Significance
Unlike other ErbB receptors, human epidermal growth factor receptor 2 (HER2) does not generally become internalized after activation but, instead, remains on the cell surface to signal for prolonged periods. This property is thought to contribute to HER2’s ability to transform cells when overexpressed. The current study demonstrates that HER2’s resistance to endocytosis depends on the presence of the calcium pump, plasma membrane calcium ATPase2 (PMCA2), in specific membrane signaling domains in which intracellular calcium must be kept low to permit continued HER2 biochemical signaling. The dramatic reduction of mammary tumors in mouse mammary tumor virus (MMTV)-Neu mice in the absence of PMCA2 demonstrates its importance in supporting the development of breast tumors. Therefore, targeting interactions between PMCA2 and HER2 may offer therapeutic strategies for breast cancer.
Keywords: calcium pumps, ErbB2, receptor internalization, HSP-90, epidermal growth factor receptor
Abstract
In the lactating mammary gland, the plasma membrane calcium ATPase2 (PMCA2) transports milk calcium. Its expression is activated in breast cancers, where high tumor levels predict increased mortality. We find that PMCA2 expression correlates with HER2 levels in breast cancers and that PMCA2 interacts with HER2 in specific actin-rich membrane domains. Knocking down PMCA2 increases intracellular calcium, disrupts interactions between HER2 and HSP-90, inhibits HER2 signaling, and results in internalization and degradation of HER2. Manipulating PMCA2 levels regulates the growth of breast cancer cells, and knocking out PMCA2 inhibits the formation of tumors in mouse mammary tumor virus (MMTV)-Neu mice. These data reveal previously unappreciated molecular interactions regulating HER2 localization, membrane retention, and signaling, as well as the ability of HER2 to generate breast tumors, suggesting that interactions between PMCA2 and HER2 may represent therapeutic targets for breast cancer.
Plasma membrane calcium ATPases (PMCAs) are a family of ion pumps that transport calcium out of cells and maintain low resting intracellular calcium levels (1–3). PMCA2 (gene symbol Atp2b2) is highly expressed in the apical membrane of mammary epithelial cells only during lactation, where it has been shown to transport calcium into milk (4–6). After weaning, PMCA2 expression rapidly decreases, contributing to the initiation of programmed cell death and mammary gland involution (7, 8). PMCA2 is also expressed in breast cancers (8–10), and high levels of tumor PMCA2 expression predict increased mortality in patients (8).
Approximately 25–30% of invasive breast cancers overexpress human epidermal growth factor receptor 2 (HER2) as a result of amplification of the ERBB2 kinase gene (11–13), and overexpression of HER2 causes breast tumors in mouse mammary tumor virus (MMTV)-Neu transgenic mice (14). HER2 functions as a heterodimer with other ERBB family members, most commonly pairing with EGFR or human epidermal growth factor receptor 3 (HER3) in breast cancers (11, 13). For reasons that remain poorly understood, in contrast to other ERBB family members, which are internalized and degraded after stimulation, HER2 remains on the cell surface and continues to signal for prolonged periods (12, 15).
In this study, we describe a previously unrecognized function for PMCA2: supporting active HER2 signaling and HER2-mediated tumor formation. Our data suggest that PMCA2 interacts with HER2 within specific membrane domains and is required for HER2 expression, membrane retention, and signaling.
Results
PMCA2 and HER2 Are Coexpressed in Breast Cancers.
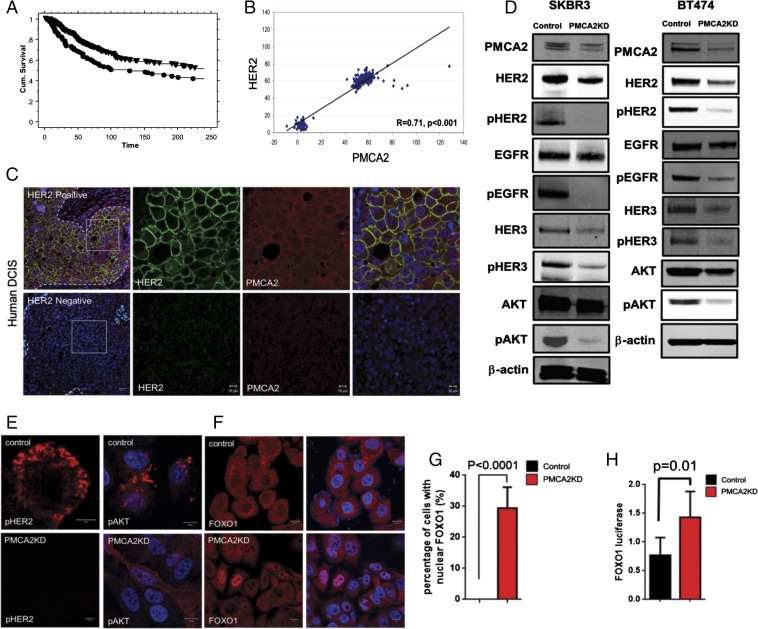
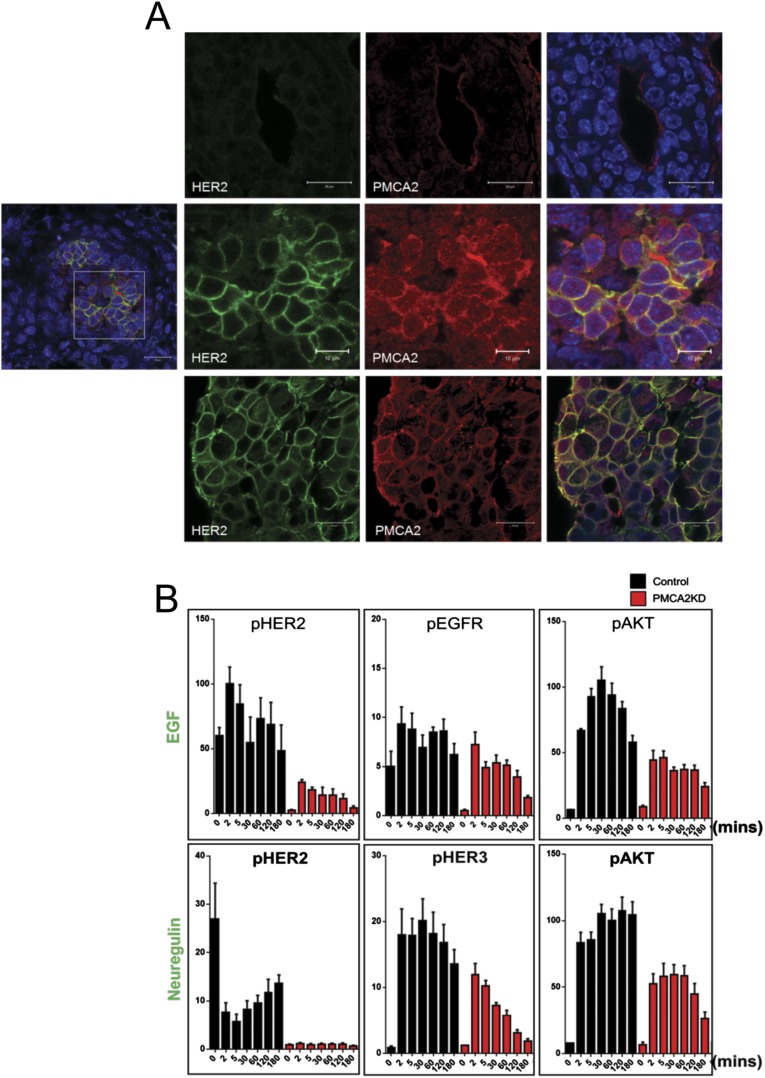
PMCA2 levels correlate with HER2 in breast tumors (8). To further explore potential interactions between PMCA2 and HER2, we analyzed their expression in a previously reported tissue microarray consisting of 652 breast cancers with a median 9 y of clinical follow-up (8, 16). Patients with the highest quartiles of both PMCA2 and HER2 expression had significantly shorter survival than patients whose tumors expressed lower levels of either protein (Fig. 1A). We also examined ATP2B2 (PMCA2) and ERBB2 (HER2) mRNA levels in a gene array study of a different cohort of 204 breast cancers of mixed subtypes (15% basal, 24% luminal A, 25% luminal B, 16% HER2, 20% normal-like) (17). As shown in Fig. 1B, there was a positive but bimodal correlation between the expression of the ATP2B2 and ERBB2 genes: one group expressed low levels of both genes, and another group had higher levels of both. We next performed immunofluorescence staining for both proteins in breast cancers. PMCA2 and HER2 were expressed at very low levels in wild-type mouse luminal epithelial cells (Fig. S1), but at much higher levels in hyperplasia and mammary tumors from MMTV-Neu mice (overexpressing HER2/Neu), where they colocalized at the cell membrane (Fig. S1). Similarly, in a series of 20 human ductal carcinoma in situ (DCIS) lesions, we found that all the HER2-positive, but none of the HER2-negative, samples expressed PMCA2. In HER2-positive DCIS, PMCA2 colocalized with HER2 at the cell membrane (Fig. 1C).
Fig. 1.
(A) Survival plots for patients whose tumors displayed the highest quartiles of both PMCA2 and HER2 immunofluorescence (Lower) compared with all other patients with lower tumor PMCA2 and/or HER2 (Upper). (B) Relationship between tumor PMCA2 and HER2 mRNA levels in 204 patients with breast cancer. (C) Representative immunofluorescence for HER2 and PMCA2 in HER2-positive (n = 16) or HER2-negative (n = 4) DCIS lesions. Boxed areas are magnified in right three panels. Panels on each end are merged images with DAPI staining. (D) Typical immunoblots for SKBR3 and BT474 cells stably transfected with a control shRNA (Control) or an shRNA directed at PMCA2 (PMCA2KD). (E) Immunostaining for pHER2 and pAKT in control or PMCA2KD SKBR3 cells. (F) Immunostaining for FOXO1 in control and PMCA2KD SKBR3 cells. (G) Quantification of nuclear staining for FOXO1 (150 cells in each of three individual experiments were scored). (H) Activity of a FOXO1-luciferase construct in control and PMCA2KD SKBR3 cells. (Scale bars, 10 μm.)
Fig. S1.
(A) Immunofluorescence costaining for HER2 (green) and PMCA2 (red) in normal murine virgin mammary ducts (Top), hyperplastic lesions from MMTV-Neu mice (Middle), and mammary tumors from MMTV-Neu mice (Bottom). Panels on far right show merged images with DAPI staining (blue). (Middle) Lower power merged image of hyperplastic regions are shown in far left panel; images to the right represent magnification of region in box in left panel. (Scale bar, 20 μm in all panels except the magnified images from boxed region of interest in hyperplastic lesions, where scale bar represents 10 μm.) (B) Time course of pHER2, pEGFR, and pAKT levels in control (black bars) and PMCA2KD (red bars) cells in response to EGF or pHER2, pHER3, and pAkt levels in response to NRG1. Cells were serum-starved for 16 h, treated with growth factors, and harvested at times listed on graphs. Each bar represents the mean ± SEM of three separate experiments.
PMCA2 Influences HER2 Signaling.
We next knocked down PMCA2 expression in SKBR3 and BT474 cells, two HER2-positive breast cancer cell lines. This reduced total HER2 levels and greatly decreased pHER2 levels (Fig. 1D). Total EGFR levels were essentially unchanged, but pEGFR, total HER3, and pHER3 levels were significantly reduced. In addition, knocking down PMCA2 reduced phosphorylated protein kinase B (pAKT) levels (Fig. 1D). Immunofluorescence staining for pHER2 and pAKT was prominent in control SKBR3 cells but greatly reduced in PMCA2KD SKBR3 cells (Fig. 1E). We measured AKT activity by assessing the localization of FOXO1 and the expression of a luciferase reporter gene regulated by FOXO1 binding (18). PMCA2KD cells displayed nuclear staining for FOXO1 and elevated FOXO1-luciferase activity, confirming reductions in AKT bioactivity (Fig. 1 F–H).
We treated SKBR3-PMCA2KD cells with epidermal growth factor (EGF) or neuregulin 1 (NRG1) to examine the effects of PMCA2 on HER2/EGFR or HER2/HER3 heterodimers, respectively. As shown in Fig. S1, basal levels of pHER2 and pEGFR were greatly reduced in PMCA2KD cells, and although they increased in response to EGF, they did not even reach the control baseline levels. In contrast, PMCA2KD cells showed a robust induction of EGFR phosphorylation, although pEGFR levels decreased more rapidly after stimulation and AKT phosphorylation was reduced. Knocking down PMCA2 also blunted responses to NRG1 (Fig. S1). As previously reported, acute treatment of the control SKBR3 cells with NRG1 actually initially decreased pHER2 levels, followed by a sustained increase between 2 and 180 min (19). Knocking down PMCA2 resulted in little change in pHER2 in response to NRG1 and blunted the increase and significantly shortened the duration of HER3 and AKT phosphorylation.
Next, we compared global changes in gene expression caused by knocking down PMCA2 with those induced by knocking down HER2 (Fig. S2; HER2KD cells). We defined 853 transcripts that were significantly altered more than twofold in either direction [P < 0.05; false discovery rate (FDR) < 0.05] in PMCA2KD cells and 840 transcripts that were changed in HER2KD cells. There was significant concordance between the changes in gene expression, with 579 (68%) of the genes altered in PMCA2KD cells also changed in HER2KD cells (Fig. S2). This is further illustrated by a heat map (Fig. S2) comparing the relative changes in all 1,127 transcripts up-regulated or down-regulated in either cell line. Functional annotation of the changes in gene expression demonstrated a strong correlation with ERBB2 signaling, and the altered genes were enriched for cancer-associated transcripts (Fig. S2). Changes in the 85 genes in the “advanced malignant tumor” category were remarkably similar between the two knockdown cell types (Fig. S2). Using quantitative reverse transcription-PCR (QPCR), we validated changes in the expression of seven cancer-associated genes that were altered in both cell lines (Fig. S2). These data support the view that PMCA2 influences HER2-dependent gene networks.
Fig. S2.
(A) Immunoblot showing reduction of total HER2 levels in HER2KD SKBR3 cells. (B) Venn diagram showing overlap between genes that were up-regulated or down-regulated by twofold or greater relative to control SKBR3 cells for HER2KD or PMCA2KD cells. The total numbers of transcripts altered in each knockdown cell line are shown in the bar graph, and the number of altered genes shared between the two knockdown cells is shown in the area of overlap on the diagram. (C) Heat map of changes in gene expression in HER2KD and PMCA2KD cells in comparison with control SKBR3 cells. Shown are the relative changes in 1,127 transcripts whose expression was altered by at least twofold in either of the cell lines. Red represents transcripts increased relative to controls, and green represents transcripts reduced relative to controls. (D) Functional annotation of the 579 genes whose expression was altered in both HER2KD and PMCA2KD cell lines. Top table includes the signaling pathways to which these genes most commonly mapped. Bottom table demonstrates the enrichment of cancer-related functional annotations in these gene sets. (E) Heat map of changes in the expression of 85 genes in the “advanced malignant tumor” cluster for HER2KD and PMCA2KD cells in comparison with control SKBR3 cells. (F) QPCR results examining the expression of seven separate genes identified as altered in the microarray dataset. Expression in PMCA2KD (red) and HER2KD (green) cells is shown relative to control (black bars) SKBR3 cells. Bars represent mean ± SEM for three separate experiments.
PMCA2 Regulates Breast Cancer Cell Growth and Tumor Formation.
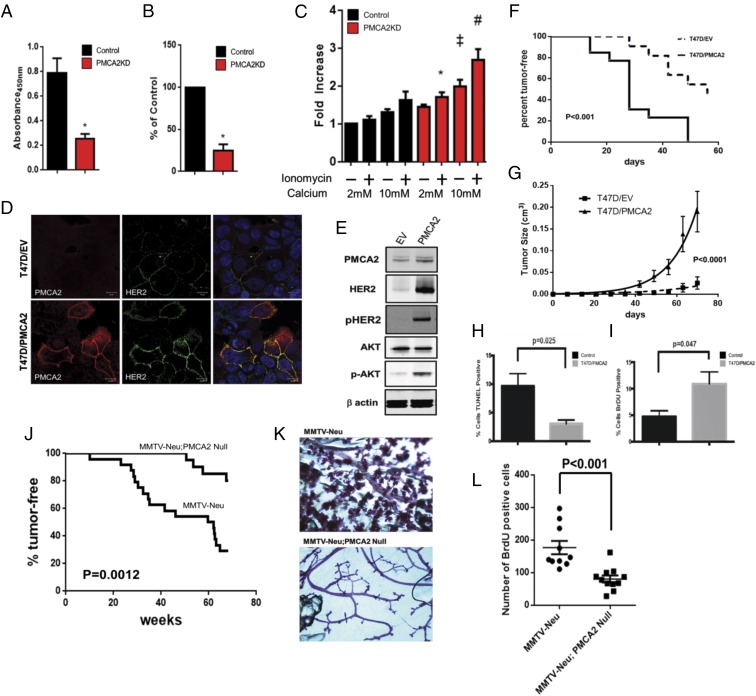
Knocking down PMCA2 inhibited the proliferation of SKBR3 cells, reducing BrdU incorporation by 62 ± 6% (Fig. 2 A and B). Baseline apoptosis tended to increase in PMCA2KD cells, although these differences were not statistically significant (Fig. 2C). However, previous studies had suggested that PMCA2 protects against calcium-induced cell death (8), and apoptosis rates were increased when PMCA2KD cells were exposed to high extracellular calcium ± ionomycin (Fig. 2C).
Fig. 2.
(A) Viable PMCA2KD and control cells as assessed by 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) assay. Asterisks denote significant difference versus control (n = 6 for each group). (B) BrdU incorporation in PMCA2KD cells relative to control cells. Asterisk denotes a significant difference with control (n = 3). (C) Apoptosis, as assessed by TUNEL assay, in PMCA2KD cells relative to controls exposed to differing concentrations of extracellular calcium ± ionomycin. *Significant difference with 2 mM calcium + ionomycin in control cells. ‡Significant difference with 10 mM calcium in control cells. #Significant difference with 10 mM calcium + ionomycin in control cells (n = 3). (D) Immunofluorescence for PMCA2 (red) and HER2 (green) in T47D cells transfected with empty vector (T47D/EV; Top) or overexpressing PMCA2 (T47D/PMCA2; Bottom). (Right) Merged images with DAPI staining. (Scale bars, 10 μm.) (E) Typical immunoblot from control T47D cells (EV) or T47D cells overexpressing PMCA2 (PMCA2). (F) Kaplan-Meier plot showing tumor latency and incidence for control T47D/EV cells (n = 11) and T47D/PMCA2 cells (n = 13) grown as xenografts. (G) Rate of tumor enlargement for the same mice described in F. (H) Apoptosis in xenograft tumors from control T47D/EV cells or T47D/PMCA2 cells (n = 4). (I) Cell proliferation in same xenograft tumors as in H (n = 4). (J) Kaplan-Meier plot showing tumor latency and incidence in MMTV-Neu mice (n = 24) versus MMTV-Neu;PMCA2-null mice (n = 20). (K) Typical whole-mount histology of non-tumor-containing mammary glands from MMTV-Neu (Top) or MMTV-Neu;PMCA2-null (Bottom) mice. (L) BrdU incorporation in MMTV-Neu versus MMTV-Neu;PMCA2-null tumors (n = 16; four histological sections from each of four tumors for each genotype).
We overexpressed PMCA2 in T47D cells, which normally display low levels of PMCA2 and HER2. This substantially increased HER2, pHER2, and pAKT levels (Fig. 2 D and E). When we grew T47D cells as xenografts in immunocompromised mice, overexpression of PMCA2 increased the incidence and shortened the latency of tumor development (Fig. 2F). The growth rate of tumors overexpressing PMCA2 was increased in comparison with control tumors, which correlated with an increase in BrdU incorporation and a decrease in apoptosis in PMCA2-overexpressing T47D tumors (Fig. 2 G–I).
Next, we crossed MMTV-Neu mice with Dfw-2J mice, which harbor a null mutation of the Atp2b2 (PMCA2) gene (6, 8, 20). The loss of PMCA2 significantly reduced tumor incidence and prolonged tumor latency (Fig. 2J). Furthermore, loss of PMCA2 inhibited the development of premalignant alveolar hyperplasia in MMTV-Neu;PMCA2-null mice compared with MMTV-Neu mice, in which it was universally present (Fig. 2K). Tumors that eventually developed in the MMTV-Neu;PMCA2-null mice demonstrated lower rates of BrdU incorporation (Fig. 2L). Apoptosis, as assessed by TUNEL staining, was negligible in both tumor types.
PMCA2 Regulates HER2 Localization and Membrane Retention.
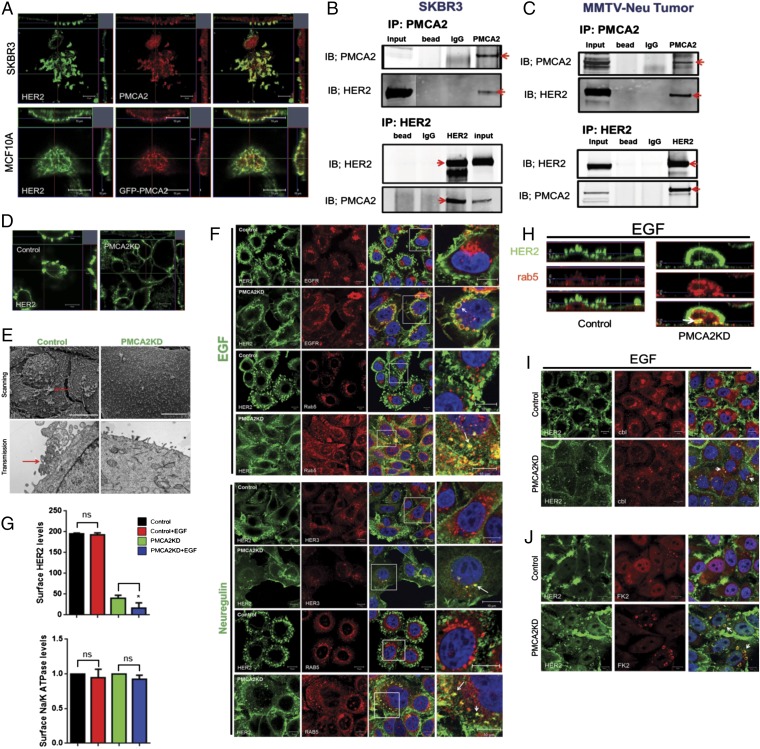
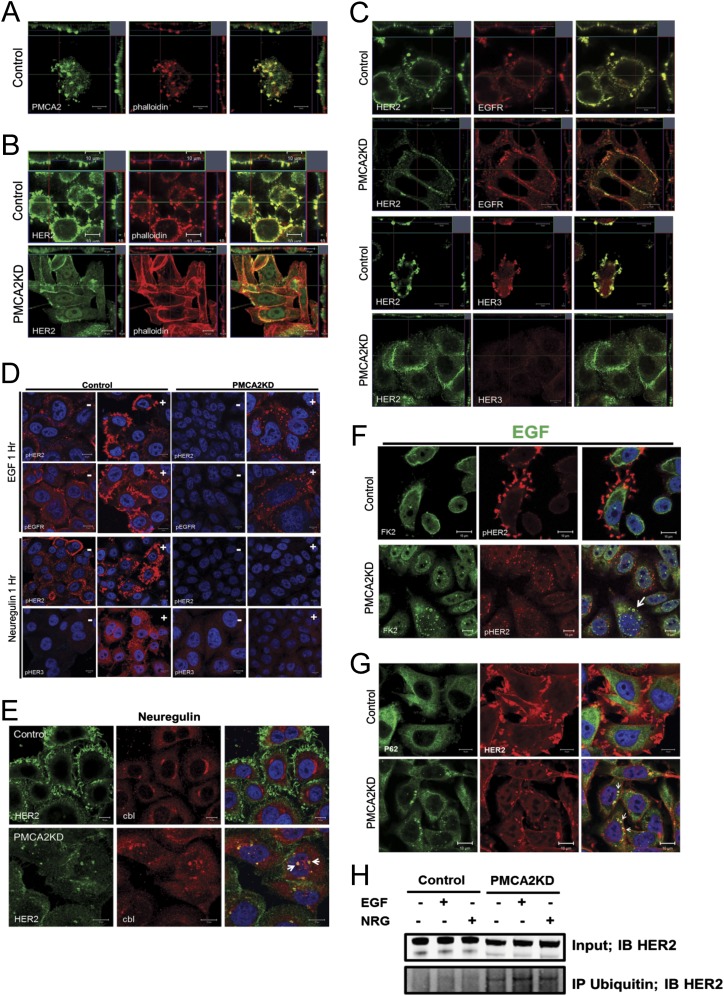
Confocal microscopy demonstrated that PMCA2 and HER2 colocalize within membrane structures protruding from the apical aspect of SKBR3 cells (Fig. 3A). A similar, although not as pronounced, pattern of colocalization was noted when we transiently expressed GFP-tagged PMCA2 in MCF10A cells that overexpressed wild-type HER2 (Fig. 3A). Costaining for actin (phalloidin) with HER2 and PMCA2 demonstrated that these structures were actin-rich (Fig. S3). EGFR and HER3 also colocalized with HER2 within similar structures, suggesting they are nodes for HER2 signaling (Fig. S3). To determine whether PMCA2 and HER2 interact within a common complex, we performed coimmunoprecipitation (IP) experiments. As shown in Fig. 3B, IP of PMCA2 from SKBR3 cells pulled down HER2 and IP of HER2 pulled down PMCA2. Similar results were obtained when we performed co-IP experiments using protein extracts prepared from MMTV-Neu tumors (Fig. 3C).
Fig. 3.
(A) Confocal images of costaining for HER2 (green) and PMCA2 (red) in SKBR3 cells (Top) or in MCF10A-HER2 cells transiently transfected with GFP-PMCA2 (Bottom). (Right) Merged images. (Insets) Z-stacks in two different orientations: apical side facing down (Top) and to left (Right). (B) Co-IP for PMCA2 and HER2 in SKBR3 cells. (Top) IP for PMCA2 and immunoblotting for PMCA2 and HER2. (Bottom) IP for HER2 and immunoblotting for HER2 and PMCA2. Red arrows point to specific bands. (C) Typical co-IP for PMCA2 and HER2, using lysates from MMTV-Neu tumors. (D) Confocal images of staining for HER2 in control or PMCA2KD cells. (Insets) Z-stacks in two different orientations, as earlier. (E) Scanning (Top) and transmission (Bottom) electron microscopy of control and PMCA2KD cells showing loss of larger membrane protrusions and ruffles (red arrows) in knockdown cells. (F) Costaining for HER2 with EGFR or Rab5 in control and PMCA2KD cells after 2 h of EGF treatment, and HER2 with HER3 or Rab5 after 2 h of NRG1 treatment. Panels on far right represent magnification of boxed areas in third panels. Arrows point to colocalization of EGFR and HER2, HER3 and HER2, or Rab5 and HER2 within cytoplasmic vesicles. (G) Quantification of surface, biotinylated HER2, or Na/K ATPase in control and PMCA2KD cells at baseline or after EGF treatment. Asterisks denote statistically significant differences compared with control. (H) Optical z-stack sections through control or PMCA2KD cells costained for HER2 (green) and Rab5 (red). Arrow points to colocalization of HER2 and Rab5 within the cytoplasm in PMCA2KD cells. (I) Costaining for HER2 and c-Cbl in control (Top) and PMCA2KD (Bottom) after EGF treatment with EGF. Arrows point to intracellular colocalization of HER2 with c-Cbl only in PMCA2KD cells. (J) Costaining for HER2 with FK2 in control (Top) and PMCA2KD (Bottom) after EGF treatment. Arrows point to intracellular colocalization of HER2 with FK2 in knockdown cells. (Scale bars, 10 μm in all panels except E, where they represent 20 μm.)
Fig. S3.
(A) Immunofluorescence confocal images for costaining for PMCA2 (green) and actin (phalloidin, red) in SKBR3 cells. (Right) Merged images. (Insets) Z-stacks of the cells in two different orientations. Note that PMCA2 and actin colocalize within apical membrane protrusions. (B) Immunofluorescence confocal images for costaining for HER2 (green) and actin (phalloidin, red) in control and PMCA2KD SKBR3 cells. (Right) Merged images. (Insets) Z-stacks of the cells in two different orientations. In control cells, HER2 is localized within actin-rich membrane domains protruding from the apical surface of the cell. However, knocking down PMCA2 leads to reorganization of the actin cytoskeleton, flattening of the cells, and more diffuse membrane staining for HER2. (C) Immunofluorescence confocal images of costaining for HER2 (green) and EGFR (red) in the top two rows and HER2 (green) and HER3 (red) in the bottom two rows. (Right) Merged images. (Insets) Z-stacks as before. Note colocalization of HER2 with EGFR or HER3 in membrane protrusions in control SKBR3 cells (rows 1 and 3). However, in PMCA2 KD cells, staining for HER2 and EGFR becomes more diffusely distributed and less well colocalized in the membrane (row 2). Furthermore, in PMCA2 KD cells, HER3 staining is much reduced and no longer colocalizes with HER2 (row 4). (D) Staining for pHER2 or pEGFR in serum-free media (−) or after 1 h treatment with EGF (+) (top two rows). In control cells, treatment with EGF leads to prominent localization of pHER2 and pEGFR within membrane protrusions. However, in PMCA2KD cells, treatment with EGF leads to internalization of both pHER2 and pEGFR. Bottom two rows: Staining for pHER2 or pHER3 in serum-free media (−) or after 1 h treatment with neuregulin (+). In control cells, treatment with neuregulin leads to prominent localization of pHER2 within membrane protrusions, although pHER3 becomes mostly cytoplasmic within 1 h of stimulation. In PMCA2KD cells, treatment with neuregulin causes almost no stimulation of pHER3. (E) Costaining for HER2 and c-Cbl in control (Top) and PMCA2KD (Bottom) cells after 2 h treatment with neuregulin. Arrows point to intracellular colocalization of HER2 with c-Cbl only in knock-down cells. (F) Costaining for pHER2 with FK2 in control (Top) and PMCA2KD (Bottom) cells after 2 h treatment with EGF. (Right) Merged images. Arrows point to intracellular colocalization of pHER2 with FK2 in knockdown cells after stimulation with EGF. (G) Costaining for HER2 with p62 in control (Top) and PMCA2KD (Bottom) cells after 2 h treatment with EGF. (Right) Merged images. Arrows point to intracellular colocalization of HER2 with p62 in knockdown cells treated with EGF. (H) Co-IP of polyubiquitin complexes with HER2 in control and PMCA2KD cells in serum-free media or after 2 h of treatment with EGF or NRG. HER2 is pulled down with antibody against polyubiquitin only in the knock-down cells. (Scale bars, 10 μm.)
We next examined immunofluorescence for HER2, EGFR, and HER3 in PMCA2KD cells. HER2 and EGFR became more diffusely distributed over the surface of these cells, whereas HER3 staining was dramatically reduced (Fig. 3D and Fig. S3). Knocking down PMCA2 also caused effacement of the actin-rich protrusions, although HER2 still appeared to colocalize with actin (Fig. 3D and Fig. S3). The change in the membrane structures was obvious using scanning and transmission electron microscopy. As shown in Fig. 3E, we observed complex membrane structures protruding from the surface of control cells, but not from the knockdown cells. Thus, PMCA2 is necessary for the organization of actin-rich membrane domains that include HER2, EGFR, and HER3.
Unlike other ErbB family members, HER2 remains on the cell surface after activation (12). Consistent with this, acute treatment of control SKBR3 cells with EGF or NRG1 increased the localization of HER2 to membrane protrusions, but caused internalization of EGFR and HER3 (Fig. 3F). In contrast, treatment of PMCA2KD cells with EGF or NRG1 caused internalization of HER2 and its colocalization with EGFR, HER3, and the endosomal marker rab5 within intracellular vesicles (Fig. 3 F and H). This was also true for the phosphorylated receptors (Fig. S3). To quantify cell surface HER2, we biotinylated cell surface proteins, IPed biotin, and measured HER2 in the immunoprecipitate from control and PMCA2KD cells. As shown in Fig. 3G, there was a dramatic reduction in biotinylated cell surface HER2 in knockdown cells at baseline or in response to acute treatment with EGF. However, this was not a general effect on all membrane proteins, as there was no change in surface-labeled Na/K ATPase.
Internalization and degradation of HER2 can occur after its polyubiquitination by the E3 ubiquitin ligase, c-Cbl (21–23). In control cells, c-Cbl and HER2 do not colocalize after stimulation with EGF or NRG1, but in PMCA2KD cells, c-Cbl becomes colocalized with HER2 at the cell surface and within the cytoplasm after receptor stimulation (Fig. 3I and Fig. S3). Using a monoclonal antibody (FK2) that recognizes polyubiquitin complexes, we also costained for HER2, pHER2, and polyubiquitin residues. FK2 staining colocalized with HER2 and pHER2 in perinuclear vesicles after EGF treatment in the knockdown cells, but not in control cells (Fig. 3J and Fig. S3). We observed similar patterns for p62, which binds to polyubiquitinated proteins localized to autophagosomes (Fig. S3). Last, IP for polyubiquitin complexes and blotting for HER2 demonstrated increased ubiquitination of HER2 in the knockdown cells (Fig. S3). These data suggest that PMCA2 prevents ubiquitination and removal of HER2 from the cell surface.
Increased Intracellular Calcium Inhibits HER2 Signaling and Stimulates HER2 Internalization.
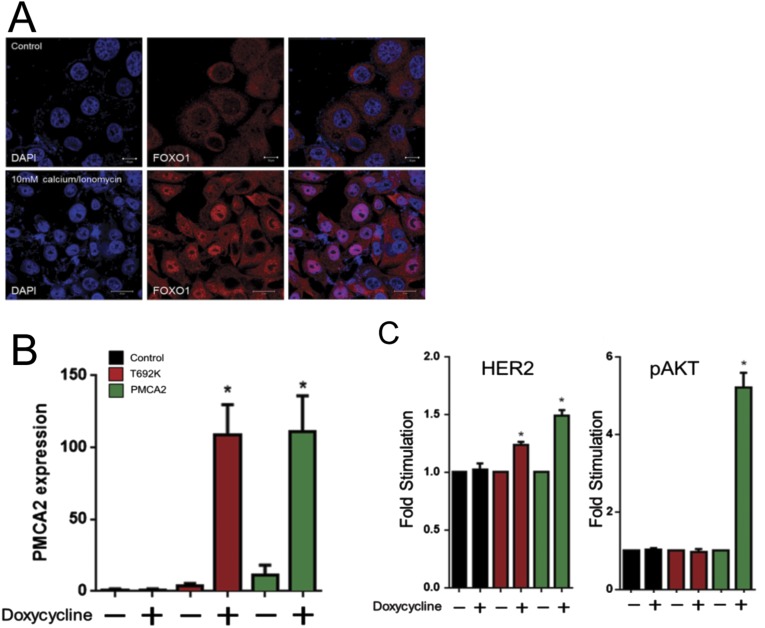
Intracellular calcium levels in PMCA2KD cells were 5.4-fold higher than in control cells (Fig. 4A), suggesting that PMCA2 might support HER2 signaling by lowering cytoplasmic calcium. To test this idea, we exposed SKBR3 cells to high extracellular calcium concentrations and ionomycin to increase intracellular calcium levels. This antagonized interactions between HER2 and PMCA2, as reflected by a reduction in the amount of PMCA2 pulled down by IP of HER2 (Fig. 4B). Treatment with calcium and ionomycin also reduced pHER2, pEGFR, pHER3, and pAKT levels (Fig. 4C) and caused nuclear translocation of FOXO1 (Fig. S4). Ionomycin and extracellular calcium flattened the actin-rich membrane protrusions (Fig. 4D) and led to costaining for HER2 and actin within the cell (Fig. 4D). Internalization of HER2 was more obvious when SKBR3 cells were pretreated with 10 mM calcium and ionomycin followed by 2 h exposure to EGF. As seen in Fig. 4E, this led to internalization and colocalization of HER2 with EGFR, and HER2 with FK2 staining in intracellular vesicles (Fig. 4E). HER2 and HER3 were also internalized in response to NRG1, but there was less colocalization of HER2 with HER3 (Fig. 4E). Finally, acute (16 h) exposure to calcium and ionomycin reduced membrane HER2 levels, as assessed by the cell surface biotinylation assay, although not to the same degree as chronic knock down of PMCA2 (Figs. 4F and 3G).
Fig. 4.
(A) Intracellular calcium measurements in PMCA2KD cells relative to control. Numbers are mean calcium concentrations estimated by FURA2 measurements. Asterisk denotes statistically significant difference. (B) Co-IP for HER2 and PMCA2 from SKBR3 cells exposed to 2 mM calcium or 10 mM calcium + ionomycin for 16 h. (C) Typical immunoblot for SKBR3 cells exposed to 2 or 10 mM calcium in the absence (−) or presence (+) of ionomycin for 16 h. (D) Confocal images of immunofluorescence for HER2 (green) and phalloidin (actin, red) in control SKBR3 cells exposed to 2 mM calcium (Top) or 10 mM calcium + ionomycin (Bottom) for 16 h. (Insets) Z-stack images in two different orientations; dotted line represents the apical surface of the cell. (E) Costaining for HER2 and EGFR, HER2 and FK2, or HER2 and HER3 in cells exposed to 10 mM calcium + ionomycin in serum-free media (SFM) for 24 h followed by 2 h of EGF. Arrows note intracellular costaining for HER2 and EGFR, HER2 and FK2, or HER2 and HER3. (F) Quantification of cell surface, biotinylated HER2 in control cells exposed to differing concentrations of extracellular calcium ± ionomycin. Values are expressed relative to control conditions at 2 mM calcium. Asterisks denote statistically significant differences compared with control at 2 mM calcium. (G) Diagram illustrating PMCA2T692K mutation. (H) Immunoblot for control T47D cells and T47D cells overexpressing PMCA2T692K or wtPMCA2 in response to doxycycline. (I) Intracellular calcium in T47D cells expressing PMCA2T692K or wtPMCA2 relative to controls (n = 3). Numbers refer to the mean calcium concentrations. Asterisks denote statistically significant differences relative to control cells. (J) BrdU incorporation in T47D cells overexpressing PMCA2T692K or wtPMCA2 relative to controls (n = 3). (K) Apoptosis in control T47D cells and T47D cells overexpressing PMCA2T692K or wtPMCA2 exposed to varying calcium concentrations ± ionomycin relative to control cells at 0.1 mM calcium. Within the same treatment group, * denotes significant differences with PMCA2-T692K; # denotes significant differences with control (n = 3). (Scale bars, 10 μm.)
Fig. S4.
(A) Staining for FOXO1 in SKBR3 cells exposed to 2 mM calcium (Top) or 10 mM calcium and ionomycin (Bottom). (B) PMCA2 mRNA expression assayed by QPCR in control T47D cells and T47D cells induced to overexpress mutant PMCA2T692K or wtPMCA2 in response to treatment with doxycycline. Bars represent the mean ± SEM for three experiments. Asterisks denote significant differences compared with controls. (C) Quantification of HER2 and pAKT expression assayed by immunoblots in control T47D cells (black bars) and T47D cells induced to overexpress mutant PMCA2T692K (red bars) or wtPMCA2 (green bars) in response to treatment with doxycycline. Bars represent the mean ± SEM for three experiments Asterisks denote significant differences compared with controls.
To test whether the calcium pumping function of PMCA2 is required for HER2 signaling, we used a doxycycline-inducible system to overexpress, in T47D cells, equal amounts of wild-type PMCA2 or a mutant form (PMCA2T692K) that has been shown to traffic normally, but that cannot pump calcium (24) (Fig. 4 G and H and Fig. S4). As expected, overexpression of wtPMCA2, but not PMCA2T692K, reduced intracellular calcium concentrations (Fig. 4I). Expression of wtPMCA2 increased HER2, pHER2, and pAKT levels, and although expression of PMCA2T692K increased total HER2 levels, it did not activate HER2/AKT signaling (Fig. 4H and Fig. S4). Overexpression of wtPMCA2, but not PMCA2T692K, stimulated proliferation and inhibited apoptosis in T47D cells (Fig. 4 J and K). Thus, PMCA2 must transport calcium to influence HER2 signaling.
PMCA2 Promotes Interactions Between HER2 and HSP-90.
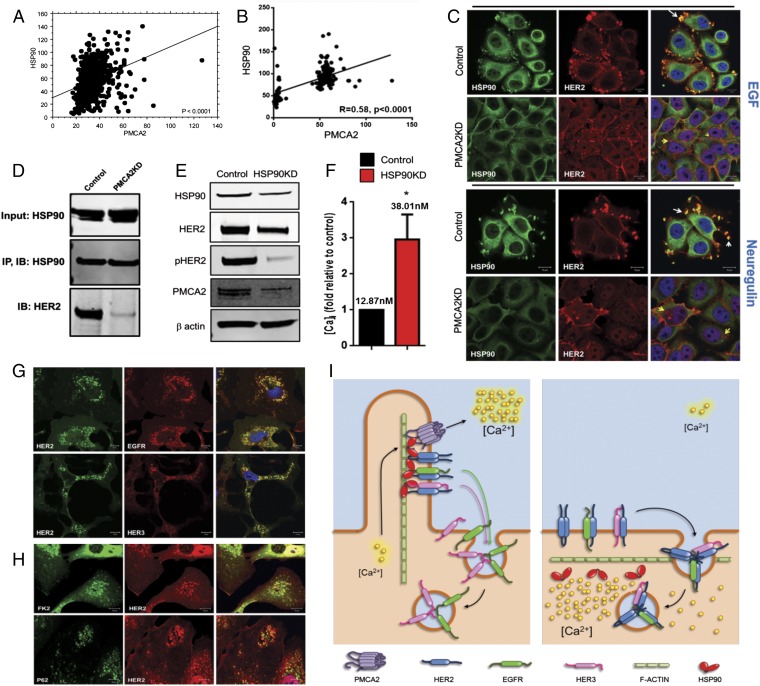
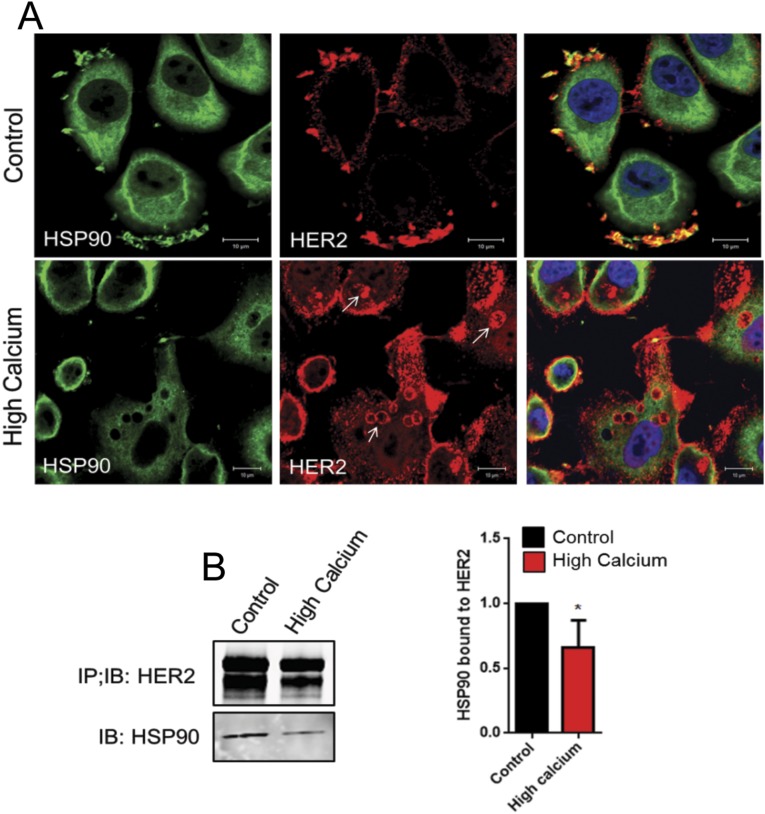
Interactions with HSP-90 stabilize HER2 at the cell membrane, prevent its internalization, and facilitate HER2 signaling (12, 25–28). Therefore, we examined the previously described tissue microarray (TMA) and microarray databases and found that HSP-90 expression correlates with PMCA2 expression in human breast cancers at both a protein (Fig. 5A) and mRNA (Fig. 5B) level. Similar to PMCA2 and HER2 (Fig. 1B), PMCA2 and HSP-90 mRNA levels were distributed in a bimodal pattern, suggesting tumors tended to have either low or high levels of both transcripts. Likewise, a χ2 analysis of the TMA data demonstrated a statistically significant clustering of tumors into groups with either high or low levels of both HSP-90 and PMCA2 staining. We found HER2 and HSP-90 colocalized in membrane protrusions in SKBR3 cells (Fig. 5C) (12, 26), but their colocalization was abolished in PMCA2KD cells (Fig. 5C). Stimulation with EGF or NRG1 accentuated colocalization between HSP-90 and HER2 in control cells, but caused internalization of HER2 and loss of colocalization with HSP-90 in PMCA2KD cells (Fig. 5C). In control cells, IP of HSP-90 pulled down HER2 (Fig. 5D). However, although total HSP-90 levels did not change in the PMCA2KD cells, the ability to co-IP HER2 with HSP-90 was greatly diminished (Fig. 5D). Interactions between HSP-90 and HER2 were also sensitive to intracellular calcium, as treatment with 10 mM calcium and ionomycin led to loss of their colocalization by immunofluorescence and reduced their co-IP (Fig. S5). Finally, knocking down HSP-90 in SKBR3 cells reduced HER2 and pHER2 levels as anticipated (Fig. 5E). However, it also reduced PMCA2 expression and increased intracellular calcium levels (Fig. 5F). Reductions in HSP-90 led to internalization and ubiquitination of HER2 and its intracellular colocalization with EGFR and HER3 after stimulation with EGF or NRG1 (Fig. 5 G and H). These data suggest that PMCA2 is required for HSP-90 to interact with cell surface HER2.
Fig. 5.
(A) Immunofluorescence automated quantitative analysis (AQUA) scores for PMCA2 versus HSP90 in breast cancer TMA. (B) PMCA2 versus HER2 mRNA levels in tumors from 204 patients with breast cancer. (C) Costaining of control and PMCA2KD cells for HSP-90 (green) and HER2 (red). (Right) Merged images. The top two rows show cells treated with EGF; the bottom two rows show cells treated with NRG1. White arrows point to colocalization of HER2 and HSP-90 in membrane protrusions in control cells. Yellow arrows point to internalized HER2, which no longer colocalizes with HSP-90 in PMCA2KD cells. (D) Co-IP of HSP-90 and HER2 in control and PMCA2KD cells. (E) Immunoblot for HSP90KD and control cells. (F) Intracellular calcium measurements in HSP90KD cells relative to controls. The numbers indicate the mean calcium measurements. *Statistical significance. (G) Costaining for HER2 and EGFR, and HER2 and HER3, in HSP90KD cells. (H) Costaining for HER2 and FK2, and HER2 and P62, in HSP90KD cells. (Scale bars, 10 μm.) (I) Model for interactions between PMCA2, HER2, and HSP-90 (Left) and consequences of loss of PMCA2 (Right).
Fig. S5.
(A) Immunofluorescence costaining for HSP90 (green) and HER2 (red). (Right) Merged images, with DAPI staining to highlight nuclei (blue). (Top) Cells at 2 mM calcium. (Bottom) Cells at 10 mM calcium + ionomycin for 16 h. Note loss of colocalization between HSP90 and HER2 after exposure to high calcium and ionomycin. Arrows in the middle panel in the bottom row highlights intracellular staining for HER2. (Scale bars, 10 μm.) (B) Co-IP of HER2 with HSP90 from SKBR3 cells at control 2 mM calcium or after exposure to 10 mM calcium + ionomycin for 16 h. B, Right shows the quantification of three co-IP experiments between HER2 and HSP90 at low or high calcium and ionomycin. Results were normalized for variation in HER2 levels. Bars represent the mean ± SEM. *Significant differences between control and high calcium conditions.
Discussion
We find that PMCA2 colocalizes with HER2 in human DCIS lesions and that expression of PMCA2 and HER2 correlate with each other in breast cancers from patients. In breast cancer cells, we find that PMCA2 interacts with HER2 within actin-rich membrane microdomains, and that PMCA2 is required for proper maintenance of these domains, localization of HER2 to these structures, interactions between HER2 with HSP-90, and the ability of HER2 to remain on the cell surface and signal after activation by either EGF or NRG1 (Fig. 5I). Finally, we find that manipulating PMCA2 levels alters the proliferation and apoptosis of breast cancer cells and that knocking out PMCA2 inhibits the development of hyperplasia and tumors in MMTV-Neu mice. Therefore, we propose that interactions between PMCA2 and HER2 regulate the cell surface stability of HER2, its biochemical signaling, and its ability to promote malignant behavior.
Transformed cells remodel intracellular calcium homeostasis to support malignant behavior, and our experiments highlight the importance of PMCAs in this process (29–31). We find that PMCA2 regulates HER2 function, in part, through its ability to lower intracellular calcium. Knocking down PMCA2 raised intracellular calcium levels and inhibited HER2 signaling in SKBR3 cells, whereas the expression of wild-type PMCA2, but not a mutant PMCA2 incapable of transporting calcium, lowered intracellular calcium and activated HER2 signaling in T47D cells. Furthermore, increasing intracellular calcium with a calcium ionophore inhibited HER2 signaling and promoted its internalization and ubiquitination. Therefore, PMCA2 appears to support HER2-mediated transformation by maintaining low intracellular calcium concentrations in the microenvironment surrounding a membrane-signaling complex that includes HER2, HER3, EGFR, and HSP-90.
Receptor internalization followed by recycling or degradation is important for modulating receptor tyrosine kinases (12, 15). Unlike other ErbB family receptors, HER2 resists internalization and degradation and remains at the cell surface, where it can continue to signal for prolonged periods (12, 32). Several groups have reported that HER2 resides within microvillus-like membrane protrusions, and Hommelgaard and colleagues hypothesized that these structures support the retention of HER2 on the cell surface (33–36). Previous experiments also showed that interactions with HSP-90 are important for HER2 stabilization within membrane protrusions and its resistance to internalization (12, 26). Thus, it is noteworthy that PMCA2 levels correlate with HSP-90 levels in human tumors and that knockdown of PMCA2 is associated with decreased interactions between HSP-90 and HER2, the internalization of HER2, and the disruption of the membrane protrusions. HER2 localization and signaling appear to be intimately related to the presence of protruding membrane microdomains, and our results do not fully distinguish whether internalization of HER2 is caused directly from loss of its interactions with PMCA2, or secondarily from elevated intracellular calcium and/or the disruption of the membrane domains. We envision that HER2 is actively retained within membrane signaling domains as a result of complex interactions with PMCA2, scaffolding proteins, and the actin cytoskeleton (37, 38). Furthermore, PMCA2 may be recruited to these signaling domains to prevent their disruption by intracellular calcium and/or to inhibit calcium-dependent movement of HER2 out of these macromolecular assemblies and into an endocytic pathway normally used by other ErbB family members (Fig. 5I) (38–40).
In summary, our findings demonstrate that PMCA2 is vital for the localization of HER2 and its partners, EGFR and HER3, to active membrane signaling domains. PMCA2 helps prevent HER2 internalization after receptor stimulation and sustains downstream signal transduction. Genetic manipulation of PMCA2 regulates the growth of breast cancers in vivo, and PMCA2 predicts outcome in human patients (8). Targeting PMCA2-HER2 interactions may offer novel opportunities for therapeutic development.
Methods
Knockdown Cell Lines.
Stable cell lines expressing shRNA directed against ATP2B2, HSP-90, and ERBB2 were generated by transducing cells with commercially prepared lentiviruses: PMCA2 (sc-42598) and HSP-90α/β (SC-35608-V) from Santa Cruz and HER2 (318-328) from AMSbio.
PMCA2 Overexpression.
T47D cell lines overexpressing wtPMCA2 were previously described (8). The Quick Change Site-Directed Mutagenesis Kit (Agilent) was used to change nucleotide 2075 of the mouse pT-REx-PMCA2w/b construct (8) from C to A, creating the T692K mutation (24). The sequence of the mutant construct was verified on both strands, and T47D cells were sequentially transfected with pcDNA6-TR (tetracycline-regulated repressor; Life Technologies) and the tet-regulated WT PMCA2, T692KPMCA2, or control expression vector. Expression was induced with 50 μg/mL doxycycline.
Tissue Microarray.
The breast carcinoma tissue microarray (YTMA49) consisted of 652 primary breast cancer specimens retrospectively obtained from 1953 to 1983, as previously described (8).
Transgenic and Mutant Mice.
We generated MMTV-Neu/PMCA2dfw-2J/dfw-2J and control MMTV-Neu/PMCA2wt/wt mice and followed them weekly for the presence of tumors. MMTV-Neu mice were obtained from Jackson Laboratory (FVB/N-Tg(MMTVneu)202Mul/J, stock number 002376), as were PMCA2wt/dfw-2J mice (CByJ.A-Atp2b2dfw-2J/J, stock number 002894). All animal experiments were approved by the Yale Institutional Animal Care and Use Committee.
Statistics.
Statistical analyses were performed with Prism 6.0 (GraphPad Software).
SI Methods
Cell Culture.
The human cell lines, SKBR3, BT474, and T47D, were obtained from ATCC and maintained in culture in DMEM +GlutaMAX-1 (Gibco-Life Technologies) containing 10% (vol/vol) FBS and pen/strep (Gibco-Life Technologies) at 37 °C in 5% (vol/vol) CO2. MCF10A-HER2 cells (gift of the D.F.S. laboratory) were cultured in DMEM/F12 (Gibco-Life Technologies) containing 5% (vol/vol) horse serum, EGF (100 μg/mL), hydrocortisone (1 mg/mL), cholera toxin (1 mg/mL), insulin (10 mg/mL), and pen/strep (Gibco-Life Technologies) at 37 °C in 5% (vol/vol) CO2. In some experiments, cells were cultured as earlier, but in media without FBS for 16 h, and then were treated with 100 ng/mL EGF (Cell Signaling) or 50 ng/mL NRG1 (Cell Signaling) for 1 or 2 h.
Knockdown Cell Lines.
Cells were cultured in 12-well plates and infected by adding the various shRNA lentiviral particles to the culture for 48 h, as per the manufacturer’s instructions. Stable clones expressing the specific shRNAs were selected using 5 μg/mL puromycin (Gibco-Life Technologies) (PMCA2 and HSP-90) or 5 μg/mL Blasticidin S HCl (Gibco-Life Technologies) (HER2).
Immunofluorescence.
Cells were grown on coverslips, fixed in 4% (wt/vol) paraformaldehyde for 20 min, permeabilized with 0.2% Triton ×100 for 10 min, washed three times with PBS, and incubated with primary antibody overnight at 4 °C. The cells were then washed three times with PBS and incubated with secondary antibody for 1 h at room temperature. After washing, coverslips were mounted using Prolong Gold antifade reagent with DAPI (Invitrogen). Paraffin-embedded tissue sections were cleared with histoclear (National Diagnostics) and graded alcohol, using standard techniques. Antigen retrieval was performed using 7 mM citrate buffer at pH 6.0 under pressure. Sections were incubated with primary antibody overnight at 4 °C and with secondary antibody for 1 h at room temperature. Coverslips were mounted using Prolong Gold antifade reagent with DAPI (Invitrogen). All images were obtained using a Zeiss 780 confocal microscope. Primary antibodies included those against HER2 (sc-284), phospho-EGFR (sc-12351), FOXO1 (sc-11350), Cbl (sc-170), and ubiquitin (sc-8017) from Santa Cruz; PMCA2 (PA1-915) and HER2 (MA1-35720) from Thermo Scientific; phospho-HER2 (2243S), phospho-AKT (4060S), AKT (4691S), EGFR (4267S), HER3 (12708P), phospho-HER3 (4791S), ERK (4695S), phospho-ERK (4370S), and anti-Rab5 (3547s) from Cell Signaling; FK2 (2325026) from Millipore; bromodeoxyuridine (M20107S) from Meridian; P62 (610832) from BD Transduction Laboratories; and HSP-90 (379400) from Invitrogen. We also stained for actin using phalloidin-Atto 488 (49409) from Sigma.
Immunoblotting.
Protein samples were prepared from cells, using standard methods, and were subjected to SDS/PAGE and transferred to a nitrocellulose membrane by wet Western blot transfer (Bio-Rad). The membrane was blocked in TBST buffer (TBS + 1% Tween) containing 5% (wt/vol) powdered milk for 1 h at room temperature. The blocked membranes were incubated overnight at 4 °C with specific primary antibodies (Odyssey blocking buffer, 927–40,000). The membranes were washed three times with TBST buffer and then incubated with specific secondary antibodies provided by LI-COR for 2 h at room temperature. After three washes with TBST buffer, the membranes were analyzed using the ODYSSEY Infrared Imaging system (LI-COR). Samples were then analyzed for specific proteins, using the antibodies listed earlier. All immunoblot experiments were performed at least three times, and representative blots are shown in the figures.
Breast Cancer Tissue Microarray.
The breast carcinoma tissue microarray (YTMA49) contains 652 primary breast cancer specimens retrospectively obtained from 1953 to 1983. Cases are evenly divided between lymph node-positive and lymph node-negative, with a median follow-up of 8.9 y. Clinicopathologic data were extracted from Yale and Connecticut Tumor Registries, including disease-specific survival. Details of staining, automated image acquisition, and analysis using the semiautomated AQUA system have been described previously (16). Briefly, slides were scanned and high-resolution images taken of each TMA spot (histospot), using different wavelength emission/excitation filters for each of the cytokeratin (tumor masking), DAPI (nuclei), and target (PMCA2, HER2, or HSP-90) stains. Immunofluorescence for each target was performed on separate sections cut from the same tumor blocks. Staining images were then read into the AQUA system for analysis. Histospots were excluded if the tumor mask represented less than 5% of the whole histospot area. The analysis of combined PMCA2 and HER2 AQUA scores was based on a total of 524 of the 652 tumors that had adequate histospots for both targets, whereas the analysis examining HSP-90 and PMCA2 was based on 540 tumors with adequate spots. The cytokeratin and DAPI images were used to assign each pixel under the tumor mask into nonoverlapping nonnuclear (membrane/cytoplasmic) and nuclear locales. AQUA scores for the target were then calculated that correspond to the average signal intensity divided by the locale area. This allowed for scores to be calculated for total under the tumor mask, as well as in the nonnuclear and nuclear compartments alone. The analyses reported were based on total tumor mask areas. The AQUA score is thus proportional to the total protein concentration averaged across the entire region of the tumor, as defined by the cytokeratin-staining mask. This produces a single score for each histospot representing individual tumor samples.
Co-IP.
Cells were lysed with RIPA buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 20 mM Tris⋅HCl, and 150 mM NaCl), and cell extracts were incubated overnight at 4 °C with protein A/G beads (sc-2003; Santa Cruz) and the specific antibody. After separating the beads by centrifugation, the immunoprecipitated proteins were eluted with NuPAGE LDS sample buffer (4X) (NP0008, Thermofisher Scientific) containing 10% (vol/vol) beta-mercaptoethanol. The resulting samples were then analyzed by Western blot, as described earlier. Analysis of biotinylated cell surface proteins was performed using the Pierce Cell Surface Protein Isolation Kit (89881) from Thermo Scientific. Briefly, cells were labeled with Sulfo-NHS-SS-Biotin, a thiol-cleavable amine-reactive biotinylation reagent, and then lysed with mild detergent. Biotinylated surface proteins were isolated with Avidin Agarose and eluted using SDS/PAGE sample buffer containing 50 mM DTT. Co-IP experiments were repeated at least three times, and representative experiments are shown in the figures.
Cell Proliferation and Apoptosis.
Cell proliferation was assessed by measuring BrdU incorporation using the cell proliferation ELISA kit (11647229001) from Roche. Apoptosis was measured by TUNEL assay, using the cell death detection ELISA Kit (11544675001) from Roche. Cell viability was quantified using the XTT cell viability assay (9095) from Cell Signaling.
Gene Array.
Total RNA was prepared from control SKBR3 cells, PMCA2KD, and HER2KD cells using TRIzol reagent (Invitrogen) and purified using the Qiagen RNeasy cleanup kit. RNA was reverse transcribed and hybridized to the Illumina HumanHT-12 v3 Expression BeadChip (Illumina) in the Yale Center for Genomic Analysis and then scanned using the Illumina BeadArray reader. The images were analyzed by Beadstudio software, and Illumina provided instructions for quality control and data analysis.
Intracellular Calcium Measurements.
Ratiometric intracellular calcium imaging was performed using 5 μM fura-2-AM (Life Technologies), as previously described (41). Cells were loaded with 5 μM fura-2-AM (Life Technologies) for 30 min at 37 °C and then imaged at a frequency of 1 Hz on a Zeiss Axiovert 100 microscope. Intracellular calcium concentrations were calculated from the background-subtracted fluorescent ratio (R) at 340 and 380 nm of Fura 2-AM-loaded cells, using the formula Kd × (R – Rmin)/(Rmax – R) × (Ff380/Fb380), where Kd is the dissociation constant of Fura 2 for calcium (225 nM), Rmin and Rmax are the empirically determined minimum and maximum fluorescent ratios, and Ff380/Fb380 is the fluorescence intensity at 380 nm in calcium-free conditions divided by the fluorescence intensity at 380 nm in saturating calcium concentrations (bound).
Xenograft Experiments.
Slow-release 17β-estradiol pellets (1.7 mg, 90-d release; Innovative Research of America) were implanted s.c. into 4–6-wk-old female nude mice (Nu/Nu; Charles River). We then injected the mice with 1 × 106 PMCA2-overexpressing cells (T47D/PMCA2) or control T47D cells s.c. Mice were monitored twice weekly for tumor development and/or growth. Tumors were measured with calipers, and the tumor volume was calculated using the equation 0.5 × length × width2 (42).
Acknowledgments
We thank Dr. Emmanuel Strehler for the kind gift of PMCA2 expression vectors, Dr. David Rimm for assistance with TMA and AQUA analysis, Dr. Sihem Khelif for providing access to human DCIS samples, and Dr. Barbara Ehrlich for help with calcium imaging. This work was supported by the following grants: NIH CA153702, NIH HD076248, and a pilot translational research award from the Yale Department of Internal Medicine (to J.J.W.), as well as Komen for the Cure Award llR12224446 (to L.L. and V.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516138113/-/DCSupplemental.
References
- 1.Brini M, Carafoli E. Calcium pumps in health and disease. Physiol Rev. 2009;89(4):1341–1378. doi: 10.1152/physrev.00032.2008. [DOI] [PubMed] [Google Scholar]
- 2.Strehler EE. Plasma membrane calcium ATPases as novel candidates for therapeutic agent development. J Pharm Pharm Sci. 2013;16(2):190–206. doi: 10.18433/j3z011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopreiato R, Giacomello M, Carafoli E. The plasma membrane calcium pump: New ways to look at an old enzyme. J Biol Chem. 2014;289(15):10261–10268. doi: 10.1074/jbc.O114.555565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faddy HM, et al. Localization of plasma membrane and secretory calcium pumps in the mammary gland. Biochem Biophys Res Commun. 2008;369(3):977–981. doi: 10.1016/j.bbrc.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinhardt TA, Lippolis JD, Shull GE, Horst RL. Null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2 impairs calcium transport into milk. J Biol Chem. 2004;279(41):42369–42373. doi: 10.1074/jbc.M407788200. [DOI] [PubMed] [Google Scholar]
- 6.VanHouten JN, Neville MC, Wysolmerski JJ. The calcium-sensing receptor regulates plasma membrane calcium adenosine triphosphatase isoform 2 activity in mammary epithelial cells: A mechanism for calcium-regulated calcium transport into milk. Endocrinology. 2007;148(12):5943–5954. doi: 10.1210/en.2007-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reinhardt TA, Lippolis JD. Mammary gland involution is associated with rapid down regulation of major mammary Ca2+-ATPases. Biochem Biophys Res Commun. 2009;378(1):99–102. doi: 10.1016/j.bbrc.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 8.VanHouten J, et al. PMCA2 regulates apoptosis during mammary gland involution and predicts outcome in breast cancer. Proc Natl Acad Sci USA. 2010;107(25):11405–11410. doi: 10.1073/pnas.0911186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee WJ, Roberts-Thomson SJ, Monteith GR. Plasma membrane calcium-ATPase 2 and 4 in human breast cancer cell lines. Biochem Biophys Res Commun. 2005;337(3):779–783. doi: 10.1016/j.bbrc.2005.09.119. [DOI] [PubMed] [Google Scholar]
- 10.Lee WJ, et al. Expression of plasma membrane calcium pump isoform mRNAs in breast cancer cell lines. Cell Signal. 2002;14(12):1015–1022. doi: 10.1016/s0898-6568(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 11.Arteaga CL, Engelman JA. ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25(3):282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertelsen V, Stang E. The Mysterious Ways of ErbB2/HER2 Trafficking. Membranes (Basel) 2014;4(3):424–446. doi: 10.3390/membranes4030424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21(2):177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Guy CT, et al. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89(22):10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315(4):683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 16.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8(11):1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 17.Mu L, et al. Favorable outcome associated with an IGF-1 ligand signature in breast cancer. Breast Cancer Res Treat. 2012;133(1):321–331. doi: 10.1007/s10549-012-1952-5. [DOI] [PubMed] [Google Scholar]
- 18.Yang JY, Hung MC. A new fork for clinical application: Targeting forkhead transcription factors in cancer. Clin Cancer Res. 2009;15(3):752–757. doi: 10.1158/1078-0432.CCR-08-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R, et al. ErbB2 dephosphorylation and anti-proliferative effects of neuregulin-1 in ErbB2-overexpressing cells; re-evaluation of their low-affinity interaction. Sci Rep. 2013;3:1402. doi: 10.1038/srep01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Street VA, McKee-Johnson JW, Fonseca RC, Tempel BL, Noben-Trauth K. Mutations in a plasma membrane Ca2+-ATPase gene cause deafness in deafwaddler mice. Nat Genet. 1998;19(4):390–394. doi: 10.1038/1284. [DOI] [PubMed] [Google Scholar]
- 21.Klapper LN, Waterman H, Sela M, Yarden Y. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res. 2000;60(13):3384–3388. [PubMed] [Google Scholar]
- 22.Marx C, Held JM, Gibson BW, Benz CC. ErbB2 trafficking and degradation associated with K48 and K63 polyubiquitination. Cancer Res. 2010;70(9):3709–3717. doi: 10.1158/0008-5472.CAN-09-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vuong TT, et al. Preubiquitinated chimeric ErbB2 is constitutively endocytosed and subsequently degraded in lysosomes. Exp Cell Res. 2013;319(3):32–45. doi: 10.1016/j.yexcr.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Giacomello M, et al. Mutations in PMCA2 and hereditary deafness: A molecular analysis of the pump defect. Cell Calcium. 2011;50(6):569–576. doi: 10.1016/j.ceca.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Citri A, Kochupurakkal BS, Yarden Y. The achilles heel of ErbB-2/HER2: Regulation by the Hsp90 chaperone machine and potential for pharmacological intervention. Cell Cycle. 2004;3(1):51–60. [PubMed] [Google Scholar]
- 26.Lerdrup M, Hommelgaard AM, Grandal M, van Deurs B. Geldanamycin stimulates internalization of ErbB2 in a proteasome-dependent way. J Cell Sci. 2006;119(Pt 1):85–95. doi: 10.1242/jcs.02707. [DOI] [PubMed] [Google Scholar]
- 27.Yan YY, et al. Blockade of Her2/neu binding to Hsp90 by emodin azide methyl anthraquinone derivative induces proteasomal degradation of Her2/neu. Mol Pharm. 2011;8(5):1687–1697. doi: 10.1021/mp2000499. [DOI] [PubMed] [Google Scholar]
- 28.Zagouri F, et al. Hsp90 inhibitors in breast cancer: A systematic review. Breast. 2013;22(5):569–578. doi: 10.1016/j.breast.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Chen YF, Chen YT, Chiu WT, Shen MR. Remodeling of calcium signaling in tumor progression. J Biomed Sci. 2013;20:23. doi: 10.1186/1423-0127-20-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monteith GR, Davis FM, Roberts-Thomson SJ. Calcium channels and pumps in cancer: Changes and consequences. J Biol Chem. 2012;287(38):31666–31673. doi: 10.1074/jbc.R112.343061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roderick HL, Cook SJ. Ca2+ signalling checkpoints in cancer: Remodelling Ca2+ for cancer cell proliferation and survival. Nat Rev Cancer. 2008;8(5):361–375. doi: 10.1038/nrc2374. [DOI] [PubMed] [Google Scholar]
- 32.Stern DF, Heffernan PA, Weinberg RA. p185, a product of the neu proto-oncogene, is a receptorlike protein associated with tyrosine kinase activity. Mol Cell Biol. 1986;6(5):1729–1740. doi: 10.1128/mcb.6.5.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carraway CA, Carvajal ME, Li Y, Carraway KL. Association of p185neu with microfilaments via a large glycoprotein complex in mammary carcinoma microvilli. Evidence for a microfilament-associated signal transduction particle. J Biol Chem. 1993;268(8):5582–5587. [PubMed] [Google Scholar]
- 34.Hommelgaard AM, Lerdrup M, van Deurs B. Association with membrane protrusions makes ErbB2 an internalization-resistant receptor. Mol Biol Cell. 2004;15(4):1557–1567. doi: 10.1091/mbc.E03-08-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanova JL, et al. Application of fusion protein 4D5 scFv-dibarnase:barstar-gold complex for studying P185HER2 receptor distribution in human cancer cells. Biochimie. 2012;94(8):1833–1836. doi: 10.1016/j.biochi.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Mori S, et al. Light and electron microscopical demonstration of c- erB-2 gene product-like immunoreactivity in human malignant tumors. Virchows Arch B Cell Pathol Incl Mol Pathol. 1987;54(1):8–15. doi: 10.1007/BF02899192. [DOI] [PubMed] [Google Scholar]
- 37.Borg JP, et al. ERBIN: A basolateral PDZ protein that interacts with the mammalian ERBB2/HER2 receptor. Nat Cell Biol. 2000;2(7):407–414. doi: 10.1038/35017038. [DOI] [PubMed] [Google Scholar]
- 38.Padányi R, et al. Apical scaffolding protein NHERF2 modulates the localization of alternatively spliced plasma membrane Ca2+ pump 2B variants in polarized epithelial cells. J Biol Chem. 2010;285(41):31704–31712. doi: 10.1074/jbc.M110.164137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bose R, et al. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc Natl Acad Sci USA. 2006;103(26):9773–9778. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardone RA, et al. The NHERF1 PDZ2 domain regulates PKA-RhoA-p38-mediated NHE1 activation and invasion in breast tumor cells. Mol Biol Cell. 2007;18(5):1768–1780. doi: 10.1091/mbc.E06-07-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boehmerle W, et al. Chronic exposure to paclitaxel diminishes phosphoinositide signaling by calpain-mediated neuronal calcium sensor-1 degradation. Proc Natl Acad Sci USA. 2007;104(26):11103–11108. doi: 10.1073/pnas.0701546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986;31(4):229–234. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]