Abstract
In contrast to mammals, adult zebrafish have a high capacity to regenerate damaged or lost myocardium through proliferation of cardiomyocytes spared from damage. The epicardial sheet covering the heart is activated by injury and aids muscle regeneration through paracrine effects and as a multipotent cell source, and has received recent attention as a target in cardiac repair strategies. Although it is recognized that epicardium is required for muscle regeneration and itself has high regenerative potential, the extent of cellular heterogeneity within epicardial tissue is largely unexplored. Here, we performed transcriptome analysis on dozens of epicardial lineage cells purified from zebrafish harboring a transgenic reporter for the pan-epicardial gene tcf21. Hierarchical clustering analysis suggested the presence of at least three epicardial cell subsets defined by expression signatures. We validated many new pan-epicardial and epicardial markers by alternative expression assays. Additionally, we explored the function of the scaffolding protein and main component of caveolae, caveolin 1 (cav1), which was present in each epicardial subset. In BAC transgenic zebrafish, cav1 regulatory sequences drove strong expression in ostensibly all epicardial cells and in coronary vascular endothelial cells. Moreover, cav1 mutant zebrafish generated by genome editing showed grossly normal heart development and adult cardiac anatomy, but displayed profound defects in injury-induced cardiomyocyte proliferation and heart regeneration. Our study defines a new platform for the discovery of epicardial lineage markers, genetic tools, and mechanisms of heart regeneration.
KEY WORDS: Heart regeneration, Epicardium, Single-cell sequencing, Caveolin-1, Zebrafish
Highlighted article: Gene expression analyses reveal that zebrafish epicardial cells are heterogeneous and identify many new epicardial markers, including Caveolin 1, which is shown to be essential for heart regeneration.
INTRODUCTION
Adult zebrafish regenerate lost cardiomyocytes (CMs) with high efficiency (Poss et al., 2002). Pre-existing CMs are the primary cellular source of new muscle (Jopling et al., 2010; Kikuchi et al., 2010), while the epicardium, a thin mesothelial cell layer covering the chambers, is activated to express embryonic markers, proliferate, and colonize the injury site (Lepilina et al., 2006). Studies over the past decade have identified the epicardium as a crucial player in heart regeneration and cardiac repair. During heart regeneration, epicardial cells provide paracrine signals, including retinoic acid (RA), Neuregulin 1 (Nrg1) and extracellular matrix (ECM) components such as fibronectin, while also functioning in vasculogenesis (Lepilina et al., 2006; Kikuchi et al., 2011a,b; Wang et al., 2013; Gemberling et al., 2015). During mammalian cardiac repair, epicardial cells also support cardiac cell survival and vascularization and are a major supply of cardiac fibroblasts, which have the capacity to be directly reprogrammed into CM-like cells (Smart et al., 2007, 2011; Ieda et al., 2010; Zhou et al., 2011; Huang et al., 2012; Qian et al., 2012; Song et al., 2012).
A recent report demonstrated that the epicardial cell population of adult zebrafish vigorously regenerates after targeted genetic ablation, explaining in part its dynamism upon myocardial injury (Wang et al., 2015). One of several key remaining questions in adult epicardial biology is to what extent the epicardium is heterogeneous at the cellular and molecular levels, and whether potential subpopulations of epicardial cells play distinct roles in regeneration. The definition of specific epicardial subsets would lead to the definition of new molecular markers, the development of genetic tools involving subset-specific regulatory sequences, and the identification of candidate genes that underlie epicardial cell responses to injuries. In zebrafish, the transcription factor tcf21 is specifically expressed in epicardial cells and epicardial-derived cells (EPDCs) (as well as in other mesothelial tissues) throughout development and regeneration, whereas two other epicardial markers widely used in murine research, tbx18 and wt1b, lack such epicardial specificity (Kikuchi et al., 2011a). In adult ventricles, tcf21 expression domains include the outermost epicardial cell layer, as well as inner EPDCs, many of which associate with coronary vessels (Kikuchi et al., 2011a). There is a clear need for a more sophisticated understanding of the diversity and molecular signature of the epicardium beyond this information.
Many recent studies have exploited technologies to acquire transcriptome data from single purified cells (Navin et al., 2011; Shalek et al., 2013; Yan et al., 2013; Knouse et al., 2014). In this study, we generated single-cell transcriptomes from the tcf21+ population isolated from adult zebrafish ventricles, and from these data we obtained evidence of at least three cell subsets defined by expression signatures. Several new markers that were present in all epicardial cells, or specific to apparent subsets, were visualized by in situ hybridization. We pursued one gene with strong pan-epicardial expression, caveolin 1 (cav1), by creating a transgenic reporter line and targeted genetic mutants. Our findings reveal a requirement for cav1 in heart regeneration and indicate a strategy for functional exploration of the adult epicardium.
RESULTS AND DISCUSSION
Single-cell transcriptome sequencing of adult epicardial cells reveals subpopulations of tcf21+ cells
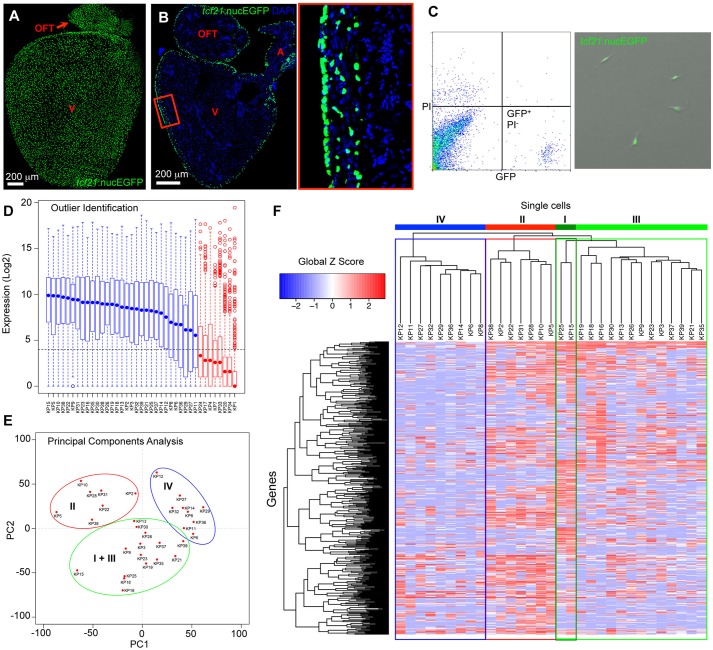
We dissociated epicardial cells from uninjured adult hearts of tcf21:nucEGFP fish and isolated EGFP-positive live cells by FACS (Fig. 1A-C). Thirty-nine single cells were captured via the Fluidigm C1 chip. Transcriptome analysis was performed with 3.6-8.8 million 50 nt read pairs obtained for each cell library. A pooled sample of ∼5000 cells was also analyzed, with sequencing yielding 10 million read pairs. To identify outliers, the Fluidigm SINGuLAR package first identified a set of core genes detected in at least half of the samples. Eight samples had median expression values below the fifteenth percentile for these core genes, suggesting technical failure. These samples were removed prior to further analysis (Fig. 1D) (Pollen et al., 2014). We detected 12,903 genes (expected count ≥1) in pooled samples, with 2826-6001 of these genes (average 3879) in each of the remaining single cells. Additionally, 15,147 unique genes (expected count ≥1 in at least one cell) were detected by single-cell sequencing. Comparison of pooled and single-cell data demonstrated a strong correlation in detected genes. Principal components analysis (PCA) and hierarchical clustering were performed using the Fluidigm SINGuLAR package for the remaining 31 single-cell sequencing libraries (Pollen et al., 2014). Both PCA and clustering indicated three main subpopulations (Fig. 1E,F). One subpopulation (Fig. 1E,F, green circle or frame) was further divided into two groups with apparent differential gene expression (Fig. 1F, dark green and light green frames). These results suggest heterogeneity within tcf21+ cells in the uninjured adult zebrafish ventricle, and further suggest at least three subtypes of tcf21+ cells.
Fig. 1.
Single-cell transcriptome sequencing of adult zebrafish epicardial cells. (A,B) Whole-mount (A) and section (B) images of an adult tcf21:nucEGFP heart. The boxed region is enlarged to the right. OFT, outflow tract; A, atrium; V, ventricle. Blue, DAPI. (C) FACS isolation of GFP-positive, propidium iodide (PI)-negative, epicardial cells from uninjured hearts of tcf21:nucEGFP adult fish. Drops of isolated cells were plated into a culture dish with DMEM plus 10% FBS for 24 h, to examine survival, morphology and GFP signals. (D) Identification of outliers using the Fluidigm SINGuLAR analysis package. Box plot of the expression levels of a set of genes detected in at least half of the samples. Eight samples (red) with median expression (blue or red dot in the box plot) values below the fifteenth percentile (dashed horizontal line) for these genes were removed before further analysis. (E) Principal components analysis (PCA) performed on the 31 cells using the Fluidigm SINGuLAR analysis package. Three main clusters were identified as marked by the ellipses. (F) Hierarchical clustering of the top 500 PCA genes across 31 cells performed in the Fluidigm SINGuLAR package. The three main clusters are labeled in the same color as in E. The green cluster was further divided into two groups with apparent differential gene expression (dark green and light green). Labels I, II, III and IV denote groups.
New markers for epicardial cells and EPDCs
To identify markers representing these potential subpopulations, we filtered our dataset for transcription factors (or nuclear proteins), secreted factors (or membrane proteins) and other genes differentially expressed across subsets, and also included published epicardial-related genes. Upon gross inspection, genes expressed in group IV (Fig. 1F, blue frame) were also strongly expressed in group II (Fig. 1F, Table S1), making it difficult to differentiate groups II and IV without combinatorial expression patterns. From these filtered genes and known expression domains, we designated the remaining three tcf21+ subpopulations as representing the outermost (epithelial) epicardial cells (I, dark green frame), internal cells (middle of the ventricular wall) likely to contain perivascular components (II, red frame), and an innermost layer (inner part of the wall) of EPDCs (III, light green frame) (Fig. 1F, Fig. 2A). Based on these designations, we could further divide the expressed genes into pan-epicardial and subset-specific markers (Table S1, Fig. 2).
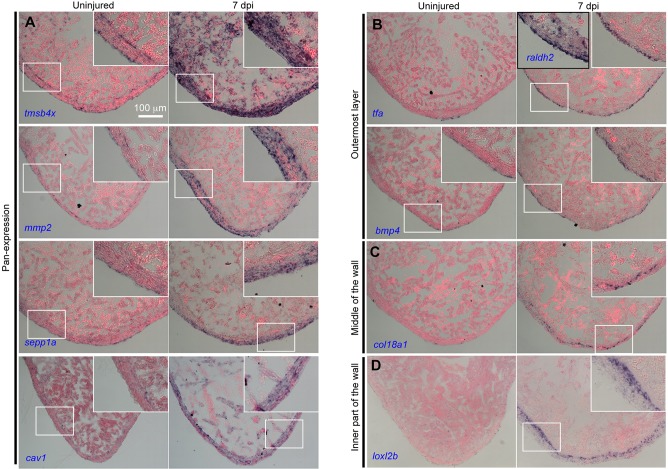
Fig. 2.
New markers of epicardial cells. In situ hybridization for selected genes on sections from regenerating ventricles at 7 days post injury (dpi) in cmlc2:CreER; bactin2:loxP-mCherry-STOP-loxP-DTA animals treated with tamoxifen, as compared with ventricles from bactin2:loxP-mCherry-STOP-loxP-DTA animals treated with tamoxifen (uninjured). raldh2 served as a 7 dpi marker for endocardium and the outermost layer of epicardium. The selected genes show: (A) pan-epicardial expression; (B) exclusive expression in the outermost layer of the ventricle wall; (C) expression within the wall; and (D) expression in the inner part of the wall but absent from the outermost layer. Insets are high-magnification views of the boxed areas. All of the genes were tested on sections from at least four uninjured hearts and five injured hearts. Each heart was represented by at least six sections. When similar patterns were detected in most of the six sections of each heart, and in all of the examined hearts, a conclusion of positive expression was made.
Among these selected genes, several have been described previously as epicardial markers. tcf21 was strongly expressed in all cells that we assessed, whereas tbx18 and wt1b were detected in a portion of these cells (Table S1) (Kikuchi et al., 2011a). The two fibronectin paralogs fn1a and fn1b, which are induced by injury in epicardial cells, were both detected in our profile. fn1b is highly expressed in all cells, whereas fn1a was detected at lower levels in a portion of cells; this is consistent with the finding that fn1a is injury induced (Table S1) (Wang et al., 2013). The epicardial (and endocardial) marker raldh2 (aldh1a2 – Zebrafish Information Network) is detected only in the outermost epithelial layer (Kikuchi et al., 2011a), and high-level detection of raldh2 was a signature of group I (Fig. 2B, Table S1). nrg1, which is reported to be expressed in tcf21+ perivascular cells, was detected in one cell of group II (Table S1) (Gemberling et al., 2015). cxcl12 encodes a CXC-motif chemokine ligand that is expressed in both epicardium and epicardial-derived smooth muscle surrounding coronary blood vessels during mouse heart development (Cavallero et al., 2015). A recent report in zebrafish described upregulation of cxcl12 in epicardial-derived perivascular cells, where it aids formation of the coronary vasculature (Harrison et al., 2015). Group III has high cxcl12a expression, whereas groups I and II have no detectable and low expression, respectively (Table S1). Further visual confirmation is required to definitively assign groups II and III. Other genes reported to have epicardial expression are igf2b, with expression in epicardial and endocardial cells in the injury site by 7 days post amputation (dpa) (Choi et al., 2013), and zTB2, with expression around the wound and surrounding compact myocardium at 3 and 7 dpa (Lien et al., 2006). These genes are predicted to be pan-epicardial expression markers by our transcriptome data.
To examine our dataset using a different technique, we made probes representing 40 genes and performed in situ hybridization to detect cardiac expression, both in uninjured ventricles and in ventricles 7 days after induced genetic ablation of ∼50% of CMs (Wang et al., 2011). We detected expression signals representing 24 genes in or around the epicardium, often representing different apparent domains. Interestingly, many genes were only detected in injured ventricles, suggesting possible roles in heart regeneration (Fig. 2, Fig. S1). Fifteen of these 24 expression domains were indicative of pan-epicardial expression. Among these genes, mmp2 exhibited a multilayer expression pattern in the walls of uninjured and injured ventricles, resembling the tcf21 reporter expression pattern (Fig. 2A). tmsb4x (zTB4), assessed for roles in epicardial cell activation in mammals (Smart et al., 2007, 2011), also had a clear two-layer expression domain in the uninjured ventricle wall, and it was strongly induced after injury in multiple cardiac cell types (Fig. 2A). zTB2 displayed weak signals in the ventricular wall after injury (Fig. S1). Other apparent pan-epicardial expression genes revealed by our dataset included cav1, p4hb, sepp1a, vmo1, mmp14a, timp2a, cyr61, hspa5, pcolce2b, lxn, mdkb, twist1 and id3. The genes cav1, p4hb, vmo1, cyr61, mdkb and hspa5 were expressed in additional cardiac tissues, such as endocardium (Fig. S1).
Some other genes, including bmp4 and tfa, were detected only in the outermost layer, both before (very weak) and after injury, resembling the epicardial expression of raldh2 (Fig. 2B). A third gene, clu, also showed weak expression in the outer epithelial layer before injury, with enhanced expression in this layer upon injury. col18a1 and ltbp1, which labeled a cluster of cells within the ventricular wall, are likely to mark the group II cells as identified by transcriptome data. Three other genes, loxl2a, loxl2b and timp2b, were induced within the ventricular wall after injury, with no obvious expression on the outer edge, are in domains consistent with group II and III markers. Importantly, several of these markers are induced by injury, and several appear to be expressed in cell types in addition to epicardium. This latter point is not surprising, as single-cell profiling approaches do not select for restricted expression. In several cases, in situ hybridization signals were weak and could not definitively specify the expressing epicardial cell group.
During heart regeneration, adult epicardial cells can reactivate expression of genes induced in epicardial cells of developing embryos (Lepilina et al., 2006). Also, a recent publication indicated that adult murine epicardial cells restore elements of embryonic signatures (Bollini et al., 2014). To test if the markers we describe are expressed in zebrafish embryos, we performed whole-mount in situ hybridization on embryos at 3 and 4 days post fertilization (dpf). We observed signals indicative of cardiac expression for several genes, including tmsb4x, col18a1 and mdkb (Fig. S2).
In total, we have identified several new epicardial markers that appear to have different expression domains. The groupings indicated by cluster analysis are largely supported by in situ hybridization. These new markers will need further assessment to probe the identities of subsets, especially groups II and III. We speculate that these apparent subsets have distinct functions in cardiac homeostasis and/or regeneration, an idea that can be explored in future functional studies.
A BAC reporter strain indicates strong pan-epicardial expression of cav1
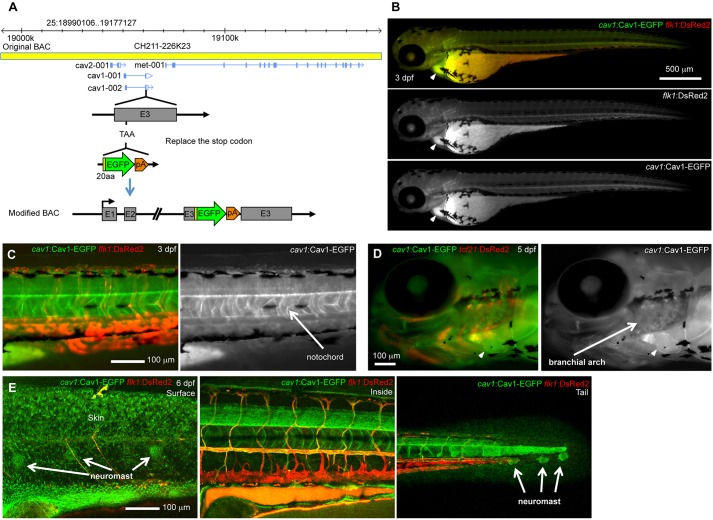
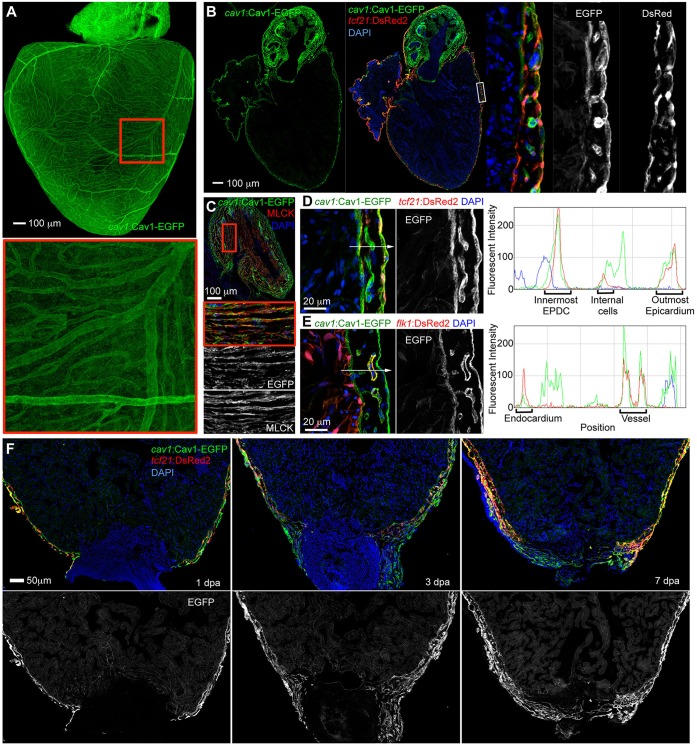
Among the listed genes, cav1 was identified as highly expressed in each epicardial group (Fig. 2A, Table S1). cav1 encodes a scaffolding protein and main component of caveolae, which are specialized membrane domains that function in vascular transport, signal transduction and membrane trafficking events (Parton and del Pozo, 2013; Shvets et al., 2014). To test cav1 expression in heart, we generated a BAC transgenic construct with EGFP fused in frame to the C-terminus of Cav1, driven by the endogenous cav1 regulatory sequences (Fig. 3A). We established a stable transgenic line, TgBAC(cav1:cav1-EGFP)pd1096 (referred to hereafter as cav1:cav1-EGFP) and examined its expression in embryos and adult heart. After crossing with tcf21:DsRed2 or flk1:DsRed2 reporter strains to label epicardial or vascular tissue, we identified conspicuous Cav1-EGFP signals in larval heart, vasculature, notochord, branchial arches, skin and neuromasts (Fig. 3B-E), largely recapitulating what has been described for endogenous cav1 expression (Fang et al., 2006; Nixon et al., 2007). Whole-mount views of adult heart revealed that the entire cardiac surface and coronary vessels showed Cav1-EGFP fluorescence (Fig. 4A). In tissue sections, EGFP signals were strong in all three suggested groups of tcf21+ cells, overlapping with tcf21:DsRed2 reporter fluorescence, which is consistent with our single-cell analysis (Fig. 2A, Fig. 4B,D). Cav1-EGFP was also detected in smooth muscle cells of the outflow tract (Fig. 4B,C), in coronary vascular endothelium, and faintly in the endocardium (Fig. 4E). Upon resection of the ventricular apex, cav1-driven epicardial EGFP expression was strong in epicardial cells colonizing the injury site (Fig. 4F). We did not detect Cav1-EGFP in cardiac muscle. Thus, although cav1 expression is also present in other selected cardiac cell types, it is a prominent pan-epicardial marker.
Fig. 3.
Expression of the cav1:cav1-EGFP reporter. (A) Generation of the cav1:cav1-EGFP BAC reporter. The original 187 kb BAC CH211-226K23 covers genes cav2, cav1 and met. The Cav1 translational stop codon (TAA) in exon 3 was replaced with an EGFP-SV40 poly(A) cassette with a 20 amino acid spacer, resulting in a C-terminal fusion protein. (B-E) Visualization of Cav1-EGFP reporter expression in embryos at 3 dpf (B,C), 5 dpf (D) and 6 dpf (E), assessed in the flk1:DsRed2 (B,C,E) or tcf21:DsRed2 (D) background. (B-D) Stereofluorescence scope images; (E) confocal microscopy images. EGFP expression was detected in heart (arrowheads in B,D), blood vessels (B,C,E), notochord (C), branchial arch (D), skin (E), neuromast (E) and other tissues.
Fig. 4.
cav1 displays pan-epicardial expression. (A) Cav1-EGFP reporter expression in a whole-mount adult heart, showing labeling of the entire cardiac surface, including coronary vessels. The boxed region is magnified beneath. (B) Tissue sections indicating that the Cav1-EGFP reporter is expressed in epicardial cells, colocalizing with tcf21:DsRed2 signals. Insets show a higher magnification view of the framed region. (C) Section of outflow tract of cav1:cav1-EGFP reporter fish, stained for the smooth muscle marker Myosin light chain kinase (MLCK) (red), indicating smooth muscle expression of the reporter. The boxed region is magnified beneath. (D,E) Ventricular sections showing Cav1-EGFP reporter expression (green) together with tcf21:DsRed2 or flk1:DsRed2 reporter expression, respectively (red). Nuclei are stained with DAPI (blue). Red/blue/green (RGB) plots along the arrowed lines were generated in ImageJ. Green signals colocalized with red signals in the outermost epicardium, internal cells (likely to be perivascular cells), vascular endothelial cells, innermost EPDCs and endocardium. (F) Upon resection of the ventricular apex, cav1-driven Cav1-EGFP expression (green) is prominent in epicardial cells near the injury site at 1 and 3 dpa, and covering the wound at 7 dpa.
cav1 mutants are defective in myocardial regeneration
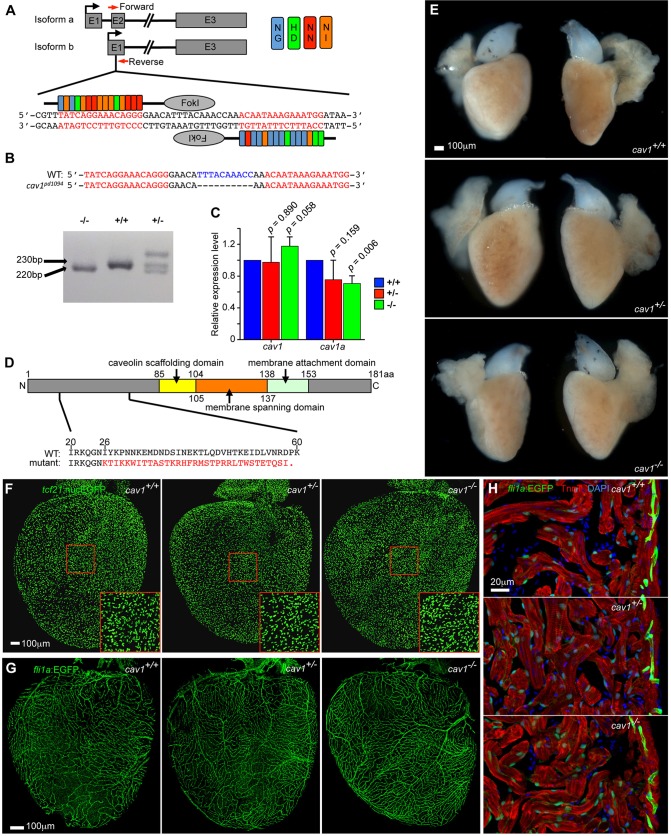
To assess requirements for cav1 in heart regeneration, we generated a mutant using TALENs. We identified a founder containing a 10 nt deletion from nucleotides 77-86 of the open reading frame (ORF) of isoform a, producing a truncated protein lacking the conserved Caveolin domains that are required for caveolae formation (Fig. 5A,B,D) (Fang et al., 2006; Parton et al., 2006). Isoform b, with an alternative translational start site generating a protein truncated by 33 amino acids, should not be affected, as the 10 nt deletion is in the 5′ UTR. The two isoforms are reported to perform nonredundant roles in zebrafish early development. Morpholino depletion of either isoform disrupts embryonic development, and each is incapable of substituting for the other (Fang et al., 2006; Nixon et al., 2007). Isoform a drives caveolae formation more efficiently than isoform b (Fujimoto et al., 2000). Also, these two isoforms have different interaction partners and effects on signaling pathways (Nohe et al., 2005; Gabor et al., 2013). Through quantitative RT-PCR using primers pairs targeting the same region of both isoforms or isoform a-specific sequences, we found that the mRNA level of cav1a, not total cav1, is slightly reduced in homozygous mutants. mRNA levels are more variable in heterozygotes (Fig. 5C).
Fig. 5.
cav1 mutants show normal heart development. (A) Strategy for generating a cav1 mutant using TALENs. The TALEN pair targets the common sequences in exon 2 of isoform a and exon 1 of isoform b. Genotyping primers are indicated by red arrows, spanning intron-exon boundaries. (B) A 10 nt deletion mutant, cav1pd1094, was identified. Genotyping results using the PCR primer pair in A, showing a 230 bp product in wild type and a 220 bp product in homozygous mutants. Samples from heterozygotes showed a higher molecular weight heterodimer product in addition to the two expected bands. (C) Quantitative RT-PCR of cav1 and cav1a in adult hearts of three genotypes, showing slightly reduced RNA levels of cav1a. Fish from three clutches served as three biological replicates, and five hearts per genotype per replicate were used. Mean±s.d., Student's t-test. (D) Sequence features of Cav1 isoform a. The mutation leads to a truncated protein lacking all the important domains for caveolae formation. (E) Whole-mount adult hearts from the three cav1 genotypes were morphologically indistinguishable. (F) Whole-mount images of tcf21:nucEGFP hearts in the three cav1 genotype backgrounds, showing no differences in epicardial cell density and distribution. (G) Whole-mount images of fli1a:EGFP hearts in the three cav1 genotype backgrounds, showing normal coronary vasculature in mutants. (H) Section images of fli1a:EGFP ventricles in the three cav1 genotype backgrounds, from samples co-stained for troponin T (Tnnt) to indicate cardiac muscle (red) and with DAPI for DNA (blue). Endocardium and muscle sarcomere appeared grossly normal in mutants. Twelve hearts per group were examined for E, and six hearts per group were examined for each of F-H.
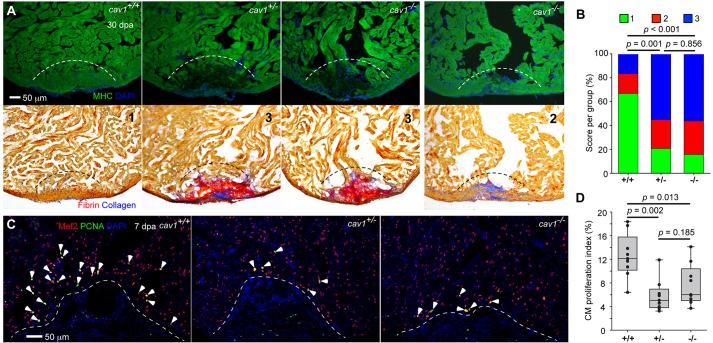
cav1 homozygous mutants survived normally through embryogenesis and adulthood and were fertile. No differences were apparent in adult cardiac size and morphology, epicardial cell density, or vascularization among wild types, heterozygotes and homozygous mutants (Fig. 5E-H). To test for function during heart regeneration, we resected ventricles of 4- to 6-month-old animals and allowed 30 days for repair. Strikingly, injured ventricles of both homozygous and heterozygous animals commonly displayed severely reduced muscularization, with gaps in the regenerated wall (22 of 29 for heterozygous mutants, and 27 of 32 for homozygotes); by contrast, control animals consistently regenerated a contiguous wall of heart muscle by this period (22 of 30) (Fig. 6A,B, Fig. S3). Instead of visible regeneration, both heterozygous and homozygous mutant animals retained pronounced fibrin and/or collagen deposits at the injured apex (Fig. 6A, Fig. S3). These defects were confirmed semi-quantitatively using a blinded scoring system (Fig. 6B).
Fig. 6.
Cav1 is required for zebrafish heart regeneration. (A) cav1 heterozygous and homozygous mutants displayed massive defects in muscle regeneration (top) and fibrin/collagen retention (bottom). Section images of ventricles at 30 dpa are stained for Myosin heavy chain (MHC, green) to indicate cardiac muscle. Dashed line indicates plane of resection. The same sections as in the top panels are stained with Acid Fuchsin-Orange G to characterize non-muscle components in the injuries (bottom; blue for collagen, red for fibrin). Twenty-two of 29 ventricles from cav1 heterozygotes and 27 of 32 from cav1 homozygotes showed obvious areas of missing myocardium and prominent scar deposition, compared with 8 of 30 ventricles from wild-type siblings. P<0.001 (Fisher's exact test) for both heterozygous and homozygous hearts. The numbers 1-3 indicates scores as in B. (B) Quantification of regeneration in cav1 mutants and wild-type siblings at 30 dpa. Representative hearts, such as those shown in A, were scored for regeneration: 1, complete regeneration of a new myocardial wall; 2, partial regeneration; 3, a strong block in regeneration. Data indicate the percentage of total hearts represented by each score for each genotype, and were analyzed by Chi-square tests. (C) Section images of injured ventricles from the three cav1 genotypes at 7 dpa. Sections are stained for Mef2 (red) and PCNA (green). Arrowheads indicate Mef2+ PCNA+ nuclei. Dashed line indicates plane of resection. (D) Quantification of CM proliferation indices in 7 dpa ventricles. For each group, nine fish were assessed, and data were analyzed using Mann–Whitney tests.
Proliferation of existing CMs is the primary, if not exclusive, cellular source mechanism for heart regeneration in zebrafish (Jopling et al., 2010; Kikuchi et al., 2010). To assess CM proliferation in cav1 mutants, we co-stained sections of 7 dpa ventricles with antibodies for the nuclear marker Mef2 and the proliferation marker PCNA (Fig. 6C) (Wills et al., 2008; Kikuchi et al., 2011b; Wang et al., 2011). CM proliferation indices were reduced by 54% in heterozygous fish (5.8% versus 12.6%) and by 39% in homozygous animals (7.7% versus 12.6%) (Fig. 6C,D). There was no significant difference between heterozygous and homozygous mutants.
To further test requirements of cav1, we made a second allele, cav1pd1104, which disrupts both isoforms and is homozygous adult viable (Fig. S4A; data not shown). Heterozygous cav1pd1104 mutants also displayed defective heart regeneration (Fig. S4B,C). The fact that zebrafish heterozygous for strong cav1 alleles have a phenotype suggests that the expression level of cav1 is important. We speculate that there may be activation in cav1 homozygous mutants of compensatory networks, as was reported recently in zebrafish (Rossi et al., 2015), that assist embryo survival and might affect injury-induced phenotypes. By this reasoning, heterozygous mutations would activate these compensatory responses to a lesser extent. These results demonstrate that cav1 is essential for injury-induced CM proliferation and natural heart muscle regeneration.
Fibronectin deposition, Tgfβ signaling activation, coronary neovascularization and epicardial regeneration in cav1 mutants
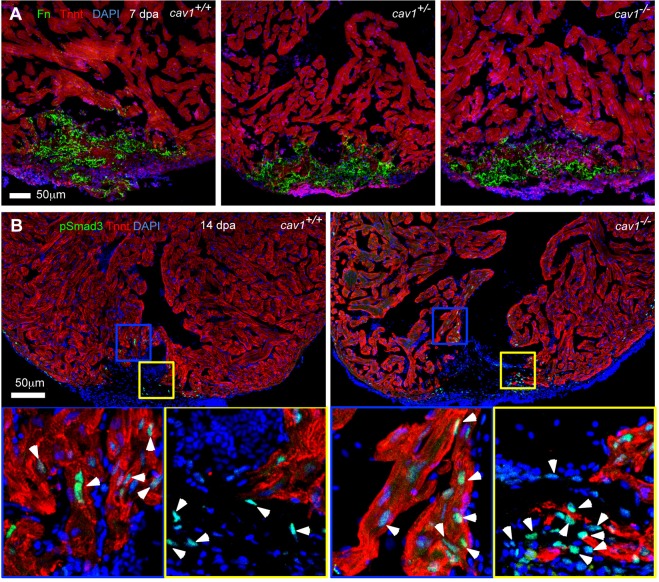
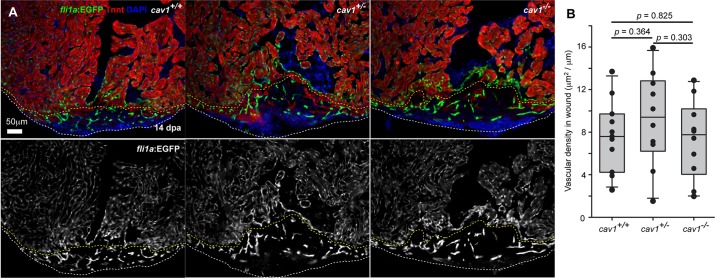
Cav1 has been reported to negatively regulate Tgfβ signaling through interaction with Tgfβ receptors, modulation of receptor expression and receptor complex turnover, and prevention of downstream Smad phosphorylation (Razani et al., 2001; Di Guglielmo et al., 2003; Wang et al., 2006; Lee et al., 2007; Tourkina et al., 2008; Chen, 2009; Miyasato et al., 2011). Cav1 was also reported to be involved in ECM remodeling and turnover (Wang et al., 2006; Tourkina et al., 2008; Miyasato et al., 2011), cell proliferation (Razani et al., 2001; Cho et al., 2004), cell migration and adhesion (Echarri et al., 2007; Grande-Garcia and del Pozo, 2008). Both Tgfβ signaling and ECM remodeling have been suggested to be indispensable for zebrafish heart regeneration (Chablais and Jazwinska, 2012; Wang et al., 2013). To test possible mechanisms of regenerative defects, we assessed fibronectin (Fn) and phospho-Smad3 (pSmad3; the antibody recognizes both Smad2 and Smad3 when phosphorylated at the equivalent sites) presence in cav1 mutants. Injured 7 dpa cav1 hearts displayed similar Fn deposition as hearts from wild-type siblings (Fig. 7A). Similarly, 14 dpa cav1 mutant and wild-type sibling hearts displayed pSmad3 signals in both CMs and non-CM cells, mainly adjacent to the wound area, indicating activation of signaling. As cav1 is expressed in coronary vessels, and coronary neovascularization is important for myocardial regeneration (Lepilina et al., 2006), we also assessed the presence of the endothelial cell marker fli1a:EGFP in all three cav1 genotypes at 14 dpa. Interestingly, wound areas of all three genotypes displayed similar vascular densities (Fig. 8A,B).
Fig. 7.
Fibronectin deposition and Tgfβ signaling activation in cav1 mutants. (A) Section images of injured ventricles from the three cav1 genotypes at 7 dpa. Sections are stained for Tnnt (red) and fibronectin (Fn, green). (B) Sections of injured ventricles at 14 dpa are stained for Tnnt (red) and pSmad3 (green). The boxed regions are enlarged to show details. Arrowheads indicate pSmad3-positive CMs (blue box, Tnnt+) and non-CMs (yellow box, Tnnt−).
Fig. 8.
Cardiac injury vascularization in cav1 mutants. (A) Sections of injured hearts of fli1a:EGFP fish in three cav1 genotypes at 14 dpa, stained for TnnT (red). White dashed lines indicate wound edges, and yellow dashed lines delineate vascularized area from endocardial area. (B) Quantification of EGFP+ vascular endothelial area in wounds with respect to wound edge lengths. Student's t-test; n=11 for wild type, n=10 for heterozygotes, and n=10 for homozygotes.
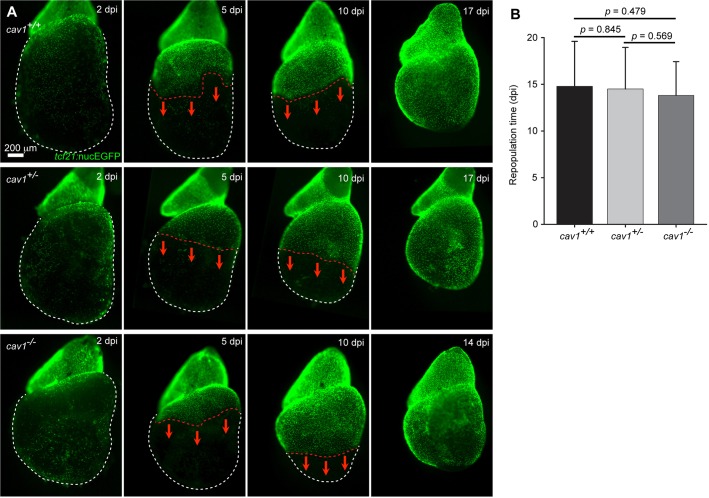
As cav1 is predominantly expressed in epicardial cells, we were curious whether the regenerative capacity of epicardial cells themselves is affected in cav1 mutants. To test this, we crossed tcf21:NTR and tcf21:nucEGFP transgenes into the cav1 homozygous mutant background, of which tcf21:NTR enables inducible genetic ablation of ∼90% of the adult tcf21+ cardiac lineage (Wang et al., 2015). We then examined epicardial regeneration in cardiac explants by treating freshly extracted and cultured hearts with 1 mM metronidazole (Mtz) for 24 h, before daily imaging (Fig. 9A). We observed similar extents and dynamics of epicardial cell repopulation between all three cav1 genotypes, suggesting that epicardial regenerative capacity is not affected (Fig. 9B).
Fig. 9.
Normal ex vivo epicardial regeneration in cav1 mutants. (A) Adult hearts of tcf21:NTR; tcf21:nucEGFP fish in different cav1 backgrounds were cultured as explants and tested for epicardial cell regeneration after at least 90% genetic ablation. White dashed lines delineate ventricles and red dashed lines delineate the epicardial sheet edge. Red arrows indicate the direction of epicardial regeneration. (B) Quantification of days to repopulate the ventricle after 90% or greater epicardial cell ablation. Mean±s.d., Student's t-test; n=18 for wild type, n=26 for heterozygotes and n=21 for homozygotes. dpi, days post Mtz incubation.
Conclusions
It is likely that the current binning of cardiac cells into designations such as myocardium, endocardium and epicardium greatly underrepresents the actual cellular diversity of the heart. Here, we have applied single-cell transcriptome profiling toward a better understanding of this diversity in adult epicardial cells, from which our results suggest at least three epicardial subsets. We identified several new epicardial markers with apparently different expression domains, many of which were upregulated after cardiac injury, suggesting possible roles in regeneration. It will also be interesting to examine the expression or function of mammalian orthologs in murine models of cardiac repair. Our single-cell transcriptomes represented 31 epicardial cells from uninjured hearts, but the analysis of hundreds or thousands of individual cells under different injury contexts would unquestionably provide higher-resolution information regarding epicardial heterogeneity. These types of analyses can be applied to other cardiac cell types, such as CMs and endocardium, and ultimately promise to radially expand the genetic toolbox and our molecular understanding of the heart and its regenerative capacity.
Cav1 functions in many contexts by the spatiotemporal regulation of signaling molecules, either by direct interaction with the signaling molecule, by compartmentalization of signaling platforms in caveolae, or by caveolar endocytosis (Gvaramia et al., 2013). The precise mechanistic basis for the CM proliferation and myocardial regeneration defects in cav1 mutants requires further investigation. These functions might overlap with the reported roles for murine Cav1 in mounting responses to myocardial injury that limit heart failure (Zhao et al., 2002; Cohen et al., 2003; Miyasato et al., 2011; Shivshankar et al., 2014).
MATERIALS AND METHODS
Zebrafish and cardiac injuries
Adult zebrafish of the Ekkwill strain were maintained as described, and resection injuries and CM ablation were performed as described (Poss et al., 2002; Wang et al., 2011). Animals between 4 and 12 months of age of both sexes were used. All surgeries, histological analyses and proliferation analyses of cav1 mutants were performed in a blinded fashion, and experiments were repeated using different clutches of animals. Transgenic lines used in this study were Tg(tcf21:mCherry-NTR)pd108 (Wang et al., 2015), Tg(tcf21:nucEGFP)pd41 (Wang et al., 2011), Tg(tcf21:DsRed2)pd37 (Kikuchi et al., 2011a), Tg(cmlc2:CreER)pd10 (Kikuchi et al., 2010), Tg(bactin2:loxP-mCherry-STOP-loxP-DTA176)pd36 (Wang et al., 2011), Tg(flk1:DsRed2)pd27 (Kikuchi et al., 2011b) and Tg(fli1a:EGFP)y1 (Lawson and Weinstein, 2002). Newly constructed strains are described below. Genetic CM ablation injuries were performed as described (Wang et al., 2011). All transgenic strains were analyzed as hemizygotes. Animal procedures were performed in accordance with Duke University guidelines.
Single-cell RNA sequencing
Ventricles were collected from adult tcf21:nucEGFP fish at 6 months of age. Ventricular nucEGFP+ epicardial cells were isolated as described previously (Zhou and Pu, 2012; Wang et al., 2015). Briefly, ventricles were collected on ice and washed several times to remove blood cells. Ventricles were digested in an Eppendorf tube with 0.5 ml HBSS plus 0.13 U/ml Liberase DH (Roche) and 1% sheep serum at 37°C, while stirring gently with a Spinbar magnetic stirring bar (Bel-Art Products). Supernatants were collected every 5 min and neutralized with sheep serum. Dissociated cells were spun down (300 g) and resuspended in DMEM plus 10% fetal bovine serum (FBS) medium with 1.5 µg/ml propidium iodide (PI), and sorted using a BD FACSVantage SE sorter for EGFP-positive and PI-negative cells. Some sorted cells were plated into a culture dish for 24 h to show cell survival, morphology and EGFP signals. Single epicardial cells were captured in a Fluidigm C1 platform. A pool of ∼5000 cells was also captured as a control. Libraries were prepared according to Fluidigm protocols, and sequencing was performed using an Illumina HiSeq 2000 with 50 nt paired-end reads. Bioinformatics analyses were performed with sequences aligned to the zebrafish genome (Zv9) using Bowtie 2 and transcript expressions were quantified using RSEM 1.2.12 (Li and Dewey, 2011; Langmead and Salzberg, 2012). Outlier identification, PCA and hierarchical clustering of genes were performed in the Fluidigm SINGuLAR package, using default settings. The tool package chose 500 genes with the most differential expression patterns across single cells for PCA and hierarchical clustering (Pollen et al., 2014). RNA sequencing data have been deposited at GEO under accession number GSE75583.
cav1:cav1-EGFP
An EGFP-SV40 poly(A) cassette with a 20 amino acid spacer was recombined into the translational stop codon of cav1 in the BAC clone CH211-226K23 using SW105 cells, to generate a C-terminal fusion protein as previously reported (Navis et al., 2013). For transgenesis, inverted Tol2 sites were inserted into the BAC backbone (Suster et al., 2009). The primers for recombination were (5′-3′): forward, ccatggggaaatgctttagtaacgtccgggtcactgctactaaggtggtgGATCTCCCCGCCGAACAGAAA; reverse, cattcccccatcccacccgatccctctggctcttctctttcactctgtccATTGGAGCTCCACCGCGGTG (lowercase, homologous to the BAC; uppercase, homologous to the EGFP cassette construct). The full name of this transgenic line is TgBAC(cav1:cav1-EGFP)pd1096.
cav1 mutants and genotyping
cav1 mutants were generated using TALENs targeting exon 2 of isoform a (exon 1 of isoform b; Fig. 5A). The TALEN was designed using TALEN Targeter (Doyle et al., 2012); the sequences are: left, NG NI NG HD NI NN NN NI NI NI HD NI NN NN NN; right, HD HD NI NG NG NG HD NG NG NG NI NG NG NN NG. The TALENs were assembled using Golden Gate assembly (Cermak et al., 2011) into destination vectors expressing heterodimeric FokI nuclease, pCS2TAL3DD and pCS2TAL3RR, optimized for production of in vitro RNA (Dahlem et al., 2012). Zebrafish embryos were injected at the one-cell stage with 100 pg total TALEN RNA. Genotyping was performed using primers: forward, AGTCTCCCACTCCACGCTCAT; reverse, TGTCCTACCTTCACCACATCGT. The wild-type product is 230 bp, whereas the mutant is 220 bp (Fig. 5B). This mutant allele designation is cav1pd1094. The second allele was generated using the following TALEN pairs to target to the common sequence of both isoforms: left, NN NN NI NG NI NI HD NN NI HD NI NN HD NI NG; right, HD NG HD HD NG NG NN NN NG NN NG NN NN NI HD NI NG. A mutant with a 23 nt deletion, cav1pd1104, was identified. Genotyping was performed using primers: forward, CTTTGGAACCGCAGGAATAC; reverse, CCATGCAGTTAAACAAATGTCC. The wild-type product is 282 bp, whereas the mutant is 259 bp (Fig. S4A).
Histology
In situ hybridization (ISH) was performed on 10 µm cryosections of paraformaldehyde-fixed hearts using digoxigenin-labeled cRNA probes as described (Poss et al., 2002) with the aid of an InSituPro robot (Intavis). Whole-mount in situ hybridization of 3 and 4 dpf embryos was also performed with the aid of the robot. The raldh2 probe was described previously (Lepilina et al., 2006). twist1 ORF was obtained from Open Biosystems and cloned into pBlueScript II vector (Agilent Technologies), and the probe was generated using T3 RNA polymerase. All other probes were cloned from 2 dpf zebrafish cDNA using the primers listed in Table S2.
Primary and secondary antibody staining were performed as previously described (Lepilina et al., 2006; Kikuchi et al., 2011b). Primary antibodies were mouse anti-Myosin heavy chain (MHC; F59, Developmental Studies Hybridoma Bank; 1:100), mouse anti-troponin T (Neomarkers, MS-295-PABX; 1:100), rabbit anti-Mef2 (Santa Cruz, sc-313; 1:100), mouse anti-PCNA (Sigma, P8825; 1:100), rabbit anti-fibronectin (Sigma, F3648; 1:200) and rabbit anti-phospho-Smad3 (Abcam, ab52903; 1:200). Secondary antibodies (Invitrogen; 1:200) were Alexa Fluor 488 or Alexa 594 goat anti-rabbit and goat anti-mouse. Acid Fuchsin-Orange G staining was performed as described (Poss et al., 2002).
Whole-mount and sectioned ventricular tissues were imaged using a Zeiss LSM 700 confocal microscope, a Zeiss Axio Zoom V16 stereomicroscope, or a Leica DM6000 compound microscope. Quantification of CM proliferation was performed as described previously, by counting a defined region typically including almost all Mef2+ PCNA+ CMs near the injury (314 µm in the y-dimension of the image). The percentages of Mef2+ PCNA+ cells from three sections showing the largest wounds were averaged to determine a proliferation index for each heart (Kikuchi et al., 2011b). The vascular density was quantified as described previously (Wang et al., 2015).
RT-qPCR
Quantitative RT-PCR was carried out in biological triplicate using a Roche LightCycler 480 II system with LightCycler 480 Probes Master Mix and the Universal Probe Library (UPL) (Roche). ef1a (eef1a1l1) served as the control. Primers (f, forward; r, reverse) and the UPL# were: ef1a-f CCTCTTTCTGTTACCTGGCAAA and ef1a-r CTTTTCCTTTCCCATGATTGA, UPL #73; cav1-f ATGACGATGTGGTGAAGGTG and cav1-r TGGTTACTGTGAAGGTGGTGA, UPL #97; cav1a-f TTTCCTCACACTCATCTGATCG and cav1a-r GGCGAGTGAGCGTATTCCT, UPL #124.
Ex vivo epicardial regeneration
Heart explant culture and ex vivo epicardial ablation were performed as described previously (Wang et al., 2015). Briefly, adult hearts were cultured in DMEM medium plus 10% fetal bovine serum, 1% non-essential amino acids, 100 U/ml penicillin, 100 µg/ml streptomycin, and 50 µM 2-mercaptoethanol, while rotating at 150 rpm. For epicardial ablation, dissected hearts were bathed for 24 h in 1 mM Mtz before washout. Explants were imaged using a Leica MZ05FA stereofluorescence microscope.
Acknowledgements
We thank J. Burris, N. Lee, S. Davies, T. Hume Thoren and N. Benkaci for fish care; D. Corcoran for bioinformatic analysis; and J. Wang and J. A. Goldman for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
J.C. and K.D.P. designed the experimental strategy, analyzed data and prepared the manuscript. B.D.C. helped with cell dissociation and proteomic analysis. A.L.D. performed surgeries and helped with proliferation analysis. A.N. and M.B. generated the cav1 transgenic and mutant animals and analyzed embryos. M.G. and R.K. provided unpublished reagents. All authors commented on the manuscript.
Funding
This work was funded by fellowships from the American Heart Association to J.C. (postdoctoral) and M.G. (predoctoral); a National Science Foundation predoctoral fellowship to B.D.C.; a National Institutes of Health (NIH) Mentored Clinical Scientist Award [K08-HL116485] to R.K.; and grants from the NIH to M.B. [R01AR065439-02] and K.D.P. [HL081674]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.130534/-/DC1
References
- Bollini S., Vieira J. M. N., Howard S., Dubè K. N., Balmer G. M., Smart N. and Riley P. R. (2014). Re-activated adult epicardial progenitor cells are a heterogeneous population molecularly distinct from their embryonic counterparts. Stem Cells Dev. 23, 1719-1730. 10.1089/scd.2014.0019 [DOI] [PubMed] [Google Scholar]
- Cavallero S., Shen H., Yi C., Lien C.-L., Kumar S. R. and Sucov H. M. (2015). CXCL12 signaling is essential for maturation of the ventricular coronary endothelial plexus and establishment of functional coronary circulation. Dev. Cell 33, 469-477. 10.1016/j.devcel.2015.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J. A., Somia N. V., Bogdanove A. J. and Voytas D. F. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 10.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chablais F. and Jazwinska A. (2012). The regenerative capacity of the zebrafish heart is dependent on TGFbeta signaling. Development 139, 1921-1930. 10.1242/dev.078543 [DOI] [PubMed] [Google Scholar]
- Chen Y.-G. (2009). Endocytic regulation of TGF-beta signaling. Cell Res. 19, 58-70. 10.1038/cr.2008.315 [DOI] [PubMed] [Google Scholar]
- Cho K. A., Ryu S. J., Oh Y. S., Park J. H., Lee J. W., Kim H.-P., Kim K. T., Jang I. S. and Park S. C. (2004). Morphological adjustment of senescent cells by modulating caveolin-1 status. J. Biol. Chem. 279, 42270-42278. 10.1074/jbc.M402352200 [DOI] [PubMed] [Google Scholar]
- Choi W.-Y., Gemberling M., Wang J., Holdway J. E., Shen M.-C., Karlstrom R. O. and Poss K. D. (2013). In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 140, 660-666. 10.1242/dev.088526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A. W., Park D. S., Woodman S. E., Williams T. M., Chandra M., Shirani J., Pereira de Souza A., Kitsis R. N., Russell R. G., Weiss L. M. et al. (2003). Caveolin-1 null mice develop cardiac hypertrophy with hyperactivation of p42/44 MAP kinase in cardiac fibroblasts. Am. J. Physiol. Cell Physiol. 284, C457-C474. 10.1152/ajpcell.00380.2002 [DOI] [PubMed] [Google Scholar]
- Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., Locke A. S., Weis A. M., Voytas D. F. and Grunwald D. J. (2012). Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8, e1002861 10.1371/journal.pgen.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Guglielmo G. M., Le Roy C., Goodfellow A. F. and Wrana J. L. (2003). Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat. Cell Biol. 5, 410-421. 10.1038/ncb975 [DOI] [PubMed] [Google Scholar]
- Doyle E. L., Booher N. J., Standage D. S., Voytas D. F., Brendel V. P., Vandyk J. K. and Bogdanove A. J. (2012). TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117-W122. 10.1093/nar/gks608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echarri A., Muriel O. and Del Pozo M. A. (2007). Intracellular trafficking of raft/caveolae domains: insights from integrin signaling. Semin. Cell Dev. Biol. 18, 627-637. 10.1016/j.semcdb.2007.08.004 [DOI] [PubMed] [Google Scholar]
- Fang P.-K., Solomon K. R., Zhuang L., Qi M., McKee M., Freeman M. R. and Yelick P. C. (2006). Caveolin-1alpha and -1beta perform nonredundant roles in early vertebrate development. Am. J. Pathol. 169, 2209-2222. 10.2353/ajpath.2006.060562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T., Kogo H., Nomura R. and Une T. (2000). Isoforms of caveolin-1 and caveolar structure. J. Cell Sci. 113, 3509-3517. [DOI] [PubMed] [Google Scholar]
- Gabor K. A., Stevens C. R., Pietraszewski M. J., Gould T. J., Shim J., Yoder J. A., Lam S. H., Gong Z., Hess S. T. and Kim C. H. (2013). Super resolution microscopy reveals that caveolin-1 is required for spatial organization of CRFB1 and subsequent antiviral signaling in zebrafish. PLoS ONE 8, e68759 10.1371/journal.pone.0068759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M., Karra R., Dickson A. L. and Poss K. D. (2015). Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife 4, e05871 10.7554/eLife.05871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande-Garcia A. and del Pozo M. A. (2008). Caveolin-1 in cell polarization and directional migration. Eur. J. Cell Biol. 87, 641-647. 10.1016/j.ejcb.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Gvaramia D., Blaauboer M. E., Hanemaaijer R. and Everts V. (2013). Role of caveolin-1 in fibrotic diseases. Matrix Biol. 32, 307-315. 10.1016/j.matbio.2013.03.005 [DOI] [PubMed] [Google Scholar]
- Harrison M. R. M., Bussmann J., Huang Y., Zhao L., Osorio A., Burns C. G., Burns C. E., Sucov H. M., Siekmann A. F. and Lien C.-L. (2015). Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev. Cell 33, 442-454. 10.1016/j.devcel.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G. N., Thatcher J. E., McAnally J., Kong Y., Qi X., Tan W., DiMaio J. M., Amatruda J. F., Gerard R. D., Hill J. A. et al. (2012). C/EBP transcription factors mediate epicardial activation during heart development and injury. Science 338, 1599-1603. 10.1126/science.1229765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M., Fu J.-D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G. and Srivastava D. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375-386. 10.1016/j.cell.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Martí M., Raya A. and Izpisua Belmonte J. C. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606-609. 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Werdich A. A., Anderson R. M., Fang Y., Egnaczyk G. F., Evans T., MacRae C. A., Stainier D. Y. R. and Poss K. D. (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601-605. 10.1038/nature08804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Gupta V., Wang J., Holdway J. E., Wills A. A., Fang Y. and Poss K. D. (2011a). tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 138, 2895-2902. 10.1242/dev.067041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Major R. J., Blum N., Dahn R. D., Begemann G. and Poss K. D. (2011b). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397-404. 10.1016/j.devcel.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse K. A., Wu J., Whittaker C. A. and Amon A. (2014). Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc. Natl. Acad. Sci. USA 111, 13409-13414. 10.1073/pnas.1415287111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N. D. and Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307-318. 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- Lee E. K., Lee Y. S., Han I.-O. and Park S. H. (2007). Expression of Caveolin-1 reduces cellular responses to TGF-beta1 through down-regulating the expression of TGF-beta type II receptor gene in NIH3T3 fibroblast cells. Biochem. Biophys. Res. Commun. 359, 385-390. 10.1016/j.bbrc.2007.05.121 [DOI] [PubMed] [Google Scholar]
- Lepilina A., Coon A. N., Kikuchi K., Holdway J. E., Roberts R. W., Burns C. G. and Poss K. D. (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607-619. 10.1016/j.cell.2006.08.052 [DOI] [PubMed] [Google Scholar]
- Li B. and Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien C.-L., Schebesta M., Makino S., Weber G. J. and Keating M. T. (2006). Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 4, e260 10.1371/journal.pbio.0040260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasato S. K., Loeffler J., Shohet R., Zhang J., Lindsey M. and Le Saux C. J. (2011). Caveolin-1 modulates TGF-beta1 signaling in cardiac remodeling. Matrix Biol. 30, 318-329. 10.1016/j.matbio.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N., Kendall J., Troge J., Andrews P., Rodgers L., McIndoo J., Cook K., Stepansky A., Levy D., Esposito D. et al. (2011). Tumour evolution inferred by single-cell sequencing. Nature 472, 90-94. 10.1038/nature09807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navis A., Marjoram L. and Bagnat M. (2013). Cftr controls lumen expansion and function of Kupffer's vesicle in zebrafish. Development 140, 1703-1712. 10.1242/dev.091819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon S. J., Carter A., Wegner J., Ferguson C., Floetenmeyer M., Riches J., Key B., Westerfield M. and Parton R. G. (2007). Caveolin-1 is required for lateral line neuromast and notochord development. J. Cell Sci. 120, 2151-2161. 10.1242/jcs.003830 [DOI] [PubMed] [Google Scholar]
- Nohe A., Keating E., Underhill T. M., Knaus P. and Petersen N. O. (2005). Dynamics and interaction of caveolin-1 isoforms with BMP-receptors. J. Cell Sci. 118, 643-650. 10.1242/jcs.01402 [DOI] [PubMed] [Google Scholar]
- Parton R. G. and del Pozo M. A. (2013). Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 14, 98-112. 10.1038/nrm3512 [DOI] [PubMed] [Google Scholar]
- Parton R. G., Hanzal-Bayer M. and Hancock J. F. (2006). Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J. Cell Sci. 119, 787-796. 10.1242/jcs.02853 [DOI] [PubMed] [Google Scholar]
- Pollen A. A., Nowakowski T. J., Shuga J., Wang X., Leyrat A. A., Lui J. H., Li N., Szpankowski L., Fowler B., Chen P. et al. (2014). Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 32, 1053-1058. 10.1038/nbt.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K. D., Wilson L. G. and Keating M. T. (2002). Heart regeneration in zebrafish. Science 298, 2188-2190. 10.1126/science.1077857 [DOI] [PubMed] [Google Scholar]
- Qian L., Huang Y., Spencer C. I., Foley A., Vedantham V., Liu L., Conway S. J., Fu J.-D. and Srivastava D. (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593-598. 10.1038/nature11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B., Engelman J. A., Wang X. B., Schubert W., Zhang X. L., Marks C. B., Macaluso F., Russell R. G., Li M., Pestell R. G. et al. (2001). Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J. Biol. Chem. 276, 38121-38138. [DOI] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M. and Stainier D. Y. R. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230-233. 10.1038/nature14580 [DOI] [PubMed] [Google Scholar]
- Shalek A. K., Satija R., Adiconis X., Gertner R. S., Gaublomme J. T., Raychowdhury R., Schwartz S., Yosef N., Malboeuf C., Lu D. et al. (2013). Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236-240. 10.1038/nature12172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivshankar P., Halade G. V., Calhoun C., Escobar G. P., Mehr A. J., Jimenez F., Martinez C., Bhatnagar H., Mjaatvedt C. H., Lindsey M. L. et al. (2014). Caveolin-1 deletion exacerbates cardiac interstitial fibrosis by promoting M2 macrophage activation in mice after myocardial infarction. J. Mol. Cell Cardiol. 76, 84-93. 10.1016/j.yjmcc.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvets E., Ludwig A. and Nichols B. J. (2014). News from the caves: update on the structure and function of caveolae. Curr. Opin. Cell Biol. 29, 99-106. 10.1016/j.ceb.2014.04.011 [DOI] [PubMed] [Google Scholar]
- Smart N., Risebro C. A., Melville A. A. D., Moses K., Schwartz R. J., Chien K. R. and Riley P. R. (2007). Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature 445, 177-182. 10.1038/nature05383 [DOI] [PubMed] [Google Scholar]
- Smart N., Bollini S., Dubé K. N., Vieira J. M., Zhou B., Davidson S., Yellon D., Riegler J., Price A. N., Lythgoe M. F. et al. (2011). De novo cardiomyocytes from within the activated adult heart after injury. Nature 474, 640-644. 10.1038/nature10188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Nam Y.-J., Luo X., Qi X., Tan W., Huang G. N., Acharya A., Smith C. L., Tallquist M. D., Neilson E. G. et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599-604. 10.1038/nature11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suster M. L., Sumiyama K. and Kawakami K. (2009). Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics 10, 477 10.1186/1471-2164-10-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourkina E., Richard M., Gooz P., Bonner M., Pannu J., Harley R., Bernatchez P. N., Sessa W. C., Silver R. M. and Hoffman S. (2008). Antifibrotic properties of caveolin-1 scaffolding domain in vitro and in vivo. Am. J. Physiol. Lung Cell Mol. Physiol. 294, L843-L861. 10.1152/ajplung.00295.2007 [DOI] [PubMed] [Google Scholar]
- Wang X. M., Zhang Y., Kim H. P., Zhou Z., Feghali-Bostwick C. A., Liu F., Ifedigbo E., Xu X., Oury T. D., Kaminski N. et al. (2006). Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J. Exp. Med. 203, 2895-2906. 10.1084/jem.20061536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Panakova D., Kikuchi K., Holdway J. E., Gemberling M., Burris J. S., Singh S. P., Dickson A. L., Lin Y.-F., Sabeh M. K. et al. (2011). The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 138, 3421-3430. 10.1242/dev.068601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Karra R., Dickson A. L. and Poss K. D. (2013). Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev. Biol. 382, 427-435. 10.1016/j.ydbio.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cao J., Dickson A. L. and Poss K. D. (2015). Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 522, 226-230. 10.1038/nature14325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills A. A., Holdway J. E., Major R. J. and Poss K. D. (2008). Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development 135, 183-192. 10.1242/dev.010363 [DOI] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. et al. (2013). Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131-1139. 10.1038/nsmb.2660 [DOI] [PubMed] [Google Scholar]
- Zhao Y.-Y., Liu Y., Stan R.-V., Fan L., Gu Y., Dalton N., Chu P.-H., Peterson K., Ross J. Jr and Chien K. R. (2002). Defects in caveolin-1 cause dilated cardiomyopathy and pulmonary hypertension in knockout mice. Proc. Natl. Acad. Sci. USA 99, 11375-11380. 10.1073/pnas.172360799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. and Pu W. T. (2012). Isolation and characterization of embryonic and adult epicardium and epicardium-derived cells. Methods Mol. Biol. 843, 155-168. 10.1007/978-1-61779-523-7_15 [DOI] [PubMed] [Google Scholar]
- Zhou B., Honor L. B., He H., Ma Q., Oh J.-H., Butterfield C., Lin R.-Z., Melero-Martin J. M., Dolmatova E., Duffy H. S. et al. (2011). Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Invest. 121, 1894-1904. 10.1172/JCI45529 [DOI] [PMC free article] [PubMed] [Google Scholar]