Significance
We combine genetics, biochemistry, cell biology, and proteomics to define Bardet–Biedl Syndrome complex (BBSome) composition, location, and function in the deadly parasite Trypanosoma brucei. BBSome mutants have reduced infectivity in mice, and quantitative proteomics identified parasite surface proteome changes that may underlie reduced virulence. To our knowledge, this work presents the first comprehensive study of the BBSome in any microbial pathogen. T. brucei is also among the earliest organisms to have diverged from other eukaryotes, showing deep evolutionary origins of the BBSome. Localization to membranes and vesicles at the flagellar pocket, together with functional analyses and interaction with clathrin and ubiquitin, supports a model whereby the BBSome functions in postendocytic sorting of select surface proteins.
Keywords: BBSome, virulence, cilium, clathrin, ubiquitin
Abstract
Cilia (eukaryotic flagella) are present in diverse eukaryotic lineages and have essential motility and sensory functions. The cilium’s capacity to sense and transduce extracellular signals depends on dynamic trafficking of ciliary membrane proteins. This trafficking is often mediated by the Bardet–Biedl Syndrome complex (BBSome), a protein complex for which the precise subcellular distribution and mechanisms of action are unclear. In humans, BBSome defects perturb ciliary membrane protein distribution and manifest clinically as Bardet–Biedl Syndrome. Cilia are also important in several parasites that cause tremendous human suffering worldwide, yet biology of the parasite BBSome remains largely unexplored. We examined BBSome functions in Trypanosoma brucei, a flagellated protozoan parasite that causes African sleeping sickness in humans. We report that T. brucei BBS proteins assemble into a BBSome that interacts with clathrin and is localized to membranes of the flagellar pocket and adjacent cytoplasmic vesicles. Using BBS gene knockouts and a mouse infection model, we show the T. brucei BBSome is dispensable for flagellar assembly, motility, bulk endocytosis, and cell viability but required for parasite virulence. Quantitative proteomics reveal alterations in the parasite surface proteome of BBSome mutants, suggesting that virulence defects are caused by failure to maintain fidelity of the host–parasite interface. Interestingly, among proteins altered are those with ubiquitination-dependent localization, and we find that the BBSome interacts with ubiquitin. Collectively, our data indicate that the BBSome facilitates endocytic sorting of select membrane proteins at the base of the cilium, illuminating BBSome roles at a critical host–pathogen interface and offering insights into BBSome molecular mechanisms.
Cilia, also called eukaryotic flagella, are emblematic organelles for which functional and structural similarities across diverse lineages indicate they have existed since the emergence of eukaryotes (1, 2). Although historically considered as machines for cell locomotion and movement of fluids across epithelia, cilia are now recognized as signaling platforms that sense and transduce environmental stimuli to drive cellular responses (3). As such, cilia constitute a critical cell–environment interface that is paramount for development and physiology of ciliated organisms. In vertebrates, cilium-dependent signaling orchestrates important developmental pathways, such as limb development and kidney morphogenesis, and is required for vision, hearing, and smell (4). In free-living protists, cilium signaling controls cell motility, mating, and response to extracellular cues (5).
The cilium is anchored to the cell surface membrane and protrudes into the extracellular milieu. At the core of the organelle is a microtubule-based axoneme that originates at the basal body in the cytoplasm. The axoneme is encased by a ciliary membrane that is contiguous with the plasma membrane but constitutes a specialized domain with distinct protein and lipid composition (6). The base of the cilium is not entirely delimited by membrane, and the ciliary matrix (soluble fraction within the cilium) is, thus, topologically contiguous with the cytoplasm. Organellar identity is maintained by a diffusion barrier that bona fide ciliary proteins must traverse; traversal of this barrier in and out of the cilium is critical for cilium function (7).
In keeping with their critical motility and sensory functions, defective cilia cause a wide range of inherited human diseases, termed ciliopathies, which exhibit diverse clinical manifestations and molecular etiologies (8). Bardet–Biedl Syndrome (BBS) is a ciliopathy that is mainly characterized by retinopathy, obesity, polydactyly, cognitive impairment, renal abnormalities, and hypogonadism; we currently know of 19 genes (bbs1–bbs19) that, when mutated, can cause BBS (9). Interestingly, eight bbs genes (bbs1, bbs2, bbs4, bbs5, bbs7, bbs8, bbs9, and bbs18) encode proteins that assemble into a complex termed the Bardet–Biedl Syndrome complex (BBSome) (10, 11). While cilium assembly is generally unaffected, BBSome mutants in vertebrates and protists exhibit sensing defects resulting from abnormal localization of select ciliary proteins (12–18). Although the BBSome is necessary for dynamic trafficking of these membrane-associated proteins through the ciliary compartment, its precise location and exact function remain enigmatic.
Ciliated pathogens cause tremendous human suffering worldwide and limit economic development in some of the world’s poorest regions (19). Despite broad awareness of the cilium’s role in the pathology of inherited human diseases, the contribution of cilia and ciliary modules, such as the BBSome, to infection by eukaryotic parasites is mostly unknown (20). Parasite survival and virulence depend on successful interaction with the host environment, and this interaction is mediated, at least in part, by cilia and ciliary proteins (21). This paradigm applies to the unicellular parasite Trypanosoma brucei, which causes African sleeping sickness in humans and Nagana in cattle. Sleeping sickness is endemic to sub-Saharan Africa, is almost always fatal if untreated, and remains one of the world’s most neglected diseases (22). T. brucei has a single flagellum, which emerges from the cytoplasm through the flagellar pocket at the posterior end of the cell (23). The flagellar pocket is a pronounced invagination of the plasma membrane that marks the boundary between the flagellar membrane and the rest of the plasma membrane. The trypanosome flagellar pocket is a key host–parasite portal, because it is the sole site of endocytosis, mediates uptake of growth factors, and is a necessary transit point for proteins en route to or from the cell surface (24).
Given the role of the BBSome in controlling delivery of ciliary proteins important for interaction with the external environment, we examined BBSome functions in mammalian-infectious, bloodstream-stage T. brucei. We report that T. brucei BBS proteins assemble into a BBSome [T. brucei Bardet–Biedl Syndrome complex (TbBBSome)] that localizes to membranes of the flagellar pocket and adjacent cytoplasmic vesicles. BBS gene knockouts (KOs) show the TbBBSome is dispensable for flagellum assembly and parasite viability but required for virulence in a mouse infection model. Quantitative proteomics and biochemical analysis suggest that the TbBBSome interacts with clathrin and ubiquitin to facilitate endocytic trafficking of select cell surface proteins and that defects in these processes underlie the virulence defect of BBSome mutants. Our combined studies offer insights into both parasite biology and the pathophysiology of human ciliopathies.
Results and Discussion
T. brucei BBS Proteins Assemble into a BBSome.
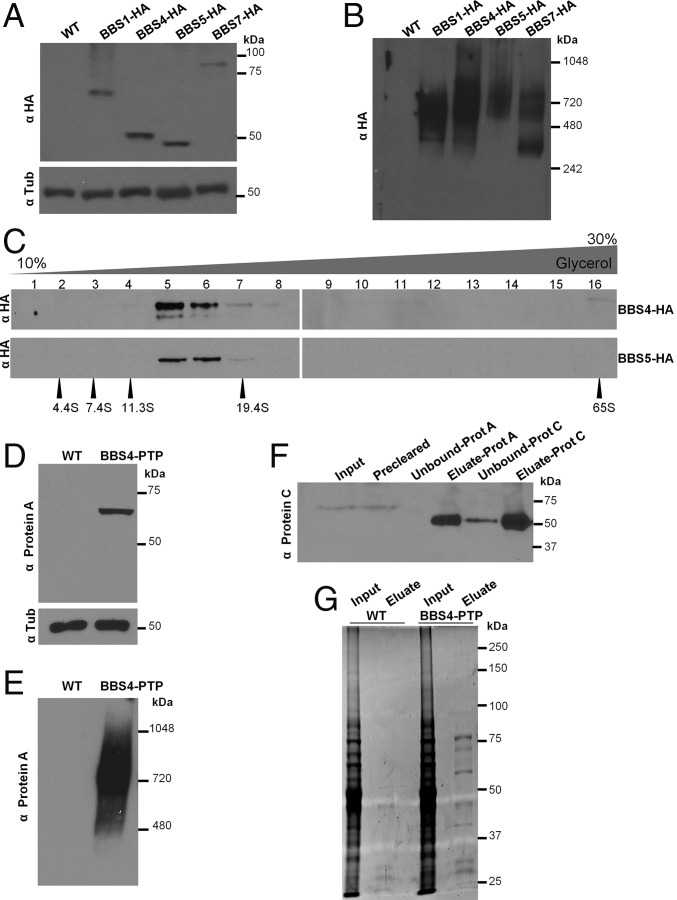
Similar to the majority of ciliated eukaryotes, T. brucei encodes all eight BBSome proteins, but unlike vertebrates, it lacks the canonical BBS chaperonins BBS6, BBS10, and BBS12, which mediate assembly of the BBSome (11, 25, 26). Therefore, we sought to determine whether T. brucei Bardet–Biedl Syndrome (TbBBS) proteins assemble into a complex in mammalian-infectious T. brucei cells. To this end, we tagged four BBSome proteins (TbBBS1, TbBBS4, TbBBS5, and TbBBS7) with the triple-HA tag at the endogenous loci and verified expression of fusion proteins in TbBBS-HA/− background (Fig. 1A). Blue native electrophoresis showed that all four fusion proteins migrate as part of an ∼700-kDa complex, whereas TbBBS7-HA is also part of a 242- to 480-kDa subcomplex (Fig. 1B). Furthermore, TbBBS4-HA and TbBBS5-HA sediment within ∼14.5S particles in glycerol gradient centrifugation (Fig. 1C). The size of the complex observed by blue native electrophoresis is consistent with the size predicted from subunit molecular masses (487 kDa) and the size reported in other organisms (10, 26). Moreover, the ∼14.5S particle observed in glycerol gradient sedimentation is in close agreement with the ∼14S value reported for the BBSome in mammals (10, 27). A smaller particle (∼12S) was reported for Chlamydomonas (18). Monomeric TbBBS proteins were not detected, indicating that the vast majority of the TbBBS protein pool is part of the BBSome.
Fig. 1.
TbBBS proteins assemble into the TbBBSome. (A) Western blot for 3× HA epitope-tagged TbBBS proteins. Wild type (WT) was used as negative control. Tubulin (Tub) blot served as loading control. (B) Western blot for HA-tagged TbBBS proteins by blue native PAGE. (C) Western blot for TbBBS4-HA and TbBBS5-HA in 10–30% (vol/vol) glycerol gradient centrifugation. Numbers 1–16 indicate gradient fractions. Protein standards of known S values are shown at the bottom. (D) Western blot for PTP epitope-tagged TbBBS4. WT was used as negative control. Tub blot served as loading control. (E) Western blot for TbBBS4-PTP by blue native PAGE. (F) Western blot for tandem purification of TbBBS4-PTP probed with anti-Protein C. TEV protease was used for elution before Protein C purification, hence the reduction in size. (G) SYPRO Ruby protein stain for tandem purification of TbBBS4-PTP. Eluates were used for shotgun proteomics (SI Appendix, Tables S1 and S5).
We next affinity-purified the TbBBSome and determined its composition by shotgun proteomics. To this end, we fused TbBBS4 to a dual tag consisting of Protein C and Protein A epitopes [Protein C-tobacco etch virus (TEV) protease site-Protein A (PTP) tag] and verified that TbBBS4-PTP is expressed in TbBBS4-PTP/− background (Fig. 1D) and remains part of an ∼700-kDa complex (Fig. 1E) as seen for TbBBS-HA proteins. We performed tandem affinity purification (Fig. 1 F and G) and detected all eight predicted BBSome subunits as the most abundant hits in the TbBBS4-PTP eluate (SI Appendix, Table S1). These proteins were not recovered in parallel purifications using control cell extracts.
Collectively, our data show presence of the BBSome in a divergent eukaryotic supergroup and make T. brucei one of a few systems in which an actual BBSome has been experimentally shown (10, 17, 18, 26). Trypanosomes are members of the Excavates, which are early branching unicellular organisms suggested to be a sister group to all other eukaryotes (28). Presence of an intact BBSome in this divergent lineage supports an early emergence in eukaryotic evolution (10, 25). Formation of the TbBBSome, despite the lack of canonical BBS chaperonins, suggests that BBSome assembly may occur intrinsically or rely on alternative chaperonins in T. brucei and other nonvertebrate organisms.
TbBBSome Associates with Membranes and Vesicles at the Flagellar Pocket.
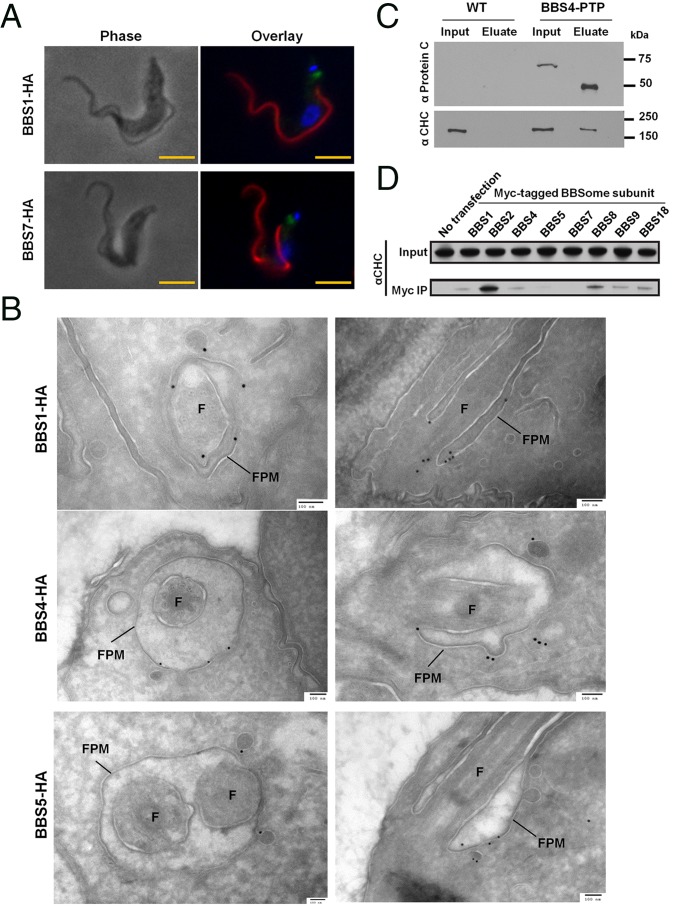
Despite its importance to cilium biology, the precise subcellular location of the BBSome is unclear (29). We assessed subcellular localization of TbBBS-HA proteins by immunofluorescence (IF) and detected all fusion proteins near the base of the flagellum (Fig. 2A and SI Appendix, Fig. S4). To date, most studies have relied on IF for determining BBSome localization. To determine TbBBSome location at higher resolution, we performed immuno-EM on three TbBBS-HA proteins. In agreement with IF localizations (Fig. 2A and SI Appendix, Fig. S4) and ref. 30, we did not detect any specific labeling within the flagellum proper. Instead, we detected gold particles on the flagellar pocket membrane and neighboring cytoplasmic vesicles for all three TbBBS-HA proteins examined (Fig. 2B and SI Appendix, Table S2). For TbBBS1-HA, we also detected a few gold particles at the basal body (Fig. 2B and SI Appendix, Table S2). Such localizations were rarely observed in control samples (SI Appendix, Table S2). Our results provide the first demonstration, to our knowledge, of BBSome localization to flagellar pocket and vesicle membranes in vivo. This finding extends in vitro observations that the BBSome forms a coat on liposomes and supports the paradigm that the BBSome regulates trafficking of membrane proteins at the cilium base (14).
Fig. 2.
The TbBBSome localizes to the flagellar pocket membrane and interacts with clathrin. (A) IF for TbBBS-HA proteins: TbBBS-HA (green), PFR2 (red), and DAPI (blue). (Scale bar: 5 µm.) (B) Immunogold labeling for TbBBS-HA proteins. Cross-sections and longitudinal sections of the flagellar pocket are shown in Left and Right, respectively. Flagellar pocket membrane (FPM) and flagellum (F) are indicated. (Scale bar: 100 nm.) Immunogold quantification is provided in SI Appendix, Table S2. (C) Western blot of TbBBS4-PTP immunoprecipitation by Protein A probed with anti-Protein C and anti-CHC. TEV protease was used to elute TbBBS4-PTP, hence the reduced size of TbBBS4 in the eluate. Input (1×) and eluate (8×) in anti-Protein C blot; input (1×) and eluate (100×) in anti-CHC blot. (D) Western blot for CHC in immunoprecipitation (IP) of Myc-tagged BBSome proteins from HEK293 cells.
In vertebrates, nematodes, and algae, the major pool of BBSome is near the cilium base, but it is also observed to travel with intraflagellar transport particles along the axoneme (18, 31–34). Absence of detectable TbBBSome in the flagellum itself might reflect technical limitations owing to low abundance of BBSome proteins (32, 33). Alternatively, it might reflect T. brucei-specific adaptations given the extensive elaboration of the flagellar pocket as the sole site of endocytosis in these organisms (24). In either case, our data suggest a TbBBSome role in vesicle trafficking at the flagellar pocket, a function in agreement with the phenotypes of Leishmania, nematode, and vertebrate BBS mutants (20, 35–38).
Association of TbBBS proteins with vesicles at the flagellar pocket points to a role in endocytosis and prompted us to ask whether the TbBBSome interacts with clathrin. Clathrin-mediated endocytosis at the flagellar pocket is considered the sole means of endocytosis in T. brucei (39) and depends on conserved and lineage-specific proteins but not AP-2 adaptin (40). Interestingly, we detected clathrin heavy chain (CHC) in TbBBS4-PTP purifications but not in control eluates (Fig. 2C). To determine whether this is a trypanosome-specific feature, we asked whether the mammalian BBSome also interacts with clathrin. We found that human BBSome proteins associated with CHC (Fig. 2D), leading us to speculate that the BBSome may function in concert with clathrin across all BBS-encoding lineages. Intriguingly, the BBSome exhibits similarities to clathrin adaptor protein complexes, such as common structural domains, membrane recruitment by small GTPases, and the ability to bind lipids (10, 14). Moreover, clathrin-mediated endocytosis is thought to operate at the base of the cilium to control cilium-mediated signaling in multiple organisms (36, 41). It is possible that endocytosis and vesicular transport were ancestral functions of the BBSome (for example, participating in phagocytosis in a protoeukaryote) and that these functions were later repurposed into a dedicated ciliary trafficking machinery on emergence of the cilium (1, 2, 25).
TbBBSome Is Required for Parasite Virulence.
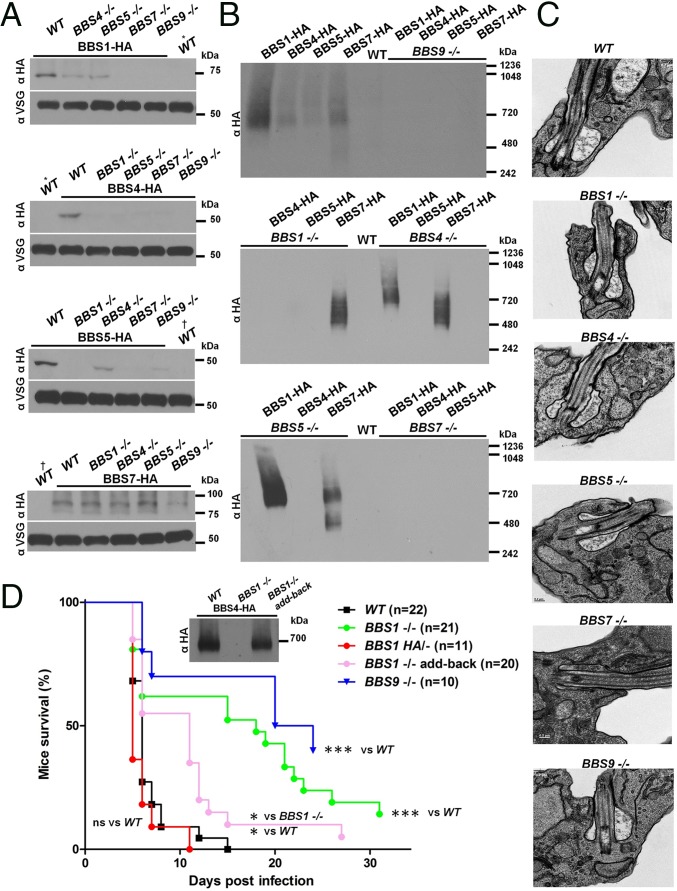
To gain insight into TbBBSome functions, we generated gene KOs of TbBBS1, TbBBS4, TbBBS5, TbBBS7, and TbBBS9. TbBBS KOs were generated in cells expressing HA-tagged BBS proteins to enable systematic analysis of the impact on BBS proteins and the BBSome. TbBBS KOs were verified by Southern blotting (SI Appendix, Fig. S1A). Expression levels of TbBBS proteins and assembly of the TbBBSome were differentially compromised in individual KOs (Fig. 3 A and B). Ablation of TbBBS4 or TbBBS5 had modest effects on the rest of the complex, whereas loss of TbBBS1 resulted in a partial TbBBSome that contains at least TbBBS7 (Fig. 3 A and B). In contrast, deletion of TbBBS7 or TbBBS9 seemed to disrupt TbBBSome altogether (Fig. 3 A and B). Together, these results indicate that TbBBS7 and TbBBS9 are core subunits, whereas TbBBS1, TbBBS4, and TbBBS5 are in the periphery of the complex. Our results show that the hierarchy of BBSome assembly in the evolutionarily divergent T. brucei is conserved with that observed in mammals (10, 42, 43).
Fig. 3.
The TbBBSome is dispensable for viability but required for parasite virulence. (A) Western blot for TbBBS-HA proteins in TbBBS KO mutants or WT background as a positive control. Lysates from WT cells (i.e., without an HA tag) served as negative control. WT lanes are the same for BBS1-HA/BBS4-HA blots (*) and BBS5-HA/BBS7-HA blots (†). VSG was used as loading control. (B) Western blot for TbBBS-HA proteins in TbBBS KO mutants by blue native PAGE. WT lysates served as negative control. The same lysates were run in parallel and blotted for VSG on SDS/PAGE (A) to verify equal total protein. (C) Transmission electron micrographs of WT and TbBBS mutants at the flagellar pocket region. (D) Survival curve for mice infected with TbBBS mutants. Only mice in which parasitemia was detected are shown. Inset shows the Western blot for TbBBS4-HA by blue native PAGE. ns, Not significant. *P < 0.05 by Mantel–Cox test; ***P < 0.001 by Mantel–Cox test.
The BBSome is not generally required for cilium morphogenesis (12, 18). However, in certain cases, loss of BBSome proteins leads to severe defects in cilium assembly (12, 17, 38, 44) or vesicle accumulation at the cilium base (20, 35), suggesting organism- and cell-specific requirements for the complex. Depletion of the TbBBSome had no discernible effect on parasite motility (SI Appendix, Fig. S1B), had only a minor effect on in vitro growth rate (SI Appendix, Fig. S1C), and did not cause conspicuous defects in axoneme structure (SI Appendix, Fig. S1D). Given the localization of the TbBBSome (Fig. 2 A and B and SI Appendix, Fig. S4), we also examined ultrastructure of the flagellar pocket and did not detect any alterations in TbBBS KOs (Fig. 3C). The TbBBSome is, thus, dispensable for parasite viability and motility as well as flagellar and cellular morphogenesis.
Having established that the TbBBSome is dispensable for parasite viability, we asked whether it is required for virulence in a mouse infection model. We used mutants that exhibit substantial loss of the complex, such as TbBBS1 KO and TbBBS9 KO. Control parasites mount a lethal mouse infection within 5–14 days (Fig. 3D). Strikingly, both TbBBS mutants exhibited decreased virulence as judged by prolonged mouse survival, including some animals that cleared detectable parasites from the bloodstream after an established infection and survived to the end of the experimental timeframe (Fig. 3D). Reintroduction of WT BBS1 restored the BBSome and suppressed the virulence defect of TbBBS1 KOs (Fig. 3D), showing that loss of the complex is responsible for the virulence defect. Moreover, BBS1-HA/− virulence was indistinguishable from control parasites, indicating that the HA tag does not affect TbBBS1 function (Fig. 3D).
TbBBSome Regulates Endocytic Trafficking of Select Membrane Proteins.
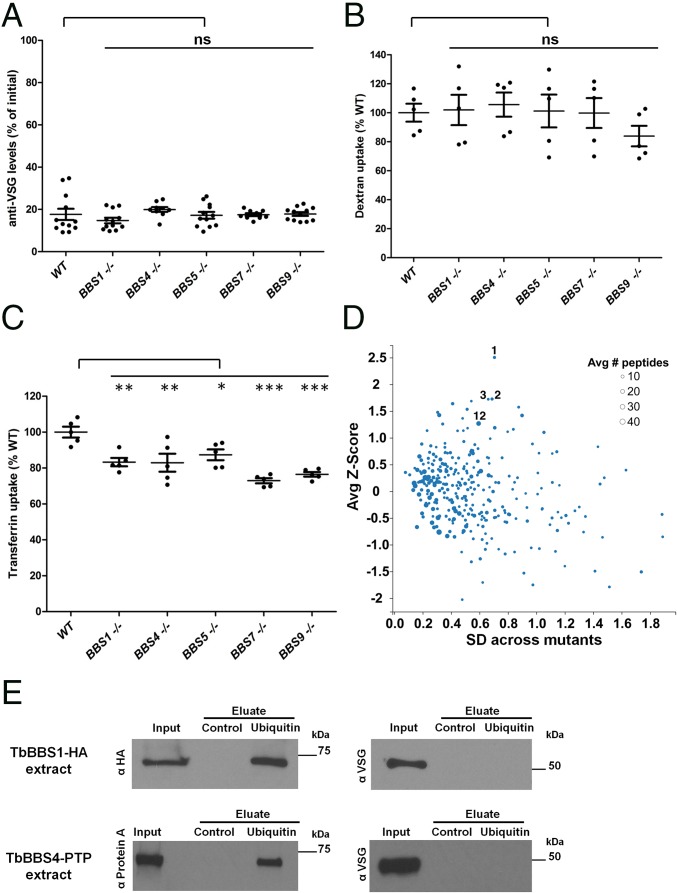
KO of the BBS1 gene in the parasite Leishmania major reduced virulence in a mouse footpad model of infection (20), but underlying mechanisms and presence of a BBSome were not examined. Given the localization of the TbBBSome, we hypothesized that virulence defects in T. brucei stem from failure to orchestrate trafficking events at the flagellar pocket, a critical host–parasite interface responsible for endocytosis of host antibodies and uptake of host-derived growth factors (24). Host antibodies bind to parasite Variant Surface Glycoproteins (VSGs) that cover the cell surface, mask other epitopes, and switch stochastically (45). VSG switching and rapid endocytic clearance of VSG–Ig complexes through the flagellar pocket enable the parasite population to avoid destruction by the host immune system (46). Therefore, we asked whether TbBBS KOs have defects in clearance of VSG–Ig complexes from the parasite surface, but mutants were similar to WT (Fig. 4A). Dextran uptake was also unaffected, indicating the TbBBSome is not required for fluid-phase endocytosis (Fig. 4B). We then gauged receptor-mediated endocytosis by assessing transferrin uptake. In T. brucei, transferrin uptake is mediated by the parasite’s heterodimeric transferrin receptor (TfR) that comprises the products of expression site-associated gene 6 (ESAG6) and ESAG7 and localizes to the flagellar pocket (47, 48). TfR binds transferrin at the flagellar pocket and delivers it to acidic endosomes, where transferrin is released and sorted to lysosomes for degradation; TfR then gets recycled to the cell surface (49). Interestingly, we found that transferrin uptake is impaired in all TbBBS KOs, with TbBBS7 and TbBBS9 mutants showing the most severe defects (Fig. 4C). Impaired uptake is not caused by lower total abundance of TfR or CHC (SI Appendix, Fig. S2A). Moreover, TfR itself is functional in the TbBBS mutants, because in conditions that block endocytosis, transferrin is bound at the cell surface at levels roughly equivalent to those in WT cells (SI Appendix, Fig. S2B). Notably, TbBBS4-HA/− and TbBBS4-PTP/− had no defects in transferrin uptake (SI Appendix, Fig. S2C), further showing that tagged TbBBS proteins are functional (Fig. 3D).
Fig. 4.
The TbBBSome regulates endocytic trafficking of select membrane proteins. (A) Anti-VSG levels at 5-min clearance as percentages of levels at 0 min. (B) Dextran uptake at 15 min as percentage of WT uptake. (C) Transferrin uptake at 15 min as percentage of WT uptake. In A–C, data are represented as means ± SEMs. ns, Not significant. *P < 0.05 by one-way ANOVA with Dunnett’s test; **P < 0.01 by one-way ANOVA with Dunnett’s test; and ***P < 0.001 by one-way ANOVA with Dunnett’s test. (D) Plot comparing cell surface protein abundance between WT and BBS mutants by stable isotope labeling by amino acids in cell culture (SILAC) proteomics. Only proteins detected in WT and all five BBS mutants are shown. The average BBS mutant to WT Z score of log2(fold change) is plotted against the SD across all five mutants. The size of each dot corresponds to the number of identified peptides per protein. Dots labeled 1, 2, 3, and 12 correspond to proteins in SI Appendix, Table S3 sorted by descending Z score (VSG-related, MBAP1, ESAG2, and ISG75, respectively). (E) Western blot for protein pulldowns. TbBBS1-HA and TbBBS4-PTP cell extracts were incubated with control beads and ubiquitin beads. Samples were blotted with anti-HA and anti-Protein A to detect tagged TbBBS proteins as well as anti-VSG as control for nonspecific pulldown. Input (1×) and eluate (320×) in anti-HA and anti-Protein A blots; Input (1×) and eluate (8,000×) in anti-VSG blots.
Defects in transferrin uptake have been reported for a plethora of T. brucei mutants and are typically accompanied by general defects in endocytosis, flagellar pocket enlargement, and cell lethality (50, 51), which are not observed in TbBBS mutants (Fig. 3C and SI Appendix, Fig. S1C). Therefore, we postulate that, in TbBBS mutants, transferrin-bound TfR is endocytosed but not properly sorted within the endosomal system; defective sorting results in decreased transferrin release within the cell and increased recycling of transferrin-bound TfR to the cell surface. Taken together with the absence of gross morphological or functional aberrations to the flagellar pocket, our results indicate that the TbBBSome is dispensable for general clathrin-mediated endocytosis (39) and that it controls the postendocytic fate of TfR and possibly, other membrane-associated surface proteins. This idea is consistent with the notion that surface protein uptake in T. brucei is nonselective and that sorting takes place at postendocytic steps (24).
We suspected that a subset of surface proteins beyond TfR depends on TbBBSome for trafficking and that these will be over- or underrepresented on the cell surface of TbBBS mutants compared with control cells. To test this hypothesis, we used an unbiased quantitative proteomic analysis to identify changes in the cell surface proteome of TbBBS KOs. Stable isotope labeling by amino acids in cell culture (SILAC) quantitative proteomics revealed that, although most proteins were of similar abundance in TbBBS KO and control cells, a few were consistently dysregulated in all five TbBBS KOs (Fig. 4D and SI Appendix, Tables S3 and S4 and Fig. S3). Among the most overabundant proteins were T. brucei membrane-bound acidic phosphatase (TbMBAP), ESAG2, two VSG-related proteins, and three members of the invariant surface glycoprotein (ISG) family (Fig. 4D and SI Appendix, Table S3). TbMBAP traffics between the flagellar pocket and endosomes and normally, is barely detectable at the cell surface but accumulates there on overexpression (52), indicating that saturable sorting mechanisms control its surface distribution. ESAG2 and ISG75 are also known surface proteins (53).
ISG75 is ubiquitinated, and mutation of the ISG75 ubiquitination motif results in overaccumulation at the cell surface, whereas loss of an ubiquitin hydrolase reduces ISG75 abundance (54, 55). The combined results indicate that ubiquitination promotes ISG75 endocytic trafficking and delivery to lysosomes for degradation (54, 55). Intriguingly, we noticed in our proteomics data that ubiquitin was consistently found to copurify with TbBBS4-PTP (SI Appendix, Table S5). To independently test whether the TbBBSome interacts with ubiquitin, we incubated parasite cell extracts with ubiquitin beads, and we, indeed, detected specific pulldown of TbBBS1-HA and TbBBS4-PTP but not of an abundant control protein (Fig. 4E). Thus, it is possible that TbBBSome interacts with ubiquitinated proteins, such as ISG75, and regulates their endocytic trafficking, although additional experiments are needed to test this directly. Nonetheless, our results present the first demonstration, to our knowledge, that BBSome proteins interact with ubiquitin. Although BBSome proteins lack canonical ubiquitin-binding domains, BBS5 contains PH-like domains (10); such domains in other proteins have been shown to bind not only phosphoinositides but also, ubiquitin (56). Thus, it is conceivable that BBS5 binds ubiquitin through its PH-like domains. In other organisms, the BBSome has been implicated in ubiquitin-mediated pathways, and many BBS mutant phenotypes could be explained by a BBSome–ubiquitin interaction. For instance, BBSome proteins interact with the proteasome and are required for degradation of signaling molecules (57). Moreover, the Notch receptor is normally sorted to lysosomes on ubiquitination but accumulates in late endosomes in BBSome mutants (37). Similarly, ubiquitination of Patched1 and Smoothened is thought to promote trafficking out of cilia (58, 59), and both proteins fail to exit cilia in the absence of the BBSome (16). Interestingly, BBS11, a vertebrate-specific BBS protein that is not part of the BBSome, is an E3 ubiquitin ligase that acts on dysbindin, a regulator of endosomal–lysosomal trafficking (60). Finally, a recent study provided evidence that the BBSome is required for trafficking of ubiquitin-conjugated membrane proteins out of cilia and subsequent lysosomal degradation (61). Thus, it seems that recognizing an ubiquitin moiety is a conserved and integral element of BBSome-dependent protein trafficking.
In summary, we provide the first demonstration, to our knowledge, of a BBSome in a member of the Excavate eukaryotic supergroup. We show the TbBBSome is localized to the flagellar pocket membrane and neighboring vesicles, suggesting a role in endocytic trafficking that is further supported by interaction with clathrin and ubiquitin. Loss of the TbBBSome has pleiotropic effects on the fidelity of the cell surface proteome, and these effects likely underlie the attenuated virulence of TbBBSome mutants. Together, our results expand the realm of BBSome-dependent functions to novel host–parasite interactions and suggest that defective endocytosis of ubiquitinated ciliary proteins might contribute to the pathobiology of human ciliopathies.
Experimental Procedures
Detailed methods are provided in SI Appendix, SI Experimental Procedures. Bloodstream-form T. brucei cells were used, and cell culture, cell fractionation, uptake assays, mouse infection, molecular biology, affinity purification, stable isotope labeling by amino acids in cell culture proteomics, tandem MS, immunostaining, and microscopy procedures were done according to standard methods. All animal experiments strictly complied with the Institutional Animal Care and Use Committee of University of California, Los Angeles (approved protocol permit ARC 2001-065).
Supplementary Material
Acknowledgments
We thank Eva Ng, Selena Zhou, Ankeet Vakharia, and Frederique de Lame for technical assistance. We thank Sofia Gkountela for assistance with flow cytometry, Wandy Beatty (Washington University) for immuno-EM, and Chantal Allamargot and Randy Nessler (University of Iowa) for transmission EM. We also thank Robert Sabatini (University of Georgia) for plasmids and James Bangs (University at Buffalo, SUNY), Mark Field (University of Dundee), and Piet Borst (The Netherlands Cancer Institute) for antibodies. Funding was provided by NIH Ruth L. Kirschstein NRSA Grant AI094750 (to M.M.S) and NIH Grants GM089778 (to J.A.W.) and AI052348 (to K.L.H.) as well as the Burroughs Wellcome Fund PATH Award (to K.L.H.). G.L. was supported by a UCLA Dissertation Year Fellowship and a UCLA Warsaw Fellowship. E.A.S. was supported by NIH-USPHS-NRSA Grant GM07104. R.S. acknowledges support from a QCB Collaboratory Postdoctoral Fellowship and the QCB Collaboratory Community directed by Matteo Pellegrini. A.R.N. was supported by Fayez Sarofim Fellowship of the Damon Runyon Cancer Research Foundation DRG 2160-13. W.D.B. was supported by CMB Ruth L. Kirschstein NRSA Training Grant GM007185.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.B.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518079113/-/DCSupplemental.
References
- 1.Jékely G, Arendt D. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. BioEssays. 2006;28(2):191–198. doi: 10.1002/bies.20369. [DOI] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life. Cold Spring Harb Perspect Biol. 2014;6(9):a016006. doi: 10.1101/cshperspect.a016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloodgood RA. Sensory reception is an attribute of both primary cilia and motile cilia. J Cell Sci. 2010;123(Pt 4):505–509. doi: 10.1242/jcs.066308. [DOI] [PubMed] [Google Scholar]
- 4.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginger ML, Portman N, McKean PG. Swimming with protists: Perception, motility and flagellum assembly. Nat Rev Microbiol. 2008;6(11):838–850. doi: 10.1038/nrmicro2009. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Gonzalo FR, Reiter JF. Scoring a backstage pass: Mechanisms of ciliogenesis and ciliary access. J Cell Biol. 2012;197(6):697–709. doi: 10.1083/jcb.201111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: How to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: An emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 9.Novas R, Cardenas-Rodriguez M, Irigoín F, Badano JL. Bardet-Biedl syndrome: Is it only cilia dysfunction? FEBS Lett. 2015;589(22):3479–3491. doi: 10.1016/j.febslet.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 10.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129(6):1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 11.Loktev AV, et al. A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell. 2008;15(6):854–865. doi: 10.1016/j.devcel.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Mykytyn K, et al. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc Natl Acad Sci USA. 2004;101(23):8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci USA. 2008;105(11):4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin H, et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141(7):1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loktev AV, Jackson PK. Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Reports. 2013;5(5):1316–1329. doi: 10.1016/j.celrep.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q, Seo S, Bugge K, Stone EM, Sheffield VC. BBS proteins interact genetically with the IFT pathway to influence SHH-related phenotypes. Hum Mol Genet. 2012;21(9):1945–1953. doi: 10.1093/hmg/dds004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valentine MS, et al. Paramecium BBS genes are key to presence of channels in Cilia. Cilia. 2012;1(1):16. doi: 10.1186/2046-2530-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lechtreck KF, et al. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187(7):1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart K, et al. Kinetoplastids: Related protozoan pathogens, different diseases. J Clin Invest. 2008;118(4):1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price HP, et al. The Leishmania major BBSome subunit BBS1 is essential for parasite virulence in the mammalian host. Mol Microbiol. 2013;90(3):597–611. doi: 10.1111/mmi.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmon D, et al. Adenylate cyclases of Trypanosoma brucei inhibit the innate immune response of the host. Science. 2012;337(6093):463–466. doi: 10.1126/science.1222753. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy PG. Clinical features, diagnosis, and treatment of human African trypanosomiasis (sleeping sickness) Lancet Neurol. 2013;12(2):186–194. doi: 10.1016/S1474-4422(12)70296-X. [DOI] [PubMed] [Google Scholar]
- 23.Langousis G, Hill KL. Motility and more: The flagellum of Trypanosoma brucei. Nat Rev Microbiol. 2014;12(7):505–518. doi: 10.1038/nrmicro3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Field MC, Carrington M. The trypanosome flagellar pocket. Nat Rev Microbiol. 2009;7(11):775–786. doi: 10.1038/nrmicro2221. [DOI] [PubMed] [Google Scholar]
- 25.van Dam TJ, et al. Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc Natl Acad Sci USA. 2013;110(17):6943–6948. doi: 10.1073/pnas.1221011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seo S, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci USA. 2010;107(4):1488–1493. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo S, et al. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet. 2011;7(11):e1002358. doi: 10.1371/journal.pgen.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He D, et al. An alternative root for the eukaryote tree of life. Curr Biol. 2014;24(4):465–470. doi: 10.1016/j.cub.2014.01.036. [DOI] [PubMed] [Google Scholar]
- 29.Smith TS, et al. Light-dependent phosphorylation of Bardet-Biedl syndrome 5 in photoreceptor cells modulates its interaction with arrestin1. Cell Mol Life Sci. 2013;70(23):4603–4616. doi: 10.1007/s00018-013-1403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price HP, et al. A role for the vesicle-associated tubulin binding protein ARL6 (BBS3) in flagellum extension in Trypanosoma brucei. Biochim Biophys Acta. 2012;1823(7):1178–1191. doi: 10.1016/j.bbamcr.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams CL, et al. Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat Commun. 2014;5:5813. doi: 10.1038/ncomms6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eguether T, et al. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev Cell. 2014;31(3):279–290. doi: 10.1016/j.devcel.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liew GM, et al. The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev Cell. 2014;31(3):265–278. doi: 10.1016/j.devcel.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei Q, et al. The BBSome controls IFT assembly and turnaround in cilia. Nat Cell Biol. 2012;14(9):950–957. doi: 10.1038/ncb2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilliam JC, et al. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012;151(5):1029–1041. doi: 10.1016/j.cell.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan OI, et al. Endocytosis genes facilitate protein and membrane transport in C. elegans sensory cilia. Curr Biol. 2012;22(6):451–460. doi: 10.1016/j.cub.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leitch CC, Lodh S, Prieto-Echagüe V, Badano JL, Zaghloul NA. Basal body proteins regulate Notch signaling through endosomal trafficking. J Cell Sci. 2014;127(Pt 11):2407–2419. doi: 10.1242/jcs.130344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen HJ, et al. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer’s vesicle cilia function. Hum Mol Genet. 2006;15(5):667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 39.Allen CL, Goulding D, Field MC. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J. 2003;22(19):4991–5002. doi: 10.1093/emboj/cdg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adung’a VO, Gadelha C, Field MC. Proteomic analysis of clathrin interactions in trypanosomes reveals dynamic evolution of endocytosis. Traffic. 2013;14(4):440–457. doi: 10.1111/tra.12040. [DOI] [PubMed] [Google Scholar]
- 41.Clement CA, et al. TGF-β signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Reports. 2013;3(6):1806–1814. doi: 10.1016/j.celrep.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, Yu D, Seo S, Stone EM, Sheffield VC. Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable bardet-biedl syndrome protein complex, the BBSome. J Biol Chem. 2012;287(24):20625–20635. doi: 10.1074/jbc.M112.341487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh Y, Nozaki S, Hartanto D, Miyano R, Nakayama K. Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J Cell Sci. 2015;128(12):2351–2362. doi: 10.1242/jcs.168740. [DOI] [PubMed] [Google Scholar]
- 44.Blacque OE, et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18(13):1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horn D. Antigenic variation in African trypanosomes. Mol Biochem Parasitol. 2014;195(2):123–129. doi: 10.1016/j.molbiopara.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engstler M, et al. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell. 2007;131(3):505–515. doi: 10.1016/j.cell.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 47.Salmon D, et al. A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell. 1994;78(1):75–86. doi: 10.1016/0092-8674(94)90574-6. [DOI] [PubMed] [Google Scholar]
- 48.Steverding D, Stierhof YD, Fuchs H, Tauber R, Overath P. Transferrin-binding protein complex is the receptor for transferrin uptake in Trypanosoma brucei. J Cell Biol. 1995;131(5):1173–1182. doi: 10.1083/jcb.131.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabiri M, Steverding D. Studies on the recycling of the transferrin receptor in Trypanosoma brucei using an inducible gene expression system. Eur J Biochem. 2000;267(11):3309–3314. doi: 10.1046/j.1432-1327.2000.01361.x. [DOI] [PubMed] [Google Scholar]
- 50.Hall B, Allen CL, Goulding D, Field MC. Both of the Rab5 subfamily small GTPases of Trypanosoma brucei are essential and required for endocytosis. Mol Biochem Parasitol. 2004;138(1):67–77. doi: 10.1016/j.molbiopara.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Demmel L, et al. The endocytic activity of the flagellar pocket in Trypanosoma brucei is regulated by an adjacent phosphatidylinositol phosphate kinase. J Cell Sci. 2014;127(Pt 10):2351–2364. doi: 10.1242/jcs.146894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engstler M, et al. The membrane-bound histidine acid phosphatase TbMBAP1 is essential for endocytosis and membrane recycling in Trypanosoma brucei. J Cell Sci. 2005;118(Pt 10):2105–2118. doi: 10.1242/jcs.02327. [DOI] [PubMed] [Google Scholar]
- 53.Gadelha C, et al. Architecture of a host-parasite interface: Complex targeting mechanisms revealed through proteomics. Mol Cell Proteomics. 2015;14(7):1911–1926. doi: 10.1074/mcp.M114.047647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leung KF, Riley FS, Carrington M, Field MC. Ubiquitylation and developmental regulation of invariant surface protein expression in trypanosomes. Eukaryot Cell. 2011;10(7):916–931. doi: 10.1128/EC.05012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alsford S, et al. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482(7384):232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slagsvold T, et al. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem. 2005;280(20):19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- 57.Liu YP, et al. Ciliopathy proteins regulate paracrine signaling by modulating proteasomal degradation of mediators. J Clin Invest. 2014;124(5):2059–2070. doi: 10.1172/JCI71898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J, et al. The role of ciliary trafficking in Hedgehog receptor signaling. Sci Signal. 2015;8(379):ra55. doi: 10.1126/scisignal.aaa5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia R, Jia H, Fan J, Liu Y, Jia J. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS Biol. 2012;10(1):e1001238. doi: 10.1371/journal.pbio.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Locke M, Tinsley CL, Benson MA, Blake DJ. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum Mol Genet. 2009;18(13):2344–2358. doi: 10.1093/hmg/ddp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu Q, et al. BBS4 and BBS5 show functional redundancy in the BBSome to regulate the degradative sorting of ciliary sensory receptors. Sci Rep. 2015;5:11855. doi: 10.1038/srep11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.