Abstract
Background: Low blood vitamin D concentration is a concern for people living in circumpolar regions, where sunlight is insufficient for vitamin D synthesis in winter months and the consumption of traditional dietary sources of vitamin D is decreasing.
Objective: The objective was to characterize the effects of diet, genetic variation, and season on serum 25-hydroxycholecalciferol [25(OH)D3] concentrations in Yup’ik Alaska Native people living in rural southwest Alaska.
Methods: This study was a cross-sectional design that assessed the associations of traditional diet (via a biomarker, the RBC δ15N value), age, gender, body mass index (BMI), community location, and genotype of select single nucleotide polymorphisms (SNPs) in cytochrome P450 family 2, subfamily R, peptide 1 (CYP2R1), 7-dehydrocholesterol reductase (DHCR7), and vitamin D binding protein (GC) with serum 25(OH)D3 concentrations in 743 Yup’ik male and female participants, aged 14–93 y, recruited between September 2009 and December 2013.
Results: Yup’ik participants, on average, had adequate concentrations of serum 25(OH)D3 (31.1 ± 1.0 ng/mL). Variations in diet, BMI, age, gender, season of sample collection, and inland or coastal community geography were all significantly associated with serum 25(OH)D3 concentration. In models not adjusting for other covariates, age, diet, and seasonal effects explained 33.7%, 20.7%, and 9.8%, respectively, of variability in serum 25(OH)D3 concentrations. Of the 8 SNPs interrogated in CYP2R1 and DHCR7, only rs11023374 in CYP2R1 was significantly associated with serum 25(OH)D3, explaining 1.5% of variability. The GC haplotype explained an additional 2.8% of variability. Together, age, diet, gender, season of sample collection, BMI, geography of the community, and genotype at rs11023374 explained 52.5% of the variability in serum 25(OH)D3 concentrations.
Conclusions: Lower consumption of the traditional diet was associated with lower serum concentrations of 25(OH)D3. Younger adults and youth in this community may be at increased risk of adverse outcomes associated with vitamin D insufficiency compared with older members of the community, especially during seasons of low sunlight exposure, because of lower consumption of dietary sources of vitamin D.
Keywords: Alaska Native, American Indian, vitamin D, public health, cholecalciferol, polyunsaturated fatty acid
Introduction
Cholecalciferol (vitamin D3) deficiency is linked to increased risk of multiple illnesses, including bone demineralization, rickets, multiple sclerosis, cardiovascular disease, colon cancer, some types of breast and prostate cancers, type 1 and 2 diabetes, respiratory infections, influenza, active tuberculosis, and depression (1–4). The concentration of 25-hydroxyvitamin D [25(OH)D9] in serum or plasma is used as the primary indicator of vitamin D sufficiency (5, 6). Although there is some controversy (7), the Institute of Medicine considers a serum concentration of <12 ng/mL (30 nM) to be deficient, 12–20 ng/mL to be insufficient, and >20 ng/mL (50 nM) to be sufficient (8). Sun exposure, diet, age, gender, BMI, disease status, and use of some drugs have been associated with serum/plasma 25(OH)D concentrations (5, 9). The heritability of vitamin D concentrations has been estimated at 29–80%, with known genetic variants explaining only 1–4% of that variation (5, 10–13). Specifically, common noncoding variants in cytochrome P450 family 2, subfamily R, peptide 1 (CYP2R1), which encodes the enzyme that hydroxylates vitamin D into 25(OH)D; in 7-dehydrocholesterol reductase (DHCR7), which encodes the enzyme that modulates the amount of vitamin D precursor in the skin for synthesis with sunlight; and in vitamin D binding protein (GC), which encodes the vitamin D binding protein, have been associated with serum 25-hydroxycholecalciferol [25(OH)D3] concentration (1, 14, 15). Indeed, a haplotype of DHCR7 that is thought to reduce enzyme function has been found more commonly in northern latitudes and is suggested to confer an evolutionary advantage (15).
Vitamin D deficiency is a public health concern in Alaska and is increasingly prevalent among Alaska Native (AN) infants and children. Hospitalization due to rickets occurs more frequently than in the general United States and at a rate of 2.23/100,000 children per year (16, 17). Cancer, also associated with vitamin D deficiency, is the leading cause of death among AN people (4, 18). Colon cancer is of particular concern to the Yup’ik people in the Yukon-Kuskokwim River Delta (Y-K Delta) and had an overall incidence in AN people of 102.6 per 100,000 for all years combined between 1999 and 2004 (19), with a relative rate of incidence of 2.03, compared with an overall incidence of 50.6 per 100,000 for non-Hispanic white people living in the same regions (19).
For the Yup’ik people who live in the Y-K Delta of rural southwestern Alaska, adequate vitamin D may be obtained from the traditional diet, including fish, marine mammals, liver, and other organ meats (20). In a study of Yup’ik communities in the Y-K Delta conducted between 2003 and 2005, the mean serum 25(OH)D3 concentration was twice the threshold for sufficiency, and it was estimated that 90% of vitamin D came from traditional food sources (20). However, reduced consumption of locally harvested foods may lead to increased rates of 25(OH)D deficiency in these communities (20–22). To better understand dietary, genetic, seasonal, and demographic factors that affect circulating 25(OH)D3 concentrations, we conducted a cross-sectional study in this population that included analysis of variants in CYP2R1, DHCR7, and GC; serum 25(OH)D3 concentrations and the RBC δ15N value, a validated biomarker of the marine-based diet in this population (23–25). Of particular interest for potential public health utility was the contribution of diet, genetic variation, and season on differences in serum 25(OH)D3 concentrations.
Methods
Research approval.
This study emerged from a partnership between the University of Washington (UW), the University of Alaska Fairbanks Center for Alaska Native Health Research, the Yukon Kuskokwim Health Corporation (YKHC), and Yup’ik communities in the Y-K Delta. The research questions were developed with Y-K Delta communities in partnership with the Center for Alaska Native Health Research under a community-based participatory research framework (26). Approval for research was received from the YKHC Executive Board of Directors and the University of Alaska Fairbanks institutional review board, as well as the UW institutional review board. This study was granted a Certificate of Confidentiality by the National Institute of General Medical Sciences to protect participant information.
Study population.
The Y-K Delta in southwestern Alaska is home to ∼23,000 people, 85% of whom are self-identified AN, and who live predominantly in remote communities and obtain health care through the YKHC (27). A total of 743 male and female research participants aged ≥14 y were recruited for the study between September 2009 and December 2013 through written and oral advertisement. They represent convenience sampling from 10 communities in the Y-K Delta.
Sample collection and processing.
Fasting venous blood samples were collected from each participant for isolation of RBCs, plasma, serum, and DNA. Blood was collected into silica-coated K2 EDTA tubes (BD Vacutainer) and centrifuged (900 × g, 15 min) at room temperature. Buffy coats were incubated with Puregene RBC Lysis Solution for 10 min and centrifuged again (1800 × g, 10 min) at room temperature. White blood cells were then resuspended in 10 mL Puregene Cell Lysis Solution until DNA purification by using the Gentra Puregene kit (Qiagen). Serum was isolated from blood collected in a BD red-top Vacutainer and transferred to amber tubes for 25(OH)D3 analysis (Becton Dickinson). All samples collected in the field were stored in aliquots at −15°C in a portable freezer while on site and then shipped to the University of Alaska Fairbanks within 7 d and stored at −80°C. Isolated DNA and serum were subsequently sent to UW for genetic and vitamin D analysis.
DNA isolation and genetic analysis.
The CYP2R1 single nucleotide polymorphisms (SNPs) genotyped were rs2060793, rs10741657, rs1993116, and rs11023374. The 4 SNPs informing DHCR7 haplotypes were rs12785878, rs3794060, rs12800438, and rs4944957. The 2 SNPs informing GC haplotypes were rs4588 and rs7041. These SNPs were chosen on the basis of previous association with 25(OH)D concentrations (10–13, 15). For CYP2R1 genotyping, DNA samples were analyzed on 96.96 Dynamic Genotyping Arrays (Fluidigm) according to the manufacturer’s established protocol for BioMark 96.96 genotyping, as described in Fohner et al. (28).
For determining DHCR7 and GC haplotypes, DNA samples were genotyped by using predesigned 5′-nuclease SNP genotyping assays (Applied Biosystems/Life Technologies), which use specific fluorogenic probes. The fluorescent 5′-nuclease assays were performed and analyzed on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). The specific PCR reaction conditions were based on the general guidelines provided by the manufacturer and incorporated 10–25 ng genomic DNA template. Thermocycling parameters consisted of an initial incubation at 95°C for 10 min, followed by 40 cycles of 92°C for 15 s and of 60°C for 1 min. Haplotypes in GC were calculated by using PHASE software version 2.1 (29, 30). Haplotype nomenclature was based on Powe et al. (31).
Pairwise r2 linkage disequilibrium (LD) patterns between the CYP2R1 SNPs and the DHCR7 SNPs were calculated by using Haploview 4.2 software (32). All SNPs were tested for deviations from Hardy-Weinberg equilibrium by using a chi-square test, with a significance level of 0.05. To calculate population frequencies of the SNPs in CYP2R1, DHCR7, and GC, genealogic information contained in pedigrees was used to create a matrix of pairwise kinship coefficients between each participant, as described in Bourgain and Zhang (33). Estimates of population frequency and variance with adjustment for relatedness between sample individuals were obtained by using the best linear unbiased estimator for allele frequency (34). Statistical analysis for allele frequency estimation was performed by using R statistical computing language (35).
RBC nitrogen isotope ratio analysis.
RBC nitrogen isotope ratios were prepared as described in O’Brien and colleagues (23, 36) and analyzed at the Alaska Stable Isotope Facility at University of Alaska Fairbanks by continuous-flow isotope ratio MS with the use of a Costech ECS4010 Elemental Analyzer (Costech Scientific) interfaced with a Delta V Plus isotope ratio mass spectrometer (Thermo Fisher Scientific) via the Conflo IV Interface (Thermo Fisher Scientific). Nitrogen isotope ratios are represented in δ notation as “permil” abundance of heavy isotope relative to an international standard: δ15N value = [(Rsample − Rstandard)/Rstandard] × 1000‰, where R is the ratio of heavy to light isotope and the standard is atmospheric nitrogen. The RBC δ15N value is a validated biomarker of RBC EPA and DHA n−3 FA content and their intake in the diet, because both δ15N values and n–3 FAs are elevated in the fish and marine mammals that are a staple of the traditional Yup’ik diet (23–25). The δ15N value of RBCs reflects intake over approximately the previous 2 mo (37).
Serum 25(OH)D3 measurement.
The total (unbound and bound) concentrations of 25(OH)D3 in serum samples were determined following a validated LC–tandem MS assay, as described in Wang et al. (38). Analytical standards were compared with reference 25(OH)D3 National Institute of Standards and Technology standards (39) and found to be within 15% of the reference concentration across the standard curve range.
Preliminary analysis and assessment of nonnormality of samples.
The distributions of both serum 25(OH)D3 concentrations and δ15N values were assessed for normality. Log transformation of δ15N values, but not serum 25(OH)D3 concentrations, improved the normality of the distribution. Accordingly, serum 25(OH)D3 concentrations were not transformed and δ15N values were log transformed for further analysis (Supplemental Figure 1).
Regression of serum 25(OH)D3 concentration against each genotype at each SNP was performed to determine which, if any, CYP2R1 or DHCR7 SNPs or whether GC haplotype or specific SNPs should be included in association analysis as an additive or recessive variant model. Each SNP was evaluated separately in a linear regression model and by an ANOVA test with serum 25(OH)D3 concentration. Genotypes were also tested in each season to assess any seasonal variation in genetic contribution. Only the genotype at rs11023374 in CYP2R1 was included for analysis, following a recessive inheritance model of the variant allele. SNPs in GC were analyzed independently and as haplotypes for their association with serum 25(OH)D3 concentration. Both GC SNPs at rs7041 and rs4588 were found to be significant predictors of serum 25(OH)D3 concentrations in other populations and so were included in regression analysis as haplotypes. Because of low DNA quantities for GC genotyping, some samples are missing GC genotype. To avoid losing power in analysis of other covariates, GC haplotypes were analyzed separately from the full regression model.
Kinship adjustment and summary statistics.
A kinship correlation matrix based on pairwise familial relationships was used to adjust for correlated serum 25(OH)D3 concentrations among sample individuals in an association analysis with a linear mixed-effects model. The pedigree information on the Yup’ik participants was used to create a kinship correlation matrix with the Coxme R package (40), and the kinship matrix was incorporated in a linear mixed-effects regression analysis that was performed by using the lmekin function from the same package.
Correlations with serum 25(OH)D3 concentration.
To determine associations with serum 25(OH)D3 concentration, a maximum likelihood mixed-effects multiple linear regression analysis was performed. To avoid spurious associations, the kinship correlation matrix was included to account for the random effects of the nonindependence of samples resulting from any familial relationships. The fixed effects of the model included cofactors found to be significantly associated with serum 25(OH)D3 concentration in previous studies (14, 20, 41). These included age (stratified by NHANES age categories), gender (binary), BMI (continuous), yearly quarter of sample collection (factored), recessive CYP2R1 rs11023374 genotype (binary), and log10 (δ15N value) (continuous, by a unit of 0.1). Inland or coastal geography of each community was also included as a binary variable. The previously mentioned lmekin function in the Coxme R package was used to evaluate the mixed-effects model with the use of maximum likelihood analysis (40). For this exploratory analysis, covariates were determined as predictors of serum 25(OH)D3 concentration if the significance of its coefficient surpassed P < 0.05. Samples that were missing data were excluded.
Sinusoidal model analysis.
A sinusoidal model previously developed to fit the seasonal pattern of 25(OH)D3 concentration (42, 43) was used to model the mean serum 25(OH)D3 concentration over the course of the year. The model was developed in a population of persons aged 45–84 y living in 6 communities across the lower 48 states of the United States (42). The model was fit to the overall Yup’ik data and adjusted for age or diet. Sinusoidal model analysis was performed with the cosinor.lm function of the Cosinor R package (44).
Linear regression with a subset of unrelated participants.
A subset of 526 unrelated participants was selected by removing individuals from the kinship matrix who were related to others by the third degree or closer. Simple linear regression with only these participants was performed by using R (35) to determine variability in serum 25(OH)D3 concentrations attributable to covariates. t tests were used to compare the demographic characteristics of the unrelated subset to the full data set.
Results
Population demographic characteristics and summary statistics.
The complete data set included 743 individuals; their demographic characteristics and summary statistics are described in Table 1. The distributions of serum 25(OH)D3 concentrations and δ15N values, both untransformed and transformed, are shown in Supplemental Figure 1. Serum 25(OH)D3 concentrations ranged from 6.0 to 68.7 ng/mL.
TABLE 1.
Demographic characteristics of Alaska Native (Yup’ik) participants1
| Time of collection2 | n | Age, y | BMI, kg/m2 | RBC log10(δ15N value) | Serum 25(OH)D33, ng/mL | Males, n | Females, n | Inland villages, n | Coastal villages, n |
| August–October | 167 | 40.9 ± 20.2 | 25.8 ± 5.3 | 0.934 ± 0.053 | 36.7 ± 12.1 | 79 | 88 | 117 | 50 |
| November–January | 310 | 35.5 ± 17.1 | 26.2 ± 5.8 | 0.939 ± 0.066 | 30.9 ± 13.0 | 171 | 139 | 161 | 149 |
| February–April | 48 | 35.1 ± 20.4 | 26.3 ± 6.0 | 0.945 ± 0.058 | 25.6 ± 12.9 | 19 | 29 | 0 | 48 |
| May–July | 218 | 34.7 ± 16.5 | 26.6 ± 6.2 | 0.935 ± 0.057 | 27.4 ± 10.6 | 121 | 97 | 63 | 155 |
| Total | 743 | 36.4 ± 18.0 | 26.2 ± 5.8 | 0.937 ± 0.060 | 30.8 ± 12.6 | 390 | 353 | 341 | 402 |
Values are means ± SDs unless otherwise indicated. Study participants all self-identified as Yup’ik. The δ15N value is a biomarker of traditional food intake as defined in Methods.
Participants were stratified by seasonal quarter of data collection. Seasons, as defined by the range of 3 mo listed, were determined by sunlight exposure and lag time of changes in serum concentrations after vitamin D synthesis.
25(OH)D3, 25-hydroxycholecalciferol.
Adjusted statistics and analyses.
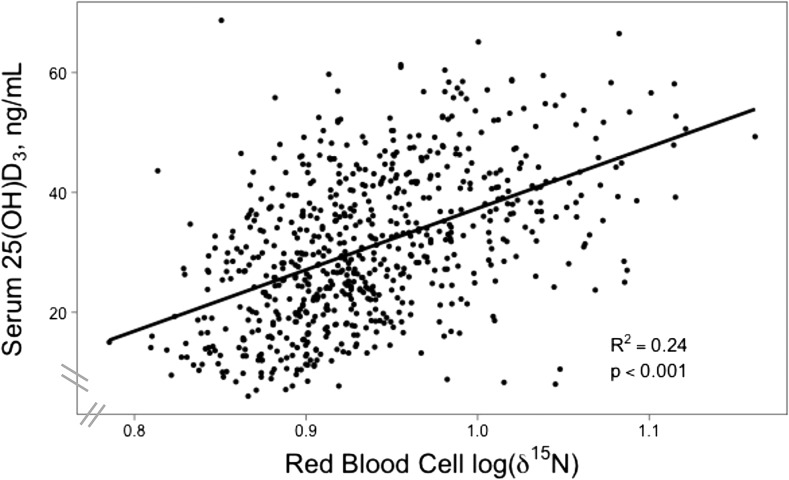
Summary statistics of the sample, adjusted for kinship coefficients between participants, are presented in Table 2. Overall, participants with dietary marker RBC δ15N values less than the median 0.927 log10(δ15N value) had significantly lower BMI, age, and serum 25(OH)D3 concentrations than participants with dietary marker values greater than or equal to the median (P < 0.05). Younger participants, compared with older participants, stratified by age <40 y and ≥40 y had significantly lower BMI, δ15N values, and serum 25(OH)D3 concentrations. A regression of the serum concentration of 25(OH)D3 by log10(δ15N value) is presented in Figure 1 and indicates a positive linear relation (R2 = 0.24, P < 0.0001). Plots of the serum concentration of 25(OH)D3 and log10(δ15N value) by decade of age are presented in Supplemental Figure 2. The mean serum concentration of 25(OH)D3 showed significant differences by season of sample collection (P < 0.0001), but the log10(δ15N value) did not vary by season of sample collection (P = 0.69).
TABLE 2.
Summary of demographic characteristics and comparative analyses adjusted for kinship correlation among Alaska Native (Yup'ik) participants1
| n | Age, y | Age-adjusted P2 | B MI, kg/m2 | BMI-adjusted P | RBC log10(δ15N value) | RBC log10(δ15N value)−adjusted P | Serum 25(OH)D3, ng/mL | 25(OH)D3-adjusted P | |
| RBC log10 (δ15N value) | <0.0001 | 7.3 × 10−10 | <0.0001 | <0.0001 | |||||
| <0.927 | 353 | 25.7 ± 0.6 | 24.8 ± 0.3 | 0.890 ± 0.002 | 25.7 ± 0.6 | ||||
| ≥0.927 | 355 | 36.5 ± 0.9 | 27.5 ± 0.4 | 0.984 ± 0.003 | 36.5 ± 0.9 | ||||
| Community location | 2.2 × 10−11 | 0.39 | 1.0 × 10−11 | 3.3 × 10−11 | |||||
| Coastal | 402 | 28.1 ± 0.7 | 26.0 ± 0.3 | 0.951 ± 0.003 | 28.1 ± 0.7 | ||||
| Inland | 341 | 34.4 ± 1.0 | 26.4 ± 0.2 | 0.920 ± 0.005 | 34.4 ± 0.9 | ||||
| rs11023374 genotype | 6.7 × 10−3 | 0.78 | 0.39 | 6.7 × 10−3 | |||||
| Recessive | 32 | 25.0 ± 2.3 | 26.5 ± 1.0 | 0.929 ± 0.011 | 25.0 ± 2.2 | ||||
| Dominant | 698 | 31.3 ± 0.5 | 26.2 ± 0.4 | 0.938 ± 0.019 | 31.3 ± 0.5 | ||||
| Age | <0.0001 | 2.9 × 10−9 | <0.0001 | ||||||
| <40 y | 441 | 23.4 ± 0.4 | 25.2 ± 0.3 | 0.913 ± 0.003 | <0.0001 | 25.7 ± 0.5 | |||
| ≥40 y | 302 | 32.2 ± 0.7 | 27.7 ± 0.4 | 0.853 ± 0.004 | 39.1 ± 0.8 | ||||
| Gender | 1.3 × 10−2 | <0.0001 | <0.0001 | 1.6 × 10−2 | |||||
| Male | 390 | 30.0 ± 0.7 | 24.9 ± 0.4 | 0.919 ± 0.004 | 30.1 ± 0.9 | ||||
| Female | 353 | 32.4 ± 0.9 | 27.7 ± 0.3 | 0.957 ± 0.003 | 32.3 ± 0.7 | ||||
| Overall | 743 | 31.0 ± 0.5 | 26.2 ± 0.2 | 0.937 ± 0.002 | 31.1 ± 0.5 |
Values are means ± SEs of the estimate unless otherwise indicated. Mixed-effects model linear regression was performed to assess differences between subsets of participants, adjusted for a kinship matrix that reflects pedigree information relating all participants. The full set of participants (n = 743) was stratified by participant characteristics (as listed): RBC log10(δ15N value) split at the mean, inland or coastal geography of the community of participant residence, homozygous recessive genotype at rs11023374 in cytochrome P450 family 2, subfamily R, peptide 1 (CYP2R1), or at least 1 reference allele, age split at age 40 y, and gender. The δ15N value is a biomarker of traditional food intake as defined in Methods. 25(OH)D3, 25-hydroxycholecalciferol.
z Scores are based on mixed-effects model linear regressions with outcomes data being mean values of age, BMI, log10(δ15N value), and serum 25(OH)D3 concentration between subsets of participants. P values were derived from a likelihood ratio test.
FIGURE 1.
Concentration of serum 25(OH)D3 by RBC log10(δ15N value) in adult Alaska Native (Yup’ik) people [n = 689 participants with data for both serum 25(OH)D3 concentration and RBC log10(δ15N value). Least-squares linear regression, with goodness of fit indicated by the R2 regression coefficient, was used. The P value, as calculated by a likelihood ratio test, was <0.0001. The δ15N value represents a biomarker of traditional food intake as defined in Methods. 25(OH)D3, 25-hydroxycholecalciferol.
Overall, 22.9% of participants had serum 25(OH)D3 concentrations below the 20-ng/mL threshold for insufficiency, as defined by the Institute of Medicine. Among participants in the lower half of nitrogen isotope ratios, 34.8% had insufficient serum 25(OH)D3 concentrations, whereas among participants in the upper half of nitrogen isotope ratios, only 8.7% were insufficient.
CYP2R1, DHCR7, and GC population genotyping.
The frequencies of minor alleles at each SNP in this Yup’ik study population are listed in Supplemental Table 1, adjusted for kinship between study participants. The association of each SNP with serum 25(OH)D3 concentration is also shown. The SNP rs11023374 in CYP2R1 was significantly associated with serum 25(OH)D3 concentrations (P = 0.009), as were both the rs4588 (P < 0.0001) and the rs7041 (P = 0.011) SNPs in GC. No other SNPs were significantly associated with serum 25(OH)D3 concentration. Season did not modify the association of any SNP with serum 25(OH)D3 concentration. All of the SNPs were in Hardy-Weinberg equilibrium.
Three SNPs in CYP2R1—rs10741657, rs2060793, and rs1993116—were in high LD in this Yup’ik study population (r2 = 0.97). The same 3 SNPs were in moderate LD with the fourth SNP, rs11023374 (r2 = 0.35) (Supplemental Figure 3A). An LD block reported for other populations (15) was also seen with the 4 SNPs informing the DHCR7 haplotypes: r2 between 0.94 and 0.97 for all pairwise comparisons of all 4 SNPs (Supplemental Figure 3B). The haplotype associated with lower DHCR7 activity was identified at 48.3% in the sample (Supplemental Table 2). The GC SNPs at rs7041 and rs4588 were in moderate LD (r2 = 0.30), but the GA haplotype (alleles found at those 2 positions) was identified in only one individual.
Modeled annual fluctuation in 25(OH)D3 concentrations.
On the basis of a sinusoidal model describing the seasonal fluctuations in serum 25(OH)D3 concentrations (42) (Supplemental Figure 4), the predicted mean serum 25(OH)D3 concentration for Yup’ik participants was 30.3 ng/mL (95% CI: 29.5, 31.2 ng/mL) over the course of the year, with a peak to trough difference of 15.9 ng/mL (95% CI: 12.5, 19.3 ng/mL). This model predicted a mean minimum concentration of 22.4 ng/mL and an average maximum concentration of 38.3 ng/mL throughout the year. The R2 of the fit was 0.11, indicating other significant sources of variation, but was consistent with the fit found in populations used in the development and validation of the model (43). Including diet as a covariate in the model improved the fit, with R2 = 0.29. Including age as a covariate in the model improved the fit, with R2 = 0.45. Twenty-four samples were excluded from the model due to missing 25(OH)D3 data.
Higher vitamin D intake, as evaluated by splitting the data set at the median n–3 FA biomarker value [log10(δ15N value)] and older age, as evaluated by splitting the data set at 40 y, were both significantly associated with higher 25(OH)D3 concentrations over the course of the year [P < 0.0001 for log10(δ15N value) split; P < 0.0001 for age split] in a model that did not adjust for the other covariates. The predicted annual mean 25(OH)D3 concentration for individuals with a log10(δ15N value) ≥0.927 was 35.6 ng/mL (95% CI: 31.0, 37.2 ng/mL) with a peak to trough difference of 12.8 ng/mL (95% CI: 8.4, 17.0 ng/mL).
Age and log10(δ15N value) were independently associated with the fit of the sinusoidal model (P < 0.0001), because both covariates were significant in the regression model that included both variables as predictors. Gender (P = 0.10) was not significantly associated with serum 25(OH)D3 concentration, but recessive genotype at rs11027334 (P = 0.03), coastal compared with inland geographic location of the community (P = 9 × 10−3), rs4588 genotype (P < 0.0001), and rs7041 genotype (P = 6 × 10−3) were significantly associated with lower yearly mean serum 25(OH)D3 concentrations in the sinusoidal model.
Seasonal analysis.
Participants were divided into seasonal quartiles on the basis of the distribution of serum 25(OH)D3 concentration over the course of a year. Peak serum 25(OH)D3 concentrations occurred in September and the trough was in March, as determined from a fit of the sinusoidal model. This result yielded a seasonal breakdown of low serum 25(OH)D3 concentrations in February, March, and April; increasing concentrations in May, June, and July; high concentrations in August, September, and October; and decreasing concentrations in November, December, and January. These divisions reflect the 6-wk lag time after seasonal patterns of sunlight exposure (45, 46).
Associations of covariates with serum 25(OH)D3 concentration.
On the basis of the significance threshold of P < 0.05 in a linear mixed-model regression, log10(δ15N value), age, gender, BMI, community location, homozygosity of the variant allele at CYP2R1 rs11023374, and season of blood draw were all significantly associated with serum 25(OH)D3 concentration after accounting for the kinship coefficients between participants (Table 3). On the basis of proportional variability explained by the kinship matrix, the heritability of serum 25(OH)D3 concentration in these study participants was estimated to be 0.46 after adjusting for age, diet, BMI, season, and community effects.
TABLE 3.
Associations of serum 25(OH)D3 concentration in Alaska Native (Yup’ik) people1
| Full model |
|||
| Characteristic | n | β-Coefficient | P2 |
| Age3 | |||
| <20 y | 157 | Ref | |
| 20–39 y | 247 | 2.7 ± 0.9 | 3.0 × 10−3 |
| 40–59 y | 177 | 8.7 ± 1.0 | <0.0001 |
| >59 y | 96 | 12.1 ± 1.4 | <0.0001 |
| Season4 | |||
| May–July | 208 | −4.1 ± 1.0 | 5.0 × 10−5 |
| August–October | 151 | Ref | |
| November–January | 283 | −3.0 ± 0.9 | 9.7 × 10−4 |
| February–April | 34 | −5.6 ± 1.7 | 6.8 × 10−4 |
| RBC log10(δ15N value)5 | 5.6 ± 0.5 | <0.0001 | |
| Gender | |||
| Male | 358 | 1.9 ± 0.7 | 6.5 × 10−3 |
| Female | 320 | Ref | |
| Village location6 | |||
| Coastal | 365 | Ref | |
| Inland | 312 | 7.8 ± 0.8 | <0.0001 |
| BMI7 | −1.1 ± 0.4 | 2.5 × 10−3 | |
| CYP2R1 rs110233748 | |||
| Homozygous variant | 28 | −3.8 ± 1.7 | 2.3 × 10−2 |
| At least 1 reference allele | 649 | Ref | |
Values are β-coefficients ± SEs from a linear mixed-effects model adjusted for a kinship matrix reflecting pedigree relationships of all participants with values for all included variables; n = 677. The numbers of participants with each predictor characteristic are listed. The outcome variable was serum 25(OH)D3 concentration (ng/mL). The significance threshold for inclusion in the model was set as P < 0.05. β-Coefficients for categorical data represent change in serum 25(OH)D3 concentration between a category and its reference group. β-Coefficients for continuous data represent change in serum 25(OH)D3 concentration for a change of 1 SD in the continuous variable. The δ15N value is a biomarker of traditional food intake as defined in Methods. CYP2R1, cytochrome P450 family 2, subfamily R, peptide 1; Ref, reference category for all discrete variables; 25(OH)D3, 25-hydroxycholecalciferol.
P values were derived from a likelihood ratio test.
Age grouping based on NHANES age groups.
Seasons of sample collection based on fluctuations of vitamin D exposure.
Values for RBC log10(δ15N value) reflect change in serum 25(OH)D3 concentration per 1 SD of log10(δ15N values). The SD of the log10(δ15N values) is 0.0605.
Community geography was determined by inland or coastal geography of the community of participant residence.
Values for BMI reflect change in serum 25(OH)D3 concentration per 1 SD of BMI. The SD of the BMI is 5.84.
CYP2R1 genotype determined as a homozygous variant allele or having at least 1 reference allele at single nucleotide polymorphism rs11023374.
Associations with serum 25(OH)D3 concentration based on maximum unrelated subjects.
In multiple linear regression that included only the 526 unrelated participants, age, log10(δ15N value), season, gender, BMI, community location, and genotype at CYP2R1 rs11023374 were found to be significantly associated with serum 25(OH)D3 concentration. These unrelated participants are representative of the entire data set (Supplemental Table 3). Together, the variables included in the regression explained 52.5% of the variability in serum 25(OH)D3 concentration of this sample set. Considered individually, season of blood draw accounted for 9.8%, age stratified by NHANES categories accounted for 33.7%, and log10(δ15N value) accounted for 20.7% of the variability (Table 4). In a combined analysis, age stratified by NHANES categories and log10(δ15N value) accounted for 36.8% of variability in 25(OH)D3.
TABLE 4.
Associations of serum 25(OH)D3 concentration in an unrelated subset of Alaska Native (Yup’ik) people1
| Full model |
||||
| Characteristic | n | β-Coefficient2 | P | Variability explained,3R2 |
| Fully adjusted model4 | 526 | (0.53) | ||
| Age5 | (0.34) | |||
| <20 y | Ref | |||
| 20–39 y | 2.7 ± 1.1 | 7.1 × 10−3 | ||
| 40–59 y | 8.6 ± 1.3 | 2.7 × 10−10 | ||
| >59 y | 12.5 ± 1.6 | 6.6 × 10−13 | ||
| Season | (0.10) | |||
| May–July | 150 | −4.9 ± 1.2 | 9.9 × 10−6 | |
| August–October | 136 | Ref | ||
| November–January | 207 | −4.1 ± 1.0 | 2.6 × 10−5 | |
| February–April | 33 | −6.9 ± 1.9 | 8.9 × 10−4 | |
| RBC log10(δ15N value)6 | 5.0 ± 0.5 | <2 × 10−16 | (0.21) | |
| Gender | (0.00) | |||
| Male | 282 | 2.3 ± 0.9 | 7.1 × 10−3 | |
| Female | 244 | Ref | ||
| Village location | (0.06) | |||
| Coastal | 270 | 7.6 ± 0.9 | 1.6 × 10−15 | |
| Inland | 256 | Ref | ||
| BMI7 | −0.6 ± 0.5 | 0.13 | (0.01) | |
| CYP2R1 rs11023374 | (0.02) | |||
| Homozygous variant | 23 | −4.5 ± 2.0 | 1.5 × 10−2 | |
| At least 1 reference allele | 503 | Ref | ||
| Age and RBC log10(δ15N value)8 | (0.37) | |||
Variability in 25(OH)D3 concentration is explained by predictors in a multiple linear regression. Participants included in this regression represent an unrelated subset (n = 526) of the overall data set (n = 743). This subset included only individuals unrelated to the third degree, as determined by using pedigree information. The significance of the association of each variable with 25(OH)D3 concentration is indicated by the P value and determined by using a likelihood ratio test. β-Coefficients for categorical data represent change in serum 25(OH)D3 concentration between a category and its reference group. β-Coefficients for continuous data represent change in serum 25(OH)D3 concentration for a change of 1 SD in the continuous variable. The δ15N value is a biomarker of traditional food intake as defined in Methods. CYP2R1, cytochrome P450 family 2, subfamily R, peptide 1; Ref, reference category for all discrete variables; 25(OH)D3, 25-hydroxycholecalciferol.
Values are β-coefficients ± SEs from a multiple linear regression model. β-Coefficients are determined as change in serum 25(OH)D3 concentration (ng/mL) per unit of change in the predictor variable, when all other predictor variables are held constant.
The goodness of fit of a multiple linear regression, on a scale of 0 to 1, as reflected in the R2 value.
The fully adjusted model of serum 25(OH)D3 concentration as the outcome variable and including all predictors in the table: age (NHANES grouping), season of sample collection, RBC log10(δ15N value), gender, village location, BMI, and genotype at CYP2R1 rs11023374.
Linear regression was used to regress each listed predictor individually with outcome data of serum 25(OH)D3 concentration. These predictors in each model are listed.
Values for the RBC log10(δ15N value) reflect change in serum 25(OH)D3 concentration per 1 SD of log10(δ15N values). The SD of the log10(δ15N values) is 0.0595.
Values for BMI reflect change in serum 25(OH)D3 concentration per 1 SD of BMI. The SD of the BMI is 5.67.
Multiple regression model of 25(OH)D3 concentration outcome and including 2 predictors: RBC log10(δ15N value) and age, divided into NHANES categories.
Repeating the full model linear regression with a subset of unrelated samples that had genotypes for the 2 SNPs in GC (n = 438), the TA haplotype was significantly associated with serum 25(OH)D3 concentration (P = 1.4 × 10−4) compared with the reference haplotype GC, but the other 2 haplotypes (GA and TC) were not significantly associated with serum 25(OH)D3 concentration. GC haplotype explained 2.8% of variability in serum 25(OH)D3 concentrations in this subset.
Discussion
We characterized serum 25(OH)D3 concentrations in a cross-section of Yup’ik people living in the Y-K Delta to better understand the role of diet, genetic variation, and seasonal sun exposure in determining vitamin D concentrations in that population. The mean (±SD) serum 25(OH)D3 concentration among the Yup’ik study participants (31.0 ± 0.1 ng/mL) was higher than the mean reported for many populations (47), including healthy individuals living at lower latitudes (25.1 ± 0.4 ng/mL) (43), who would arguably have greater opportunity to synthesize vitamin D from sunlight throughout the year. A greater intake of traditional foods rich in fats from marine mammals and fish, as measured by log10(δ15N value) validated with 24-h diet recalls, was associated with a higher serum 25(OH)D3 concentration in the Yup’ik study population (25, 48, 49). Not surprisingly, diet appears to be a more important source of vitamin D than the seasonal contribution of sunlight in the Yup’ik population, with older participants consuming more of the traditional foods rich in vitamin D (50). Although the direction of the association varies by region (47), serum 25(OH)D3 concentration has been associated with age in other populations and has been linked to age-related lifestyle differences that affect the amount of time outside and exposed to sunlight (47). Because the association with age is found in the Yup’ik study participants, even after adjusting for diet, there are likely age-related differences in lifestyle and diet that are not being captured in this study. Age may track aspects of the traditional diet that are not captured well by δ15N, such as consumption of seal oil, which contains no nitrogen.
Although SNPs in CYP2R1 have been associated with serum 25(OH)D3 concentrations in other populations, only rs11023374 was associated with serum 25(OH)D3 concentrations in the Yup’ik study participants. The SNP rs11023374 was in weaker LD compared with the other SNPs, suggesting that it may be more closely linked with the causal variant. The strong LD patterns between the other SNPs are similar to what has been found in other populations (1, 13). Even so, genotype at rs11023374 accounted for only 1.5% of variability in serum 25(OH)D3 concentrations. Furthermore, the DHCR7 haplotype associated with increased production of vitamin D from sunlight was found at a lower frequency in the Yup’ik participants than in other populations living at northern latitudes (15), and none of the SNPs informing DHCR7 haplotypes were associated with serum 25(OH)D3 concentration. These results suggest that the haplotype does not confer a particular advantage for maintaining vitamin D concentrations in this population and that dietary sources were sufficient historically, or that LD with causal SNPs was broken. The largest genetic contribution to serum 25(OH)D3 concentration was the haplotype from the 2 nonsynonymous coding SNPs in GC (encodes the plasma vitamin D binding protein), which explained an additional 2.8% of variability. As in other populations, only 3 of the 4 possible haplotypes were found in the Yup’ik study participants (31). Although the contribution of these 10 genetic variants was found to be minor, the heritability estimate of 0.46 for serum 25(OH)D3 concentration suggests that uncharacterized genetic variation should still be considered for associations with disease risk and vitamin D status. Although the heritability estimate must be interpreted with caution, because it does not account for the shared environment of participants, it suggests that CYP2R1, DHCR7, GC, and other candidate genes in the vitamin D pathway (10), should be sequenced to identify any novel variation or LD patterns in the Yup’ik population.
Consistent with the results of the regression analysis, modeling results indicated that greater consumption of the traditional diet provides sufficient vitamin D concentrations over the entire year. Although the specific source may vary by season, traditional marine foods are available year round, either fresh, frozen, or dried, as indicated by the consistency of the dietary marker throughout the year. However, the traditional diet of fish, marine mammals, birds, land mammals, and berries is being replaced by nutrient-poor imported foods in many indigenous arctic communities, especially among youth and young adults. Studies in Inuit communities in Nunavut, Canada, found similar patterns of decreased consumption of the traditional diet among younger generations and linked this transition with lower vitamin D concentrations and increased incidence of illnesses associated with vitamin D deficiency (4, 21). Deviation from the traditional diet among younger individuals is prompting public health concern, because it is thought to increase the risk of illnesses associated with vitamin D deficiency. Future studies are needed to understand the health impacts of these dietary patterns and low vitamin D concentrations in Yup’ik communities.
In contrast to findings from other populations (51), female Yup’ik participants had higher serum 25(OH)D3 concentrations than did male participants, likely reflecting differences in traditional food intake estimated from δ15N values (24). After adjustment for diet, however, male participants had higher concentrations of serum 25(OH)D3, likely because of spending more time outside. Whereas nitrogen isotope ratios were higher in coastal communities and in women, suggesting greater intakes of traditional marine foods, participants from inland communities had higher concentrations of serum 25(OH)D3 and community location explained 6.4% of serum 25(OH)D3 concentration variability. One possible explanation is that certain foods, such as lush fish (a white fish, also known as Burbot, found in local rivers), have higher vitamin D content in relation to δ15N value and are consumed more frequently in inland communities.
Strengths of this study are that it combined dietary, genetic, seasonal, and demographic measures to more completely characterize sources of variability in serum 25(OH)D3 concentrations and risk of vitamin D deficiency in a population with highly variable serum 25(OH)D3 concentrations and who are experiencing a transition in dietary patterns. Moreover, it used an objective measurement of dietary intake of vitamin D and sampled concentrations throughout the year in a region with drastic changes in sunlight availability. A limitation of this study is that it did not evaluate sources of vitamin D2, such as supplementation in market foods, which would contribute to overall vitamin D status and to the risk of deficiency and insufficiency. Other important limitations of these data are that they are cross-sectional and that we have not directly measured sunlight exposure. A longitudinal study is needed to confirm the seasonal variation in serum 25(OH)D3 concentration within individuals, although this variation would be expected on the basis of studies in other populations (42, 43). Furthermore, due to convenience sampling, the data set contains an abundance of samples collected in transition seasons between maximum and minimum annual serum 25(OH)D3 concentrations (May–July and November–January) and has especially undersampled the low season (February–April). As a result, mean serum 25(OH)D3 concentrations may be skewed high.
In summary, mean serum 25(OH)D3 concentrations were high relative to other populations and were highly correlated with dietary sources of vitamin D; however, they were also highly variable, with a larger portion of younger people insufficient and deficient for vitamin D concentrations compared with older participants. A public health campaign to raise awareness about potential health benefits of a traditional diet could possibly increase mean serum 25(OH)D3 concentrations and maintain stability during periods of low sunlight exposure. Efforts to increase serum 25(OH)D3 concentrations could also help reverse recent increases in the incidence of vitamin D deficiency, such as among young pregnant women in AN communities, who are seeing a re-emergence of illnesses associated with vitamin D deficiency (16). Although supplementation and fortification of foods with vitamin D may reduce the risk of vitamin D deficiency in the Yup’ik population (4), the promotion of a traditional diet could be more accessible and affordable, as well as have positive health impacts that go beyond increasing vitamin D concentrations in these communities.
Acknowledgments
We thank Barbara Kavanaugh, Program Manager for the Northwest-Alaska Pharmacogenomics Research Network, for directing programmatic collaborations. We also thank Joseph Klejka, Medical Director of the Yukon-Kuskokwim Health Corporation, for his input and guidance. JY, TAT, BBB, and KET designed the research; ZW, DMO, SEH, JB, JP, PLS, JMT, and BBB conducted the research; HWW and HKT provided the essential materials; AEF analyzed the data and wrote the manuscript; and KET had primary responsibility for the final content. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: AN, Alaska Native; CYP2R1, cytochrome P450 family 2, subfamily R, peptide 1; DHCR7, 7-dehydrocholesterol reductase; GC, vitamin D binding protein; LD, linkage disequilibrium; SNP, single nucleotide polymorphism; UW, University of Washington; Y-K Delta, Yukon-Kuskokwim River Delta; YKHC, Yukon Kuskokwim Health Corporation; 25(OH)D, 25-hydroxyvitamin D; 25(OH)D3, 25-hydroxycholecalciferol.
References
- 1.Ramos-Lopez E, Bruck P, Jansen T, Herwig J, Badenhoop K. CYP2R1 (vitamin D 25-hydroxylase) gene is associated with susceptibility to type 1 diabetes and vitamin D levels in Germans. Diabetes Metab Res Rev 2007;23:631–6. [DOI] [PubMed] [Google Scholar]
- 2.Levin GP, Robinson-Cohen C, Boer IHD, Houston DK, Lohman K, Liu Y, Kritchevsky SB, Cauley JA, Tanaka T, Ferrucci L, et al. . Genetic variants and associations of 25-hyroxyvitamin D concentrations with major clinical outcomes. JAMA 2012;308:1898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, He JW, Fu WZ, Zhang CQ, Zhang ZL. An analysis of the association between the vitamin D pathway and serum 25-hydroxyvitamin D levels in a healthy Chinese population. J Bone Miner Res 2013;28:1784–92. [DOI] [PubMed] [Google Scholar]
- 4.Sharma S, Barr AB, Macdonald HM, Sheehy T, Novotny R, Corriveau A. Vitamin D deficiency and disease risk among aboriginal Arctic populations. Nutr Rev 2011;69:468–78. [DOI] [PubMed] [Google Scholar]
- 5.Berry D, Hypponen E. Determinants of vitamin D status: focus on genetic variations. Curr Opin Nephrol Hypertens 2011;20:331–6. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann B, Meurer M. Vitamin D metabolism. Dermatol Ther (Heidelb) 2010;23:2–12. [DOI] [PubMed] [Google Scholar]
- 7.Holick MF, Binkley NC, Bischoff-Ferrari HA, Gordon CM, Hanley DA, Heaney RP, Murad MH, Weaver CM; Endocrine Society. Evaluation, treatment, and prevention of vitamin D deficiency: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab 2011;96:1911–30. [DOI] [PubMed] [Google Scholar]
- 8.Institute of Medicine. Dietary Reference Intakes for calcium and vitamin D. Washington (DC): National Academies Press; 2011. [PubMed] [Google Scholar]
- 9.Bolland MJ, Grey AB, Ames RW, Mason BH, Horne AM, Gamble GD, Reid IR. The effects of seasonal variation of 25-hydroxvitamin D and fat mass on a diagnosis of vitamin D sufficiency. Am J Clin Nutr 2007;86:959–64. [DOI] [PubMed] [Google Scholar]
- 10.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, Jacobs EJ, Ascherio A, Helzlsouer K, Jacobs K, et al. . Genome-wide association study of circulating vitamin D levels. Hum Mol Genet 2010;19:2739–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dastani Z, Li R, Richards B. Genetic regulation of vitamin D levels. Calcif Tissue Int 2013;92:106–17. [DOI] [PubMed] [Google Scholar]
- 12.Hiraki LT, Major JM, Chen C, Cornelis MC, Hunter DJ, Rimm EB, Simon KC, Weinstein SJ, Purdue MP, Yu K, et al. . Exploring the genetic architecture of circulating 25-hydroxyvitamin D. Genet Epidemiol 2013;37:92–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang TJ, Zhang F, Richards JB. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet 2010;376:9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman CD, Meyers KJ, Iyengar SK, Liu Z, Karki CK, Igo RP Jr, Truitt B, Robinson J, Sarto GE, Wallace R, et al. . Vitamin D intake and season modify the effects of the GC and CYP2R1 genes on 25-hydroxyvitamin D concentrations. J Nutr 2013;143:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuan V, Martineau AR, Griffiths CJ, Hypponen E, Walton R. DHCR7 mutation linked to higher vitamin D status allowed early human migration to northern latitudes. BMC Evol Biol 2013;13:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singleton R, Lescher R, Gessner BD, Benson M, Bulkow L, Rosenfeld J, Thomas T, Holman RC, Haberling D, Bruce M, et al. . Rickets and vitamin D deficiency in Alaska Native children. J Pediatr Endocrinol Metab 2015;28:815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gessner BD, Plotnik J, Muth PT. 25-Hydroxyvitamin D levels among healthy children in Alaska. J Pediatr 2003;143:434–7. [DOI] [PubMed] [Google Scholar]
- 18.Yin L, Grandi N, Raum E, Haug U, Arndt V, Brenner H. Meta-analysis: longitudinal studies of serum vitamin D and colorectal cancer risk. Aliment Pharmacol Ther 2009;30:113–25. [DOI] [PubMed] [Google Scholar]
- 19.Perdue DG, Perkins C, Jackson-Thompson J, Coughlin SS, Ahmed F, Haverkamp DS, Jim MA. Regional differences in colorectal cancer incidence, stage, and subsite among American Indians and Alaska Natives, 1999–2004. Cancer 2008;113:1179–90. [DOI] [PubMed] [Google Scholar]
- 20.Luick B, Bersamin A, Stern JS. Locally harvested foods support serum 25-hydroxyvitamin D sufficiency in an indigenous population of Western Alaska. Int J Circumpolar Health 2014 Mar 20;73. doi: 10.3402/ijch.v73.22732. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frost JT, Hill L. Vitamin D deficiency in a nonrandom sample of southeast Alaska Natives. J Acad Nutr Diet 2008;108:1508–11. [DOI] [PubMed] [Google Scholar]
- 22.Kenny DE, O’Hara TM, Chen TC, Lu Z, Tian X, Holick MF. Vitamin D content in Alaskan Arctic zooplankton, fishes, and marine mammals. Zoo Biol 2004;23:33–43. [Google Scholar]
- 23.O’Brien DM, Kristal AR, Jeannet MA, Wilkinson MJ, Bersamin A, Luick B. Red blood cell delta15N: a novel biomarker of dietary eicosapentaenoic acid and docosahexaenoic acid intake. Am J Clin Nutr 2009;89:913–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nash SH, Bersamin A, Kristal AR, Hopkins SE, Church RS, Pasker RL, Luick BR, Mohatt GV, Boyer BB, O’Brien DM. Stable nitrogen and carbon isotope ratios indicate traditional and market food intake in an indigenous circumpolar population. J Nutr 2012;142:84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bersamin A, Zidenberg-Cherr S, Stern JS, Luick BR. Nutrient intakes are associated with adherence to a traditional diet among Yup’ik Eskimos living in remote Alaska Native communities: the CANHR study. Int J Circumpolar Health 2007;66:62–70. [DOI] [PubMed] [Google Scholar]
- 26.Boyer BB, Mohatt GV, Lardon C, Plaetke R, Luick BR, Hutchison SH, Antunez de Mayolo G, Ruppert E, Bersamin A. Building a community-based participatory research center to investigate obesity and diabetes in Alaska Natives. Int J Circumpolar Health 2005;64:281–90. [DOI] [PubMed] [Google Scholar]
- 27.Boedeker B, Foster S. 2010 Census counts: American Indians/Alaska Natives alone or in combination with one or more other races, Alaska. Alaska Area Native Health Service, Anchorage (AK); 2011. [Google Scholar]
- 28.Fohner AE, Robinson R, Yracheta J, Dillard DA, Schilling B, Khan B, Hopkins S, Boyer BB, Black J, Wiener H, et al. . Variation in genes controlling warfarin disposition and response in American Indian and Alaska Native people: CYP2C9, VKORC1, CYP4F2, CYP4F11, GGCX. Pharmacogenet Genomics 2015;25:343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens M, Smith NW, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001;68:978–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 2003;73:1162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powe CE, Evans MK, Wenger J, Zonderman AB, Berg AH, Nalls M, Tamez H, Zhang D, Bhan I, Karumanchi SA, et al. . Vitamin D-binding protein and vitamin D status of black Americans and white Americans. N Engl J Med 2013;369:1991–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett J, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–5. [DOI] [PubMed] [Google Scholar]
- 33.Bourgain C, Zhang Q. Kinship and inbreeding coefficients computation in general pedigrees. 1.1 ed Boston: Free Software Foundation; 2009. [Google Scholar]
- 34.McPeek MS, Wu X, Ober C. Best linear unbiased allele-frequency estimation in complex pedigrees. Biometrics 2004;60:359–67. [DOI] [PubMed] [Google Scholar]
- 35.Team RCR. A language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2014. [Google Scholar]
- 36.O’Brien DM. Stable isotope ratios as biomarkers of diet for health research. Annu Rev Nutr 2015;35:565–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nash SH, Kristal AR, Boyer BB, King IB, Metzgar JS, O’Brien DM. Relation between stable isotope ratios in human red blood cells and hair: implications for using the nitrogen isotope ratio of hair as a biomarker of eicosapentaenoic acid and docosahexaenoic acid. Am J Clin Nutr 2009;90:1642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Senn T, Kalhorn T, Zheng XE, Zheng S, Davis CL, Hebert MF, Lin YS, Thummel KE. Simultaneous measurement of plasma vitamin D(3) metabolites, including 4beta,25-dihydroxyvitamin D(3), using liquid chromatography-tandem mass spectrometry. Anal Biochem 2011;418:126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phinney KW, Bedner M, Tai SS, Vamathevan VV, Sander LC, Sharpless KE, Wise SA, Yen JH, Schleicher RL, Chaudhary-Webb M, et al. . Development and certification of a standard reference material for vitamin D metabolites in human serum. Anal Chem 2012;84:956–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Therneau T. Coxme: mixed effects Cox models. 2012 [cited 2014 Oct 10]. Available from: http://CRAN.R-project.org/package=coxme.
- 41.Chen TC, Chimeh F, Lu Z, Mathieu J, Person KS, Zhang A, Kohn N, Martinello S, Berkowitz R, Holick MF. Factors that influence the cutaneous synthesis and dietary sources of vitamin D. Arch Biochem Biophys 2007;460:213–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sachs MC, Shoben A, Levin GP, Robinson-Cohen C, Hoofnagle AN, Swords-Jenny N, Ix JH, Budoff M, Lutsey PL, Siscovick DS, et al. . Estimating mean annual 25-hydroxyvitamin D concentrations from single measurements: the Multi-Ethnic Study of Atherosclerosis. Am J Clin Nutr 2013;97:1243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoben AB, Kestenbaum B, Levin G, Hoofnagle AN, Psaty BM, Siscovick DS, de Boer IH. Seasonal variation in 25-hydroxyvitamin D concentrations in the cardiovascular health study. Am J Epidemiol 2011;174:1363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sachs M, (2014). Package “Cosinor”—tools for estimating and predicting the Cosinor model [cited 2014 Oct 10]. Available from: http://github.com/sachsmc/cosi.
- 45.Lucas RM, Ponsonby AL, Dear K, Valery PC, Taylor B, van der Mei I, McMichael AJ, Pender MP, Chapman C, Coulthard A, et al. . Vitamin D status: multifactorial contribution of environment, genes and other factors in healthy Australian adults across a latitude gradient. J Steroid Biochem Mol Biol 2013;136:300–8. [DOI] [PubMed] [Google Scholar]
- 46.Nair-Shalliker V, Clements M, Fenech M, Armstrong BK. Personal sun exposure and serum 25-hydroxy vitamin D concentrations. Photochem Photobiol 2013;89:208–14. [DOI] [PubMed] [Google Scholar]
- 47.Hilger J, Friedel A, Herr R, Rausch T, Roos F, Wahl DA, Pierroz DD, Weber P, Hoffmann K. A systematic review of vitamin D status in populations worldwide. Br J Nutr 2014;111:23–45. [DOI] [PubMed] [Google Scholar]
- 48.Ryman TK, Boyer BB, Hopkins S, Philip J, O’Brien D, Thummel K, Austin MA. Characterising the reproducibility and reliability of dietary patterns among Yup’ik Alaska Native people. Br J Nutr 2015;113:634–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryman TK, Austin MA, Hopkins S, Philip J, O’Brien D, Thummel K, Boyer BB. Using exploratory factor analysis of FFQ data to identify dietary patterns among Yup’ik people. Public Health Nutr 2014;17:510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bersamin A, Luick BR, Ruppert E, Stern JS, Zidenberg-Cherr S. Diet quality among Yup’ik Eskimos living in rural communities is low: the Center for Alaska Native Health Research Pilot Study. J Am Diet Assoc 2006;106:1055–63. [DOI] [PubMed] [Google Scholar]
- 51.Holick MF. High prevalence of vitamin D inadequacy and implications for health. Mayo Clin Proc 2006;81:353–73. [DOI] [PubMed] [Google Scholar]