Significance
In the assisted reproductive technology (ART) clinic, pregnancy is defined by the rise of human chorionic gonadotropin upon embryo implantation. Achieving embryo implantation is a major roadblock to the success of ART; it is estimated that only 50% of transferred embryos implant in patients seeking ART, and that half of these embryos are subsequently lost. Thus, understanding the molecular pathways during the window of implantation will improve ART success. In this study, we conditionally deleted activin-like kinase 3 (ALK3) in mice and demonstrate that bone morphogenetic protein (BMP) signaling via ALK3 defines uterine receptivity. This mouse model will be a valuable research tool for studying implantation failure in women, and the results herein will contribute to our knowledge regarding female infertility.
Keywords: BMP signaling pathway, endometrium, implantation failure, infertility, progesterone receptor
Abstract
The window of implantation is defined by the inhibition of uterine epithelial proliferation, structural epithelial cell remodeling, and attenuated estrogen (E2) response. These changes occur via paracrine signaling between the uterine epithelium and stroma. Because implantation defects are a major cause of infertility in women, identifying these signaling pathways will improve infertility interventions. Bone morphogenetic proteins (BMPs) are TGF-β family members that regulate the postimplantation and midgestation stages of pregnancy. In this study, we discovered that signaling via activin-like kinase 3 (ALK3/BMPR1A), a BMP type 1 receptor, is necessary for blastocyst attachment. Conditional knockout (cKO) of ALK3 in the uterus was obtained by producing Alk3flox/flox-Pgr-cre–positive females. Alk3 cKO mice are sterile and have defects in the luminal uterine epithelium, including increased microvilli density and maintenance of apical cell polarity. Moreover, Alk3 cKO mice exhibit an elevated uterine E2 response and unopposed epithelial cell proliferation during the window of implantation. We determined that dual transcriptional regulation of Kruppel-like factor 15 (Klf15), by both the transforming growth factor β (TGF-β) transcription factor SMAD family member 4 (SMAD4) and progesterone receptor (PR), is necessary to inhibit uterine epithelial cell proliferation, a key step for embryo implantation. Our findings present a convergence of BMP and steroid hormone signaling pathways in the regulation of uterine receptivity.
Drastic structural and molecular changes are necessary for the luminal uterine epithelium to achieve a receptive status. Because it is the first site of contact between the mother and the invading blastocyst, remodeling of the luminal epithelium is crucial for establishing a successful pregnancy (1, 2). The steroid hormones, estrogen (E2) and progesterone (P4), coordinate many of these changes; however, many other growth factors and cytokines are involved (3, 4). The bone morphogenetic proteins (BMPs) are a class of growth factors that are important during pregnancy. BMPs are members of the transforming growth factor β (TGF-β) signaling pathway that regulate many cellular pathways, including processes related to normal reproductive function and pregnancy (5–13). BMPs transmit their signals by binding to a cell surface heterodimeric receptor complex that is composed of a combination of two type 1 receptors [activin-like kinase 2 (ALK2), ALK3, or ALK6] and two type 2 receptors [activin A receptor type II A (ACVR2A), ACVR2B, or BMP type 2 receptor (BMPR2)] (14–16). Upon ligand binding, the heterodimeric BMP receptor complex signals by phosphorylating and activating SMAD1/5/8, which associate with SMAD family member 4 (SMAD4) and translocate to the nucleus to regulate gene expression (17).
BMPs are expressed in a temporal manner throughout early and late pregnancy (18), and conditional deletion of BMP2 results in female infertility due to failed decidualization of the endometrial stroma (12, 19). Conditional deletion of the BMPR1, ALK2, results in female infertility due to impaired endometrial stromal cell decidualization that abrogates pregnancy in the postimplantation period (11). Conditional deletion of the BMPR2 also results in female infertility; these mice have defective spiral artery remodeling during the midgestation period that results in intrauterine growth restriction; implantation site hemorrhage; and, ultimately, fetal death (13). Despite the crucial roles for ALK2 and BMPR2 during the postimplantation stages, the specific BMP ligands and receptors that coordinate blastocyst attachment are not yet known.
Decreased uterine response to E2 determines receptivity during the window of implantation. Because tightly regulated levels of E2 are required for successful implantation (20), dynamic epithelial-stromal communication is crucial during this period. Paracrine signaling pathways, such as the paracrine signaling pathways regulated by Indian hedgehog (IHH)/chicken ovalbumin upstream promoter transcription factor II (COUP-TFII) (21, 22) or heart and neural crest derivatives expressed 2 (HAND2) (23), suppress E2-mediated actions in the luminal epithelium during the process of implantation (21–23). The proliferative response to E2 is also regulated by the transcription factors Kruppel-like factor 4 (KLF4) and KLF15 (24). The KLFs are evolutionary conserved DNA binding proteins with diverse roles in development and physiology (25). Previous studies revealed that Klf15 is a direct target of progesterone receptor (PR), whereas Klf4 expression is up-regulated by E2 in the mouse uterus (24, 26). During proestrus, the elevated levels of circulating E2 maintain uterine epithelial cell proliferation through the activation of KLF4. KLF4 binds to the promoter of Minichromosome maintenance-2 (Mcm2) and induces its expression, resulting in cell cycle progression from G1 to S phase. Following the postcopulatory surge of P4, KLF15 expression is induced and inhibits uterine epithelial proliferation by repressing Mcm2 expression (24). Thus, the uterine response to E2 and P4 is regulated by a host of transcription factors that are activated spatiotemporally during the reproductive cycle.
Highly coordinated molecular pathways are necessary to achieve normal blastocyst attachment. Previous studies from our laboratory and other laboratories indicate that the BMP signaling pathway is a major regulator of the pathways that control pregnancy (11–13). Here, we demonstrate that ALK3, a BMPR1, is a key regulator of blastocyst attachment. Because Alk3 null mice are embryonic lethal at embryonic day 9.5 due to failed mesoderm formation (27), we conditionally deleted Alk3 in PR-expressing tissues by mating Alk3flox/flox females (28) to males with a Pgr-cre (29) knock-in allele to produce, eventually, Alk3 conditional knockout (cKO) females. Compared with control females, we discovered that Alk3 cKO females are sterile due to failed blastocyst attachment. Molecular analyses during the window of implantation reveal that Alk3 cKO mice have severe defects in the uterine luminal epithelium that abrogate blastocyst attachment.
Although the use of assisted reproductive technology (ART) has greatly increased since the birth of the first child was achieved through in vitro fertilization in 1978, the success rate of live births using ART remains low (30). Several factors contribute to female infertility, yet it is thought that failed implantation accounts for ∼75% of failed pregnancies (31, 32). Our current examination of the Alk3 cKO mice indicates that characterization and understanding of the BMP ligands, BMPRs, and downstream gene targets hold the potential for novel therapeutic targets and strategies for female infertility.
Results
Conditional Deletion of Alk3 Results in Female Infertility.
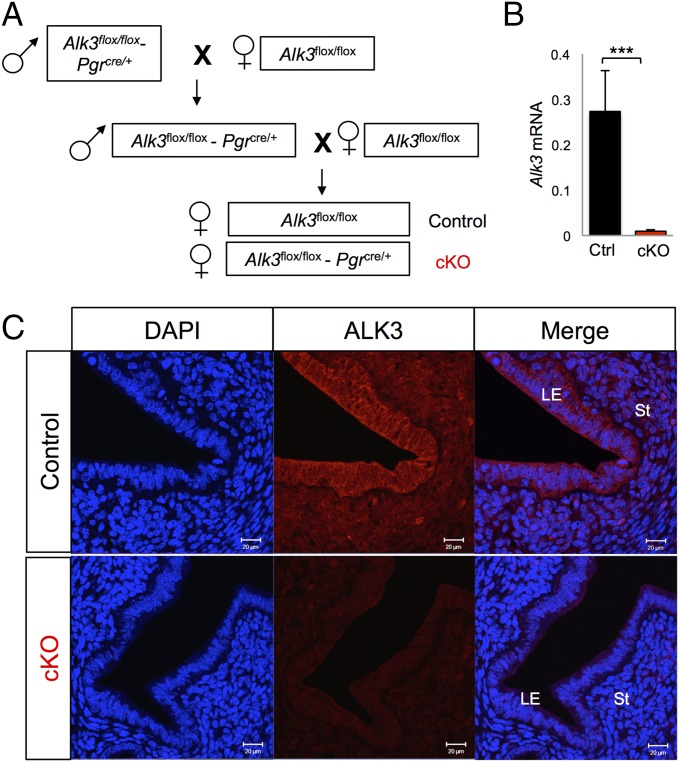
We determined that ALK3 is expressed throughout early pregnancy in the mouse uterus and that its expression peaks at 3.5 d postcoitus (dpc) (Fig. S1). Given that Alk3 null mice are embryonically lethal (27, 28), we generated conditional mutant mice by mating Alk3flox/flox females (28) to Alk3flox/flox-Pgr-cre males (Fig. 1A). This breeding strategy allowed us to study the role of ALK3 in the Pgr-expressing tissues of the female reproductive tract (ovaries, uterus, and oviduct) (29). We first assessed the effectiveness of ALK3 recombination in the uterus by immunofluorescence and quantitative RT-PCR (qRT-PCR). Alk3 expression was significantly decreased in the uterus of Alk3 cKO mice (Fig. 1B), indicating successful ALK3 deletion. Fig. 1C demonstrates that although Alk3 was readily detected in the uterine stroma and luminal epithelium of the control mice, ALK3 immunoreactivity was absent in the Alk3 cKO mice. We performed a 6-mo fertility trial, which indicated that although each control female gave birth to 58 ± 8.1 offspring, Alk3 cKO female mice were sterile (Table 1). We concluded that ALK3 is essential for female reproductive function.
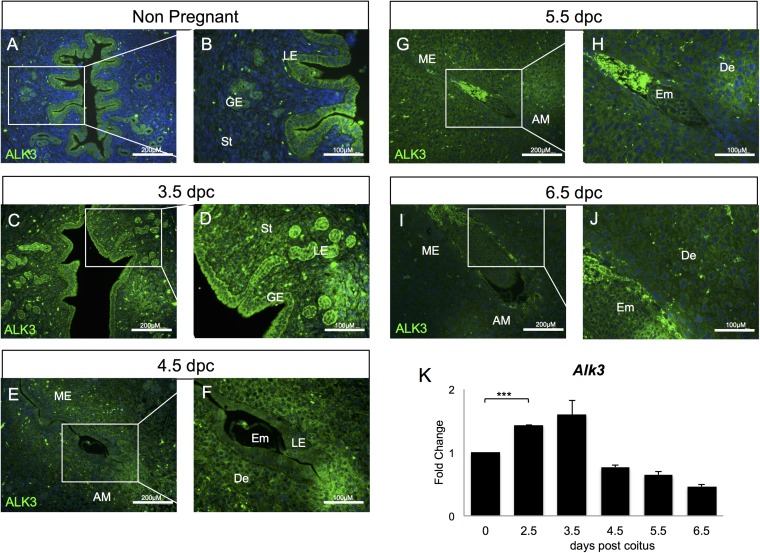
Fig. S1.
ALK3 is dynamically expressed in the uterus throughout early pregnancy. (A and B) In the nonpregnant uterus, ALK3 is confined to the glandular and luminal epithelium. (C and D) At 3.5 dpc, ALK3 expression is more intense in all uterine compartments. (E and F) At 4.5 dpc, ALK3 expression surrounds the embryo. (G–J) ALK3 is located in the decidual cells surrounding the embryo at 5.5 dpc and 6.5 dpc. (K) qRT-PCR analysis of Alk3 expression in the uteri of WT mice (n = 6). AM, antimesometrial; De, decidua; Em, embryo; GE, glandular epithelium; LE, luminal epithelium; ME, mesometrial; St, stroma. ***P < 0.0001. (Scale bars: lower magnification, 200 μM; higher magnification, 100 μM.) Data are mean ± SEM.
Fig. 1.
Generating mice with conditional deletion of ALK3. (A) Breeding scheme used to generate Alk3 cKO female mice. (B) Expression of uterine Alk3 was quantified using qRT-PCR in the uteri of control (Ctrl) and Alk3 cKO mice (n = 4). (C) Immunofluorescence of ALK3 in the uterus of control (Top) and Alk3 cKO (Bottom) mice at 3.5 dpc. (Scale bars: 20 μM.) LE, luminal epithelium; St, stroma. ***P < 0.0001. Data are mean ± SEM.
Table 1.
Six-month fertility trial demonstrates that Alk3 cKO females are sterile
| Genotype | No. of mice, n | Pups per litter | Pups born per female in 6 mo |
| Alk3flox/flox | 10 | 9.0 ± 0.8 | 58 ± 8.1 |
| Alk3flox/flox-Pgrcre | 10 | 0 | 0 |
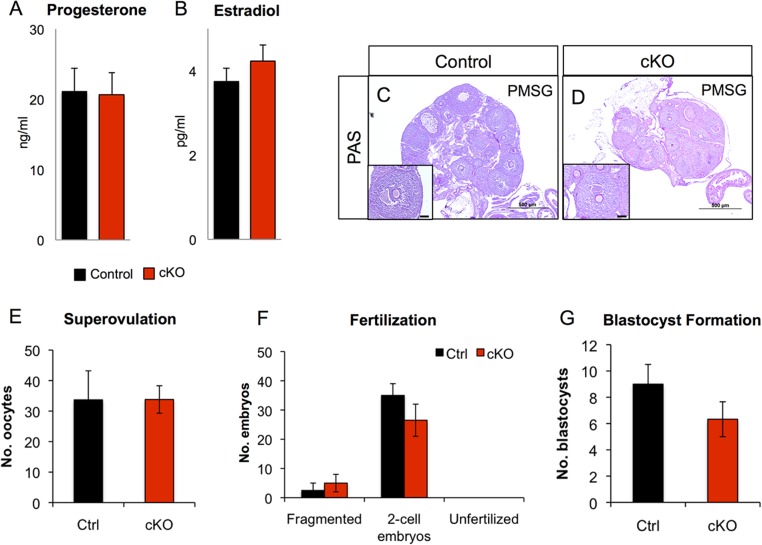
To determine whether the infertility of Alk3 cKO mice was the result of ovarian defects, we assessed ovarian function using various parameters. Ovarian transplants were performed between WT-WT, WT-cKO, and cKO-WT mice (Table S1). WT recipients with WT or Alk3 cKO donor ovaries delivered pups; however, none of the cKO recipient mice with WT ovaries delivered pups, indicating normal ovarian function in Alk3 cKO mice. Likewise, serum hormone levels collected from mice at 3.5 dpc indicated no significant differences in the circulating levels of estradiol (3.75 pg/mL ± 0.31 in controls vs. 4.23 pg/mL ± 0.38 in Alk3 cKO mice) or P4 (21.11 ng/mL ± 3.24 in controls vs. 20.65 ng/mL ± 3.07 in Alk3 cKO mice) (Fig. S2 A and B). Gonadotropin stimulation of sexually immature mice with pregnant mare serum gonadotropin (PMSG) induced follicle maturation equally in Alk3 controls and Alk3 cKO mice, indicating normal ovarian follicle maturation in the cKO mice (Fig. S2 C and D). These results indicated that the infertility observed in Alk3 cKO mice was due to uterine or oviductal defects.
Table S1.
Ovarian transplants between WT and Alk3 cKO mice demonstrate normal ovarian function in cKO mice
| Donor ovary | Recipient | No. of females that delivered pups | No. of pups per litter | No. of litters over 3 mo |
| WT | WT | 2/2 | 5.2 ± 1.4 | 3 |
| Alk3 cKO | WT | 3/3 | 3.3 ± 0.6 | 2 |
| WT | Alk3 cKO | 0/3 | 0 | 0 |
Fig. S2.
Normal ovarian function in Alk3 cKO mice. (A and B) Serum P4 and estradiol levels in control and Alk3 cKO mice at 3.5 dpc (n = 14). Superovulation was induced in control (C) and Alk3 cKO mice (D) with peritoneal injection of PMSG. (Scale bars: lower magnification, 500 μM; Inset, 100 μM.) (E) Number of oocytes was also quantified in control and Alk3 cKO mice after PMSG administration (n = 3). (F) Analysis of fertilization rate in control and Alk3 cKO mice (n = 3). (G) Analysis of blastocyst formation in control and Alk3 cKO mice (n = 3). Data are mean ± SEM.
After ovulation, unfertilized eggs enter the oviduct, where fertilization occurs if mating occurred (3). To assess whether oviduct function was compromised in the cKOs, we measured the response to superovulation, quantified the fertilization rate, and assessed blastocyst formation in control and Alk3 cKO female mice. No differences in any of these parameters were observed (Fig. S2 E–G), indicating that conditional deletion of Alk3 with Pgr-cre did not interfere with oocyte quality or fertilization. Overall, we determined that the infertility in Alk3 cKO mice did not affect ovarian or oviductal function.
Defective Embryo Attachment and Decidualization in the Uterus of Alk3 cKO Mice.
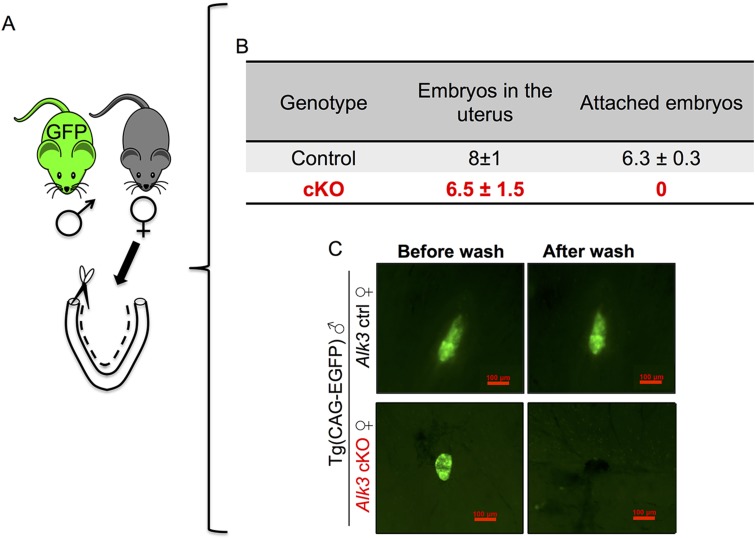
Because normal ovarian and oviductal function was observed in the Alk3 cKO mice, we hypothesized that uterine defects would be the major cause of infertility. We determined that blastocyst implantation was defective in Alk3 cKO mice by using vascular blue dye injection at 4.5 dpc to visualize implantation sites in the uterus (Fig. 2 A and B). Although numerous implantation sites were observed in the control mice (Fig. 2A), implantation sites were completely absent in the Alk3 cKO mice (Fig. 2B). In mice, blastocyst apposition and attachment occur on day 4 of pregnancy, at which point blastocysts can no longer be flushed from the uterus (33–35). We tested whether defective blastocyst attachment was occurring by mating Alk3 cKO females to males carrying a transgene of the EGFP to visualize GFP-expressing embryos (Fig. S3A). We observed that GFP-expressing embryos were entirely flushed from the uterine lumen of Alk3 cKO mice at 4.5 dpc, whereas most embryos remained attached to the uteri of the control mice (Fig. S3 B and C). We concluded that infertility in the Alk3 cKO mice was likely the result of defective blastocyst attachment.
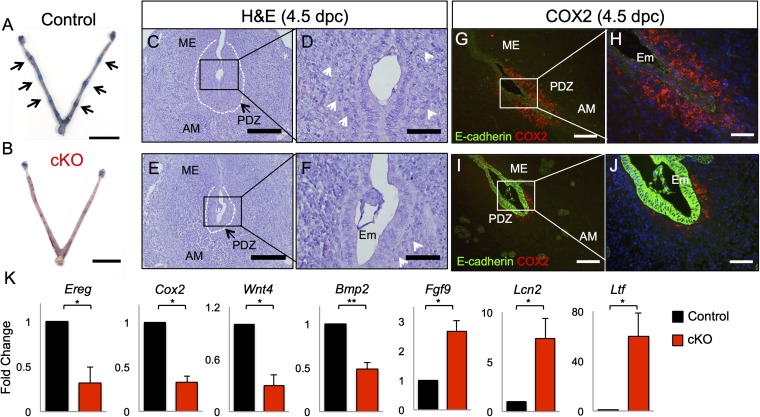
Fig. 2.
Conditional deletion of ALK3 impairs blastocyst implantation and uterine decidualization. Implantation sites at 4.5 dpc (black arrows) were visualized by Chicago Blue vascular dye injection in control (A) and Alk3 cKO (B) mice. (Scale bars: 1 cm.) H&E staining was performed in the uteri of 4.5-dpc control (C and D) and Alk3 cKO mice (E and F). (Scale bars: lower magnification, 200 μM; higher magnification, 50 μM.) Immunofluorescence of COX2 (red) and E-cadherin (green) in the 4.5-dpc uterus of control (G and H) and Alk3 cKO (I and J) mice is shown. (Scale bars: lower magnification, 100 μM; higher magnification, 50 μM.) (K) qRT-PCR analysis of implantation-related genes (Ereg, Cox2, Wnt4, and Bmp2) and E2-regulated genes (Fgf9, Lcn2, and Ltf) in control and Alk3 cKO uteri at 4.5 dpc (n = 4). AM, antimesometrial; Em, embryo; ME, mesometrial; PDZ, primary decidual zone (outlined by white dashed line). White arrowheads indicate decidual cells. *P < 0.05; **P < 0.001. Data are mean ± SEM.
Fig. S3.
Defective blastocyst attachment in Alk3 cKO mice. (A) Control and Alk3 cKO females were mated to GFP-expressing male mice. On day 4.5 of pregnancy, the uteri of pregnant females were collected and GFP-expressing embryos were visualized on the uterus using a fluorescence microscope. (B) To test embryo attachment, the uterus was then flushed and the number of attached or dislodged GFP-expressing embryos was quantified. As shown in B, the majority of control embryos remained attached to the uterine wall, whereas the Alk3 cKO embryos were all dislodged after flushing. (C) Image demonstrating the GFP-expressing embryos attached to the uterus before and after flushing. (Scale bars: 100 μM.)
We analyzed the uteri of control and Alk3 cKO mice on day 4.5 to characterize implantation. In contrast to control mice, which showed attached embryos (Fig. 2 C and D), complete luminal closure, and formation of the primary decidual zone, Alk3 cKO mice had defective embryo attachment with incomplete luminal closure (Fig. 2 E and F). We also saw fewer decidual cells in the Alk3 cKO mouse uterus compared with the controls (Fig. 2 D and F, indicated by white arrowheads), which gave rise to a reduced primary decidual zone compared with controls. This result indicated that unlike embryos in the control mice, embryos in the Alk3 cKO mice did not fully attach or penetrate the luminal uterine epithelium. Thus, in addition to inhibiting blastocyst attachment, the absence of BMP signals mediated via ALK3 blunted the decidual response in the Alk3 cKO mice.
It was previously determined that cyclooxygenase 2 (COX2) expression correlated with embryo implantation and that implantation was abrogated in Cox2 null mice (36). We assessed whether COX2 expression would be present in the uteri of control and Alk3 cKO mice at 4.5 dpc (Fig. 2 G–J). Compared with the control mice, where COX2 expression was strongly detected in the subepithelial stroma surrounding the embryo (Fig. 2 G–H), only weak COX2 expression was detected in the Alk3 cKO mice (Fig. 2 I and J). Accordingly, the expression levels of various implantation-associated genes, including epiregulin (Ereg), Cox2, wingless-type MMTV integration site family member 4 (Wnt4), and Bmp2, were significantly decreased in the Alk3 cKO uteri at 4.5 dpc (Fig. 2K). We also detected enhanced E2 response in the uteri of cKOs at 4.5 dpc (Fig. 2K), as shown by increased expression of fibroblast growth factor 9 (Fgf9), lipocalin 2 (Lcn2), and lactoferrin (Ltf). Overall, these results confirmed defective blastocyst implantation in the Alk3 cKO mice.
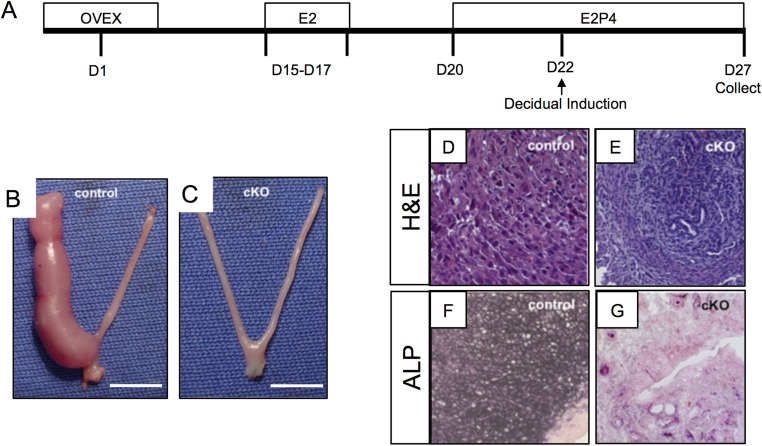
In mice, decidualization of the endometrial stroma occurs in response to mechanical and growth factor signaling elicited by the implanting embryo (37). Endometrial stromal cell decidualization can be artificially induced in mice with a mechanical trauma to the uterus (38). We tested the response of control and Alk3 cKO mice to the mechanical induction of decidualization (Fig. S4). Control and Alk3 cKO mice were ovariectomized and treated with hormones as indicated in Fig. S4A, followed by the induction of a mechanical stimulus to one uterine horn to mimic decidualization. After 5 d, uterine horns were collected and processed for histology. Compared with the enlarged uterine horns of control mice (Fig. S4B), the uterine horns of Alk3 cKO mice did not decidualize (Fig. S4C). We also observed the presence of decidual cells and alkaline phosphatase activity in the decidualized horn of control mice (Fig. S4 D and F), but not in Alk3 cKO mice (Fig. S4 E and G), indicating that ALK3 is necessary for endometrial stromal cell decidualization.
Fig. S4.
Conditional deletion of ALK3 results in impaired decidualization. (A) Experimental overview of the artificial decidualization performed in control and Alk3 cKO mice. D, day; OVEX, ovariectomy. Gross uteri were collected 5 d after mechanical induction of decidualization in control (B) and Alk3 cKO (C) mice. (Scale bars: 1 cm.) H&E staining of the decidualized horns in uteri of control (D) and Alk3 cKO (E) mice is shown. Alkaline phosphatase (ALP) activity in the decidualized uteri of control (F) and Alk3 cKO (G) mice is shown. (Magnification: D–G, 40×.)
Defective Uterine Luminal Epithelium in Alk3 cKO Mice.
To understand the role of ALK3 in the luminal uterine epithelium, we obtained the gene expression profiles of isolated luminal uterine epithelium from 3.5-dpc control and Alk3 cKO mice (Fig. S5A). We assessed the purity of the isolated epithelial cells by staining with cytokeratin 8 (KRT8) and vimentin (VIM) antibodies (Fig. S5 B and C), and by measuring Krt18 and Vim expression with qRT-PCR (Fig. S5 D and E). Microarray profiling demonstrated that 439 gene probes were differentially expressed between the control and Alk3 cKO mice (Fig. S5F; <0.7, >1.5-fold change, P < 0.05). Gene set enrichment analysis demonstrated that genes involved in mediating E2 response and formation of cell adhesions, tight junctions, and cytoskeletal dynamics were enriched in the gene expression dataset (Fig. S5 G and H). Thus, gene expression signatures were different between the control and Alk3 cKO uterine luminal epithelium during the window of implantation.
Fig. S5.
Microarray expression profiling of control and Alk3 cKO uterine epithelium. (A) Epithelial and stromal cells were isolated in control and Alk3 cKO mice at 3.5 dpc. Purity of the epithelial (B) and stromal (C) cells was verified by performing dual immunofluorescent staining of cytokeratin 8 (green) and VIM (red). qRT-PCR quantification of Krt18 (D) and Vim (E) was also performed. (F) Heat map of the microarray expression profiles showing 439 probes that were differentially expressed with P < 0.05. Gene set enrichment analysis shows that genes involved in E2 response (G) and cell adhesion/tight junctions (H) were represented in the dataset. (Scale bar: 100 μM.) *P < 0.05; **P < 0.001. Data are mean ± SEM.
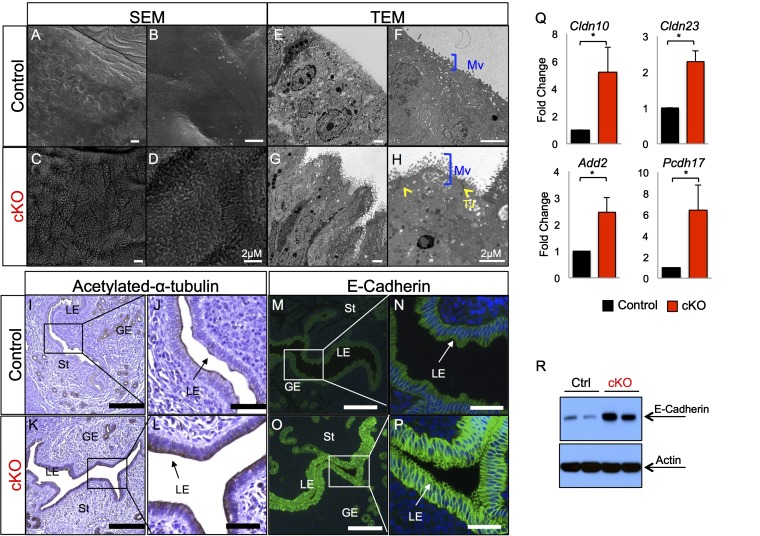
Previous studies have shown that the luminal uterine epithelium undergoes dramatic morphological remodeling into a state that is permissive of blastocyst attachment/invasion (1). These changes, described as the “plasma membrane transformation,” consist of decreased epithelial cell polarity, microvilli flattening, and desmosome/tight junction disassembly (2). To characterize the epithelial cell defects in the uterine epithelium of Alk3 cKO mice further, we performed SEM (Fig. 3 A–D) and transmission electron microscopy (TEM) (Fig. 3 E–H) in the uteri of 3.5-dpc control and Alk3 cKO mice. Both SEM and TEM analyses showed that Alk3 cKO mice had longer and more abundant microvilli than the controls; TEM also demonstrated the presence of tight junctions in the luminal epithelium (Fig. 3H, yellow arrowheads). These analyses indicated the presence of morphological defects in the luminal uterine epithelium of Alk3 cKO mice.
Fig. 3.
ALK3 regulates luminal epithelial remodeling during the window of implantation. SEM of control (A and B) and Alk3 cKO (C and D) uteri at 3.5 dpc is shown. TEM of the luminal uterine epithelium in the uterus of control (E and F) and Alk3 cKO (G and H) mice at 3.5 dpc is shown. (Scale bars: 2 μM.) (Low-power images: A, C, E, and G; high-power images: B, D, F, and H). IHC of acetylated α-tubulin in the 3.5-dpc uterus of control (I and J) and Alk3 cKO (K and L) mice is shown. (Scale bars: lower magnification, 200 μM; higher magnification, 50 μM.) E-cadherin staining on the 3.5-dpc uterus of control (M and N) and Alk3 cKO (O and P) mice is shown. (Scale bars: lower magnification, 200 μM; higher magnification, 40 μM.) (Q) qRT-PCR of genes involved in tight junction formation and cytoskeletal dynamics in the luminal epithelium of 3.5-dpc control and Alk3 cKO mice (n = 4). (R) E-cadherin immunodetection in the 4.5-dpc uterus of control and Alk3 cKO mice. GE, glandular epithelium; Mv, microvilli (blue bracket); TJ, tight junctions (yellow arrowheads). *P < 0.05. Data are mean ± SEM.
To characterize this defect further, we used markers of epithelial cell polarity in the 3.5-dpc uteri of control and Alk3 cKO mice (2, 39). We used immunohistochemistry (IHC) to detect the expression of acetylated-α-tubulin, a marker of stable microtubules, and observed stronger expression in the luminal epithelium of the cKOs (Fig. 3 I–L). Likewise, the expression of E-cadherin, a marker of cell polarity and cell junctions, was more intense in the cKOs (Fig. 3 M–P). We also observed that the expression of genes involved in cell junctions, such as claudin 10 (Cldn10) and Cldn23, and cytoskeletal dynamics, such as adducin 2 (Add2) and protocadherin 17 (Pcdh17), were more highly expressed in the luminal epithelium of the cKOs at 3.5 dpc (Fig. 3Q). A Western blot confirmed that this increase in E-cadherin persisted in the uterus at 4.5 dpc (Fig. 3R). Overall, these results demonstrated structural defects in the luminal epithelium of the Alk3 cKO mice during the window of implantation.
Enhanced E2 Receptor Signaling in the Uterus of Alk3 cKO Mice.
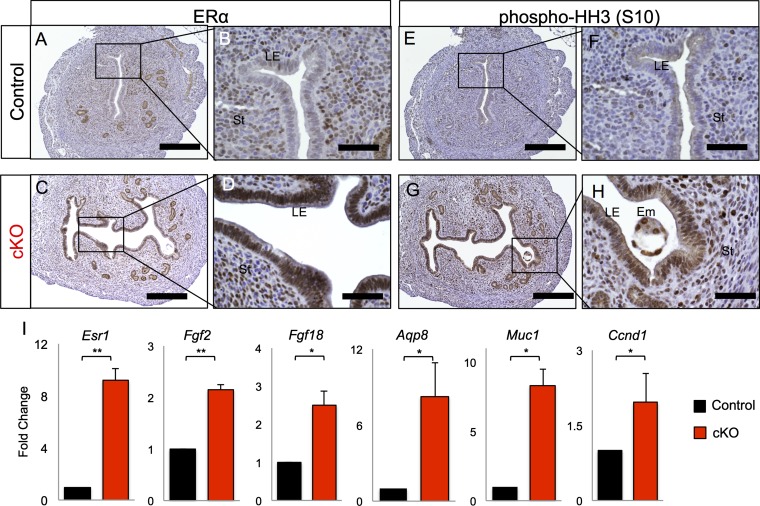
The transition of the luminal uterine epithelium from the nonreceptive phase to the receptive phase is coordinated by the action of the steroid hormones E2 and P4. Our microarray analysis indicated differences in the expression of E2-related genes between control and Alk3 cKO mice (Fig. S5 F and G). To validate this result, we analyzed the uterine expression of estrogen receptor α (ERα) and several downstream signaling targets at 3.5 dpc (Fig. 4). We detected strong ERα expression in the luminal epithelium of the Alk3 cKO mice, which was weaker in the controls (Fig. 4 A–D). Immunostaining of phosphorylated histone H3 (Ser10) demonstrated increased proliferation of the luminal epithelial cells in the uteri of the Alk3 cKO mice (Fig. 4 E–H). We also detected increased expression of Esr1 and of several E2-mediated genes, such as fibroblast growth factor 2 (Fgf2), fibroblast growth factor 18 (Fgf18), aquaporin 8 (Aqp8), and mucin 1 (Muc1), in the luminal epithelium from uteri of 3.5-dpc Alk3 cKO mice (Fig. 4I). Expression of cyclin D1 (Ccnd1) was also significantly increased in the luminal epithelium of the Alk3 cKO uteri (Fig. 4I).
Fig. 4.
Elevated E2 signaling in the uterus of Alk3 cKO mice during the window of implantation. Immunodetection of ERα in the uterus of control (A and B) and Alk3 cKO (C and D) mice at 3.5 dpc is shown. Immunodetection of phosphorylated histone H3 (phospho-HH3) in the control (E and F) and Alk3 cKO (G and H) uteri is shown. (Scale bars: lower magnification, 200 μM; higher magnification, 50 μM.) (I) qRT-PCR analysis of E2-regulated genes in the luminal uterine epithelium of 3.5-dpc control (black bars) or Alk3 cKO (red bars) mice (n = 4). *P < 0.05; **P < 0.001. Data are mean ± SEM.
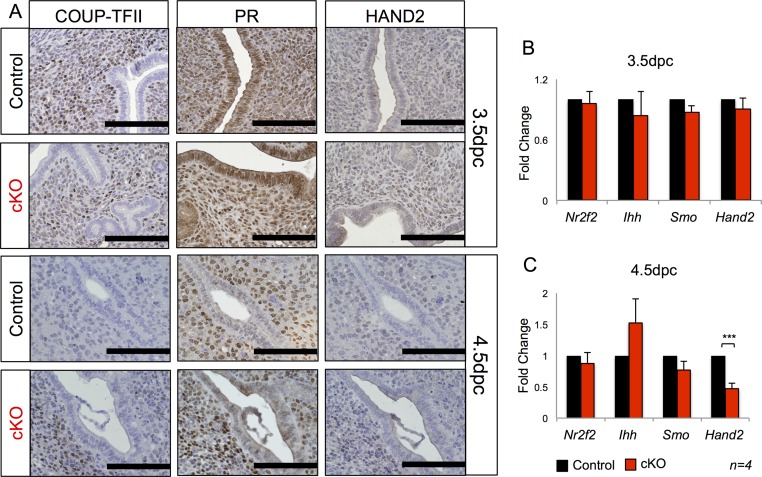
Response to E2 in the uterus is highly regulated during the window of implantation (20). Although E2 is necessary to prepare the uterus for implantation, excessive E2 response prematurely closes the window of implantation (20). COUP-TFII decreases uterine E2 response during the window of implantation by inhibiting ERα activity (22). This inhibition is driven by P4, which activates Indian hedgehog, Patched (Ptch), and Smoothened (Smo), which are upstream regulators of COUP-TFII (21). However, IHC analysis of COUP-TFII or PR and gene expression of Nr2f2 (COUP-TFII), Ihh, and Smo failed to detect significant differences between controls and cKO mouse uteri at 3.5 dpc and 4.5 dpc (Fig. S6). We also measured the gene and protein levels of HAND2, a key transcription factor that inhibits luminal epithelial proliferation during the window of implantation (23). qRT-PCR analysis of control and Alk3 cKO uteri showed a slight but significant reduction in Hand2 expression at 4.5 dpc (0.47 ± 0.08; P < 0.004) but not at 3.5 dpc (Fig. S6 B and C); however, immunostaining of HAND2 did not detect any significant differences between control and Alk3 cKO mice (Fig. S6A). These results indicated that the abnormal response to E2 was independent of COUP-TFII activity.
Fig. S6.
Analysis of the IHH/COUP-TFII and HAND2 pathways in the Alk3 cKO mice. (A) Immunostaining of COUP-TFII, PR, and HAND2 in the uterus of 3.5-dpc and 4.5-dpc control and Alk3 cKO mice. qRT-PCR expression of Nr2f2, Ihh, Smo, and Hand2 in the uterus of pregnant mice at 3.5 dpc (B) and 4.5 dpc (C) is shown (n = 4). (Scale bars: 100 μM.) ***P < 0.0001. Data are mean ± SEM.
KLF15 Expression Is Decreased in Alk3 cKO Mice During the Window of Implantation.
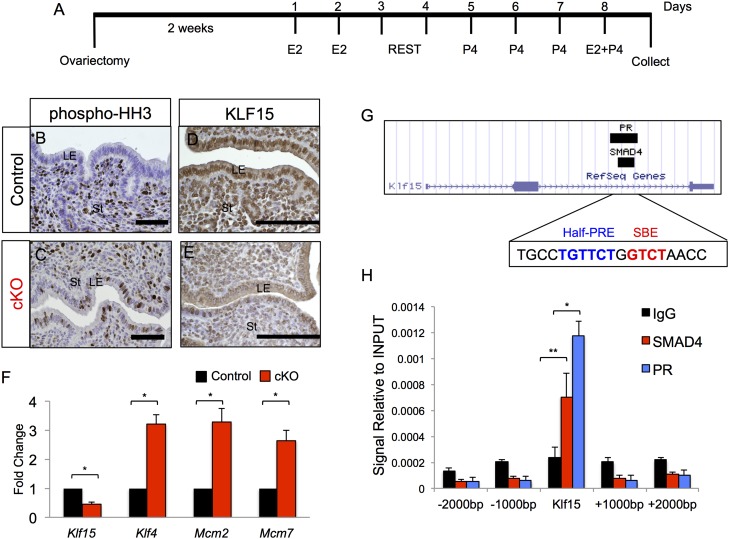
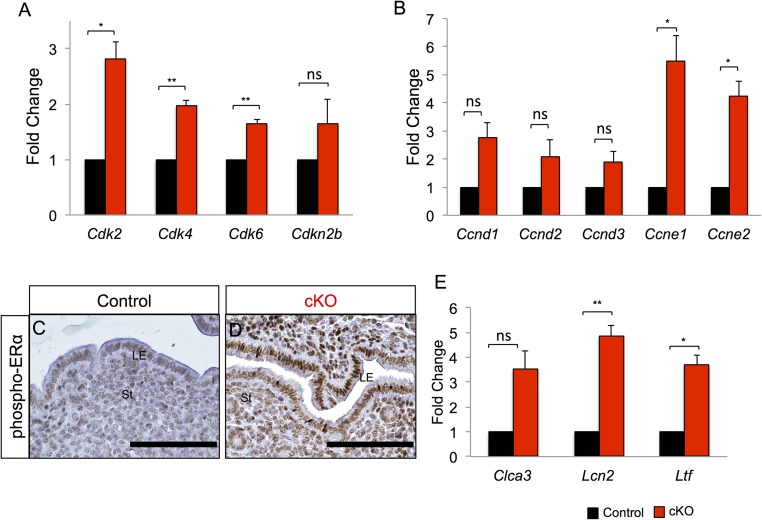
Previous studies demonstrated that proliferation of the uterine luminal epithelium is tightly regulated by the transcription factors KLF4 and KLF15 (24). KLF4 increases in response to E2, whereas KLF15 expression increases after E2 + P4 treatment. KLF4 and KLF15 regulate the transcriptional activation and repression of the Mcm2 and Mcm7 genes, respectively. MCM2 and MCM7 are part of the DNA replication licensing machinery that drives cell cycle progression from G1 to S phase. We subjected control and Alk3 cKO mice to E2 and P4 treatments to induce artificial pregnancy as described in Fig. 5A (24). Following the last E2 + P4 injection, we collected uteri and observed increased phosphorylated histone H3 (Fig. 5 B and C) and decreased KLF15 (Fig. 5 D and E) immunoreactivity in the luminal epithelium of Alk3 cKO mice. Gene expression analysis of the uteri indicated that Klf15 expression was decreased, whereas Klf4, Mcm2, and Mcm7 were significantly increased (Fig. 5F). The expression of cyclin-dependent kinase 2 (CDK2), CDK4, and CDK7 (Cdk2, Cdk4, and Cdk7) and of cyclin e1 and cyclin e2 (Ccne1 and Ccne2) was significantly increased in the luminal epithelium of Alk3 cKO mice (Fig. S7 A and B). Cyclin E/CDK2 complexes are activated during specific cell cycle phases and are required for the G1/S transition (40, 41). We also observed that phosphorylated ERα and its downstream targets, Lcn2 and Ltf, were increased in the luminal epithelium of Alk3 cKO mice (Fig. S7 C–E). Thus, in Alk3 cKO mice, P4 does not attenuate luminal epithelial cell proliferation or E2 action, likely due to decreased KLF15 expression.
Fig. 5.
SMAD4 and PR inhibit luminal epithelial cell proliferation via KLF15. (A) Experimental scheme used to induce artificial pregnancy in control and Alk3 cKO mice. Immunodetection of phospho-HH3 in control (B) and Alk3 cKO (C) mice is shown following induction of artificial pregnancy. Immunodetection of KLF15 in the uterus of control (D) and Alk3 cKO (E) mice is shown. (Scale bars: B and C, 50 μM; D and E, 100 μM.) (F) qRT-PCR analysis of luminal uterine epithelium isolated from control and Alk3 cKO mice subjected to artificial pregnancy treatments (n = 3). (G) Image obtained from the UCSC Genome Browser indicating the ChIP-Seq intervals on the Klf15 gene for SMAD4 (chr6: 90420971–90421737) and PR (chr6: 90420636–90421879). The blue sequence indicates the half-PRE (TGTTCT), and the red sequence indicates the SMAD binding element (GTCT). (H) SMAD4 and PR ChIP-qPCR performed in the uteri of 3.5-dpc WT female mice. Amplified DNA regions correspond to the predicted SMAD4 and PR binding sites on Klf15 and regions 1,000 bp and 2,000 bp upstream and downstream of Klf15. Data were normalized to INPUT (n = 5). *P < 0.05; **P < 0.001. Data are mean ± SEM.
Fig. S7.
Increased uterine epithelial proliferation and E2 response in Alk3 cKO mice. Mice were subjected to artificial pregnancy as described in Fig. 5A. qRT-PCR analysis of the CDKs (A; Cdk2, Cdk4, Cdk6, Cdkn2b) and cyclin gene expression (B; Ccdn1, Ccnd2, Ccnd3, Ccne1, Ccne2) in the uterine luminal epithelium of control and Alk3 cKO mice is shown (n = 3). Immunostaining of phosphorylated ERα in the uteri of control (C) and Alk3 cKO (D) mice after artificial induction of pregnancy is shown. (Scale bars: 100 μM.) (E) qRT-PCR expression of E2-regulated genes, Clca3, Lcn2, and Ltf, in the uterine luminal epithelium of artificially pregnant mice (n = 3). *P < 0.05; **P < 0.001. ns, not significant. Data are mean ± SEM.
SMAD4 and PR Regulate the Expression of KLF15 in the Mouse Uterus.
BMPs initiate an intracellular signaling cascade that results in the activation and nuclear translocation of SMAD1/5/8 and SMAD4 (17). SMAD1/5/8 and SMAD4 are transcription factors that are enriched at GCCT or CTCT consensus DNA sequences. We analyzed a recently published SMAD4 ChIP-sequencing (Seq) dataset (42) and identified that SMAD4 bound to the promoter and intron regions of the Klf15 gene (Fig. 5G). Because KLF15 is activated by P4, we also mined a PR ChIP-Seq dataset (43) and identified that PR and SMAD4 bound to overlapping regions on intron 2 of Klf15 (Fig. 5G). We also observed the presence of SMAD binding elements (GTCT) and a progesterone receptor element (PRE) half-site (TGTTTC) in this region (Fig. 5G). We performed PR and SMAD4 ChIP, followed by qPCR, to confirm SMAD4 and PR binding on Klf15. We used uteri from 3.5-dpc WT mice and performed immunoprecipitations with SMAD4 and PR Abs. As shown in Fig. 5H, both PR and SMAD4 were significantly enriched on intron 2 of Klf15, whereas no binding was detected in the genomic regions upstream (−1,000 bp, −2,000 bp) or downstream (+1,000 bp, +2,000 bp) of the Klf15 gene (Fig. 5H). These results indicate that both PR and SMAD4 are required for the transcriptional regulation of Klf15 during the window of implantation.
Discussion
Previous studies demonstrated that BMP signals are important during various stages of pregnancy. For example, BMP2 and ALK2 are required for normal endometrial stromal cell decidualization and during postimplantation embryo development (11, 12). BMPR2 regulates important processes at midgestation, such as spiral artery remodeling and uterine natural killer cell infiltration (13). It was recently demonstrated that conditional deletion of TGF-β type 1 receptor/ALK5 in the female reproductive tract with Pgr-cre resulted in severe pregnancy defects, including abnormal implantation, decreased uterine natural killer cell infiltration, and defective spiral artery remodeling (44). Likewise, conditional deletion of Nodal with Pgr-cre leads to intrauterine growth restriction and placental defects (45).
Here, we discovered that conditional deletion of the BMPR1, ALK3, prevented pregnancy during the early stages of embryo attachment and implantation. Our results indicated that failure of embryo attachment in Alk3 cKO mice was due to failed remodeling of the uterine epithelial cells into a state that is receptive for embryo implantation. The uterine epithelial cell defects included loss of microvilli retraction, increased epithelial cell polarity, elevated response to E2, and increased cell proliferation (Figs. 3 and 4). Ovarian E2 induces epithelial cell proliferation during proestrus, which is opposed by the rising P4 levels following copulation in preparation for embryo implantation. On day 4 of pregnancy, implantation is induced by a surge of nidatory E2 (46, 47). Previous studies demonstrated that the uterine response to E2 is highly regulated during the window of implantation (20, 22, 48). We observed elevated expression of phosphorylated ERα and its target genes in the uteri of Alk3 cKO mice during the time of implantation (Fig. 4), indicating that the uterus was nonreceptive to an implanting blastocyst.
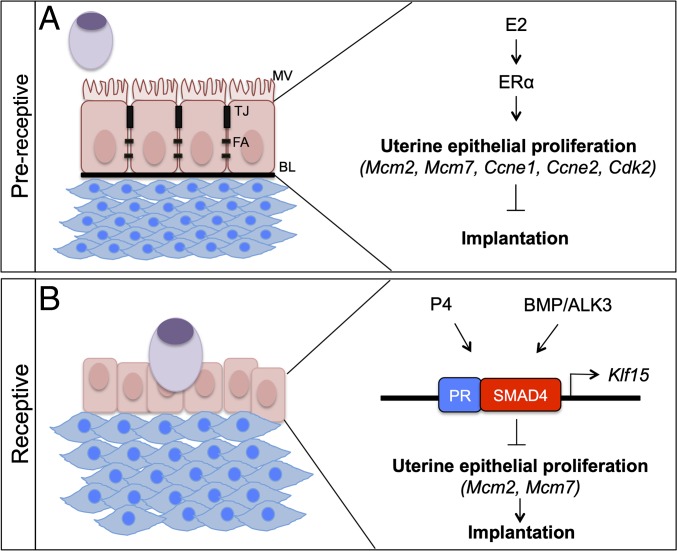
Our studies showed that the transition of a nonreceptive uterus to a receptive uterus requires ALK3. As summarized in Fig. 6, remodeling of the endometrium to support embryo attachment and implantation involves attenuation of E2 signaling and increased P4 signaling. Our results indicated that BMP signals mediated via ALK3 directed the structural remodeling of the luminal uterine epithelium, decreased E2 signaling in the luminal epithelium, and activated KLF15 expression (Fig. 6B). We also observed that coregulation of Klf15 by SMAD4 and PR is necessary to suppress uterine epithelial proliferation, a key requirement for embryo implantation.
Fig. 6.
Model describing the role of ALK3 during the window of implantation. (A) In the prereceptive state, the luminal epithelium is characterized by the presence of microvilli (MV), tight junctions (TJ), focal adhesions (FA), and an intact basal lamina (BL). During this state, E2 signaling via ERα is predominant and stimulates the proliferation of uterine luminal epithelial cells via Mcm2, Mcm7, Ccne1, Ccne2, and Cdk2. (B) During the receptive phase, the luminal uterine epithelium stops proliferating and the embryo penetrates into the endometrial stroma, where it stimulates decidualization. P4 signaling is dominant during this phase. BMP signaling via ALK3 is required for uterine receptivity. SMAD4 and PR bind to the same region of Klf15 and inhibit the DNA replication licensing genes, Mcm2 and Mcm7. Overall, coregulation by SMAD4 and PR inhibits the proliferation of the uterine epithelium, allowing embryo implantation.
KLF15 is a transcription factor that directly opposes uterine luminal epithelial cell proliferation during the window of implantation (24). KLF15 regulates the transcription of the DNA replication licensing gene Mcm2, thereby blocking DNA synthesis and inhibiting uterine epithelial cell proliferation (24). In the luminal uterine epithelium, the expression levels of KLF4 and KLF15 increase in response to E2 and P4, respectively. E2 induction of KLF4 results in KLF4 binding to the Mcm2 promoter, which leads to G1/S transition, DNA synthesis, and mitosis. Conversely, in the presence of E2 and P4, KLF15 is active, resulting in repression of the DNA licensing machinery and opposed cell proliferation. We observed that Klf15 mRNA was significantly decreased in ovariectomized Alk3 cKO mice treated with E2 and P4 to induce artificial pregnancy. This decrease in Klf15 correlated with increased epithelial cell proliferation in the luminal uterine epithelium of the Alk3 cKO mice. We also observed that Klf4, Mcm2, and Mcm7 were increased in the epithelium of the Alk3 cKO mice (Fig. 5). Thus, similar to P4, BMP signals via ALK3 to inhibit uterine epithelial cell proliferation during the window of implantation.
Using previously published SMAD4 (42) and PR (43) ChIP-Seq data, we identified that overlapping SMAD4 and PR binding sites were present in the Klf15 gene. Analysis of these binding sites indicated the presence of half-PREs (TGTTCT) and several BMP-responsive elements (GCCG) and SMAD binding elements (GTCT). We confirmed that SMAD4 and PR bound to Klf15 using ChIP-qPCR in uteri of 3.5-dpc mice, the window of implantation when uterine epithelial cell proliferation is inhibited. This result indicated an interaction between SMAD4 and PR that is necessary for the transcriptional regulation of Klf15. Further studies are necessary to characterize whether SMAD4 and PR are in direct contact, and whether they jointly regulate additional genes in the uterus during the window of implantation.
Although our study only analyzed the proximal binding of SMAD4 to the Klf15 gene, other studies suggest that the genome-wide binding patterns of SMADs include proximal promoter regions and distal enhancer regions. For example, Liu et al. (42) performed SMAD4 ChIP-Seq in mouse lungs and discovered that 25% of binding sites mapped ±1 kb of transcription start sites, but that a higher percentage (29%) of binding sites mapped to distal intergenic regions. Similarly, results from a SMAD1/5 ChIP-Seq study performed in mouse ES cells indicated that binding of a SMAD1/OCT4/SOX2 complex was located to enhancer regions (49). Another study reported similar binding patterns, in which ∼85% of all SMAD1/5 binding events occurred in distal enhancer regions in human umbilical vein endothelial cells and pulmonary arterial smooth muscle cells (50). Because genome-wide SMAD1/5 binding experiments have not been performed in uterine tissues, further studies are needed to establish the genome-wide binding patterns of SMAD1, SMAD4, and SMAD5 during the window of implantation.
Our results suggest that ALK3 is necessary during the transition from the E2-dominant phase to the P4-dominant phase, which is permissive for embryo attachment and invasion. Although ALK3 is expressed in both the endometrial epithelium and stroma during early pregnancy, we observe major defects in the luminal uterine epithelium. We believe that these pronounced luminal epithelial cell defects impair embryo attachment and stromal invasion in Alk3 cKO mice. In particular, increased ERα expression in the luminal epithelium and the elevated expression of its downstream targets are defects that contribute to the infertility of the Alk3 cKO mice. There are several possible explanations for the elevated E2 response observed in Alk3 cKO mice during pregnancy. First, it is possible that ALK3 signaling induces SMAD1/5 interaction and repression of ERα, either in the cytoplasm or at the promoter regions of its target genes. Such direct interactions between SMAD1/SMAD3 and ERα have been previously reported (51, 52). Second, as demonstrated for Klf15, it is possible that dual transcriptional regulation by SMAD4 and PR at target genes is necessary to oppose E2 action on the uterus. PR binding studies in the uterus indicate that SMAD1/5 and SMAD4 binding motifs are significantly enriched in PR target genes (43), and thus it is likely that PR and SMAD4 may frequently bind similar target genes.
Although we observed an absence of endometrial stromal cell decidualization in the Alk3 cKO mice following artificial induction of decidualization (Fig. S4), we observed a few decidual cells in the stroma adjacent to the embryo during natural pregnancy (Fig. 2 E and F). We believe that the embryo’s contact with the uterine epithelium was likely sufficient to trigger the decidual response in the Alk3 cKOs. However, the absence of embryo attachment and invasion, as well as the absence of BMP signals through ALK3, did not support further decidualization of the endometrial stroma and resulted in a blunted decidual response. Thus, the artificial decidualization model provided information regarding the decidual response of the Alk3 cKO uterus to hormones and a mechanical stimulus. However, natural pregnancy indicated that growth factor signals (e.g., BMPs) or mechanical signals elicited by the attaching embryo provide a stimulus that is not provided by the artificial model.
Previous studies demonstrated the expression of various BMPs in the uterus. BMP2 is highly expressed in the mouse endometrium surrounding the implanting embryo and in the decidual tissues (18). Although BMP2 cKO mice are infertile, embryos achieve implantation and do not phenocopy the defects observed in the Alk3 cKO mice (12, 19). This finding indicates that BMP2 is not the ligand for ALK3 during implantation or that other BMPs compensate for its absence. During implantation, BMP7 is expressed in the endometrial stroma surrounding the embryo, whereas BMP5 is weakly expressed in the uterine stroma near the myometrium (18, 53). Thus, it is plausible that BMP7, BMP5, or a combination of these ligands may drive the transition of the luminal uterine epithelium into the receptive phase. Future studies are needed to determine the physiological ligand for ALK3 during implantation.
Despite the increased use of ART in the clinic, failure rates remain high: Of the 151,923 procedures performed in 2011, only 47,818 (about 31%) resulted in live-birth deliveries (30). Implantation failure is one of the various factors contributing to the failure of ART interventions (54). The process of embryo implantation requires communication between the maternal endometrium and the implanting embryo. This bidirectional communication depends on hormones, growth factors, cytokines, and cell surface receptors (3). Improving the success of current reproductive medicine will require an understanding of the bidirectional maternal and embryonic signaling pathways. Data from our studies demonstrate the importance of signaling via ALK3 in the uterine luminal epithelium in the establishment of pregnancy. In particular, our studies demonstrate the crucial role for ALK3 in the preparation of the luminal uterine epithelium for blastocyst attachment. We believe that our data and future studies of the Alk3 cKO mice that we generated will be useful for studying the biological mechanisms in female infertility.
Methods
Mouse Strains and Breeding Scheme to Generate Alk3 cKO Female Mice.
Mice carrying the Alk3 floxed allele were previously generated by Yuji Mishina, University of Michigan, Ann Arbor, MI, and Richard Behringer, University of Texas, MD Anderson Cancer Center, Houston (28). Briefly, loxP sites were inserted into the Alk3 locus spanning exon 2, which encodes a portion of the extracellular domain. To generate Alk3 cKO mice, Alk3flox/flox mice were bred to mice carrying the Pgr-cre knock-in allele (Pgrcre/+) (29) to obtain Alk3flox/flox Pgrcre/+ females (cKO mice). Mice were genotyped by PCR analysis using genomic tail DNA with the primers listed in Table S1 using the conditions described by Mishina et al. (28) and Soyal et al. (29). The mice were maintained on a hybrid C57BL/6J and 129S5/SvEvBrd genetic background. Animal handling and studies were performed following the NIH Guide for the Care and Use of Laboratory Animals (55) and were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Fertility Analyses.
Fertility of Alk3flox/flox and Alk3 cKO mice was monitored over the course of 6 mo. The fertility studies were performed by mating Alk3 control and Alk3 cKO female mice with WT males for 6 mo. Normal mating was observed in the Alk3 cKO female mice by monitoring the cages the daily for the presence of vaginal plugs, as well as the total number of pups and litters delivered.
Histological Analyses.
Uteri were removed from mice and fixed in 10% (vol/vol) formalin overnight, followed by dehydration in 70% (vol/vol) ethanol. Tissues were processed for paraffin embedding in the Pathology and Histology Core Facility at Baylor College of Medicine. Paraffin sections were then stained with H&E (Electron Microscopy Sciences) or with periodic acid Schiff (Sigma) following the manufacturer’s conditions. To visualize implantation sites, Alk3 control and cKO females were mated to WT males and injected with Chicago Sky Blue Dye at 4.5 dpc as described (56). Implantation sites from controls or uteri from cKO females were then collected and processed for histology or mRNA analysis.
Tissue Collection for Protein or mRNA Extraction and qRT-PCR.
Uterine tissues were collected and immediately frozen in dry ice. For protein extraction, the tissue was ground with a pestle in T-PER buffer (Pierce) with a protease inhibitor mixture (Roche). RNA was extracted using the QIAGEN RNEasy Kit by homogenizing the tissue with an OMNI-TH disrupter following the manufacturer’s protocol. RNA (1–2 μg) was transcribed to cDNA using qScript cDNA Supermix (Quanta). cDNA was then used to amplify specific primers (listed in Tables S2 and S3) using SYBR Green (Life Technologies/ABI) on a Roche 480 Light Cycler II. Data were analyzed using the ΔΔCt method as described (57) and analyzed using a Student’s t test or ANOVA analyses with Tukey’s multiple comparison posttests using Prism GraphPad version 7.
Table S2.
List of primers
| Name | Forward (5′–3′) | Reverse (5′–3′) |
| Gene expression primers, 5′–3′ | ||
| Cldn10 | CTGGAAGGTCTCCACCATCG | GGATGTAACCATCCAACGCC |
| Cldn23 | CCCGACGAGTGGAACTACTTC | GGCCAGCGACGAAAAACAC |
| Bmp2 | GGGACCCGCTGTCTTCTAGT | TCAACTCAAATTCGCTGAGGAC |
| Ltf | TGAGGCCCTTGGACTCTGT | ACCCACTTTTCTCATCTCGTTC |
| Fgf9 | ATGGCTCCCTTAGGTGAAGTT | TCATTTAGCAACACCGGACTG |
| Lcn2 | GCAGGTGGTACGTTGTGGG | CTCTTGTAGCTCATAGATGGTGC |
| Fgf2 | GCGACCCACACGTCAAACTA | TCCCTTGATAGACACAACTCCTC |
| Fgf18 | AGGGGAAGCTAATTGCCAAGA | CCTTGCGGGTAAAGGCCAT |
| Muc1 | GGCATTCGGGCTCCTTTCTT | TGGAGTGGTAGTCGATGCTAAG |
| Alk3 | GTATGCTCCATGGCACTGGT | AGCATCATCTGGGCAGTGTC |
| Vim | TGCACGATGAAGAGATCCAGG | CTCCTGGAGGTTCTTGGCAG |
| Krt18 | GAGGGCTCAGATCTTTGCGA | CATGGATGTCGCTCTCCACA |
| Pcdh17 | AGCCACTCGAACAAGAACCA | TGCATTTTCAGCCTGATTGGC |
| Add2 | GAGCAGTTCCTACCTGGCTG | ATCCACTCCTGGAACCCAGT |
| Ptgs2 | AGCCAGGCAGCAAATCCTT | CAGTCCGGGTACAGTCACAC |
| Ereg | TGCTTTGTCTAGGTTCCCACC | GGCGGTACAGTTATCCTCGG |
| Wnt4 | GAGAAGTGTGGCTGTGACCGG | ATGTTGTCCGAGCATCCTGACC |
| Aqp8 | AGTACAATCTAGGGCAGCCG | ACTACACATTGGTGTCTGCTCC |
| Klf4 | GTGCCCCGACTAACCGTTG | GTCGTTGAACTCCTCGGTCT |
| Klf15 | GAGACCTTCTCGTCACCGAAA | GCTGGAGACATCGCTGTCAT |
| Mcm2 | ATCCACCACCGCTTCAAGAAC | TACCACCAAACTCTCACGGTT |
| Mcm7 | AGTATGGGACCCAGTTGGTTC | GCATTCTCGCAAATTGAGTCG |
| Ptch1 | AAAGAACTGCGGCAAGTTTTTG | CTTCTCCTATCTTCTGACGGGT |
| Smo | GAGCGTAGCTTCCGGGACTA | CTGGGCCGATTCTTGATCTCA |
| Nr2f2 | TCAACTGCCACTCGTACCTG | CCATGATGTTGTTAGGCTGCAT |
| Ihh | CTCTTGCCTACAAGCAGTTCA | CCGTGTTCTCCTCGTCCTT |
| 36B4 | AAGCAAAGGAAGAGTCGGAG | CCAGACCGGAGTTTTAAGAGAAG |
| Ccnd1 | CATCAAGTGTGACCCGGACTG | CCTCCTCCTCAGTGGCCTTG |
| Ccnd2 | GAGTGGGAACTGGTAGTGTTG | CGCACAGAGCGATGAAGGT |
| Ccnd3 | CCAGCGTGTCCTGCAGAGTT | CCTTTTGCACGCACTGGAAG |
| Ccne1 | TGTTACAGATGGCGCTTGCTC | TTCAGCCAGGACACAATGGTC |
| Ccne2 | TGCTGCCGCCTTATGTCATT | TCCGAGATGTCATCCCATTCC |
| Cdk2 | CTGCCATTCTCACCGTGTCC | AGCTTGATGGACCCCTCTGC |
| Cdk4 | CCCAATGTTGTACGGCTGA | GGAGGTGCTTTGTCCAGGTA |
| Cdk6 | GCTTCGTGGCTCTGAAGCGCG | TGGTTTCTGTGGGTACGCCGG |
| Cdkn2b | CCCTGCCACCCTTACCAGA | CAGATACCTCGCAATGTCACG |
| ChIP primers | ||
| ChIP Klf15 − 2K | ATCAAGGCAGGGTCTCTTGC | TTGCATCCCCAGCATCATGT |
| ChIP Klf15 − 1K | TCTAGACAGCTGGGGCATCT | CTGTCCAATGTCTCCCACCC |
| ChIP Klf15 + 2K | ATGGGACATTGCAGGGACAG | ACCCAGGTGATGTGCTGATG |
| ChIP Klf15 + 1K | TGGGGTAGGCCTGCTAATCT | GGTACATTCGGTCACAGGCA |
| Klf15 BS F | CACCTGTCCCAGTCCAAAGC | AGGCTGGCTTCAGTTCTTCC |
| Genotyping primers, 5′–3′ | ||
| Alk3 Fx 0 | CTCTGAATTTCTAGTCCACATCTGC | |
| Alk3 Fx 1 | GGTTTGGATCTTAACCTTAGG | |
| Alk3 Fx 2 | GCAGCTGCTGCTGCAGCCTCC | |
| Alk3 Fx 3 | AGACTGCCTTGGGAAAAGCGC | |
| Alk3 Fx 4 | TGGCTACAATTTGTCTCATGC | |
| Alk3 Fx 5 | GGACTATGGACACACAATGGC | |
| PR cre WT | CCCAAAGAGACACCAGGAAG | |
| PR cre F | TATACCGATCTCCCTGGACG | |
| PR cre R | ATGTTTAGCTGGCCCAAATG | |
F, forward; R, reverse.
Table S3.
List of antibodies
| Antibody | Company | Species | Catalog no. | IHC dilution | Western blot dilution | ChIP |
| ALK3 | Proteintech | Rabbit | 12702 | 1:100 | — | — |
| Acetylated α-tubulin | Cell Signaling | Rabbit | 5335 | 1:200 | 1:1,000 | — |
| E-cadherin | Cell Signaling | Rabbit | 3195 | 1:200 | — | — |
| ERα | Thermo Scientific | Mouse | MS-354-P0 | 1:200 | — | — |
| Phospho-ERα (S118) | Abcam | Rabbit | ab31477 | 1:750 | — | — |
| Phospho-histone H3 (S10) | Abcam | Rabbit | ab5176 | 1:200 | — | — |
| COX2 | Santa Cruz Biotechnology | Goat | sc-1747 | 1:200 | — | — |
| COX2 | Santa Cruz Biotechnology | Rabbit | sc-1747-R | 1:200 | — | — |
| Cytokeratin-8 | Hybridoma Bank | Rat | TROMA-1 | 1:50 | 1:500 | — |
| KLF15 | Abcam | Rabbit | ab2647 | 1:200 | — | — |
| PR | Santa Cruz Biotechnology | Rabbit | sc-7208 | 1:200 | — | 5 μg |
| SMAD4 | R&D Systems | Goat | AF2097 | — | 1:200 | 5 μg |
| VIM | Sigma | Mouse | V5255 | 1:200 | — | — |
Separation of Uterine Epithelial and Stromal Cells.
To obtain epithelial and stromal uterine cells, mice were mated and their uteri were collected 3.5 dpc. We used the conditions described by Clementi et al. (11) to obtain epithelial and/or stromal cells from the uterus. Briefly, uteri were collected and cut into 2- to 5-mm cross-sections, followed by incubation in 1% trypsin solution in HBSS. After a 30-min incubation at 37 °C, the epithelial cells were mechanically separated from the uterus using forceps and a mouth pipette. Purity of the epithelial cells was determined by analyzing Krt8 and Vim mRNA and protein levels using qRT-PCR and immunofluorescence, respectively.
Hormone Treatments.
Mice were subjected to artificial pregnancy by timed hormone injections as described (24). Briefly, 6-wk-old mice were ovariectomized, allowed 2 wk of recovery, primed with 100 ng of E2 (in sesame oil), rested for 2 d, and then divided into four groups and injected with the following hormone regimen for 4 d: (i) injected with oil, (ii) injected with 50 ng of E2 on day 4, (iii) injected with 1 mg of P4 on days 1–4, and (iv) injected with 1 mg of P4 on days 1–3 and then with 1 mg of P4 + 50 ng of E2 on day 4. Mouse uteri were collected 15 h following the last injection. RNA was isolated from whole uteri, fixed in formalin, or processed for epithelial/stromal cell isolation. To induce superovulation in mice, 3-wk-old female control and Alk3 cKO females were injected with 5 international units of PMSG. Forty-eight hours later, the ovaries were collected, processed for histology, and analyzed for the presence of follicles.
SI Methods
Mouse Strains Used for Embryo Attachment Studies.
For the embryo attachment experiments, we used males with a transgene containing the EGFP gene under the control of the CMV-immediate early enhancer/chicken β-actin/rabbit β-globin hybrid promoter [mouse line FVB.Cg-Tg(CAG-EGFP)B5Nagy/J].
Immunofluorescence and Immunohistochemistry.
Paraffin-embedded tissues were rehydrated in Histoclear (National Diagnostics); 100%, 95%, 80%, and 60% (vol/vol) ethanol; and water. Antigen retrieval was performed in 10 mM citric acid and 0.05% Tween-20 (pH 6) at high heat. For immunofluorescence, slides were washed in Tris-buffered saline, blocked for 1 h in 3% (wt/vol) BSA, and incubated in a humidified chamber overnight at 4 °C. Secondary Ab incubation was performed in the dark for 1 h at room temperature, followed by incubation with DAPI. Coverslips were mounted using Vectashield Fluorescence Mounting Medium (Vector Laboratories). For immunohistochemistry, sections were blocked using an Avidin/Biotin Blocking Kit (Vector Laboratories), followed by blocking in 3% (wt/vol) BSA and overnight incubation in primary Ab. The following day, sections were washed and incubated in biotinylated secondary Abs (Jackson Laboratories), followed by avidin/biotin complex formation (Vectastain ABC). Sections were then incubated with DAB Peroxidase (HRP) substrate (Vector Laboratories) and counterstained.
Mechanical Induction of Decidualization and Ovarian Transplants.
Mechanical induction of decidualization was performed as described by Finn and Martin (38). Mice were ovariectomized and allowed to recover for 2 wk, followed by three daily s.c. injections of 100 ng of E2. Following 2 d of rest, mice were injected with three daily s.c. injections of 1 mg of P4 + 6.7 ng of E2. To induce decidualization artificially, one uterine horn was scratched with a needle and the other unscratched horn served as a control. After continuous injections of 1 mg of P4 + 6.7 ng of E2 for 5 d, the mice were killed and uterine horns were collected, weighed, and processed for histological analyses. Ovarian transplants were performed as described by a protocol kindly provided by Iruela-Arispe, University of California, Los Angeles, CA.
Microarray Analysis.
For microarray analyses, labeled cRNA was hybridized to Illumina Mouse WG-6 v.2.0 by the Genomics and Proteomics Core at Texas Children’s Hospital. Array data were quantile-normalized. Differentially expressed genes were defined using a two-sided t test and fold change (on log-transformed data). The microarray data were deposited to the National Center for Biotechnology Information’s Gene Expression Omnibus and are available under accession number GSE73717.
ChIP Followed by qPCR.
For SMAD4 and PR ChIP, 3.5-dpc uteri were collected, cut into 3- to 4-mm cross-sections, and fixed in 1% paraformaldehyde in 1× PBS for 15 min, followed by quenching in 0.125 M glycine. Uteri were then washed three times in cold PBS and flash-frozen. ChIP was performed following a protocol described by Lee et al. (58). We used 5 μg of rabbit polyclonal PR Ab (H190, sc7208; Santa Cruz Biotechnology) previously used to perform ChIP-Seq in the mouse uterus (43) and 5 μg of goat polyclonal SMAD4 Ab (AF2097; R&D Systems) previously used to perform ChIP-Seq in mouse lung (42). Uteri were resuspended in lysis buffer 1 + protease inhibitors [50 mM Hepes-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% (vol/vol) glycerol, 0.5% Nonidet P-40, 0.25% Triton X-100] and pulverized in a tissue homogenizer. Lysates were centrifuged and then resuspended in lysis buffer 2 + protease inhibitors [10 mM Tris⋅HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA]. Finally, lysates were resuspended on lysis buffer 3 [10 mM Tris⋅HCl (pH 8), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine] and sonicated for 25 cycles at power 3 (10 s ON, 50 s OFF). Chromatin was incubated overnight with Abs on a rocker at 4 °C. Dynal Beads (Life Technologies) were used to capture the protein/DNA/Ab complexes for 4 h on a rocker at 4 °C. Dynal Beads were then washed with radioimmunoprecipitation assay buffer [50 mM Hepes-KOH (pH 7.5), 500 mM LiCl, 1 mM EDTA, 1% Nonidet P-40, 0.7% Na-deoxycholate], and protein/DNA complexes were eluted [50 mM Tris⋅HCl (pH 8), 10 mM EDTA, 1% SDS], followed by cross-link reversal overnight at 65 °C. RNase digest was performed, followed by column DNA purification. qPCR was performed on the chromatin-immunoprecipitated DNA using the primers listed in Tables S2 and S3. Primers were designed to amplify the following chromosomal intervals of Klf15: SMAD4 binding site (chr6: 90420971–90421737) and PR binding site (chr6: 90420636–90421879) based on mm9. Data were normalized to INPUT and expressed as signal relative to INPUT.
TEM and SEM.
To analyze the luminal uterine epithelium, 3.5-dpc control and cKO mice were collected. We verified pregnancy by flushing the uteri and determining the presence of blastocysts, as well as by epithelial quantification of the pregnancy genes, Jam2, Hdc, and Clca. For SEM, samples were fixed in Trump’s glutaraldehyde, postfixed in 2% (wt/vol) buffered osmium tetroxide, dehydrated in a graded series of ethanol, critical point-dried, mounted, and sputter-coated with gold/palladium. Samples were imaged using a JEOL 6100 scanning electron microscope at the Department of Pathology Laboratories at the Texas Children’s Hospital. For TEM, samples were fixed in 2.5% (wt/vol) glutaraldehyde and 2% (wt/vol) paraformaldehyde in 0.1 M cacodylate buffer (pH 7.4), followed by postfixation in 1% osmium tetroxide. After dehydration in ethanol and embedding in low-viscosity resin, the tissues were stained for 1 h in saturated uranyl acetate with 50% (vol/vol) ethanol during dehydration. Imaging was performed at the Integrated Microscopy Core at Baylor College of Medicine using the H7500 transmission electron microscope.
Statistical Analyses.
All experiments were performed in at least three independent samples and were analyzed using two-tailed Student’s t tests or one-way ANOVA analyses with Tukey’s multiple comparison posttests using Excel and GraphPad version 6. Data are presented as mean ± SEM.
Acknowledgments
We thank Drs. Yuji Mishina and Richard Behringer for their kind gift of the Alk3flox/flox mice. These studies were supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development Grants HD032067 (to M.M.M.) and HD042311 (to J.P.L.), Institutional Research and Academic Career Development Award K12-GM084897 (to D.M.), National Cancer Institute Grant CA125123 (to C.J.C.), and the A. I. & Manet Schepps Endowment for Discovery (to D.M. and M.M.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GEO73717).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523758113/-/DCSupplemental.
References
- 1.Kaneko Y, Lindsay LA, Murphy CR. Focal adhesions disassemble during early pregnancy in rat uterine epithelial cells. Reprod Fertil Dev. 2008;20(8):892–899. doi: 10.1071/rd08148. [DOI] [PubMed] [Google Scholar]
- 2.Kaneko Y, Day ML, Murphy CR. Uterine epithelial cells: Serving two masters. Int J Biochem Cell Biol. 2013;45(2):359–363. doi: 10.1016/j.biocel.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Dey SK. Roadmap to embryo implantation: Clues from mouse models. Nat Rev Genet. 2006;7(3):185–199. doi: 10.1038/nrg1808. [DOI] [PubMed] [Google Scholar]
- 4.Vasquez YM, DeMayo FJ. Role of nuclear receptors in blastocyst implantation. Semin Cell Dev Biol. 2013;24(10-12):724–735. doi: 10.1016/j.semcdb.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katagiri TST, Miyazono K. The Bone Morphogenetic Proteins. Cold Spring Harbor Laboratory Press; Plainview, NY: 2008. [Google Scholar]
- 6.Peng J, et al. Uterine activin receptor-like kinase 5 is crucial for blastocyst implantation and placental development. Proc Natl Acad Sci USA. 2015;112(36):E5098–E5107. doi: 10.1073/pnas.1514498112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, et al. Transforming growth factor β receptor type 1 is essential for female reproductive tract integrity and function. PLoS Genet. 2011;7(10):e1002320. doi: 10.1371/journal.pgen.1002320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pangas SA, Matzuk MM. The TGF-β Family in the Reproductive Tract. Cold Spring Harbor Laboratory Press; Plainview, NY: 2008. [Google Scholar]
- 9.Dong J, et al. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383(6600):531–535. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- 10.Li Q, et al. Redundant roles of SMAD2 and SMAD3 in ovarian granulosa cells in vivo. Mol Cell Biol. 2008;28(23):7001–7011. doi: 10.1128/MCB.00732-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clementi C, et al. Activin-like kinase 2 functions in peri-implantation uterine signaling in mice and humans. PLoS Genet. 2013;9(11):e1003863. doi: 10.1371/journal.pgen.1003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee KY, et al. Bmp2 is critical for the murine uterine decidual response. Mol Cell Biol. 2007;27(15):5468–5478. doi: 10.1128/MCB.00342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagashima T, et al. BMPR2 is required for postimplantation uterine function and pregnancy maintenance. J Clin Invest. 2013;123(6):2539–2550. doi: 10.1172/JCI65710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducy P, Karsenty G. The family of bone morphogenetic proteins. Kidney Int. 2000;57(6):2207–2214. doi: 10.1046/j.1523-1755.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- 15.Miyazono K, Kamiya Y, Morikawa M. Bone morphogenetic protein receptors and signal transduction. J Biochem. 2010;147(1):35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- 16.Matzuk MM, Burns KH. Genetics of mammalian reproduction: modeling the end of the germline. Annu Rev Physiol. 2012;74:503–528. doi: 10.1146/annurev-physiol-020911-153248. [DOI] [PubMed] [Google Scholar]
- 17.Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 18.Paria BC, et al. Cellular and molecular responses of the uterus to embryo implantation can be elicited by locally applied growth factors. Proc Natl Acad Sci USA. 2001;98(3):1047–1052. doi: 10.1073/pnas.98.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, et al. Bone morphogenetic protein 2 functions via a conserved signaling pathway involving Wnt4 to regulate uterine decidualization in the mouse and the human. J Biol Chem. 2007;282(43):31725–31732. doi: 10.1074/jbc.M704723200. [DOI] [PubMed] [Google Scholar]
- 20.Ma WG, Song H, Das SK, Paria BC, Dey SK. Estrogen is a critical determinant that specifies the duration of the window of uterine receptivity for implantation. Proc Natl Acad Sci USA. 2003;100(5):2963–2968. doi: 10.1073/pnas.0530162100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurihara I, et al. COUP-TFII mediates progesterone regulation of uterine implantation by controlling ER activity. PLoS Genet. 2007;3(6):e102. doi: 10.1371/journal.pgen.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee DK, et al. Suppression of ERalpha activity by COUP-TFII is essential for successful implantation and decidualization. Mol Endocrinol. 2010;24(5):930–940. doi: 10.1210/me.2009-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, et al. The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science. 2011;331(6019):912–916. doi: 10.1126/science.1197454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray S, Pollard JW. KLF15 negatively regulates estrogen-induced epithelial cell proliferation by inhibition of DNA replication licensing. Proc Natl Acad Sci USA. 2012;109(21):E1334–E1343. doi: 10.1073/pnas.1118515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knoedler JR, Denver RJ. Krüppel-like factors are effectors of nuclear receptor signaling. Gen Comp Endocrinol. 2014;203:49–59. doi: 10.1016/j.ygcen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wade HE, et al. Multimodal regulation of E2F1 gene expression by progestins. Mol Cell Biol. 2010;30(8):1866–1877. doi: 10.1128/MCB.01060-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9(24):3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 28.Mishina Y, Hanks MC, Miura S, Tallquist MD, Behringer RR. Generation of Bmpr/Alk3 conditional knockout mice. Genesis. 2002;32(2):69–72. doi: 10.1002/gene.10038. [DOI] [PubMed] [Google Scholar]
- 29.Soyal SM, et al. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis. 2005;41(2):58–66. doi: 10.1002/gene.20098. [DOI] [PubMed] [Google Scholar]
- 30.Sunderam S, et al. Centers for Disease Control and Prevention (CDC) Assisted reproductive technology surveillance--United States, 2011. MMWR Surveill Summ. 2014;63(10):1–28. [PubMed] [Google Scholar]
- 31.Wilcox AJ, et al. Incidence of early loss of pregnancy. N Engl J Med. 1988;319(4):189–194. doi: 10.1056/NEJM198807283190401. [DOI] [PubMed] [Google Scholar]
- 32.Norwitz ER, Schust DJ, Fisher SJ. Implantation and the survival of early pregnancy. N Engl J Med. 2001;345(19):1400–1408. doi: 10.1056/NEJMra000763. [DOI] [PubMed] [Google Scholar]
- 33.van Mourik MS, Macklon NS, Heijnen CJ. Embryonic implantation: Cytokines, adhesion molecules, and immune cells in establishing an implantation environment. J Leukoc Biol. 2009;85(1):4–19. doi: 10.1189/jlb.0708395. [DOI] [PubMed] [Google Scholar]
- 34.Achache H, Revel A. Endometrial receptivity markers, the journey to successful embryo implantation. Hum Reprod Update. 2006;12(6):731–746. doi: 10.1093/humupd/dml004. [DOI] [PubMed] [Google Scholar]
- 35.Hantak AM, Bagchi IC, Bagchi MK. Role of uterine stromal-epithelial crosstalk in embryo implantation. Int J Dev Biol. 2014;58(2-4):139–146. doi: 10.1387/ijdb.130348mb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim H, et al. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell. 1997;91(2):197–208. doi: 10.1016/s0092-8674(00)80402-x. [DOI] [PubMed] [Google Scholar]
- 37.Ramathal CY, Bagchi IC, Taylor RN, Bagchi MK. Endometrial decidualization: Of mice and men. Semin Reprod Med. 2010;28(1):17–26. doi: 10.1055/s-0029-1242989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finn CA, Martin L. Endocrine control of the timing of endometrial sensitivity to a decidual stimulus. Biol Reprod. 1972;7(1):82–86. doi: 10.1093/biolreprod/7.1.82. [DOI] [PubMed] [Google Scholar]
- 39.Daikoku T, et al. Conditional deletion of Msx homeobox genes in the uterus inhibits blastocyst implantation by altering uterine receptivity. Dev Cell. 2011;21(6):1014–1025. doi: 10.1016/j.devcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15(5):2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim S, Kaldis P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development. 2013;140(15):3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, et al. ErbB2 Pathway Activation upon Smad4 Loss Promotes Lung Tumor Growth and Metastasis. Cell Rep. 2015;10(9):1599–1613. doi: 10.1016/j.celrep.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubel CA, et al. Research resource: Genome-wide profiling of progesterone receptor binding in the mouse uterus. Mol Endocrinol. 2012;26(8):1428–1442. doi: 10.1210/me.2011-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng J, et al. Uterine activin receptor-like kinase 5 is crucial for blastocyst implantation and placental development. Proc Natl Acad Sci USA. 2015;112(36):E5098–E5107. doi: 10.1073/pnas.1514498112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park CB, DeMayo FJ, Lydon JP, Dufort D. NODAL in the uterus is necessary for proper placental development and maintenance of pregnancy. Biol Reprod. 2012;86(6):194. doi: 10.1095/biolreprod.111.098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCormack JT, Greenwald GS. Evidence for a preimplantation rise in oestradiol-17beta levels on day 4 of pregnancy in the mouse. J Reprod Fertil. 1974;41(2):297–301. doi: 10.1530/jrf.0.0410297. [DOI] [PubMed] [Google Scholar]
- 47.Finn CA, Martin L. The control of implantation. J Reprod Fertil. 1974;39(1):195–206. doi: 10.1530/jrf.0.0390195. [DOI] [PubMed] [Google Scholar]
- 48.Zhang S, et al. Uterine Rbpj is required for embryonic-uterine orientation and decidual remodeling via Notch pathway-independent and -dependent mechanisms. Cell Res. 2014;24(8):925–942. doi: 10.1038/cr.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 50.Morikawa M, et al. ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res. 2011;39(20):8712–8727. doi: 10.1093/nar/gkr572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto T, Saatcioglu F, Matsuda T. Cross-talk between bone morphogenic proteins and estrogen receptor signaling. Endocrinology. 2002;143(7):2635–2642. doi: 10.1210/endo.143.7.8877. [DOI] [PubMed] [Google Scholar]
- 52.Matsuda T, Yamamoto T, Muraguchi A, Saatcioglu F. Cross-talk between transforming growth factor-beta and estrogen receptor signaling through Smad3. J Biol Chem. 2001;276(46):42908–42914. doi: 10.1074/jbc.M105316200. [DOI] [PubMed] [Google Scholar]
- 53.Erickson GF, Fuqua L, Shimasaki S. Analysis of spatial and temporal expression patterns of bone morphogenetic protein family members in the rat uterus over the estrous cycle. J Endocrinol. 2004;182(2):203–217. doi: 10.1677/joe.0.1820203. [DOI] [PubMed] [Google Scholar]
- 54.Timeva T, Shterev A, Kyurkchiev S. Recurrent implantation failure: The role of the endometrium. J Reprod Infertil. 2014;15(4):173–183. [PMC free article] [PubMed] [Google Scholar]
- 55. Committee on Care and Use of Laboratory Animals (1996) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
- 56.Deb K, Reese J, Paria BC. Methodologies to study implantation in mice. Methods Mol Med. 2006;121:9–34. doi: 10.1385/1-59259-983-4:007. [DOI] [PubMed] [Google Scholar]
- 57.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 58.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1(2):729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]