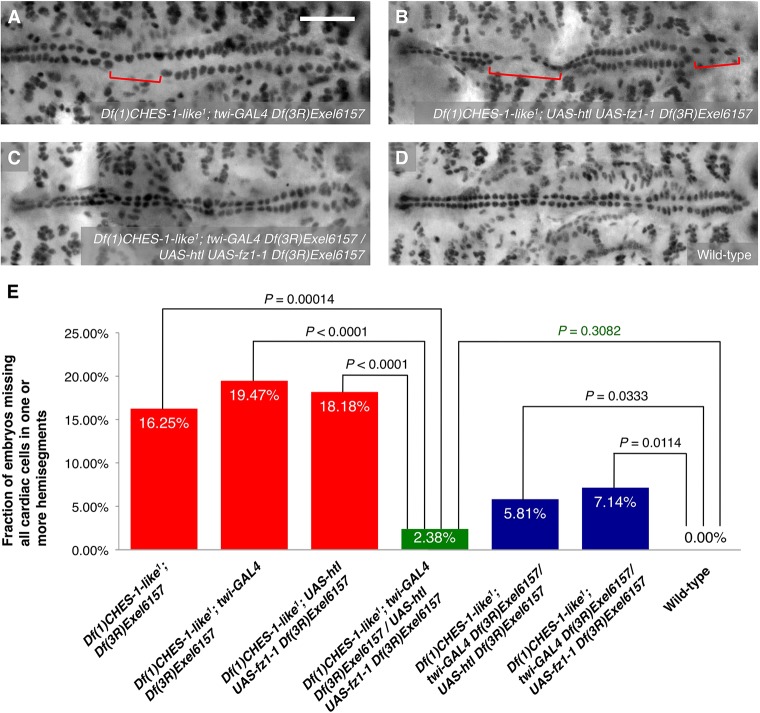
Simultaneous mesoderm-targeted ectopic expression of both Htl and Fz reduces the severity of the MCH phenotype in embryos lacking both CHES-1-like and jumu functions to wild-type levels
Finally, given that ectopic pan-mesodermal expression of either Htl or Fz receptors individually results in partial rescue of the MCH phenotype, we examined the effects of simultaneously expressing both these full-length proteins in embryos lacking both Fkh genes. Expressing both receptors under the control of the twi-GAL4 driver significantly reduces the severity of the MCH phenotype in the Fkh double mutants to a level that is even lower than those obtained when either Htl or Fz are ectopically expressed individually (Fig. 8; Table 1). Of note, although significant rescue of the MCH phenotype was indeed observed with ectopic mesoderm-targeted expression of either receptor alone (Figs 5, 7), the severity of the phenotype in each case was significantly greater than that in wild-type embryos (Fig. 8E). By contrast, expressing both Htl and Fz simultaneously in the CHES-1-like; jumu double mutants reduces the occurrence of the MCH phenotype to levels that are not significantly different from that in wild-type embryos (Fig. 8E; Table 1). Collectively, these results indicate that the two Fkh TFs, CHES-1-like and Jumu, mediate CM specification by activating the expression of receptors of both the FGF and Wg signaling pathways.
Fig. 8.
Simultaneous mesoderm-targeted ectopic expression of both Htl and Fz reduces the severity of the MCH phenotype in embryos lacking both CHES-1-like and jumu functions to wild-type levels. (A-C) Mef2 antibody staining of CCs, illustrating the presence or absence of the MCH phenotype (square brackets), in representative Stage 16 embryos lacking both functional CHES-1-like and jumu genes but possessing (A) twi-GAL4, (B) UAS-htl and UAS-fz1-1, or (C) twi-GAL4, UAS-htl and UAS-fz1-1 transgenes, and therefore simultaneously expressing Htl and Fz constitutively throughout the entire mesoderm. Scale bar: 50 μm. (D) Mef2 antibody staining of CCs in a representative Stage 16 wild-type embryo. (E) Quantification and significance of the MCH phenotypes.
DISCUSSION
In this study, we demonstrate that the two Drosophila Fkh TFs CHES-1-like and Jumu function in a redundant manner to regulate cardiac progenitor specification, with each TF being sufficient to ensure proper specification of the CM, and the MCH phenotype being detected only when the functions of both Fkh genes are eliminated. Utilizing in vivo ChiP binding data, cis-trans enhancer analyses, examination of transcript expression by in situ hybridization in appropriate mutants, phenotypic analyses of these same mutants, and genetic interaction and rescue assays, we show that one of the pathways by which this CM specification process is achieved involves the transcriptional activation of the FGFR-encoding gene htl by the Fkh TFs binding to its enhancer. Our expression profiling results combined with genetic interaction and rescue assays further indicate that the same Fkh TFs mediate an additional pathway involved in this process of cardiac progenitor specification by also activating the expression of the Wg/Wnt receptor encoding gene fz.
Regulation of cardiac progenitor specification by control of the expression of receptors of relevant signaling pathways
Studies of the signaling pathways involved in cardiac mesoderm/cardiac progenitor specification have focused largely on the signal transduction mechanisms themselves or on the mechanisms responsible for appropriate expression of the relevant signaling ligands. Our study highlights yet another mechanism by which CM specification is modulated: regulating the expression of the receptors involved in these signaling pathways by the activity of upstream TFs. Particularly in the case of the FGFR Htl, we found that the Fkh TFs transcriptionally activate its requisite expression in a tight spatiotemporal domain that corresponds to the cells destined to become cardiac progenitors in the dorsal mesoderm, independent of any effect of Htl on cell migration (Beiman et al., 1996; Gisselbrecht et al., 1996; Shishido et al., 1997).
The instructive role of FGF/FGFR signaling in specifying cardiac progenitors has been shown in mouse, chick and zebrafish (Alsan and Schultheiss, 2002; Barron et al., 2000; Crossley and Martin, 1995; Reifers et al., 2000) in addition to Drosophila. Fkh TF-mediated regulation of FGFRs in non-cardiogenic processes has also been observed in vertebrates (Coffer and Burgering, 2004; Mandinova et al., 2009; Nakada et al., 2009; Tsai et al., 2003). Given the conservation of both genes and regulatory networks in heart development in both Drosophila and vertebrates (Cripps and Olson, 2002; Olson, 2006), and the numerous cardiogenic processes in mammals in which the Fkh genes are involved (Evans-Anderson et al., 2008; Hu et al., 2004; Korver et al., 1998; Roessler et al., 2008; Wang et al., 2004; Yu et al., 2010), it will be of particular interest to see if cardiac progenitor specification in vertebrates also involves the regulation of relevant FGFRs by Fkh genes.
Although signaling through the Wg pathway in Drosophila is essential for normal heart formation, the role of Wnt signaling in specifying cardiac progenitors in vertebrates is considerably more complex: signaling through Wnt3a is required for initial mesoderm formation but inhibits later cardiac specification, whereas signaling through Wnt5a and Wnt11 stimulates cardiac progenitor specification (Gessert and Kuhl, 2010). Regardless, regulation of Fz homologs in vertebrates could provide another mechanism for modulating this process.
We had previously suggested that the Fkh gene FoxM1, which transcriptionally regulates a mammalian polo ortholog (Laoukili et al., 2005), and mutations in which exhibit cardiac progenitor cell division defects in mouse hearts (Korver et al., 1998), was the functional ortholog of CHES-1-like and jumu in mammals (Ahmad et al., 2012). Thus, it was intriguing to learn that FoxM1 also regulates at least one mammalian fz homolog, Frizzled family receptor 6 (Fzd6) (Kim et al., 2005). Given that FZD6 is also expressed in the human heart (Uhlen et al., 2010), it will be of interest to see if this Wnt receptor, or some other Fkh-regulated Fz homolog, also plays a role in cardiac specification.
Redundancy and robustness in cardiac mesoderm specification
Crucial developmental steps are frequently robust and interconnected in order to prevent damage to any one of their constituent genetic components resulting in the overall disruption of the process in which they are involved. This concept is demonstrated at multiple levels by the CM specification process examined in the present study.
One elegant illustration of this regulatory design feature is the degree of mutual redundancy exhibited by CHES-1-like and jumu, with either of these genes being able to compensate for defects in the function of the other to mediate normal heart formation. That is, each Fkh TF is individually sufficient to activate the expression of htl to bring about proper FGFR-mediated CM specification, with the absence of either the htl transcript or htl enhancer activity in a hemisegment, as well as the presence of the MCH phenotype, being detected only when both Fkh functions are eliminated in the same embryo.
Furthermore, the absence of both Fkh TFs reduces but does not completely eliminate CM specification; similarly, the expression of the htl enhancer and transcript is reduced but not completely abrogated in embryos lacking both Fkh functions. These observations indicate that additional redundant mechanisms must exist to at least partially maintain htl expression and CM specification in the absence of the Fkh TFs. A preliminary computational analysis we performed using the algorithm PhylCRM (Warner et al., 2008) identified a highly conserved Pointed binding site in the htl enhancer ChIPCRM2610 as well as several putative Twist and Tinman binding sites, suggesting that these cardiogenic TFs, plus potentially other TFs not presently known to function in heart development, might mediate redundant mechanisms for activating htl expression and thereby CM specification in embryos lacking CHES-1-like and jumu functions.
Yet another example of an interconnection among the components of the cardiac gene regulatory network is the control of not one, but two CM specification pathways – the FGF/FGFR signaling pathway and the Wnt signaling pathway – by these Fkh TFs. Although we cannot rule out the possibility that the Fkh TFs might also regulate additional CM specification pathways, the observation that ectopic expression of both Htl and Fz simultaneously can restore wild-type levels of CM specification in the CHES-1-like; jumu double mutants suggests that the Fkh-mediated regulation of only these two receptors is sufficient for wild-type function.
Finally, a fourth example of the robustness of this system is the previously known redundancy in the Wnt signaling pathway itself. Fz is not the only receptor utilized by the Wnt signaling pathway for CM specification: the functions of both Fz and Fz2 have to be eliminated before any major disruption is detected in cardiac progenitor specification (Bhanot et al., 1999; Chen and Struhl, 1999).
Pleiotropic roles of CHES-1-like and jumu in cardiogenesis
Our previous studies had suggested that the Fkh TFs might play roles in multiple cardiogenic processes. We had observed a statistically significant over-representation of primary and secondary Fkh TF binding motifs in combination with other known cardiogenic TF binding sites in putative enhancers of genes known to be expressed in the heart (Zhu et al., 2012). Machine-learning modeling of cardiac enhancers based on sequence features of known heart enhancers had also identified Fkh TFs as important regulators (Ahmad et al., 2014). Furthermore, overexpression of Jumu in the mesoderm resulted in elevated expression levels of many known cardiac genes (Ahmad et al., 2012).
With the results of this study, we now show that the same Fkh genes mediate at least two distinct cardiogenic processes (Fig. 9): (1) cardiac progenitor specification by regulating the expression of the FGFR encoded by htl and the Wnt receptor encoded by fz, and (2) the subsequent cell divisions of these cardiac progenitors by regulating the activity of Polo kinase (Ahmad et al., 2012). Such pleiotropic roles for the same TFs provide an elegant illustration of how the vast array of complex processes involved in cardiogenesis can be orchestrated by a small set of regulators.
Fig. 9.
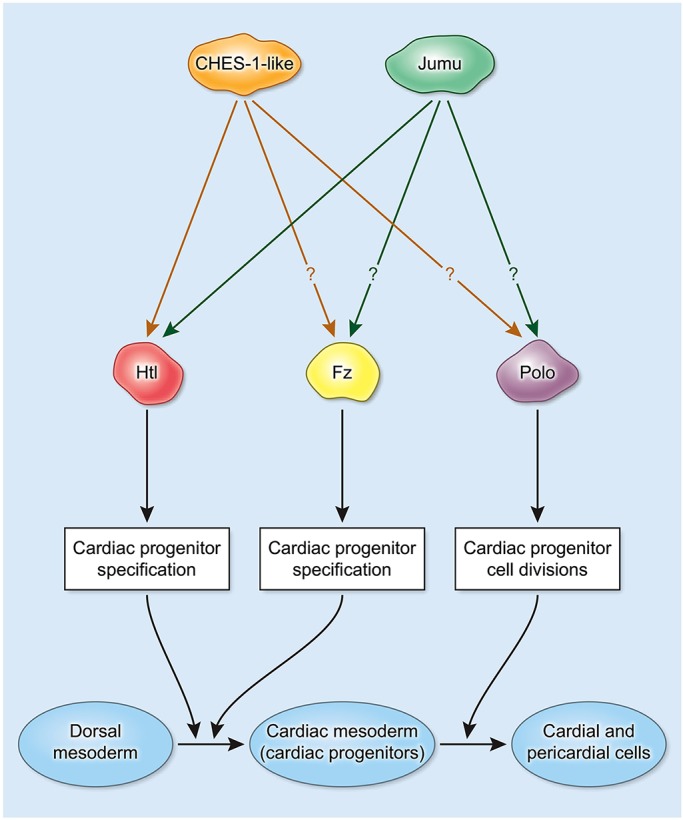
Pleiotropic roles of CHES-1-like and jumu in cardiogenesis. Schematic illustrating the roles of the Fkh TFs in multiple cardiogenic processes. Both CHES-1-like and Jumu transcriptionally activate expression of htl via its cardiac enhancer in a mutually redundant manner to achieve proper specification of cardiac progenitors from the dorsal mesoderm. Both Fkh TFs also transcriptionally activate fz expression to mediate CM specification, but it has yet to be determined if the activation is direct. Subsequently, both Fkh TFs activate Polo kinase to bring about proper cell divisions of these cardiac progenitors, but it is not yet known whether this activation is direct.
MATERIALS AND METHODS
Drosophila strains and transgenic reporter constructs
The following mutant alleles, deficiencies and transgenes were used: Df(1)CHES-1-like1, Df(3R)Exel6157 and Ubi-polo (Ahmad et al., 2012); twi-GAL4 (Greig and Akam, 1993); Mef2-GAL4 (Ranganayakulu et al., 1996); tinD-GAL4 (Yin et al., 1997); Hand-GAL4 (Han and Olson, 2005); UAS-2EGFP (Halfon et al., 2002); UAS-DN-htl (Michelson et al., 1998b); htlAB42 (Gisselbrecht et al., 1996); stumpsYY202 (Michelson et al., 1998a); UAS-htl (Michelson et al., 1998b); fzR52 (Jones et al., 1996); CHES-1-likeKK101264 (Dietzl et al., 2007); jumuGL00363 (Ni et al., 2011) and UAS-fz1-1 (Boutros et al., 2000). Wild-type and mutated ChIPCRM2610 enhancers were synthesized by Integrated DNA Technologies, cloned into the pWattB-nlacZ vector (Busser et al., 2012) and targeted to the attP40 site (Markstein et al., 2008) by phiC31-mediated integration (Groth et al., 2004) to create the ChIPCRM2610WT and ChIPCRM2610Fkh lacZ reporter-driving lines.
RNA interference assays
RNA interference (RNAi) assays targeted to CM precursors were performed as previously described (Ahmad et al., 2012). UAS-RNAi constructs from the Transgenic RNAi Project (TRiP) at Harvard Medical School (Ni et al., 2011) and the Vienna Drosophila Resource Center (VDRC) (Dietzl et al., 2007) were expressed using both the tinD-GAL4 and Hand-GAL4 drivers simultaneously. The efficiency of RNAi knockdowns was enhanced both by allowing embryos to age at 29°C and by using UAS-dcr2.
In situ hybridization, immunohistochemistry and cell counting
Embryo fixation, probe synthesis and histochemical staining were carried out as previously described (Ahmad et al., 2014; Ahmad et al., 2012; Estrada et al., 2006; Zhu et al., 2012). For all quantitative studies of gene expression, cells in >250 hemisegments were counted for each genotype examined. Because mutations in CHES-1-like and jumu cause defective cardiac progenitor cell divisions that result in certain hemisegments having atypical numbers of cells, only hemisegments with the correct number of Tin-CCs were examined and scored in Df(1)CHES-1-like1 embryos, Df(3R)Exel6157 embryos and Df(1)CHES-1-like1; Df(3R)Exel6157 embryos.
Transcriptional expression profiling
Microarray-based gene expression profiles for GFP-positive mesodermal cells isolated by fluorescence-activated sorting of single cell suspensions prepared from Stage 11-early Stage 12 twi-GAL4 UAS-2EGFP embryos, Df(1)CHES-1-like1; twi-GAL4 UAS-2EGFP embryos and twi-GAL4 Df(3R)Exel6157/UAS-2EGFP Df(3R)Exel6157 embryos were obtained by Affymetrix microarray hybridization as described previously (Ahmad et al., 2012). Array data were normalized with the rma command (with default settings) in the affy Bioconductor package (Gautier et al., 2004) and linear models were fitted using limma (Smyth, 2004). The microarray data utilized in this study are available from Gene Expression Omnibus with the accession numbers GSE3854, GSE34946 and GSE65439.
Statistical analyses
A description of the statistical analysis of genetic interaction assays, rescue assays, cis-trans enhancer assays and geneset enrichment is provided in the supplementary materials and methods.
Acknowledgements
We thank M. Bulyk for sharing data on the htl enhancer prior to publication; P. Adler, B. Paterson, J. Lipsick, M. Mlodzik, J. Skeath, the TRiP at Harvard Medical School and the Vienna Drosophila Resource Center for providing fly strains and antibodies; B. Busser, T. Tansey, S. Kumar and C. Sonnenbrot for technical assistance; and the Flow Cytometry Core Facility and the DNA Sequencing and Genomics Core Facility of the National Heart Lung and Blood Institute Division of Intramural Research for purifying mesodermal cells and for expression profiling, respectively.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
S.M.A. and A.M.M. conceived and designed the overall research project, interpreted the results, and wrote the manuscript. S.M.A. and P.B. purified mesodermal cells for expression profiling. S.M.A. performed all other experiments. S.S.G. analyzed the expression profiling data and performed geneset enrichment analysis. N.J. performed all phenotype- and enhancer expression-related statistical analyses.
Funding
This work was supported by the National Heart, Lung, and Blood Institute (NHLBI) Division of Intramural Research (A.M.M.). Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.122952/-/DC1
References
- Ahmad S. M., Tansey T. R., Busser B. W., Nolte M. T., Jeffries N., Gisselbrecht S. S., Rusan N. M. and Michelson A. M. (2012). Two forkhead transcription factors regulate the division of cardiac progenitor cells by a Polo-dependent pathway. Dev. Cell 23, 97-111. 10.1016/j.devcel.2012.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S. M., Busser B. W., Huang D., Cozart E. J., Michaud S., Zhu X., Jeffries N., Aboukhalil A., Bulyk M. L., Ovcharenko I. et al. (2014). Machine learning classification of cell-specific cardiac enhancers uncovers developmental subnetworks regulating progenitor cell division and cell fate specification. Development 141, 878-888. 10.1242/dev.101709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsan B. H. and Schultheiss T. M. (2002). Regulation of avian cardiogenesis by Fgf8 signaling. Development 129, 1935-1943. [DOI] [PubMed] [Google Scholar]
- Badis G., Berger M. F., Philippakis A. A., Talukder S., Gehrke A. R., Jaeger S. A., Chan E. T., Metzler G., Vedenko A., Chen X. et al. (2009). Diversity and complexity in DNA recognition by transcription factors. Science 324, 1720-1723. 10.1126/science.1162327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron M., Gao M. and Lough J. (2000). Requirement for BMP and FGF signaling during cardiogenic induction in non-precardiac mesoderm is specific, transient, and cooperative. Dev. Dyn. 218, 383-393. [DOI] [PubMed] [Google Scholar]
- Beiman M., Shilo B. Z. and Volk T. (1996). Heartless, a Drosophila FGF receptor homolog, is essential for cell migration and establishment of several mesodermal lineages. Genes Dev. 10, 2993-3002. 10.1101/gad.10.23.2993 [DOI] [PubMed] [Google Scholar]
- Bhanot P., Fish M., Jemison J. A., Nusse R., Nathans J. and Cadigan K. M. (1999). Frizzled and Dfrizzled-2 function as redundant receptors for Wingless during Drosophila embryonic development. Development 126, 4175-4186. [DOI] [PubMed] [Google Scholar]
- Bodmer R. and Frasch M. (2010). Development and aging of the Drosophila heart. In Heart Development and Regeneration (ed. Rosenthal N. and Harvey R. P.), pp. 47-86. London: Academic Press. [Google Scholar]
- Boutros M., Mihaly J., Bouwmeester T. and Mlodzik M. (2000). Signaling specificity by Frizzled receptors in Drosophila. Science 288, 1825-1828. 10.1126/science.288.5472.1825 [DOI] [PubMed] [Google Scholar]
- Busser B. W., Taher L., Kim Y., Tansey T., Bloom M. J., Ovcharenko I. and Michelson A. M. (2012). A machine learning approach for identifying novel cell type-specific transcriptional regulators of myogenesis. PLoS Genetics 8, e1002531 10.1371/journal.pgen.1002531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. M. and Struhl G. (1999). Wingless transduction by the Frizzled and Frizzled2 proteins of Drosophila. Development 126, 5441-5452. [DOI] [PubMed] [Google Scholar]
- Coffer P. J. and Burgering B. M. T. (2004). Forkhead-box transcription factors and their role in the immune system. Nat. Rev. Immunol. 4, 889-899. 10.1038/nri1488 [DOI] [PubMed] [Google Scholar]
- Cripps R. M. and Olson E. N. (2002). Control of cardiac development by an evolutionarily conserved transcriptional network. Dev. Biol. 246, 14-28. 10.1006/dbio.2002.0666 [DOI] [PubMed] [Google Scholar]
- Crossley P. H. and Martin G. R. (1995). The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121, 439-451. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Estrada B., Choe S. E., Gisselbrecht S. S., Michaud S., Raj L., Busser B. W., Halfon M. S., Church G. M. and Michelson A. M. (2006). An integrated strategy for analyzing the unique developmental programs of different myoblast subtypes. PLoS Genet. 2, e16 10.1371/journal.pgen.0020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans-Anderson H. J., Alfieri C. M. and Yutzey K. E. (2008). Regulation of cardiomyocyte proliferation and myocardial growth during development by FOXO transcription factors. Circ. Res. 102, 686-694. 10.1161/CIRCRESAHA.107.163428 [DOI] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B. M. and Irizarry R. A. (2004). affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307-315. 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- Gessert S. and Kuhl M. (2010). The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ. Res. 107, 186-199. 10.1161/CIRCRESAHA.110.221531 [DOI] [PubMed] [Google Scholar]
- Gisselbrecht S., Skeath J. B., Doe C. Q. and Michelson A. M. (1996). heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev. 10, 3003-3017. 10.1101/gad.10.23.3003 [DOI] [PubMed] [Google Scholar]
- Gisselbrecht S. S., Barrera L. A., Porsch M., Aboukhalil A., Estep P. W. III, Vedenko A., Palagi A., Kim Y., Zhu X., Busser B. W. et al. (2013). Highly parallel assays of tissue-specific enhancers in whole Drosophila embryos. Nat. Methods 10, 774-780. 10.1038/nmeth.2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig S. and Akam M. (1993). Homeotic genes autonomously specify one aspect of pattern in the Drosophila mesoderm. Nature 362, 630-632. 10.1038/362630a0 [DOI] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R. and Calos M. P. (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775-1782. 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon M. S., Gisselbrecht S., Lu J., Estrada B., Keshishian H. and Michelson A. M. (2002). New fluorescent protein reporters for use with the Drosophila Gal4 expression system and for vital detection of balancer chromosomes. Genesis 34, 135-138. [DOI] [PubMed] [Google Scholar]
- Han Z. and Bodmer R. (2003). Myogenic cells fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division. Development 130, 3039-3051. 10.1242/dev.00484 [DOI] [PubMed] [Google Scholar]
- Han Z. and Olson E. N. (2005). Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development 132, 3525-3536. 10.1242/dev.01899 [DOI] [PubMed] [Google Scholar]
- Hu T., Yamagishi H., Maeda J., McAnally J., Yamagishi C. and Srivastava D. (2004). Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development 131, 5491-5502. 10.1242/dev.01399 [DOI] [PubMed] [Google Scholar]
- Imam F., Sutherland D., Huang W. and Krasnow M. A. (1999). stumps, a Drosophila gene required for fibroblast growth factor (FGF)-directed migrations of tracheal and mesodermal cells. Genetics 152, 307-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. H., Liu J. and Adler P. N. (1996). Molecular analysis of EMS-induced frizzled mutations in Drosophila melanogaster. Genetics 142, 205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I.-M., Ramakrishna S., Gusarova G. A., Yoder H. M., Costa R. H. and Kalinichenko V. V. (2005). The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J. Biol. Chem. 280, 22278-22286. 10.1074/jbc.M500936200 [DOI] [PubMed] [Google Scholar]
- Korver W., Schilham M. W., Moerer P., van den Hoff M. J., Dam K., Lamers W. H., Medema R. H. and Clevers H. (1998). Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor Trident. Curr. Biol. 8, 1327-1330. 10.1016/S0960-9822(07)00563-5 [DOI] [PubMed] [Google Scholar]
- Laoukili J., Kooistra M. R., Brás A., Kauw J., Kerkhoven R. M., Morrison A., Clevers H. and Medema R. H. (2005). FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7, 126-136. 10.1038/ncb1217 [DOI] [PubMed] [Google Scholar]
- Mandinova A., Kolev V., Neel V., Hu B., Stonely W., Lieb J., Wu X., Colli C., Han R., Pazin M. J. et al. (2009). A positive FGFR3/FOXN1 feedback loop underlies benign skin keratosis versus squamous cell carcinoma formation in humans. J. Clin. Invest. 119, 3127-3137. 10.1172/JCI38543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E. and Perrimon N. (2008). Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476-483. 10.1038/ng.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson A. M., Gisselbrecht S., Buff E. and Skeath J. B. (1998a). Heartbroken is a specific downstream mediator of FGF receptor signalling in Drosophila. Development 125, 4379-4389. [DOI] [PubMed] [Google Scholar]
- Michelson A. M., Gisselbrecht S., Zhou Y., Baek K.-H. and Buff E. M. (1998b). Dual functions of the heartless fibroblast growth factor receptor in development of the Drosophila embryonic mesoderm. Dev. Genet. 22, 212-229. [DOI] [PubMed] [Google Scholar]
- Nakada C., Iida A., Tabata Y. and Watanabe S. (2009). Forkhead transcription factor foxe1 regulates chondrogenesis in zebrafish. J. Exp. Zool. B Mol. Dev. Evol. 312B, 827-840. 10.1002/jez.b.21298 [DOI] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Ma L., Bristow C. A., Miller S. W., Wagner U., Kheradpour P., Eaton M. L., Loriaux P., Sealfon R. et al. (2011). A cis-regulatory map of the Drosophila genome. Nature 471, 527-531. 10.1038/nature09990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Q., Zhou R., Czech B., Liu L. P., Holderbaum L., Yang-Zhou D., Shim H. S., Tao R., Handler D., Karpowicz P. et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405-407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E. N. (2006). Gene regulatory networks in the evolution and development of the heart. Science 313, 1922-1927. 10.1126/science.1132292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W. J., Liu J. and Adler P. N. (1994). Frizzled gene expression and development of tissue polarity in the Drosophila wing. Dev. Genet. 15, 383-389. 10.1002/dvg.1020150410 [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G., Schulz R. A. and Olson E. N. (1996). Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev. Biol. 176, 143-148. 10.1006/dbio.1996.9987 [DOI] [PubMed] [Google Scholar]
- Reifers F., Walsh E. C., Leger S., Stainier D. Y. and Brand M. (2000). Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar). Development 127, 225-235. [DOI] [PubMed] [Google Scholar]
- Robasky K. and Bulyk M. L. (2011). UniPROBE, update 2011: expanded content and search tools in the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 39, D124-D128. 10.1093/nar/gkq992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E., Ouspenskaia M. V., Karkera J. D., Vélez J. I., Kantipong A., Lacbawan F., Bowers P., Belmont J. W., Towbin J. A., Goldmuntz E. et al. (2008). Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am. J. Hum. Genet. 83, 18-29. 10.1016/j.ajhg.2008.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido E., Ono N., Kojima T. and Saigo K. (1997). Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development 124, 2119-2128. [DOI] [PubMed] [Google Scholar]
- Smyth G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Tsai P. T., Lee R. A. and Wu H. (2003). BMP4 acts upstream of FGF in modulating thymic stroma and regulating thymopoiesis. Blood 102, 3947-3953. 10.1182/blood-2003-05-1657 [DOI] [PubMed] [Google Scholar]
- Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S. et al. (2010). Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248-1250. 10.1038/nbt1210-1248 [DOI] [PubMed] [Google Scholar]
- Vincent S. D. and Buckingham M. E. (2010). How to make a heart: the origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 90, 1-41. 10.1016/S0070-2153(10)90001-X [DOI] [PubMed] [Google Scholar]
- Vincent S., Wilson R., Coelho C., Affolter M. and Leptin M. (1998). The Drosophila protein Dof is specifically required for FGF signaling. Mol. Cell 2, 515-525. 10.1016/S1097-2765(00)80151-3 [DOI] [PubMed] [Google Scholar]
- Wang B., Weidenfeld J., Lu M. M., Maika S., Kuziel W. A., Morrisey E. E. and Tucker P. W. (2004). Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development 131, 4477-4487. 10.1242/dev.01287 [DOI] [PubMed] [Google Scholar]
- Ward E. J. and Skeath J. B. (2000). Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development 127, 4959-4969. [DOI] [PubMed] [Google Scholar]
- Warner J. B., Philippakis A. A., Jaeger S. A., He F. S., Lin J. and Bulyk M. L. (2008). Systematic identification of mammalian regulatory motifs’ target genes and functions. Nat. Methods 5, 347-353. 10.1038/nmeth.1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Xu X. L. and Frasch M. (1997). Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development 124, 4971-4982. [DOI] [PubMed] [Google Scholar]
- Yu S., Shao L., Kilbride H. and Zwick D. L. (2010). Haploinsufficiencies of FOXF1 and FOXC2 genes associated with lethal alveolar capillary dysplasia and congenital heart disease. Am. J. Med. Genet. A 152A, 1257-1262. 10.1002/ajmg.a.33378 [DOI] [PubMed] [Google Scholar]
- Zhu X., Ahmad S. M., Aboukhalil A., Busser B. W., Kim Y., Tansey T. R., Haimovich A., Jeffries N., Bulyk M. L. and Michelson A. M. (2012). Differential regulation of mesodermal gene expression by Drosophila cell type-specific Forkhead transcription factors. Development 139, 1457-1466. 10.1242/dev.069005 [DOI] [PMC free article] [PubMed] [Google Scholar]