Significance
We report the identification and characterization of a family of proteins encoded by insect DNA viruses and present in the venom of parasitic wasps. These molecules are homologous to the product of the uncharacterized Drosophila gene diedel (die). We show that Diedel is an immunomodulatory cytokine, which down-regulates the evolutionarily conserved immune deficiency (IMD) pathway of host defense in flies. The importance of this factor is highlighted by the fact that die mutant flies, which express high levels of IMD-regulated immunity genes, have reduced viability. Our work provides the first characterization of virokines in insects to our knowledge, and reveals that besides RNA interference and apoptosis, two well-characterized antiviral responses, insect viruses can also suppress a major signaling pathway of the innate immune response.
Keywords: virokine, cytokine, Sindbis virus, Edin, antiviral immunity
Abstract
Viruses are obligatory intracellular parasites that suffer strong evolutionary pressure from the host immune system. Rapidly evolving viral genomes can adapt to this pressure by acquiring genes that counteract host defense mechanisms. For example, many vertebrate DNA viruses have hijacked cellular genes encoding cytokines or cytokine receptors to disrupt host cell communication. Insect viruses express suppressors of RNA interference or apoptosis, highlighting the importance of these cell intrinsic antiviral mechanisms in invertebrates. Here, we report the identification and characterization of a family of proteins encoded by insect DNA viruses that are homologous to a 12-kDa circulating protein encoded by the virus-induced Drosophila gene diedel (die). We show that die mutant flies have shortened lifespan and succumb more rapidly than controls when infected with Sindbis virus. This reduced viability is associated with deregulated activation of the immune deficiency (IMD) pathway of host defense and can be rescued by mutations in the genes encoding the homolog of IKKγ or IMD itself. Our results reveal an endogenous pathway that is exploited by insect viruses to modulate NF-κB signaling and promote fly survival during the antiviral response.
Like all animals, insects are plagued by viral infections. Insect viruses have a major economic impact, both as the result of infection of beneficial insects (e.g., silkworms, honey bees) and their applied use in insect control strategies. In addition, some insect viral pathogens, known as arboviruses (arthropod-borne viruses), can be transmitted to humans and cause serious disease. Finally, the powerful genetic tools available for the fruit fly Drosophila melanogaster can be used in conjunction with characterized insect viruses to decipher antiviral immune responses in a model system. The large number and high diversity of the RNA and DNA viruses that have been characterized in insect hosts represents an extensive set of models to investigate virus–host interactions.
The pioneering studies on insect innate immunity were focused on bacterial infections and led to the discovery of the first antimicrobial peptide (AMP), Cecropin (1). The characterization of the mechanisms regulating expression of AMP genes in Drosophila subsequently revealed two evolutionarily conserved signaling pathways, Toll and immune deficiency (IMD), controlling transcription factors of the NF-κB family (2, 3). The Toll pathway is activated following infection by fungi and bacteria containing Lysine-type peptidoglycan (PGN) in their cell wall and regulates expression of a set of AMPs, including Drosomycin, through the p65-like transcription factor DIF (4). It shares striking similarities with the Toll-like receptor/interleukin (IL)-1 receptor pathway in mammals. The IMD pathway is triggered by bacteria containing diaminopimelic acid PGN in their cell wall (e.g., Gram-negative bacteria). This pathway activates expression of a different set of AMPs (including diptericin, cecropins, drosocin, and attacins) and acts through the p105-like transcription factor Relish (3, 5). It is evocative of the tumor necrosis factor (TNF) receptor pathway from vertebrates.
Previous studies have revealed that insect susceptibility to viral infection is determined by host factors, which either facilitate (6, 7) or restrict viral replication (8–10). In addition, the bacterial flora, particularly endosymbiotic Wolbachia strains, modifies susceptibility to viral infections (11, 12). Viruses also trigger dedicated host–defense mechanisms as in other organisms. RNA interference (RNAi) mechanisms mediate a strong antiviral response against a broad range of RNA and DNA viruses in Drosophila (13–16). RNAi involves the RNaseIII enzyme Dicer-2, which produces virus-derived small interfering RNAs (siRNAs). These siRNAs are then loaded onto the Argonaute-2 (AGO2) nuclease and guide the RNA-induced silencing complex (RISC) to target complementary viral sequences for silencing (17). Apoptosis is also a major response to viral infection, initially characterized in Lepidopteran insects in the context of DNA viruses (18), and more recently in Drosophila (19). Activation of this programmed cell death pathway rapidly eliminates infected cells before completion of the viral replication cycle. Insect viruses often encode inhibitors of RNAi or apoptosis (known as VSRs or VSAs, for viral suppressors of RNAi or apotosis, respectively), attesting to the restrictive pressures exerted by these host antiviral mechanisms in insects (20–26).
Besides RNAi and apoptosis, global gene expression analyses have indicated that viral infections trigger complex transcriptional responses. These gene expression profiling studies have been supported by experiments with mutant flies, which point to the involvement of the Toll, IMD, and Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathways in the control of viral infections (reviewed in ref. 27). Although a few induced genes have been associated with resistance to Sindbis virus (SINV) (e.g., Diptericin-B, Attacin-C) (28) or Drosophila C virus (DCV) (e.g., Vago) (29), this inducible response remains poorly characterized. In particular, little is known about the function of many of the genes induced by viral immune challenge (27). Here, we investigate the function of the Drosophila gene diedel (die) that is strongly induced by some viral infections (30, 31). We report that die belongs to a larger family that includes homologs found in the genomes of members of three large classes of insect viruses, the Entomopoxvirinae, the Ascoviridae, and the Baculoviridae. We show that die and also a viral homolog can promote host survival by modulating the activation of the IMD pathway and NF-κB signaling.
Results
die Is Strongly Induced by Viral Infection and Has Several Viral Homologs.
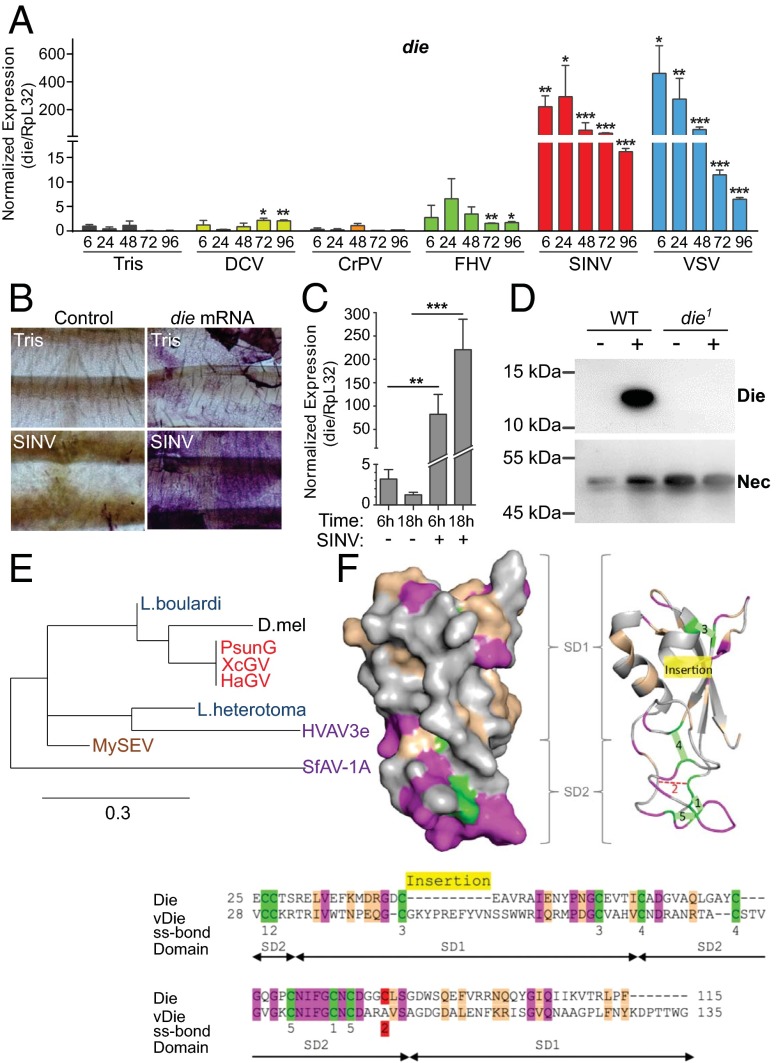
We monitored expression of die in virus-infected flies and observed transcript induction with four of the five viruses tested (see also ref. 32). By contrast, we did not observe a significant induction of die by bacteria in our infection conditions (SI Appendix, Fig. S1 and ref. 33). The die induction was particularly striking (up to 460-fold) in the case of SINV and VSV. With these viruses, the time course of induction showed highest levels during early time points (6 h, 24 h), with subsequent steadily decline (Fig. 1A). VSV and SINV are enveloped viruses, suggesting that lipids or other moieties from the envelope may contribute to the induction of die. Indeed, a significant up-regulation of die was observed when UV-inactivated SINV was injected into flies (SI Appendix, Fig. S2). We tested representative mutants of the RNAi, JAK/STAT, Toll, and IMD pathways and observed that the NF-κB transcription factor Dif was required for the up-regulation of die. However, the Toll adapter MyD88 was not required, indicating that Dif is not activated by the canonical Toll pathway in this context (SI Appendix, Fig. S3).
Fig. 1.
die is up-regulated in response to viral infection. (A) Wild-type flies were injected with Tris, DCV, CrPV, FHV, SINV, or VSV. die mRNA level was monitored by qPCR at the indicated time points. Data represent the mean ± SD of two independent experiments, each containing two groups of six flies. Student's t test: *P < 0.05, **P < 0.01, ***P < 0.001. (B) Whole-mount in situ hybridization on fat body of Tris or SINV-injected flies (18 h after injection) with Digoxigenin-labeled sense (control) or antisense die RNA probes. (C) die mRNA levels in dissected fat body extracts prepared at the indicated time points from flies injected with Tris (-) or SINV were quantified by qPCR. Data represent the mean ± SD of three independent experiments, each containing six flies. (D) Protein extract from hemolymphs of wild-type and die1 null mutant flies injected with Tris (-) or SINV were analyzed by Western blotting with Die or Necrotic (Nec)-specific antibodies. The circulating protein Nec was used as loading control. (E) Phylogeny of Die-related molecules identified in the genome of the indicated viruses (granuloviruses in red, ascoviruses in violet, and the entomopoxvirus in brown) and in the venom of Leptopilina parasitic wasps (in blue). (F) Overall structure of Die. The conserved residues between D. melanogaster Die and ORF121 from SfAV-1a (vDie) are plotted on the molecular surface (Upper Left) according to the following code: in green the conserved cysteins, in red the lacking cystein, in magenta the strictly conserved amino acid residues, and in pink the homologous amino acid residues.
In situ hybridization on whole-mount adult tissues showed expression of die in the fat body, which was strongly increased upon infection with SINV (Fig. 1B). Up-regulation of die in the fat body was confirmed by RT-PCR analysis on dissected tissue (Fig. 1C). The die transcript encodes a putative signal peptide, and the Die protein is readily secreted into the supernatant of transfected S2 cells (SI Appendix, Fig. S4A and ref. 30). We produced an antibody against the recombinant protein from S2 cells, which detects Die on Western blots from hemolymph after infection with SINV (Fig. 1D). We conclude that Die is a circulating hemolymph component, which is synthesized in the fat body and strongly up-regulated after infection by some viruses.
Homologs of die were found in three different and unrelated families of large DNA viruses that infect Lepidoptera such as Entomopoxvirinae (Mythimna separata entomopoxvirus L), Baculoviridae (Pseudaletia unipuncta granulovirus, Helicoverpa armigera granulovirus, Xestia c-nigrum granulovirus) and Ascoviridae [Spodoptera frugiperda ascovirus 1a (SfAv-1a), Heliothis virescens ascovirus], suggesting that these viruses independently hijacked this gene from an insect genome (Fig. 1E). Homologs of Die are also present in the venom of two related parasitoid wasp species, Leptopilina boulardi and Leptopilina heterotoma (Fig. 1E and SI Appendix, Fig. S4 B and C) (34, 35). This wasp venom suppresses the Drosophila larval immune response when injected together with wasp eggs (36). All these proteins contain predicted signal peptides, suggesting that they are secreted, like the Drosophila Die protein.
The crystal structure of Die has been solved in a previous study (30). Die is composed of two subdomains SD1 and SD2. SD1 is made of an antiparallel β-sheet covered by an α-helix and displays a ferredoxin-like fold. SD2 reveals a previously unidentified protein fold made of loops connected by four disulfide bridges (Fig. 1F). Die is an extracellular protein, which presents a high level of stability certainly because of the presence of five disulfide bridges. The alignment of the sequence of Die with those of its homologs found in viruses and venoms shows that the cystein array is well conserved with the presence of the CC and CXC motif in most of the sequences (SI Appendix, Fig. S4C). At the tridimensional level, the presence of five disulfide bridges in such a rather small protein (12 kDa) imposes strong structural constraints, so we can assume that the homologs display a fold similar to that of Die. Plotting the similar residues between Die and its viral homolog SfAV-1a reveals large conserved molecular surface areas (Fig. 1F), suggesting that the two proteins exert related functions.
die Promotes Survival Following SINV Infection.
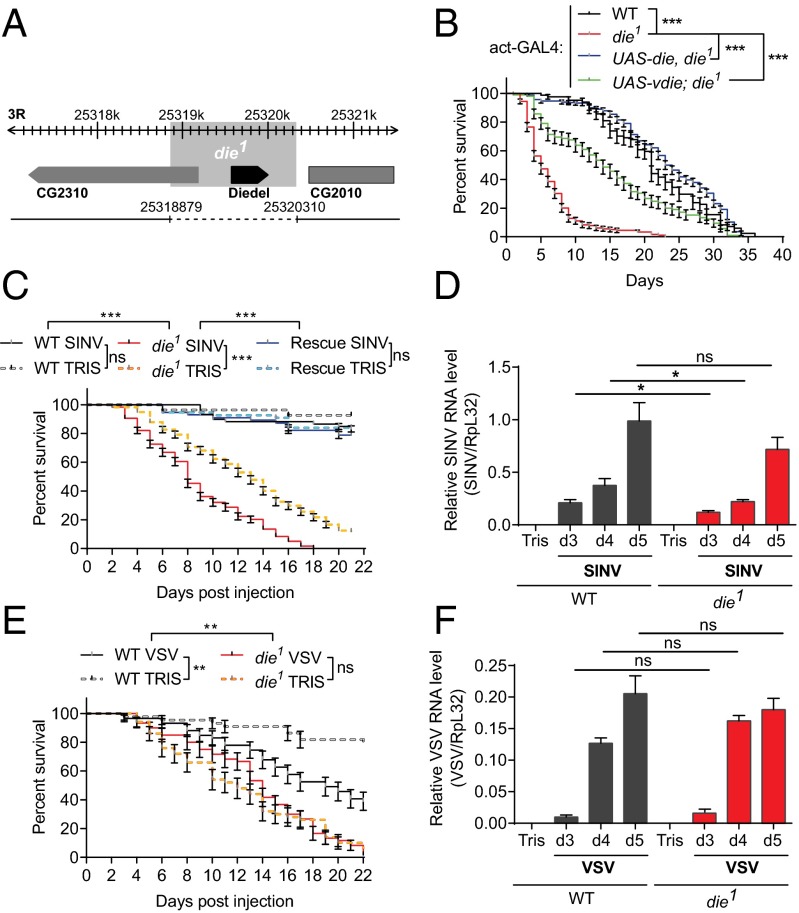
To characterize the function of die, we generated mutant flies by imprecise excision of a transposon inserted next to the die coding sequence. The die1 allele has a large deletion that includes the whole die gene plus the 5′ region of the adjacent gene, CG2310 (Fig. 2A). We were unable to detect neither mRNA (SI Appendix, Fig. S5 A and B) nor protein (Fig. 1D) in these flies, confirming that die1 is a null allele. The die1 homozygous flies show reduced viability. We assayed the ratio of die1 homozygote and heterozygote flies by using a GFP-labeled balancer chromosome and observed that the number of die1 homozygote flies steadily decreases during larval development (SI Appendix, Fig. S5B). Similarly, homozygous die1 adults have reduced viability in the absence of immune challenge at 29 °C or 25 °C (Fig. 2B and SI Appendix, Fig. S5D). Importantly, the viability of the die1 mutant chromosome was fully rescued by the expression of a transgenic die cDNA, indicating that the reduced viability resulted from the deletion of die (Fig. 2B). Altogether, these findings suggest that die promotes survival throughout development and in adult flies, although it is not absolutely required for viability. Of note, expression of the Die homolog from SfAV-1a partially rescued the lifespan of die1 mutants (Fig. 2B).
Fig. 2.
die promotes viability in uninfected and SINV-infected flies. (A) A null mutant allele for die (die1) was generated by imprecise P-element excision, which resulted in a deletion spanning the whole die gene plus the 5′ coding region of the adjacent gene CG2310. The breakpoints of the deletion are indicated. (B) The lifespan of flies of the indicated genotypes was monitored daily at 29 °C. The reduced viability of the mutant flies was rescued when a UAS-die transgene was expressed under the control of the ubiquitous actin-GAL4 driver in die1 mutants (act-GAL4; UAS-die, die1 referred to as Rescue below). (C) Flies of the indicated genotype were injected with Tris or SINV and survival was monitored daily at 29 °C. Note that all flies have the actin-GAL4 driver. (D) Quantitative RT-PCR analysis of the accumulation of SINV RNA at indicated time points after infection in wild-type and die1 mutant flies. (E) Flies of the indicated genotype were injected with Tris or VSV and survival was monitored daily at 29 °C. (F) Quantitative RT-PCR analysis of the accumulation of VSV RNA at indicated time points after infection in wild-type and die1 mutant flies. Data represent the mean ± SE (B, C, and E) or SD (D and F) of at least three independent experiments, each containing three groups of 10 (B, C, and E) or 6 (D and F) flies. Log rank test (B, C, and E) and t test (D and F): *P < 0.05, **P < 0.01, ***P < 0.001.
We next challenged die1 mutants with several viruses. Interestingly, die1 flies showed reduced survival after a challenge with a sublethal dose of SINV, without any increase in viral load (Fig. 2 C and D and SI Appendix, Fig. S5E). Increased lethality did not result from the general weakness of die1 flies, because their viability was similar to wild-type flies following injection of Tris buffer or the slowly replicating virus, VSV (Fig. 2 E and F). The die1 mutation did not affect survival following challenge with the fast replicating viruses Drosophila C virus, Cricket Paralysis virus, or Flock House virus, which kill wild-type flies within 10 days (SI Appendix, Fig. S6). Of note, induction of the JAK/STAT regulated gene Tot-M was not affected in die1 mutant flies, suggesting that die does not encode a negative regulator of the JAK/STAT pathway (37) (SI Appendix, Fig. S7). Furthermore, the die1 mutation did not increase lethality compared with controls following infection by the fungus Beauveria bassiana, the Gram-positive bacteria Enteroccocus faecalis, or the Gram-negative bacteria Enterobacter cloacae (SI Appendix, Fig. S8 A–C). Thus, die specifically promotes fly survival following SINV infection, although without directly affecting viral replication. Finally, we observed that Dif mutant flies are not susceptible to SINV infection, indicating that the basal level of die expression is sufficient to promote the survival of the flies (SI Appendix, Fig. S9).
Up-Regulated Expression of Immunity Genes in die1 Mutant Flies.
To understand the contribution of die to viability, we analyzed the transcriptome of die1 mutant flies with and without viral immune challenge. In uninfected conditions, die1 mutants showed 142 up-regulated and 69 down-regulated genes, compared with wild-type controls (cutoff of twofold change) (Fig. 3A and SI Appendix, Table S1). Four genes were up-regulated more than 40-fold in die1 mutant flies, whereas the most strongly down-regulated gene was repressed 18-fold (Fig. 3B). Significantly, Gene Ontology (GO) analysis revealed a strong enrichment for immune response genes among the up-regulated gene set (Fig. 3C). The down-regulated gene set, however, showed no GO enrichment that identified particular biological processes (SI Appendix, Fig. S10). During SINV challenge, we observed 171 up-regulated genes and 16 down-regulated genes in wild-type flies compared with 98 up-regulated and 10 down-regulated genes in die1 mutant flies (SI Appendix, Table S2). In SINV-infected conditions, 102 genes were up-regulated in die1 mutants compared with wild-type control. Of these genes, only 16 were induced by SINV infection (SI Appendix, Table S3). Strikingly, GO analysis revealed an exclusive enrichment for molecules participating in host–defense (e.g., antimicrobials peptides, PGRPs) (Fig. 3 D and E). Moreover, 15 of these 16 genes are targets of the transcription factor Relish.
Fig. 3.
die and its viral homolog suppress expression of stress and immune genes. (A) Pie charts showing the number of up (in red) and down-regulated (in blue) genes in die1 compared with wild-type flies as determined by genomewide microarray analysis in noninfected conditions. (B) Fold change of the top 40 deregulated genes in die1 null mutants in noninfected conditions (die1/WT). (C) Gene ontology analysis for biological process for the 142 up-regulated genes in noninfected conditions when die is mutated. (D) Fold change of the 16 genes up-regulated in die1 mutants compared with wild-type flies and induced 4 d after infection by SINV. (E) Gene ontology analysis for biological processes for the 16 up-regulated genes in die1 mutant infected by SINV. (F–H) mRNA levels of Cyp12d1-d (F), Metchnikowin (G), and edin (H) were monitored by qPCR in wild-type, die1, Rescue, and act-GAL4,UAS-vdie, die1 flies 4 d after injection with Tris (-) or SINV. Data represent the mean ± SD of three independent experiments, each containing three groups of six flies. t test: ***P < 0.001. For B and D, data represent the mean of two independent biological samples.
We confirmed by quantitative PCR (qPCR) that Cyp12d1-d and Metchnikowin (Mtk) are strongly up-regulated in die1 mutant compared with control flies in the absence of infection (Fig. 3 F and G). The effect is particularly striking for the expression of Cyp12d1-d, which is undetectable in wild-type flies (Fig. 3F). Importantly, the expression of a transgenic die cDNA in die1 mutant flies suppressed expression of these genes, confirming that the phenotype results from the lack of die expression (Fig. 3 F and G). In addition, elevated during infection (edin) and Attacin-A (Att-A) were strongly induced by SINV in die1 mutant flies, but not in wild-type controls or in rescue flies (Fig. 3H). Of note, neither edin nor Att-A expression was induced by VSV infection in die1 mutant flies, which may explain why die1 mutant flies do not exhibit increased lethality when challenged with VSV (SI Appendix, Fig. S11). Taken together, these data suggest that Die is an immunomodulatory cytokine dampening expression of immune response genes in the fat body. Importantly, expression of the SfAV1 homolog of die also suppressed expression of these genes in die1 mutant flies (Fig. 3 F–H). The rescue by v-die was complete for some genes (e.g., Cyp12d1-d) and partial for others (e.g., Mtk, edin), possibly explaining the partial complementation for survival (Fig. 2B).
Activation of the IMD Pathway in die1 Mutant Flies Enhances Pathogenesis.
The above results suggest a possible involvement of the IMD pathway in the phenotype of die1 mutant flies. To validate this hypothesis, we produced double mutant flies for die and kenny (key), which encodes the homolog of IKKγ, a critical component of the IMD pathway (38). In this context, expression of Cyp12d1-d was completely suppressed (Fig. 4A). Similarly, induction of Att-A and edin by SINV infection was not observed in die, IKKγ double mutant flies (Fig. 4 B and C). Mutation of the IKKγ gene also improved viability of die1 mutant flies both in the absence of immune challenge (Fig. 4D) and most strikingly after SINV infection (Fig. 4E). Similarly, an imd mutation also rescued survival of die1 mutant flies following SINV infection (Fig. 4F). By contrast, a mutation in the gene Dif, which encodes the NF-κB family transcription factor of the Toll pathway, did not improve survival of die1 mutant flies (SI Appendix, Fig. S12). Of note, antibiotic treatment to clear the bacterial flora did not improve the viability of die1 mutant flies, and these flies mount a normal immune response to bacterial infection, suggesting that receptors other than PGRPs activate the IMD pathway in die1 mutant flies (SI Appendix, Figs. S8 and S13). The expression of known inhibitors of the IMD pathway was not modified in die1 mutant flies, indicating that Die acts through a previously unidentified mechanism (SI Appendix, Fig. S14). We conclude that the reduced viability of die1 mutants results from inappropriate activation of the IMD pathway and, by extension, that both insect DNA viruses and parasitoid wasps use Die homologs to suppress the host IMD-mediated immune response.
Fig. 4.
IMD pathway mediates deregulation of gene expression and reduction of viability in die mutant flies. (A–C) mRNA levels of Cyp12d1-d (A), Attacin-A (B) and edin (C) were monitored in wild-type, keyc02831, die1, and double mutant keyc02831; die1 flies at 4 d after injection with Tris or SINV. (D and E) Wild-type, keyc02831, die1, and double null mutant keyc02831; die1 flies were injected with Tris (D) or SINV (E) and survival was monitored daily at 29 °C. (F) Wild-type, imd1, die1, and double mutant imd1, die1 flies were injected with SINV and survival was monitored daily at 29 °C. Data represent the mean ± SE (D–F) or SD (A–C) of three independent experiments, each containing three groups of 6 (A–C) or 10 flies (D–F). t test (A–C) and log rank test (D–F): *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Insect Virokines Validate Putative Regulatory Components of the IMD Pathway.
Large DNA viruses are particularly prone to hijack host genes that allow them to evade immune responses. These big viral genomes (of 100–200 kb) can be regarded as information repositories concerning the host antiviral defenses (39). Identifying viral homologs of cellular genes can therefore validate critical host gene regulatory functions and point to novel antiviral response components. For example, the characterization of the viral inhibitor of apoptosis protein (vIAP) in Baculoviridae and later Entomopoxvirinae led to the identification of the host cIAPs, which regulate caspase activity in insects and in vertebrates (22, 40). Similarly, the identification of a viral cytokine in the genome of Herpesvirus saimiri paved the way for the discovery of the mammalian cytokine IL-17 and its receptor (41). More generally, the identification of suppressors of host defenses within viral genomes provides useful indications of the restrictive pressures mounted by the host immune system.
Many insect and plant RNA and DNA viruses express VSR, which confirms the importance of RNAi in the control of viral infections. For the most part, these VSRs are structurally unrelated to each other, and antagonize antiviral RNAi at multiple levels, either binding, sequestering, and degrading long dsRNAs or siRNA duplexes or antagonizing the catalytic activity of AGO2 (20, 21, 23–26). Extensive characterization of the Lepidopteran large DNA viruses has identified a range of VSAs, targeting another prominent antiviral pathway. These VSAs include suicide substrates for caspases (e.g., AcMNPV-p35, AMP-p33), and inhibitors (vIAPs), which bind to their N terminus (reviewed in ref. 22).
Previous studies have indicated that the JAK/STAT (42, 43) and NF-κB signaling pathways may have a role in antiviral immunity, although the molecular mechanisms were not characterized (reviewed in ref. 27, see also refs. 43–46). Of note, imd or IKKγ mutant flies were not more susceptible to SINV infection than wild-type controls in our conditions (SI Appendix, Fig. S15). Nevertheless, the virus clearly induced IMD target genes in die1 mutant flies, unlike VSV for which we observed no phenotype. Another hint for a role of NF-κB signaling in the antiviral response comes from the polydnaviruses (PDVs), which have an obligate symbiotic association with parasitic wasps. The PDV genome encodes vankyrin proteins, which show homology to IκB but lack the peptide degradation motif (47, 48). Members of the vankyrin family can antagonize activation of the Toll and IMD NF-κB proteins (49, 50). In addition, the cellular response leading to wasp egg encapsulation is blocked. Given that the PDVs are obligate symbionts of Ichneumonid and Braconid parasitic wasps, it is unclear that the vankyrins are targeted to modulate the antiviral immune response. Alternatively, their major function may be to block the cellular immune response that encapsulates the wasp parasite, on which survival of the PDV depends. Therefore, viral Die represents the first example to our knowledge for a bona fide viral suppressor of inducible immune responses to be identified in insects.
Cytokines orchestrate immune responses and as such are good targets for viral evasion strategies. Indeed, several cytokines and cytokine binding proteins have been identified in the genomes of mammalian Herpesviruses and Poxviruses (39). Among these cytokines, viral IL-10 is particularly striking. IL-10 is a pleiotropic cytokine related to interferons with both immunostimulatory and immunosuppressive properties. vIL-10 orthologs have so far been found in the genomes of 12 members of the Herpesviridae family, 2 members of the Alloherpesviridae family, and 7 members of the Poxviridae family. Analysis of the evolution of these viral genes indicates that their acquisition from cellular genomes occurred independently at least eight times (51). Altogether, these data attest of the importance of IL-10 in the orchestration of the mammalian immune response (52). Similarly, viral homologs of the Drosophila Die cytokine have been identified in six DNA viruses that belong to three unrelated families (Baculoviridae, Entomopoxvirinae, Ascoviridae) and in the venom of parasitic wasps, as discussed above. This cytokine therefore reveals an important regulatory control point in Drosophila immunity.
A Regulator of Systemic Activation of the IMD Pathway.
Our data identify Die as a cytokine participating in the homeostasis of the immune response in Drosophila, by modulating expression of IMD-regulated genes. It is now well established that excessive or sustained activation of the IMD pathway has deleterious effects, and that this pathway is severely constrained by multiple negative regulators acting at several levels (5, 53). For example, Relish-dependent chronic expression of AMPs in the brain of flies mutant for dnr1 causes neurodegeneration (54).
The TNF receptor pathway in mammals, which shares many similarities with the IMD pathway, is also tightly regulated. Indeed, chronic TNF activation contributes to many disease pathologies (55). Thus, activation of this evolutionarily conserved innate immunity signaling pathway is a double-edged sword and must be regulated at multiple levels to prevent harmful consequences for the host, be it an insect or a mammal.
Whereas a number of negative regulators of the IMD pathway act locally in epithelia to modulate the expression of AMPs by the endogenous flora (53, 56, 57), Die appears to have a broader function in homeostasis, because its mutation leads to a strong phenotype in the absence of infectious challenge and even in antibiotic-treated flies. The strongly improved viability of die1; IKKγ− and die1; imd− double mutant flies attests that the die function per se is largely dispensable for tissue homeostasis, although die clearly has a major role in preventing activation of the IMD pathway. Our data are consistent with a model where chronic expression of a subset of IMD-regulated genes in the absence of immune challenge, and strong up-regulation of the pathway during SINV infection, trigger collateral damage to the host, resulting in increased lethality (SI Appendix, Fig. S16). The reported toxicity associated with ectopic AMP or Edin expression supports this model (54, 58). It is possible that homologs of die were acquired by some viruses to promote survival of the infected insects and to allow optimal viral replication. Alternatively, suppression of the expression of specific components of the host–defense program by die homologs may be beneficial to insect pathogens. In this regard, it is interesting to note that Edin was recently shown to regulate the cellular response to parasite wasp infections (59).
Methods
Drosophila Strains.
w1118 (used as wild-type flies), y1 w*; P{Act5C-GAL4}25FO1/CyO, y+ w1118; keyc02831, y1 sc* v1; P{VALIUM20-mCherry}attP2 (dsRNA against mCherry), y1 sc* v1; P{TRiP.HMS00183}attP2 (dsRNA against Myd88), y1 sc* v1; P{TRiP.HMS00084}attP2 (dsRNA against Cactus), y1 sc* v1; P{TRiP.HM05257}attP2 (dsRNA against Dif), w1118; Df(2R)BSC279/CyO (Deficiency covering Myd88), w1118; Df(2L)Exel7068/CyO (Deficiency covering Dif), w1118; Df(2R)BSC856/CyO-Df(2R)B80, and y+ (Deficiency covering key) spz4/TM3 were obtained from the Bloomington Fly Stock Center. imd1, Dcr2L811fsX, hopmsv1/M38, RelE20, Myd88c03881, key1, and Dif1 mutant flies were described previously (29). die1 flies were generated after an imprecise excision of the P element of the strain d06279. The die1 mutant chromosome was recombined with the strain w1118, and identical phenotypes were observed with two lines resulting from independent recombination events. All flies were in a PstS background and were tested for the presence of Wolbachia and persistent infection by DCV, FHV, and Nora virus, and treated when necessary. The fly stocks were raised on standard cornmeal-agar medium at 25 °C. Adult flies between 4–6 d after hatching were used in infection experiments.
Infections.
Virus infections were made by intrathoracic injection (Nanoject II apparatus; Drummond Scientific) of 4.6 nL of a viral suspension in 10 mM Tris⋅HCl, pH 7.5 [500 plaque-forming units (pfu) per fly for DCV and FHV, 5 pfu per fly for CrPV, 2,500 pfu per fly for SINV], with the exception of VSV (5,000 pfu per fly), which was injected directly in the Vero cell culture medium (DMEM). Control injections used the same volume of 10 mM Tris⋅HCl, pH 7.5. Infected flies were then incubated at 25 °C or at 29 °C. For survival experiments, three tubes of 10 flies were injected with each viral strain. In the case of bacteria, flies were pricked with a thin needle previously dipped in a concentrated overnight culture of E. cloacae, E. faecalis, or Micrococcus luteus in LB medium. For B. bassiana survival experiments, natural infection with fungal spores was used.
Quantitative RT-PCR.
RNA was isolated with TRI Reagent RT (Molecular Research Central) according to the manufacturer’s instruction. RNA was reverse transcribed by using iScriptcDNA Synthesis Kit (Bio-Rad). Analysis of RNA expression was performed by real-time quantitative RT-PCR by using iTaq SYBR Green Kit (Bio-Rad) except for Fig. 1A (see Taq-Man procedure in SI Appendix). Gene expression was normalized to the expression of the RNA encoding the ribosomal protein (RpL32). Primers used for real-time PCR were listed in SI Appendix, SI Methods.
Statistical Analysis.
An unpaired two-tailed Student's t test was used for statistical analysis of data with GraphPad Prism (GraphPad Software). Survival curves were plotted and analyzed by log-rank analysis (Kaplan–Meier method) using GraphPad Prism (GraphPad Software). P values lower than 0.05 were considered statistically significant: *P < 0.05, **P < 0.01, ***P < 0.001.
Additional methods are available in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank Estelle Santiago for expert technical assistance; Stefanie Müller and Safia Deddouche for help in the early stages of the project; Yves Bigot and Brian Federici for discussion and providing SfAV-1a DNA; and Carine Meignin, Dominique Ferrandon, and David Gubb for discussions, help with preparation of the figures, and critical reading of the manuscript. The microarray analysis were performed at the Plateforme Biopuces et Séquençage, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg, France. This work was supported by CNRS, Université de Strasbourg, Université d’Aix Marseille; NIH Grant P01 AI070167; Agence Nationale de la Recherche (ANR) Grants ANR-11-ISV3-002 and ANR-13-BSV3-0009; and Investissements d’Avenir Program Grants NetRNA ANR-10-LABX-36 and I2MC ANR-11-EQPX-0022.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The normalized and raw data for the microarray analysis of gene expression in Fig. 3 and SI Appendix, Tables S1–S3 were deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE68174).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516122113/-/DCSupplemental.
References
- 1.Steiner H, Hultmark D, Engström A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292(5820):246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426(6962):33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 3.Hultmark D. Drosophila immunity: Paths and patterns. Curr Opin Immunol. 2003;15(1):12–19. doi: 10.1016/s0952-7915(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 4.Valanne S, Wang J-H, Rämet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186(2):649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- 5.Kleino A, Silverman N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol. 2014;42(1):25–35. doi: 10.1016/j.dci.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majzoub K, et al. RACK1 controls IRES-mediated translation of viruses. Cell. 2014;159(5):1086–1095. doi: 10.1016/j.cell.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, et al. pelo is required for high efficiency viral replication. PLoS Pathog. 2014;10(4):e1004034. doi: 10.1371/journal.ppat.1004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magwire MM, et al. Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet. 2012;8(11):e1003057. doi: 10.1371/journal.pgen.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magwire MM, Bayer F, Webster CL, Cao C, Jiggins FM. Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a duplication. PLoS Genet. 2011;7(10):e1002337. doi: 10.1371/journal.pgen.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martins NE, et al. Host adaptation to viruses relies on few genes with different cross-resistance properties. Proc Natl Acad Sci USA. 2014;111(16):5938–5943. doi: 10.1073/pnas.1400378111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedges LM, Brownlie JC, O’Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322(5902):702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- 12.Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6(12):e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saleh M-C, et al. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458(7236):346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller S, et al. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc Natl Acad Sci USA. 2010;107(45):19390–19395. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Y-H, et al. RNA-based immunity terminates viral infection in adult Drosophila in the absence of viral suppression of RNA interference: Characterization of viral small interfering RNA populations in wild-type and mutant flies. J Virol. 2011;85(24):13153–13163. doi: 10.1128/JVI.05518-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bronkhorst AW, et al. The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc Natl Acad Sci USA. 2012;109(51):E3604–E3613. doi: 10.1073/pnas.1207213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding S-W. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10(9):632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 18.Clem RJ, Fechheimer M, Miller LK. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254(5036):1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 19.Liu B, et al. P53-mediated rapid induction of apoptosis conveys resistance to viral infection in Drosophila melanogaster. PLoS Pathog. 2013;9(2):e1003137. doi: 10.1371/journal.ppat.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bronkhorst AW, van Cleef KWR, Venselaar H, van Rij RP. A dsRNA-binding protein of a complex invertebrate DNA virus suppresses the Drosophila RNAi response. Nucleic Acids Res. 2014;42(19):12237–12248. doi: 10.1093/nar/gku910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Cleef KWR, et al. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res. 2014;42(13):8732–8744. doi: 10.1093/nar/gku528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clem RJ. Viral IAPs, then and now. Semin Cell Dev Biol. 2015;39:72–79. doi: 10.1016/j.semcdb.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Hussain M, Abraham AM, Asgari S. An Ascovirus-encoded RNase III autoregulates its expression and suppresses RNA interference-mediated gene silencing. J Virol. 2010;84(7):3624–3630. doi: 10.1128/JVI.02362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296(5571):1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 25.van Mierlo JT, et al. Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS Pathog. 2014;10(7):e1004256. doi: 10.1371/journal.ppat.1004256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nayak A, et al. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol. 2010;17(5):547–554. doi: 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamiable O, Imler J-L. Induced antiviral innate immunity in Drosophila. Curr Opin Microbiol. 2014;20:62–68. doi: 10.1016/j.mib.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Z, Kingsolver MB, Avadhanula V, Hardy RW. An antiviral role for antimicrobial peptides during the arthropod response to alphavirus replication. J Virol. 2013;87(8):4272–4280. doi: 10.1128/JVI.03360-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deddouche S, et al. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9(12):1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- 30.Coste F, et al. Crystal structure of Diedel, a marker of the immune response of Drosophila melanogaster. PLoS One. 2012;7(3):e33416. doi: 10.1371/journal.pone.0033416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp C, et al. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol. 2013;190(2):650–658. doi: 10.4049/jimmunol.1102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merkling SH, et al. The heat shock response restricts virus infection in Drosophila. Sci Rep. 2015;5:12758. doi: 10.1038/srep12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002;3(5):711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 34.Colinet D, et al. Extensive inter- and intraspecific venom variation in closely related parasites targeting the same host: The case of Leptopilina parasitoids of Drosophila. Insect Biochem Mol Biol. 2013;43(7):601–611. doi: 10.1016/j.ibmb.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Goecks J, et al. Integrative approach reveals composition of endoparasitoid wasp venoms. PLoS One. 2013;8(5):e64125. doi: 10.1371/journal.pone.0064125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee MJ, et al. Virulence factors and strategies of Leptopilina spp.: Selective responses in Drosophila hosts. Adv Parasitol. 2009;70:123–145. doi: 10.1016/S0065-308X(09)70005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436(7052):871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 38.Rutschmann S, et al. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat Immunol. 2000;1(4):342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- 39.Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol. 2003;3(1):36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- 40.Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67(4):2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao Z, et al. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity. 1995;3(6):811–821. doi: 10.1016/1074-7613(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 42.Dostert C, et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. 2005;6(9):946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 43.Merkling SH, et al. The epigenetic regulator G9a mediates tolerance to RNA virus infection in Drosophila. PLoS Pathog. 2015;11(4):e1004692. doi: 10.1371/journal.ppat.1004692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gammon DB, et al. A single vertebrate DNA virus protein disarms invertebrate immunity to RNA virus infection. eLife. 2014;3 doi: 10.7554/eLife.02910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreira ÁG, et al. The Toll-dorsal pathway is required for resistance to viral oral infection in Drosophila. PLoS Pathog. 2014;10(12):e1004507. doi: 10.1371/journal.ppat.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 2009;5(9):e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kroemer JA, Webb BA. Ikappabeta-related vankyrin genes in the Campoletis sonorensis ichnovirus: Temporal and tissue-specific patterns of expression in parasitized Heliothis virescens lepidopteran hosts. J Virol. 2005;79(12):7617–7628. doi: 10.1128/JVI.79.12.7617-7628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thoetkiattikul H, Beck MH, Strand MR. Inhibitor kappaB-like proteins from a polydnavirus inhibit NF-kappaB activation and suppress the insect immune response. Proc Natl Acad Sci USA. 2005;102(32):11426–11431. doi: 10.1073/pnas.0505240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bitra K, Suderman RJ, Strand MR. Polydnavirus Ank proteins bind NF-κB homodimers and inhibit processing of Relish. PLoS Pathog. 2012;8(5):e1002722. doi: 10.1371/journal.ppat.1002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gueguen G, Kalamarz ME, Ramroop J, Uribe J, Govind S. Polydnaviral ankyrin proteins aid parasitic wasp survival by coordinate and selective inhibition of hematopoietic and immune NF-kappa B signaling in insect hosts. PLoS Pathog. 2013;9(8):e1003580. doi: 10.1371/journal.ppat.1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ouyang P, et al. IL-10 encoded by viruses: A remarkable example of independent acquisition of a cellular gene by viruses and its subsequent evolution in the viral genome. J Gen Virol. 2014;95(Pt 2):245–262. doi: 10.1099/vir.0.058966-0. [DOI] [PubMed] [Google Scholar]
- 52.Banchereau J, Pascual V, O’Garra A. From IL-2 to IL-37: The expanding spectrum of anti-inflammatory cytokines. Nat Immunol. 2012;13(10):925–931. doi: 10.1038/ni.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee K-Z, Ferrandon D. Negative regulation of immune responses on the fly. EMBO J. 2011;30(6):988–990. doi: 10.1038/emboj.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao Y, Chtarbanova S, Petersen AJ, Ganetzky B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc Natl Acad Sci USA. 2013;110(19):E1752–E1760. doi: 10.1073/pnas.1306220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beutler B, Rietschel ET. Innate immune sensing and its roots: The story of endotoxin. Nat Rev Immunol. 2003;3(2):169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 56.Paredes JC, Welchman DP, Poidevin M, Lemaitre B. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity. 2011;35(5):770–779. doi: 10.1016/j.immuni.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 57.Lhocine N, et al. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4(2):147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 58.Gordon MD, Ayres JS, Schneider DS, Nusse R. Pathogenesis of listeria-infected Drosophila wntD mutants is associated with elevated levels of the novel immunity gene edin. PLoS Pathog. 2008;4(7):e1000111. doi: 10.1371/journal.ppat.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanha-Aho L-M, et al. Edin expression in the fat body is required in the defense against parasitic wasps in Drosophila melanogaster. PLoS Pathog. 2015;11(5):e1004895. doi: 10.1371/journal.ppat.1004895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.