Abstract
Exogenous bone morphogenetic proteins (Bmp) are well known to induce ectopic bone formation, but the physiological effect of Bmp signaling on normal bone is not completely understood. By deleting the receptor Bmpr1a in osteoblast lineage cells with Dmp1-Cre, we observed a dramatic increase in trabecular bone mass in postnatal mice, which was due to a marked increase in osteoblast number that was likely to be driven by hyperproliferation of Sp7+ preosteoblasts. Similarly, inducible deletion of Bmpr1a in Sp7+ cells specifically in postnatal mice increased trabecular bone mass. However, deletion of Smad4 by the same approaches had only a minor effect, indicating that Bmpr1a signaling suppresses trabecular bone formation through effectors beyond Smad4. Besides increasing osteoblast number in the trabecular bone, deletion of Bmpr1a by Dmp1-Cre also notably reduced osteoblast activity, resulting in attenuation of periosteal bone growth. The impairment in osteoblast activity correlated with reduced mTORC1 signaling in vivo, whereas inhibition of mTORC1 activity abolished the induction of protein anabolism genes by BMP2 treatment in vitro. Thus, physiological Bmpr1a signaling in bone exerts a dual function in both restricting preosteoblast proliferation and promoting osteoblast activity.
KEY WORDS: Bmp, Bmpr1a, Smad4, Osteoblast, mTORC1, Mouse
Summary: The genetic deletion of Bmpr1a in mice reveals a dual function for BMP signaling in restricting preosteoblast proliferation and promoting osteoblast activity, probably via regulation of mTORC1.
INTRODUCTION
The bone morphogenetic protein (Bmp) family includes over 30 secreted signaling molecules in humans, regulating both embryogenesis and postnatal tissue homeostasis (Moustakas and Heldin, 2009; Wu and Hill, 2009). Bmp proteins signal through serine/threonine kinase receptors known as type I and type II receptors. Four type I receptors, namely Bmpr1a (Alk3), Bmpr1b (Alk6), Acvrl1 (Alk1) and Acvr1 (Alk2), and three type II receptors, namely Bmpr2 (BMPRII), Acvr2a (ActRIIA) and Acvr2b (ActRIIB), are believed to mediate Bmp signaling (Miyazono et al., 2010). Binding of dimeric Bmp proteins leads to the assembly of a hetero-tetramer comprising two molecules of each receptor type, resulting in the phosphorylation and activation of the type I receptor by the type II receptor, which possesses constitutively active kinase activity (Wrana et al., 1994). Upon activation, the type I receptors activate receptor Smads (Smad1, 5, 8) through phosphorylation, and the activated Smads recruit the common partner Smad4 and other nuclear factors to regulate gene expression (Massague, 2012; Wharton and Derynck, 2009; Wu and Hill, 2009). In addition to Smad signaling, Bmp proteins have been shown to activate other pathways, including TAK1-p38 and PI3K-Akt signaling (Ghosh-Choudhury et al., 2002, 2013; Massague, 2012; Miyazono et al., 2010). The functional significance of Smad-dependent versus Smad-independent Bmp signaling is likely to depend on the cellular context.
Originally discovered in bone, Bmp proteins have been extensively studied in the context of skeletal cell types (Urist et al., 1979). These studies have led to the clinical use of Bmp2 and Bmp7 as bone anabolic agents in orthopedic applications. In recent years, mouse genetic studies have established the essential role of Bmp proteins in cartilage development. Individual or combinatorial deletion of Bmp2, 4 and 7 in the embryonic limb mesenchyme revealed that a threshold of combinatorial Bmp levels is necessary for chondrogenesis (Bandyopadhyay et al., 2006). Conditional deletion of the type I Bmp receptors Bmpr1a and Bmpr1b, or of Smad1 and Smad5, in chondrocytes caused severe chondrodysplasia (Retting et al., 2009; Yoon et al., 2005). Although deletion of Smad4 in chondrocytes only modestly affected cartilage development, its deletion in the limb bud mesenchyme completely abolished chondrogenesis in the mouse embryo (Benazet et al., 2012; Lim et al., 2015; Zhang et al., 2005). Thus, Smad-dependent Bmp signaling critically controls multiple steps of cartilage development.
The role of Bmp signaling in osteoblasts is less well understood. Removal of Bmpr1a in osteocalcin-positive osteoblasts suppressed osteoblast function without affecting osteoblast numbers, but also reduced bone resorption in the aged mice (Mishina et al., 2004). Deletion of Smad4 with the same Cre driver also reduced osteoblast activity and bone resorption, but additionally decreased osteoblast number (Tan et al., 2007). Both of those studies reported a lower trabecular bone mass in the young mutant mice but a reversed phenotype when the mice aged. However, more recent studies with tamoxifen-inducible Cre to delete Bmpr1a in Col1a1-positive cells either in utero or postnatally showed a marked increase in trabecular bone mass, which was attributed to suppression of osteoclastogenesis by increased Wnt signaling (Kamiya et al., 2010, 2008a,b). Deletion of Acvr1 by the same approach also increased bone mass but the cellular basis was not investigated (Kamiya et al., 2011). In addition, systemic injection of a soluble Bmpr1a receptor increased both trabecular and cortical bone mass through an early increase in osteoblast number followed by a decrease in osteoclast number (Baud'huin et al., 2012). Thus, both positive and negative roles have been described for Bmp in the regulation of bone mass, but the mechanisms for such opposing roles are not clear.
Here we have revisited the function of physiological Bmp signaling in osteoblast lineage cells. By deleting Bmpr1a with either Dmp1-Cre or doxycycline-regulated Sp7-Cre we show that Bmp signaling normally restrains osteoblast number within the trabecular bone region through suppression of preosteoblast proliferation. In addition, Bmp signaling stimulates osteoblast activity and this is likely to be through activation of the mTORC1 complex.
RESULTS
Bmpr1a deletion with Dmp1-Cre increases trabecular bone mass but reduces periosteal bone growth
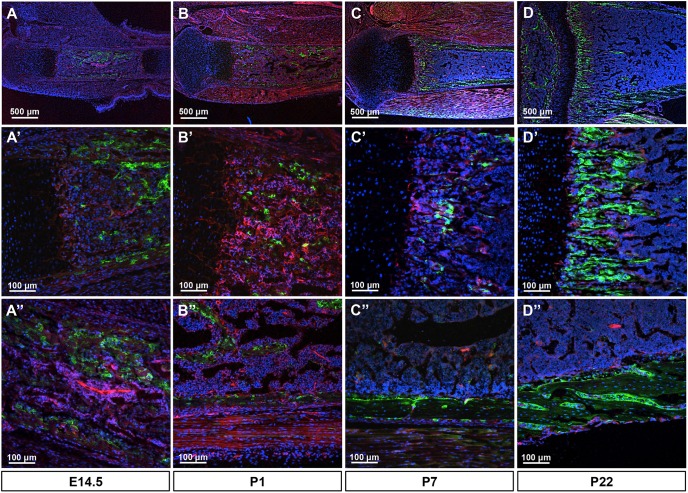
To examine the physiological role of Bmp signaling in postnatal bone formation, we conditionally removed Bmpr1a with Dmp1-Cre, which was previously reported to target mainly osteocytes (Lu et al., 2007). We first assessed the targeting specificity of Dmp1-Cre using the mT/mG reporter mouse. At E14.5, we detected Cre activity, as indicated by GFP expression, at the nascent periosteum and the invading mesenchyme, with only occasional GFP+ cells at the chondro-osseous junction (Fig. 1A-A″). At postnatal day (P) 1 or 7, GFP+ cells were detected at both the bone collar and the emerging bony trabeculae (Fig. 1B-B″). In the bone collar, both osteoblasts and osteocytes were positive but GFP was stronger in the former. By P22, GFP+ cells were not only present in osteoblasts and osteocytes of both cortical and trabecular bone, but were also abundant at the chondro-osseous junction, which is known to be enriched for osteoblast precursors (Fig. 1D-D″) (Regan et al., 2014). Thus, Dmp1-Cre targets osteoblast lineage cells at an earlier stage than previously reported.
Fig. 1.
Dmp1-Cre targets osteoblast lineage cells in mice. (A-D) Representative fluorescence images of longitudinal sections of the tibia from Dmp1-Cre; mT/mG mice at the indicated ages. (A′-D′) Representative trabecular bone regions at higher magnification. (A″-D″) Representative cortical bone regions at higher magnification. Red, mTomato; green, mGFP; blue, DAPI staining of nuclei. Red and green protein fluorescence was captured directly.
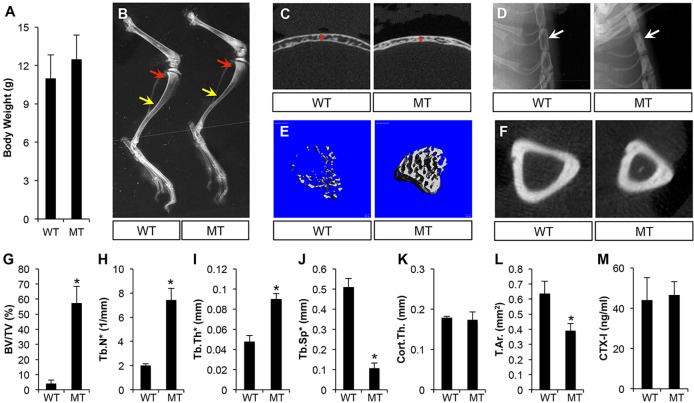
We next deleted Bmpr1a with Dmp1-Cre. The mutant mice (Dmp1-Cre; Bmpr1af/f) appeared to be normal in body size and weight (Fig. 2A). X-ray radiography at P33 revealed that the long bones of the mutant mice were of normal length, but with a greater trabecular bone mass at the metaphysis (Fig. 2B, red arrows). An increase in trabecular bone mass was also observed in the calvarium, the sternum and the vertebra of the mutant mice (Fig. 2C,D; data not shown). By contrast, both the tibia and the calvarium were thinner than normal (Fig. 2B,C). Micro-computed tomography (μCT) analyses confirmed that the mutant mice exhibited a significant increase in trabecular bone volume, trabecular number and thickness, with a corresponding decrease in trabecular spacing (Fig. 2E,G-J). By contrast, Bmpr1a deletion reduced the cortical area without altering cortical thickness (Fig. 2F,K,L). Despite the changes in bone mass, the total bone resorption activity, as indicated by serum CTX-I levels, was normal in the mutant mice (Fig. 2M). Therefore, Bmpr1a deletion with Dmp1-Cre increases trabecular bone mass but reduces periosteal bone growth.
Fig. 2.
Dmp1-Cre; Bmpr1af/f mice display higher trabecular bone mass but smaller bone diameters at P33. (A) Body weight measurements. (B-D) X-ray radiography of the hindlimb (B), calvarium (C) and sternum (D). Arrows in B denote trabecular bone region (red) or diaphysis of tibia (yellow). Red line in C denotes thickness of calvaria. Arrow in D denotes sternum. (E) μCT 3D reconstruction of trabecular bone. (F) μCT images of mid-diaphyseal region of the tibia. (G-L) μCT quantification of trabecular bone (G-J) or cortical bone (K,L). (M) Serum CTX-I measurement. WT, Bmpr1af/f; MT, Dmp1-Cre; Bmpr1af/f. *P<0.05, n=3. BV, bone volume; TV, total volume; Tb.N*, trabeculae number; Tb.Th*, trabeculae thickness; Tb.Sp*, trabeculae spacing; Cort.Th., cortical thickness; T.Ar., total cross-sectional area.
Bmpr1a restrains osteoblast numbers in trabecular bone but promotes osteoblast activity
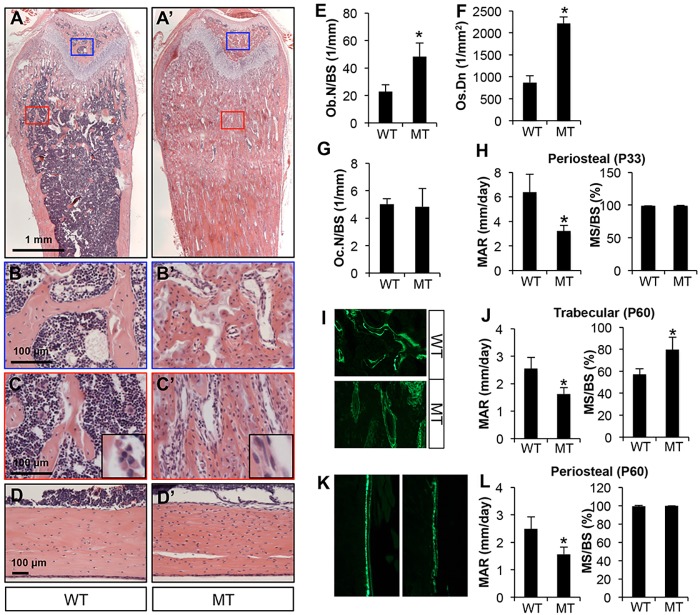
We next assessed the cellular basis for the bone phenotype in the Dmp1-Cre; Bmpr1af/f mice. Histology of the femur confirmed a significant increase in trabecular bone mass in both primary and secondary ossification centers of the mutant mice at P33 (Fig. 3A-C′). Quantification of the trabecular bone region revealed a marked increase in osteoblast number when normalized to bone surface in the mutant (Fig. 3E). Both trabecular and cortical bone also exhibited a greater osteocyte density than in the control (Fig. 3C′,D′,F). However, osteoclast number normalized to bone surface was normal (Fig. 3G). A closer examination of the trabecular bone osteoblasts revealed a flatter morphology than normal, indicating that osteoblast activity might be compromised (Fig. 3C,C′, insets). To assess this possibility, we performed calcein double-labeling experiments. At P33, quantification at the trabecular bone was difficult owing to the scarcity of discrete double-labeled surfaces, but the periosteum at the diaphysis of the mutant bone exhibited a clear reduction in the mineral apposition rate (MAR) with no change in the mineralizing bone surface (MS/BS) (Fig. 3H). At P60, in the trabecular bone MAR was decreased but MS/BS was increased in the mutant, indicating a decrease in osteoblast activity but an increase in osteoblast number (Fig. 3I,J). The periosteal surface at the diaphysis showed a similar decrease in MAR but no change in MS/BS in mutant mice (Fig. 3K,L). Thus, impaired osteoblast activity explains the decrease in periosteal growth, but the marked increase in osteoblast number is likely to drive the massive trabecular bone accrual in the Dmp1-Cre; Bmpr1af/f mice.
Fig. 3.
Bmpr1a deletion increases trabecular osteoblast number but reduces osteoblast activity. Mice were analyzed at P33 (A-H) or P60 (I-L). (A,A′) H&E staining of longitudinal sections of the femur. Blue or red boxes denote areas of secondary or primary ossification centers, respectively, shown at higher magnification beneath. (B,B′) Secondary ossification centers. (C,C′) Primary ossification centers. Insets show representative osteoblast morphology at higher magnification. (D,D′) H&E staining of longitudinal sections through the cortical bone of the femur. (E-G) Histomorphometric quantification of osteoblast number (Ob.N; E), osteocyte density (Os.Dn) in cortical bone (F) and osteoclast number (Oc.N; G), all in the femur. (H) Dynamic histomorphometry in the tibia. (I,K) Representative images of double labeling in the trabecular bone region (I) or periosteum (K) in the tibia. (J,L) Dynamic histomorphometric parameters in trabecular bone (J) or periosteum (L) in the tibia. WT, Bmpr1af/f; MT, Dmp1-Cre; Bmpr1af/f. *P<0.05, n=3. MS, mineralizing surface; BS, bone surface; MAR, mineral apposition rate.
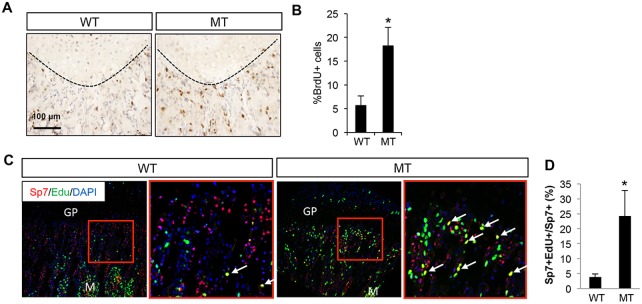
To explore the basis for the increase in trabecular osteoblast number in the Dmp1-Cre; Bmpr1af/f mice, we examined the status of proliferation and apoptosis in osteoblast lineage cells on bone sections. BrdU labeling experiments detected proliferation at the chondro-osseous junction, which was likely to be in osteoblast precursors (Fig. 4A). Importantly, the proliferation rate was significantly increased in the mutant mice, indicating that Bmpr1a may normally suppress the proliferation of osteoblast precursors (Fig. 4B). TUNEL staining detected little signal in either control or mutant mice (data not shown). To determine whether Bmpr1a deletion stimulated the proliferation of Sp7+ preosteoblasts, we performed EdU labeling experiments followed by immunofluorescence staining for Sp7. Whereas few Sp7+ cells at the chondro-osseous junction incorporated EdU in the control mouse, the number significantly increased in the mutant, resulting in a 5-fold increase in the percentage of Sp7+ EdU+ cells among total Sp7+ preosteoblasts (Fig. 4C,D). It should be noted that not all EdU+ cells at the chondro-osseous junction stained positively for Sp7 in either control or mutant mice. Nonetheless, Bmpr1a deletion with Dmp1-Cre results in hyperproliferation among the Sp7+ preosteoblasts, which might account for the increase in osteoblast number.
Fig. 4.
Bmpr1a deletion increases the proliferation of Sp7+ preosteoblasts. (A) Representative images for BrdU staining of longitudinal femur sections at P33. (B) Quantification of BrdU-positive cells (%) at the chondro-osseous junction. (C) Representative images for EdU labeling (green) and Sp7 immunostaining (red). Area in red box is shown at higher magnification to the right. Arrows indicate Sp7+ EdU+ cells (yellow). GP, growth plate; M, marrow. (D) Quantification of Sp7+ EdU+ cells among total Sp7+ cells. WT, Bmpr1af/f; MT, Dmp1-Cre; Bmpr1af/f. *P<0.05, n=3.
Postnatal deletion of Bmpr1a in Sp7+ cells increases trabecular bone mass
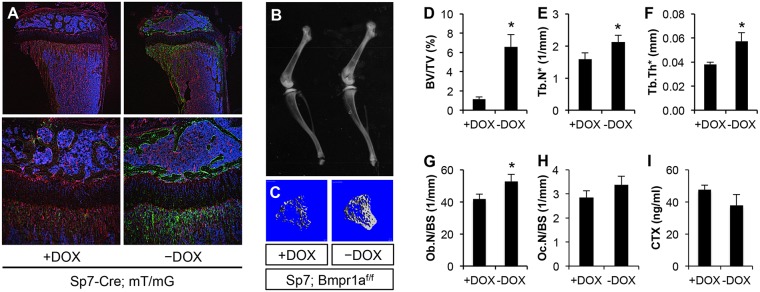
Hyperproliferation of the Sp7+ preosteoblasts in the Dmp1-Cre; Bmpr1af/f mice could be a cell-autonomous effect of Bmpr1a deletion in those cells. We therefore investigated the effect of Bmpr1a removal in Sp7+ cells in postnatal mice. We induced Bmpr1a deletion with Sp7-Cre following withdrawal of doxycycline (Dox). We first confirmed the efficacy of the strategy using the mT/mG reporter mice. After 12 days of Dox withdrawal between P21 and P33, Cre activity, as indicated by GFP expression, was detected in osteoblasts and osteocytes as well as preosteoblasts at the chondro-osseous junction (Fig. 5A, right panels). Mice of the same genotype (Sp7-Cre; mT/mG) but maintained on Dox expressed little GFP in bone (Fig. 5A, left panels). Importantly, deletion of Bmpr1a with the same Dox regimen (12 days of Dox withdrawal) notably increased the trabecular bone mass in Sp7-Cre; Bmpr1af/f mice, as detected by X-ray radiography (Fig. 5B). Analyses by μCT confirmed a marked increase in trabecular bone volume, trabecular number and thickness following Dox withdrawal, whereas the cortical diameter or thickness at the mid-diaphysis was not affected (Fig. 5C-F; data not shown). Histomorphometry revealed that Bmpr1a deletion increased osteoblast numbers without affecting osteoclasts (Fig. 5G,H). Serum CTX-I assays indicated that Dox withdrawal did not alter the total bone resorption activity in the Sp7-Cre; Bmpr1af/f mice (Fig. 5I). Thus, postnatal deletion of Bmpr1a in the Sp7+ cells greatly increases trabecular bone mass.
Fig. 5.
Postnatal deletion of Bmpr1a by Sp7-Cre increases trabecular bone formation. (A) Direct fluorescence images of longitudinal sections of the tibia from Sp7-Cre; mT/mG mice with or without doxycycline (Dox) withdrawal (P21-P33) at low (top row) or high (bottom row) magnification. Red, mTomato; green, mGFP; blue, DAPI staining of nuclei. (B) X-ray radiography of the hindlimb bones from mice of genotype Sp7-Cre; Bmpr1af/f subjected to the same Dox regimen as above. (C) μCT 3D reconstruction of trabecular bone of the tibia. (D-F) μCT quantification of trabecular bone in the tibia. (G,H) Histomorphometric analyses of osteoblast and osteoclast numbers in trabecular bone of the femur. (I) Serum CTX-I measurements. +DOX, Sp7-Cre; Bmpr1af/f mice were maintained on Dox water from conception until sacrifice at P33. −DOX, Sp7-Cre; Bmpr1af/f mice were raised on Dox water from conception until P21, when they were weaned off Dox until sacrifice at P33. *P<0.05, n=3.
Deletion of Smad4 in the osteoblast lineage has a relatively mild effect
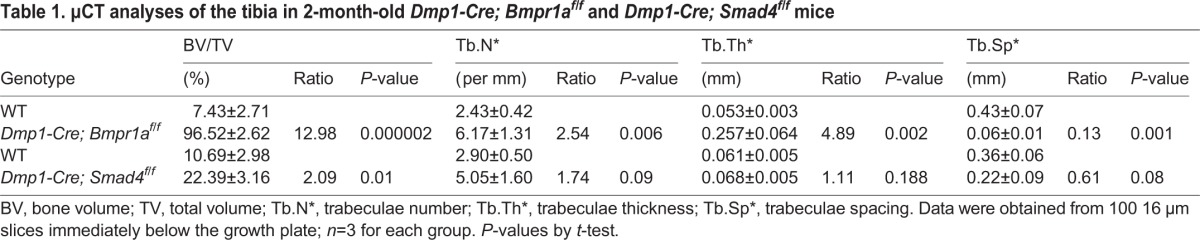
To evaluate the contribution of Smad4 to Bmpr1a signaling in the osteoblast lineage, we deleted Smad4 with Dmp1-Cre. Such deletion (Dmp1-Cre; Smad4f/f) increased trabecular bone mass only by 2-fold at 2 months of age, much less than the 13-fold increase observed in Dmp1-Cre; Bmpr1af/f mice (Table 1). According to X-ray radiography, Smad4 deletion did not notably reduce the diameter at the mid-diaphysis of the tibia. Consistent with the lesser effect on trabecular bone mass, the Smad4 mutant mice, contrary to the Bmpr1a mutant, did not exhibit a significant change in the proliferation rate of osteoblast precursors at the chondro-osseous junction (Fig. S1A). Calcein double-labeling experiments also failed to detect any significant difference in MS/BS or MAR between control and Smad4-deficient littermates (Fig. S1B,C). Thus, Smad4 plays a relatively minor role compared with Bmpr1a in regulating bone accrual.
Table 1.
μCT analyses of the tibia in 2-month-old Dmp1-Cre; Bmpr1af/f and Dmp1-Cre; Smad4f/f mice
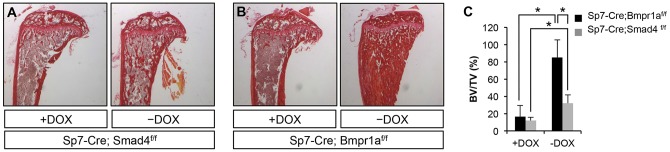
To assess the role of Smad4 in postnatal Sp7+ preosteoblasts, we deleted Smad4 with Sp7-Cre following Dox withdrawal. After Dox withdrawal from 3 through to 8 weeks of age, mice of genotype Sp7-Cre; Smad4f/f exhibited no increase in trabecular bone mass (data not shown). Even after 9 weeks of Dox withdrawal (3 through to 12 weeks of age), the Sp7-Cre; Smad4f/f mice showed only a slight increase in bone mass, much lower than that in the Sp7-Cre, Bmpr1af/f mice following the same Dox regimen (Fig. 6A,B). Quantification by μCT confirmed the significant disparity in trabecular bone volume over total volume (BV/TV) between the two mice after 9 weeks of Dox withdrawal (Fig. 6C). Thus, Bmpr1a is likely to employ additional downstream effectors besides Smad4 to suppress trabecular bone formation.
Fig. 6.
Deletion of Smad4 mildly affects trabecular bone mass. (A,B) Picrosirius Red staining of longitudinal sections of the tibia. (C) μCT quantification of trabecular bone of the tibia. −DOX, mice raised on Dox from conception until P21, and then weaned off Dox for 9 weeks before harvest. +DOX, mice maintained on Dox from conception until harvest at the same age as the −Dox mice. *P<0.05, n=3.
Bmpr1a signaling activates mTORC1
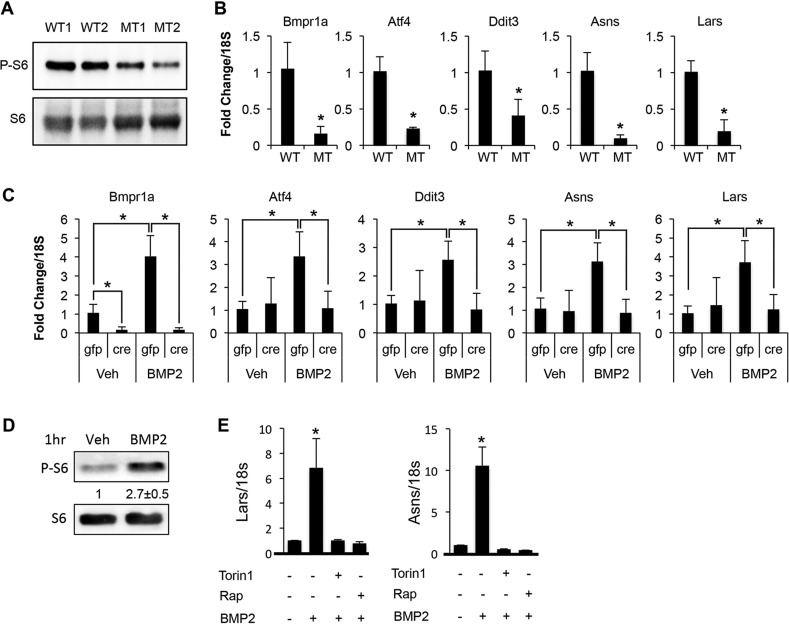
We next investigated the mechanism responsible for the stimulatory effect of Bmpr1a signaling on osteoblast activity. Because recent studies have implicated the mTORC1 complex in promoting bone formation, we hypothesized that Bmp signaling might stimulate osteoblast activity via mTORC1 signaling (Chen et al., 2014; Karner et al., 2015). Western blot analyses showed that the level of phosphorylated ribosomal protein S6 (P-S6), a common readout for mTORC1 activity, was notably reduced in bone extracts of Dmp1-Cre; Bmpr1af/f mice (Fig. 7A). By contrast, we did not detect a consistent change in the phosphorylation of p38 (Mapk1) in the mutant bones (data not shown).
Fig. 7.
Bmp-mTORC1 signaling induces expression of protein anabolism genes. (A) Western blot of protein extracts from long bones at P33. Examples are shown for two mice with each genotype. (B) qRT-PCR with RNA isolated from calvaria at P33. (C) qRT-PCR with RNA isolated from Bmpr1af/f newborn calvarial cells infected with Ad-Cre or Ad-Gfp virus followed by vehicle or BMP2 treatment for 96 h. (D) Western blot in ST2 cells following 1 h treatment with vehicle (Veh) or BMP2. Phosphorylated ribosomal protein S6 (P-S6) is normalized to S6. Quantification (average with s.d.) is shown for three independent experiments. (E) qRT-PCR with RNA from ST2 cells treated with BMP2 or vehicle for 72 h with or without chemical inhibitors. *P<0.05, n=3. WT, Bmpr1af/f; MT, Dmp1-Cre; Bmpr1af/f.
We have recently shown that mTORC1 activation induces the expression of Atf4, Ddit3 and their downstream target genes important for protein synthesis, such as asparagine synthetase (Asns) and leucyl-tRNA synthetase (Lars) (hereafter referred to collectively as protein anabolism genes) (Karner et al., 2015). Consistent with that finding, the mRNA levels of Atf4, Ddit3, Asns and Lars were markedly reduced in the calvarium of Dmp1-Cre; Bmpr1af/f mice (Fig. 7B). To confirm the direct effect of Bmpr1a on the protein anabolism genes, we deleted the receptor with adenovirus expressing Cre in primary calvarial cells isolated from the Bmpr1af/f mice. BMP2 induced an increase in the mRNA level of Bmpr1a in the calvarial cells infected with the control GFP-expressing virus, but the cells infected with the Cre-expressing virus expressed only a negligible level of Bmpr1a in either basal or BMP2 treatment conditions. Importantly, the Bmpr1a-deficient cells failed to activate the expression of Atf4, Ddit3, Asns and Lars in response to BMP2 (Fig. 7C). Overall, Bmpr1a deletion results in the diminution of Bmp-induced mTORC1 signaling, and correlates with suppression of osteoblast activity in vivo.
To test whether Bmp signaling directly activates mTORC1, we treated ST2 cells, a mouse bone marrow stromal cell line, with recombinant BMP2. ST2 cells are known to undergo osteoblastogenesis in response to BMP2 (Yamaguchi et al., 1996). BMP2 increased P-S6 levels by almost 3-fold over the control after 1 h (Fig. 7D). Moreover, chemical inhibition of mTORC1 with either Torin1 or rapamycin markedly suppressed the induction of Lars and Asns by BMP2 (Fig. 7E). Taken together, the data support the notion that Bmp signaling activates the mTORC1 pathway to enhance osteoblast activity.
DISCUSSION
We have investigated the role of physiological Bmp signaling in osteoblast lineage cells. Deletion of Bmpr1a with either Dmp1-Cre or Sp7-Cre, both of which target osteoblast precursors at the chondro-osseous junction of long bones, markedly increased trabecular bone mass, and this was likely to be through hyperproliferation of Sp7+ preosteoblasts. Deletion of Bmpr1a also results in a lower osteoblast activity, correlating with reduced mTORC1 signaling. The data support a model wherein Bmp signaling exerts a dual function in both restricting the proliferation of preosteoblasts and promoting the activity of mature osteoblasts.
The marked increase in trabecular bone mass is similar to that reported in mice in which Bmp1r was deleted with tamoxifen-inducible ColI-CreERTM in embryos, weanlings or adults (Kamiya et al., 2010, 2008a,b). Based on the decrease in osteoclast number in either the embryonic calvaria or the postnatal vertebrae, these previous studies attributed the excessive bone mass to suppression of bone resorption. Here, we did not detect a change in either osteoclast numbers in the trabecular region of long bones or the overall bone resorption activity as indicated by serum CTX-I levels. In addition, qRT-PCR analyses showed that Bmpr1a deletion reduced both Rankl (Tnfsf11) and Opg (Tnfrsf11b) to a similar extent in the calvaria of P33 mice, resulting in a normal Rankl/Opg ratio expected to maintain normal osteoclastogenesis (Fig. S2). The discrepancy regarding osteoclasts between this and the other studies could be due to differences between the Cre drivers employed or in the skeletal sites being analyzed. Our finding is consistent with previous studies showing that inducible deletion of Bmpr1a with Mx1-Cre, or inhibition of Bmp signaling with a soluble Bmpr1a receptor, increases trabecular bone volume mainly through an increase in osteoblast number (Baud'huin et al., 2012; Zhang et al., 2003).
The negative effect of Bmpr1a signaling on trabecular osteoblast number is unexpected, as Bmp has been well documented to promote osteoblast differentiation in vitro. Our results point to hyperproliferation of the Sp7+ preosteoblasts as a major reason for the increase in osteoblast number within the trabecular bone. It remains possible that osteoblast differentiation is partially compromised by Bmpr1a deletion but the deficit is overcome by the increase in preosteoblasts. The mechanism for the increased proliferation is not clear at present. Bmpr1a deletion in the Sp7+ preosteoblasts might cell-autonomously increase their proliferation. Alternatively, the increase in proliferation could be secondary to Bmpr1a deletion in the more mature cell types, such as osteoblasts or osteocytes. In this regard, it is worth noting that the protein level of sclerostin in osteocytes is markedly reduced in the Dmp1-Cre; Bmpr1af/f mouse (data not shown). This observation is consistent with a previous report and could indicate that upregulation of paracrine Wnt signaling stimulates the proliferation of Sp7+ preosteoblasts (Kamiya et al., 2008b). However, because the Bmpr1a mutant possesses more trabecular bone than the sclerostin knockout mouse, sclerostin suppression might not be solely responsible for the trabecular bone phenotype caused by Bmpr1a deletion (Li et al., 2008). Whatever the precise mechanism, it is likely to involve effectors beyond Smad4, as deletion of Smad4 does not produce a similar effect. Overall, further studies are necessary to elucidate the molecular mechanism responsible for the negative regulation of preosteoblast proliferation by Bmpr1a.
The positive regulation of osteoblast activity by Bmpr1a is consistent with several previous studies. Transgenic overexpression of noggin in osteoblasts reduced periosteal bone formation (Okamoto et al., 2006). Deletion of Bmpr1a in osteoblasts with Og2-Cre also suppressed osteoblast activity (Mishina et al., 2004). Moreover, as Bmp2 deletion in the limb mesenchyme resulted in thinner long bones, similar to those in the Bmpr1a mutants described here, Bmp2 might signal through Bmpr1a to control periosteal growth (Tsuji et al., 2006). The reduced periosteal growth appears to be due to diminution of osteoblast activity in the absence of Bmp signaling. Because the defect is not recapitulated by Smad4 deletion, additional effectors are likely to contribute to the anabolic function of Bmpr1a in osteoblasts. Our biochemical studies have identified mTORC1 as a likely candidate, which probably functions downstream of PI3K-Akt signaling previously shown to be activated by Bmp (Ghosh-Choudhury et al., 2002, 2013). In light of recent findings concerning the role of mTORC1 in bone formation and its regulation by Wnt signaling, mTORC1 might be a common mediator for multiple bone anabolic signals (Chen and Long, 2015; Chen et al., 2014).
Finally, the decrease in osteoblast activity in the Bmpr1a-deficient mice is at odds with the marked reduction of sclerostin in bone. Sclerostin knockout mice develop thicker bones due to increased osteoblast activity, and the effect is generally attributed to upregulation of Wnt signaling (Li et al., 2008). If so, our results would indicate that stimulation of periosteal bone growth by Wnt requires intact Bmpr1a in the osteoblast lineage. Alternatively, sclerostin might impede osteoblast activity through direct suppression of Bmp signaling. This notion is consistent with a recent finding that sclerostin inhibits Bmp7 secretion (Krause et al., 2010). Further studies are necessary to elucidate the potential interaction between sclerostin and Bmp signaling in regulating bone formation.
MATERIALS AND METHODS
Mouse strains
Sp7-Cre (Rodda and McMahon, 2006), Dmp1-Cre (Lu et al., 2007), Bmpr1af/f (Mishina et al., 2002), Smad4f/f (Yang et al., 2002) and mT/mG (Muzumdar et al., 2007) mouse strains were as previously described. All analyses were performed on paired littermates. The Animal Studies Committee at Washington University approved all mouse procedures in this study.
Cell culture
Primary osteoblasts were isolated from calvaria of newborn mouse pups on the day of birth. Briefly, dissected calvaria were transferred to ice-cold PBS and washed thoroughly. Tissue was digested with 2 mg/ml collagenase solution (Sigma, C0130) for 15 min at 37°C with shaking at 75 rpm to remove excess connective tissue. Cells were then collected from three consecutive digestions in 4 mg/ml collagenase solution under the same conditions. Cells were pooled and plated in αMEM with 10% FBS (Gibco). For adenovirus infections, cells were incubated with Ad-Cre or Ad-Gfp virus (MOI of 100) for 6 h. At 24 h after infection, cells were split into 24-well plates and grown to confluence before being treated with vehicle (4 μM HCl) or 300 ng/ml recombinant human BMP2 (R&D Systems) for 96 h and harvested for qRT-PCR experiments.
ST2 cells were cultured in αMEM with 10% FBS. For western blot, the cells were serum-starved for 6 h before treatment with vehicle or 300 ng/ml BMP2 for 1 h. Antibodies for P-S6 (2211S), S6 (Rps6) (2217S) and β-actin (8457S) were from Cell Signaling Technology and used at 1:1000 dilution. For qRT-PCR, the cells were treated with BMP2 for 72 h with or without 100 nM Torin1 (Tocris) or 20 nM rapamycin (Sigma).
Morphological analyses of bones
X-ray radiography was performed with a Faxitron X-ray system, with 20 s exposure at 25 kV. Micro-computed tomography (μCT 40, Scanco Medical) was performed on the tibia and the femur. μCT quantification of the trabecular bone was assessed by measuring 100 μCT slices (1.6 mm) immediately below the growth plate. For cortical bone parameters, 50 μCT slices (0.8 mm) starting from 6.4 mm below the growth plate were analyzed.
Hematoxylin and Eosin (H&E), tartrate-resistant acid phosphatase (TRAP), and Picrosirius Red staining were performed on paraffin sections, following decalcification with 14% (w/v) EDTA (pH 7.4) for 2 weeks at room temperature with daily changes of solution. For dynamic histomorphometry of postnatal mice, calcein (Sigma) was injected at 7.5 mg/kg body weight intraperitoneally at 6 and 2 days prior to sacrifice. Bones were embedded in methyl-methacrylate and sectioned. Histomorphometric parameters were acquired with Bioquant Osteo II.
In vivo assays
For serum CTX-I assays, serum was collected from mice starved for 6 h, and analyzed with the RatLaps ELISA kit (Immunodiagnostic Systems) according to the manufacturer's instructions.
For BrdU labeling assays, mice were injected intraperitoneally with BrdU (10 mg/ml; Sigma, B5002)/FdU (1.2 mg/ml; Sigma, F0503) solution at 0.1 ml/10 g body weight at 4 h prior to sacrifice. Sections were deparaffinized and stained using a BrdU staining kit (Invitrogen, 93-3943) according to the manufacturer's instructions. BrdU labeling indices were calculated as the percentage of positive cells among total cells within similar areas under the growth plate across the different mice.
EdU was injected intraperitoneally at 10 μg/g body weight at 4 h before harvest. Frozen sections were subjected to immunostaining for Sp7 (Abcam, ab22552), followed by a Click reaction performed according to the manufacturer's instructions (Click-iT EdU Alexa Fluor 488 imaging kit, Invitrogen). Images were acquired with the Nikon C-1 confocal system.
Biochemical analyses of bones
For western blots of bone extracts, femurs and tibiae from P33 mice were cleanly dissected with the epiphysis removed. After removing the marrow by centrifugation, bones were cut into small pieces and rinsed three times with ice-cold PBS. Bone pieces were snap-frozen in liquid nitrogen, pulverized at 2000 rpm for 20 s using a Mikro-Dismembrator (Sartorius) and then lysed with RIPA buffer containing protease and phosphatase inhibitors (Roche).
For qRT-PCR, calvaria from P33 mice were dissected free of connective tissue in ice-cold PBS, and then snap-frozen in liquid nitrogen and pulverized at 2000 rpm for 20 s using a Mikro-Dismembrator. 1 ml Trizol (Invitrogen) was added directly to the pulverized bone for RNA extraction. RNA was treated with DNaseI and purified with the RNeasy RNA extraction kit (Qiagen). 1 µg RNA was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad). qRT-PCR reactions were performed in biological triplicate and technical duplicate with SYBR Green Supermix (SsoAdvanced, Bio-Rad) and analyzed on an ABI StepOne Plus machine. Gene expression was normalized to 18S rRNA and relative expression was determined using the 2−(ΔΔCt) method. Primer sequences are listed in Table S1.
Statistics
Three mice per group were analyzed unless indicated otherwise. Quantitative data are presented as mean±s.d. Statistical significance was determined by Student's t-test. P≤0.05 was considered significant.
Acknowledgements
We thank Dr Jerry Feng (Baylor College of Dentistry) for the Dmp1-Cre mice and Dr Yuji Mishina (University of Michigan School of Dentistry) for the Bmpr1af/f mice. We thank Masato Hoshi of the Sanjay Jain laboratory for technical assistance with the confocal microscope.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
J.L., Y.S., C.M.K., S.-Y.L., W.-C.L. and G.H. performed experiments; J.L. and F.L. analyzed data and wrote the paper; F.L. conceived the project.
Funding
This work was supported by National Institutes of Health (NIH) grants [AR060456 and AR055923] to F.L. The confocal microscopy experiments were supported in part by the NIH-funded George O'Brien Center for Kidney Disease Research [P30 DK079333], Kidney Translational Research Core and the Renal Division at the Washington University School of Medicine. The bone morphometric studies were partly supported by a grant [P30 AR057235] to the Washington University Musculoskeletal Research Center. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.126227/-/DC1
References
- Bandyopadhyay A., Tsuji K., Cox K., Harfe B. D., Rosen V. and Tabin C. J. (2006). Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2, e216 10.1371/journal.pgen.0020216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud'huin M., Solban N., Cornwall-Brady M., Sako D., Kawamoto Y., Liharska K., Lath D., Bouxsein M. L., Underwood K. W., Ucran J. et al. (2012). A soluble bone morphogenetic protein type IA receptor increases bone mass and bone strength. Proc. Natl. Acad. Sci. USA 109, 12207-12212. 10.1073/pnas.1204929109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benazet J.-D., Pignatti E., Nugent A., Unal E., Laurent F. and Zeller R. (2012). Smad4 is required to induce digit ray primordia and to initiate the aggregation and differentiation of chondrogenic progenitors in mouse limb buds. Development 139, 4250-4260. 10.1242/dev.084822 [DOI] [PubMed] [Google Scholar]
- Chen J. and Long F. (2015). mTORC1 signaling promotes osteoblast differentiation from preosteoblasts. PLoS ONE 10, e0130627 10.1371/journal.pone.0130627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Tu X., Esen E., Joeng K. S., Lin C., Arbeit J. M., Rüegg M. A., Hall M. N., Ma L. and Long F. (2014). WNT7B promotes bone formation in part through mTORC1. PLoS Genet. 10, e1004145 10.1371/journal.pgen.1004145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Choudhury N., Abboud S. L., Nishimura R., Celeste A., Mahimainathan L. and Choudhury G. G. (2002). Requirement of BMP-2-induced phosphatidylinositol 3-kinase and Akt serine/threonine kinase in osteoblast differentiation and Smad-dependent BMP-2 gene transcription. J. Biol. Chem. 277, 33361-33368. 10.1074/jbc.M205053200 [DOI] [PubMed] [Google Scholar]
- Ghosh-Choudhury N., Mandal C. C., Das F., Ganapathy S., Ahuja S. and Ghosh Choudhury G. (2013). c-Abl-dependent molecular circuitry involving Smad5 and phosphatidylinositol 3-kinase regulates bone morphogenetic protein-2-induced osteogenesis. J. Biol. Chem. 288, 24503-24517. 10.1074/jbc.M113.455733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N., Ye L., Kobayashi T., Lucas D. J., Mochida Y., Yamauchi M., Kronenberg H. M., Feng J. Q. and Mishina Y. (2008a). Disruption of BMP signaling in osteoblasts through type IA receptor (BMPRIA) increases bone mass. J. Bone Miner. Res. 23, 2007-2017. 10.1359/jbmr.080809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N., Ye L., Kobayashi T., Mochida Y., Yamauchi M., Kronenberg H. M., Feng J. Q. and Mishina Y. (2008b). BMP signaling negatively regulates bone mass through sclerostin by inhibiting the canonical Wnt pathway. Development 135, 3801-3811. 10.1242/dev.025825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N., Kobayashi T., Mochida Y., Yu P. B., Yamauchi M., Kronenberg H. M. and Mishina Y. (2010). Wnt inhibitors Dkk1 and Sost are downstream targets of BMP Signaling Through the Type IA Receptor (BMPRIA) in osteoblasts. J. Bone Miner. Res. 25, 200-210. 10.1359/jbmr.090806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N., Kaartinen V. M. and Mishina Y. (2011). Loss-of-function of ACVR1 in osteoblasts increases bone mass and activates canonical Wnt signaling through suppression of Wnt inhibitors SOST and DKK1. Biochem. Biophys. Res. Commun. 414, 326-330. 10.1016/j.bbrc.2011.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner C. M., Esen E., Okunade A. L., Patterson B. W. and Long F. (2015). Increased glutamine catabolism mediates bone anabolism in response to WNT signaling. J. Clin. Invest. 125, 551-562. 10.1172/JCI78470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause C., Korchynskyi O., de Rooij K., Weidauer S. E., de Gorter D. J. J., van Bezooijen R. L., Hatsell S., Economides A. N., Mueller T. D., Lowik C. W. G. M. et al. (2010). Distinct modes of inhibition by sclerostin on bone morphogenetic protein and Wnt signaling pathways. J. Biol. Chem. 285, 41614-41626. 10.1074/jbc.M110.153890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ominsky M. S., Niu Q.-T., Sun N., Daugherty B., D'Agostin D., Kurahara C., Gao Y., Cao J., Gong J. et al. (2008). Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J. Bone Miner. Res. 23, 860-869. 10.1359/jbmr.080216 [DOI] [PubMed] [Google Scholar]
- Lim J., Tu X., Choi K., Akiyama H., Mishina Y. and Long F. (2015). BMP-Smad4 signaling is required for precartilaginous mesenchymal condensation independent of Sox9 in the mouse. Dev. Biol. 400, 132-138. 10.1016/j.ydbio.2015.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Xie Y., Zhang S., Dusevich V., Bonewald L. F. and Feng J. Q. (2007). DMP1-targeted Cre expression in odontoblasts and osteocytes. J. Dent. Res. 86, 320-325. 10.1177/154405910708600404 [DOI] [PubMed] [Google Scholar]
- Massague J. (2012). TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616-630. 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y., Hanks M. C., Miura S., Tallquist M. D. and Behringer R. R. (2002). Generation of Bmpr/Alk3 conditional knockout mice. Genesis 32, 69-72. 10.1002/gene.10038 [DOI] [PubMed] [Google Scholar]
- Mishina Y., Starbuck M. W., Gentile M. A., Fukuda T., Kasparcova V., Seedor J. G., Hanks M. C., Amling M., Pinero G. J., Harada S.-I. et al. (2004). Bone morphogenetic protein type IA receptor signaling regulates postnatal osteoblast function and bone remodeling. J. Biol. Chem. 279, 27560-27566. 10.1074/jbc.M404222200 [DOI] [PubMed] [Google Scholar]
- Miyazono K., Kamiya Y. and Morikawa M. (2010). Bone morphogenetic protein receptors and signal transduction. J. Biochem. 147, 35-51. 10.1093/jb/mvp148 [DOI] [PubMed] [Google Scholar]
- Moustakas A. and Heldin C.-H. (2009). The regulation of TGF beta signal transduction. Development 136, 3699-3714. 10.1242/dev.030338 [DOI] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L. and Luo L. (2007). A global double-fluorescent cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Okamoto M., Murai J., Yoshikawa H. and Tsumaki N. (2006). Bone morphogenetic proteins in bone stimulate osteoclasts and osteoblasts during bone development. J. Bone Miner. Res. 21, 1022-1033. 10.1359/jbmr.060411 [DOI] [PubMed] [Google Scholar]
- Regan J. N., Lim J., Shi Y., Joeng K. S., Arbeit J. M., Shohet R. V. and Long F. (2014). Up-regulation of glycolytic metabolism is required for HIF1alpha-driven bone formation. Proc. Natl. Acad. Sci. USA 111, 8673-8678. 10.1073/pnas.1324290111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retting K. N., Song B., Yoon B. S. and Lyons K. M. (2009). BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development 136, 1093-1104. 10.1242/dev.029926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda S. J. and McMahon A. P. (2006). Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133, 3231-3244. 10.1242/dev.02480 [DOI] [PubMed] [Google Scholar]
- Tan X., Weng T., Zhang J., Wang J., Li W., Wan H., Lan Y., Cheng X., Hou N., Liu H. et al. (2007). Smad4 is required for maintaining normal murine postnatal bone homeostasis. J. Cell Sci. 120, 2162-2170. 10.1242/jcs.03466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K., Bandyopadhyay A., Harfe B. D., Cox K., Kakar S., Gerstenfeld L., Einhorn T., Tabin C. J. and Rosen V. (2006). BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat. Genet. 38, 1424-1429. 10.1038/ng1916 [DOI] [PubMed] [Google Scholar]
- Urist M. R., Mikulski A. and Lietze A. (1979). Solubilized and insolubilized bone morphogenetic protein. Proc. Natl. Acad. Sci. USA 76, 1828-1832. 10.1073/pnas.76.4.1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K. and Derynck R. (2009). TGFbeta family signaling: novel insights in development and disease. Development 136, 3691-3697. 10.1242/dev.040584 [DOI] [PubMed] [Google Scholar]
- Wrana J. L., Attisano L., Wieser R., Ventura F. and Massagué J. (1994). Mechanism of activation of the TGF-beta receptor. Nature 370, 341-347. 10.1038/370341a0 [DOI] [PubMed] [Google Scholar]
- Wu M. Y. and Hill C. S. (2009). Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev. Cell 16, 329-343. 10.1016/j.devcel.2009.02.012 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Ishizuya T., Kintou N., Wada Y., Katagiri T., Wozney J. M., Rosen V. and Yoshiki S. (1996). Effects of BMP-2, BMP-4, and BMP-6 on osteoblastic differentiation of bone marrow-derived stromal cell lines, ST2 and MC3T3-G2/PA6. Biochem. Biophys. Res. Commun. 220, 366-371. 10.1006/bbrc.1996.0411 [DOI] [PubMed] [Google Scholar]
- Yang X., Li C., Herrera P.-L. and Deng C.-X. (2002). Generation of Smad4/Dpc4 conditional knockout mice. Genesis 32, 80-81. 10.1002/gene.10029 [DOI] [PubMed] [Google Scholar]
- Yoon B. S., Ovchinnikov D. A., Yoshii I., Mishina Y., Behringer R. R. and Lyons K. M. (2005). Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc. Natl. Acad. Sci. USA 102, 5062-5067. 10.1073/pnas.0500031102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Niu C., Ye L., Huang H., He X., Tong W.-G., Ross J., Haug J., Johnson T., Feng J. Q. et al. (2003). Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425, 836-841. 10.1038/nature02041 [DOI] [PubMed] [Google Scholar]
- Zhang J., Tan X., Li W., Wang Y., Wang J., Cheng X. and Yang X. (2005). Smad4 is required for the normal organization of the cartilage growth plate. Dev. Biol. 284, 311-322. 10.1016/j.ydbio.2005.05.036 [DOI] [PubMed] [Google Scholar]