Significance
Single-span membrane proteins (ssMPs) are anchored by single hydrophobic helices to cell surfaces, where they mediate cell–cell communications. Unfortunately, hydrophobic helices also cause aggregation in solution, rendering ssMPs nonfunctional. We discovered that in vitro-synthesized ssMPs localize on an oil drop’s surface, preventing aggregation of ssMPs in solution and promoting assembly of functional structures on the drop’s surface. We use this approach to synthesize and display apoptosis-inducing ssMPs and show that these “death drops” are functional and can kill cultured cancer cells. Our results illustrate a one-pot method for rapid synthesis and assembly of functional ssMPs, which is facilitated by the hydrophobic interaction rather than limited by it. Such functionalized oil drops represent a platform to communicate with cells.
Keywords: biophysics, hydrophobic, self-assembly, membrane protein, cell-free protein synthesis
Abstract
Single-span membrane proteins (ssMPs) represent approximately one-half of all membrane proteins and play important roles in cellular communications. However, like all membrane proteins, ssMPs are prone to misfolding and aggregation because of the hydrophobicity of transmembrane helices, making them difficult to study using common aqueous solution-based approaches. Detergents and membrane mimetics can solubilize membrane proteins but do not always result in proper folding and functionality. Here, we use cell-free protein synthesis in the presence of oil drops to create a one-pot system for the synthesis, assembly, and display of functional ssMPs. Our studies suggest that oil drops prevent aggregation of some in vitro-synthesized ssMPs by allowing these ssMPs to localize on oil surfaces. We speculate that oil drops may provide a hydrophobic interior for cotranslational insertion of the transmembrane helices and a fluidic surface for proper assembly and display of the ectodomains. These functionalized oil drop surfaces could mimic cell surfaces and allow ssMPs to interact with cell surface receptors under an environment closest to cell–cell communication. Using this approach, we showed that apoptosis-inducing human transmembrane proteins, FasL and TRAIL, synthesized and displayed on oil drops induce apoptosis of cultured tumor cells. In addition, we take advantage of hydrophobic interactions of transmembrane helices to manipulate the assembly of ssMPs and create artificial clusters on oil drop surfaces. Thus, by coupling protein synthesis with self-assembly at the water–oil interface, we create a platform that can use recombinant ssMPs to communicate with cells.
Membrane proteins populate the surfaces of cells and allow cells to sense and interact with their external environments. Membrane proteins constitute 25–30% of all proteins identified in sequenced genomes, and understanding their functions is important, because they represent the majority of current drug targets (1). Conventionally, function is inferred from structural studies of membrane proteins; these studies involve expression and purification of natural or recombinant membrane proteins followed by crystallization. However, membrane proteins are notoriously hard to work with because of their hydrophobic domains that can cause misfolding and aggregation (2). Consequently, detergents and membrane mimetics (3) are used to solubilize membrane proteins. To avoid cytotoxicity caused by in vivo expression and enable high-throughput production, cell-free systems are used to express membrane proteins in vitro (4–7). Despite recent progresses, understanding the functions of membrane proteins is still a lengthy and difficult process.
Here, we develop a one-pot approach for synthesis, assembly, and display of single-span membrane proteins (ssMPs) for direct functional studies. ssMPs contain a single transmembrane (TM) helix that anchors the hydrophilic ectodomains on the membrane surface; ssMPs represent approximately one-half of all membrane proteins and are involved in important cellular functions, such as cell–cell communication and cell adhesion (1). We synthesize ssMPs in a reconstituted cell-free protein synthesis system in the presence of oil drops (8, 9). We believe that the interior of oil drops could provide the hydrophobic environment for the single TM helix, whereas the fluidic surface of oil drops mimics the cell membrane to display the ectodomain and allows lateral movement. Using a reconstituted rather than a cell-extract–based cell-free system, we synthesize ssMPs in a chemically defined mixture (10, 11). Using oil drops to anchor ssMPs, we further remove most of the soluble components of the cell-free system. We expect that these oil drops can be directly used without additional purification for functional studies, such as cell-based assays. We apply our one-pot approach to produce natural apoptosis-inducing receptor ligands and show that these single-span membrane receptor ligands efficiently killed cultured cancer cells. Oil drops have been used to interact with cells (12, 13) and study protein interactions (14–17); here, we produce oil drops coated with membrane proteins. By in vitro-synthesizing proteins from DNA templates, we can also rapidly engineer synthetic ssMPs to modulate their assembly and function (18) on oil drops and therefore, show a unique way to manipulate aggregation-prone hydrophobic proteins.
Results and Discussion
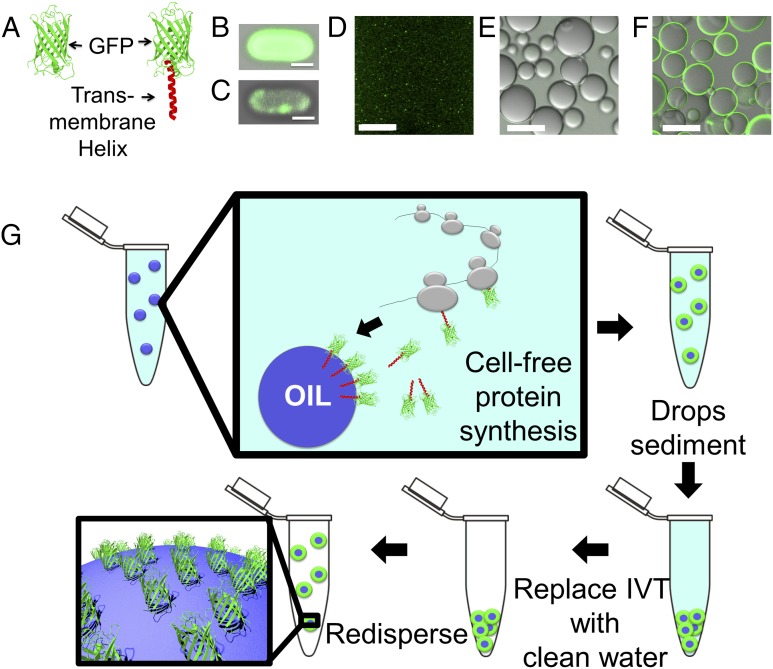
To conveniently visualize and monitor the synthesized protein, we genetically fused GFP with the TM helix (GFP-TM) from human programmed cell death protein 1 (PD1) (19–21). We used this fusion protein as an artificial ssMP (GFP-TM) (Fig. 1A). When synthesized in Escherichia coli, GFP-TM exhibits weak fluorescence and appears to aggregate and clump against the cell membrane (Fig. 1C), whereas GFP exhibits strong fluorescence and appears to be soluble and disperse uniformly inside the cell (Fig. 1B). When synthesized in vitro in a cell-free system, GFP-TM again forms aggregates (Fig. 1D), whereas GFP disperses uniformly in solution (2) (SI Materials and Methods). The formation of GFP-TM aggregates is similar to the behavior of natural TM proteins, which are known to be aggregation-prone during protein synthesis in E. coli and in vitro.
Fig. 1.
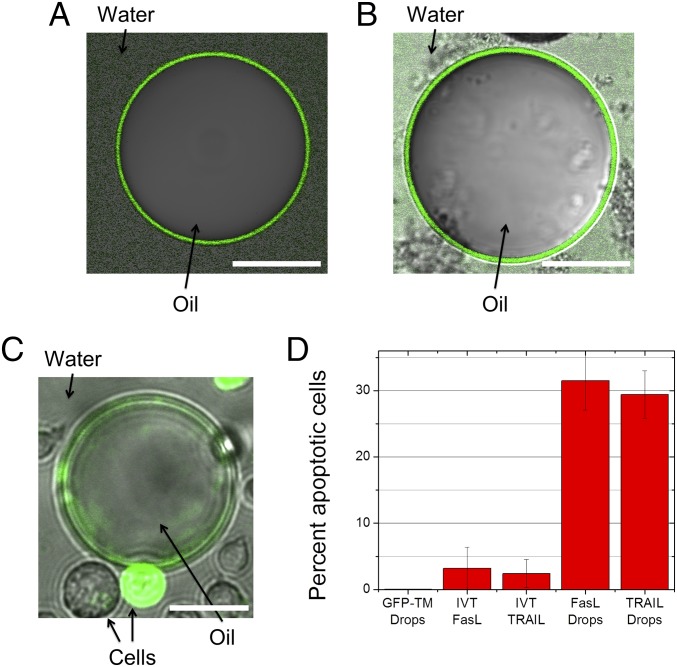
(A) Cartoon depiction of (Left) GFP and (Right) GFP-TM. (B) Confocal fluorescent microscope image of an E. coli cell producing GFP. (Scale bar: 1 μm.) (C) Confocal fluorescent microscope image of an E. coli cell producing GFP-TM. (Scale bar: 1 μm.) (D) Experimental image of GFP-TM synthesized in aqueous solution in vitro. (Scale bar: 80 μm.) (E) Experimental image of GFP synthesized in the presence of oil drops after washing. GFP does not readily anchor to the oil drop surface and is washed away. (Scale bar: 250 μm.) (F) Experimental image of GFP-TM–coated oil drops after washing. GFP-TM anchors to the oil drop surface and is not washed away. (Scale bar: 50 μm.) (G) Cartoon depiction of TM synthesis, assembly on oil drop surface, and subsequent purification.
Interestingly, when synthesized in vitro in the presence of oil drops (HFE-7500), GFP-TM is localized at the water–oil interface and cannot be easily washed away (Figs. 1F and 2 and Fig. S1). In fact, a very simple procedure was followed: oil was added to the cell-free synthesis solution at room temperature without surfactant, and the combination was then vortex-mixed. The resultant cell-free solution and oil drops were then incubated at 37 °C for 90 min, producing oil drops coated with GFP-TM. Under the same conditions, GFP disperses evenly in solution, does not significantly localize on the drop surface, and is easily washed away (Figs. 1E and 2). GFP-TM is very stably localized on the drop surface; it takes many days for the signal from GFP-TM to completely disappear (Fig. S1). These results suggest that the localization of GFP-TM on the drop surface is dependent on the TM helix and that oil drops provide a hydrophobic environment for the TM helix, capturing newly synthesized GFP-TM before the TM helix causes aggregation. We speculate that the TM helix may enter the interior of the oil drop, leaving the hydrophilic GFP domain in the aqueous solution (Fig. 1G) (8). However, we cannot rule out the possibility that the TM helix is localized at the water–oil interface without entering the interior of the oil drop (see Fig. S2). In either case, the interaction between GFP-TM and the oil drop seems to be sufficient to prevent some GFP-TM from aggregation in solution. Because the fluorinated oil used here is denser than water, sedimentation is sufficient to separate protein-coated drops from their surrounding solution that contains soluble components of in vitro protein synthesis (Fig. 1G). By repeated washing of oil drops, we can further purify the surface-bound proteins. To explore the generality of our method, we used two other oils, Silicon 1000 and Miglyol 812, as well as four additional TM helices. In all cases, GFP-TM proteins are strongly localized on oil surfaces, even after extensive washing, suggesting that our method can potentially be applied to any helix and many oil types (SI Materials and Methods and Fig. S3).
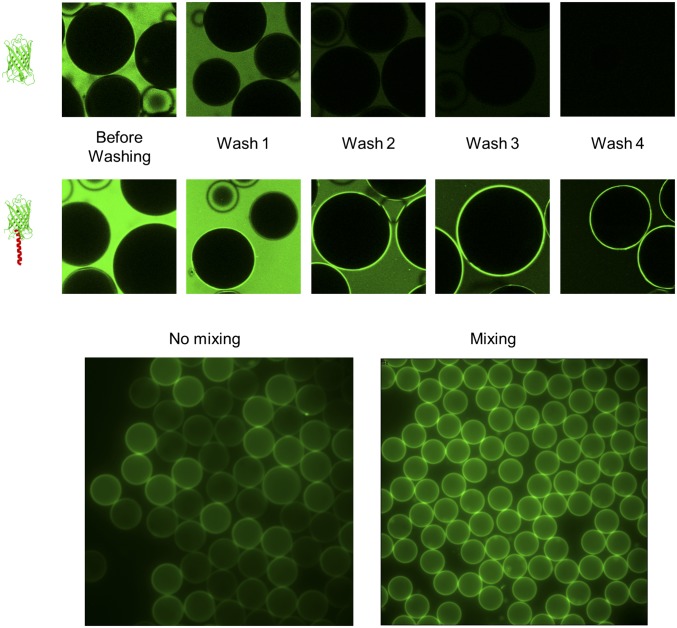
Fig. 2.
Images of (Top and Middle) GFP and (Bottom) GFP-TM synthesized in the presence of oil drops after multiple stages of washing. GFP-TM–coated drops made (Bottom Left) without stirring and (Bottom Right) with stirring. (Bottom Right) The drops made with stirring are brighter and more uniformly coated.
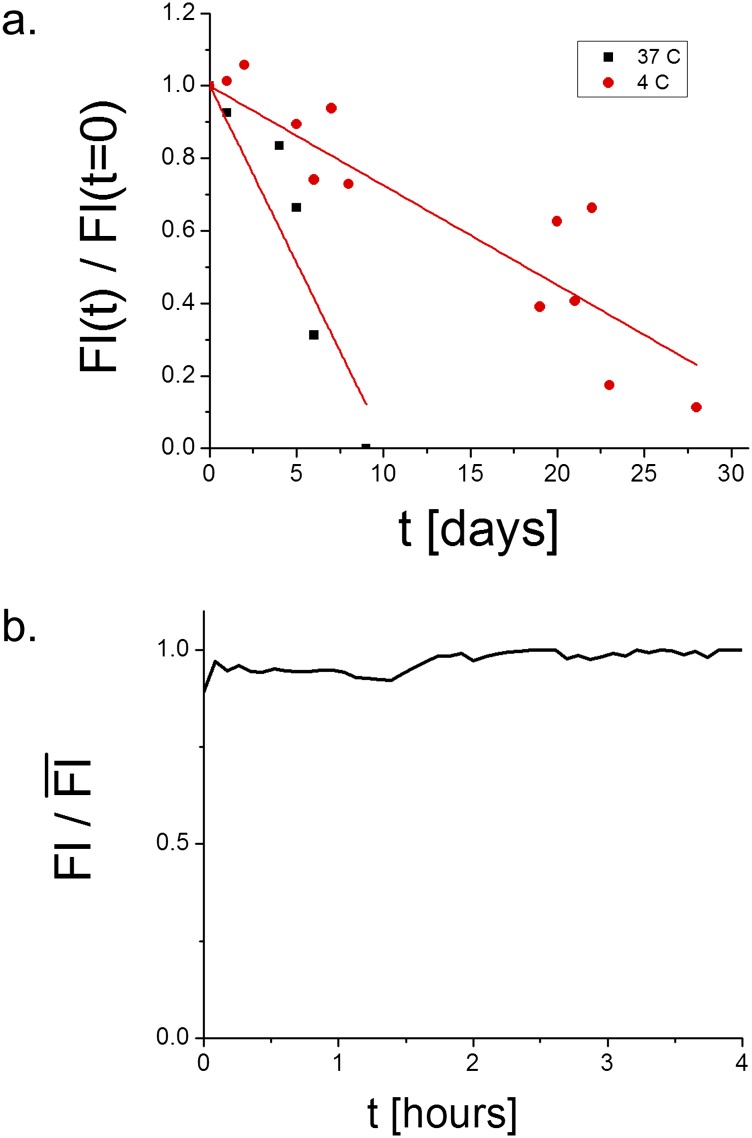
Fig. S1.
(A) Fl plotted as a function of time, t, for GFP-TM–coated drops stored at 4 °C (black squares) and 37 °C (red circles). (B) Fl normalized by the maximum of Fl, , of a GFP-TM–coated drop in a microfluidic device with water flowing past it. Fl is plotted as a function of time.
Fig. S2.
GFP-TM was synthesized absent oil drops. Then, oil drops were added at room temperature. After 4 h, the GFP-TM coated the drops as seen here.
Fig. S3.
Three different types of oil are coated with GFP-TM after in vitro synthesis.
Like all membrane proteins, ssMPs tend to self-aggregate rapidly after in vitro synthesis. Even in the presence of oil drops, only a small fraction of newly synthesized ssMPs are localized on the drop surface (Figs. S4, S5, and S6). Simply incubating the cell-free solution with oil drops without stirring captures only 1–3% of synthesized ssMPs on oil drops, leaving most ssMPs aggregated in bulk, and results in an uneven coverage of drops (Fig. 2). We found that gently stirring oil drops during in vitro synthesis reactions made the drop coverage density much more uniform (Fig. 2). Additional optimization of the in vitro synthesis conditions may further increase the amounts of surface-bound ssMPs.
Fig. S4.
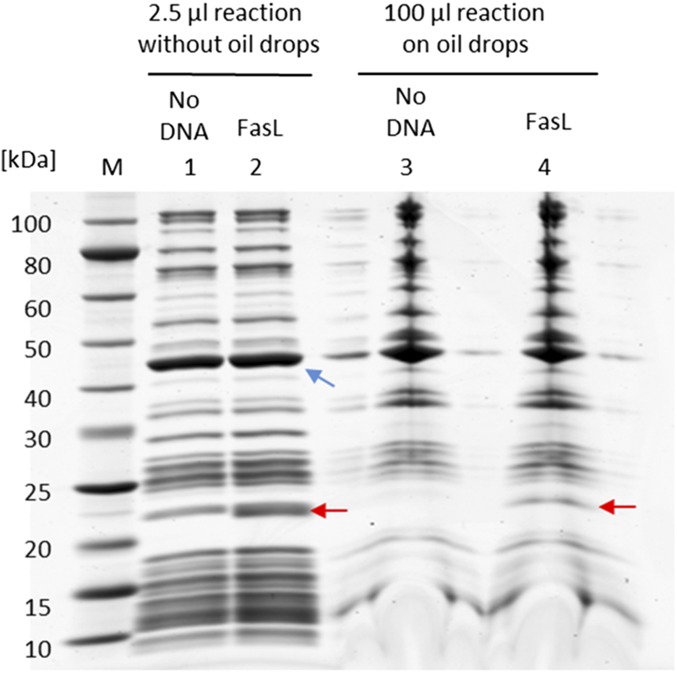
SDS/PAGE analysis of in vitro synthesis of FasL in the absence or presence of oil drops. Lanes 1 and 2 show in vitro reactions (2.5 μL per lane) without oil drops. Lanes 3 and 4 show proteins that remain on oil drops after oil drops are incubated with 100-μL reactions and washed. The synthesized FasL is indicated by red arrows. The EF-Tu protein of the cell-free system, indicated by the blue arrow, is used as the internal standard (0.32 mg/mL) for the quantification of the synthesized protein.
Fig. S5.
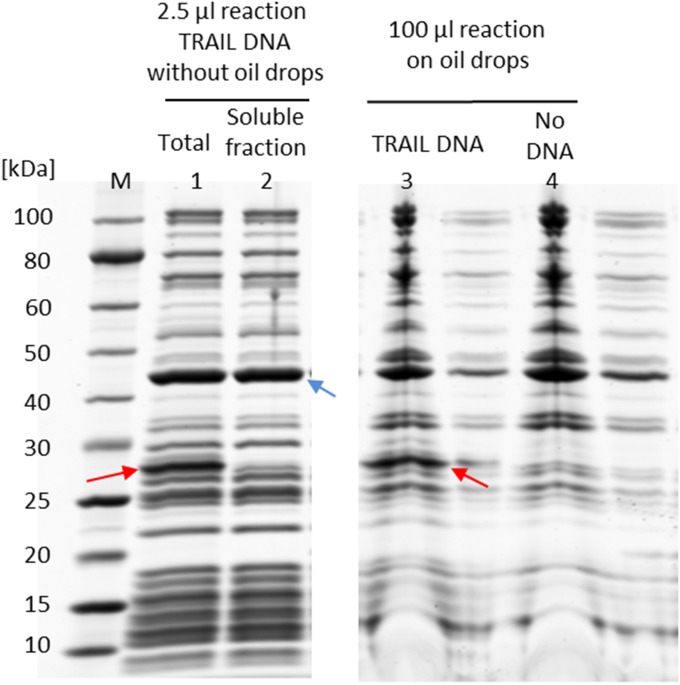
SDS/PAGE analysis of in vitro synthesis of TRAIL in the absence or presence of oil drops. Lane 1 shows the in vitro reaction (2.5 μL) expressing TRAIL without oil drops. Lane 2 shows the soluble fraction from the in vitro reaction (2.5 μL) expressing TRAIL without oil drops after the reaction is centrifuged at 3,000 × g for 5 min at room temperature. As separate experiments, lane 3 shows proteins that remain on oil drops after oil drops are incubated with 100-μL reactions with TRAIL DNA and washed. Lane 4 shows proteins that remain on oil drops after oil drops are incubated with 100-μL reactions without TRAIL DNA and washed. The synthesized TRAIL is indicated by red arrows. The EF-Tu protein of the cell-free system, indicated by the blue arrow, is used as the internal standard (0.32 mg/mL) for the quantification of the synthesized protein.
Fig. S6.
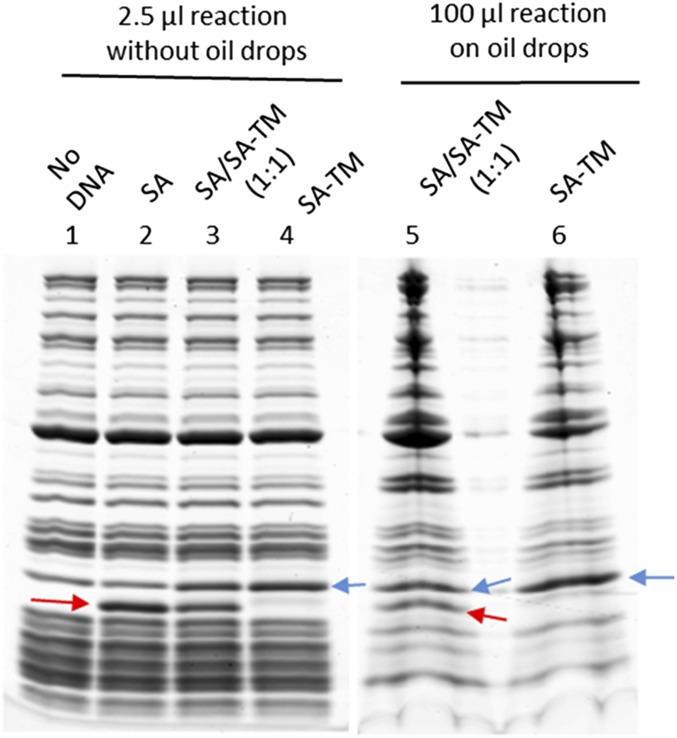
SDS/PAGE analysis of in vitro synthesis of SA, SA/SA-TM hybrid, and SA-TM in the absence or presence of oil drops. Lanes 1–4 show in vitro reactions (2.5 μL) with no DNA, SA DNA, SA:SA-TM DNA mix (1:1 ratio), and SA-TM DNA, respectively. Lanes 5 and 6 show proteins that remain on oil drops after oil drops are incubated with 100-μL reactions with SA:SA-TM DNA mix (1:1 ratio) or only SA-TM DNA and washed. The synthesized SA is indicated by red arrows. The synthesized SA-TM is indicated by blue arrows.
To apply the same approach to natural ssMPs, we synthesize human apoptosis-inducing death ligands, FasL and TRAIL (22–25). We omit the cytoplasmic domain of FasL and TRAIL and express the portions consisting of the extracellular ectodomain and the TM helix. Based on the SDS/PAGE analyses of in vitro synthesis reactions, we estimate that the yields of FasL and TRAIL are 100 and 200 μg/mL, respectively (Figs. S4 and S5). In the absence of oil drops, the synthesized TRAIL is completely aggregated, because the TRAIL band disappears from the soluble fraction after centrifugation (Fig. S5). In the presence of oil drops, TRAIL is found among proteins that bind to oil drops and represents ∼3% of the total synthesized TRAIL (Fig. S5). The presence of FasL and TRAIL on the surface of oil drops is further visualized by fluorescent anti-FasL antibody and fluorescent anti-TRAIL antibodies, respectively (Fig. 3 A and B). These data suggest that oil drops can be used to localize ssMPs at the water–oil interface, thus solubilizing them.
Fig. 3.
(A) Fas ligands and (B) TRAIL on the oil drop surface visualized by using FITC-conjugated anti-FasL or anti-TRAIL antibody, respectively. Representative individual drops are shown, because the fluorescent signal is weaker on these drops. (Scale bar: 100 μm.) (C) Confocal fluorescent microscope image of Fas ligand-coated oil drops inducing apoptosis in a Jurkat cell (green). (Scale bar: 20 μm.) (D) Percentage of apoptotic Jurkat cells during a 90-min incubation period while exposed to oil drops coated with FasL, oil drops coated with TRAIL, TRAIL synthesized with IVT-absent oil drops, FasL synthesized with IVT-absent oil drops, or oil drops coated with GFP-TM. Error bars represent SDs.
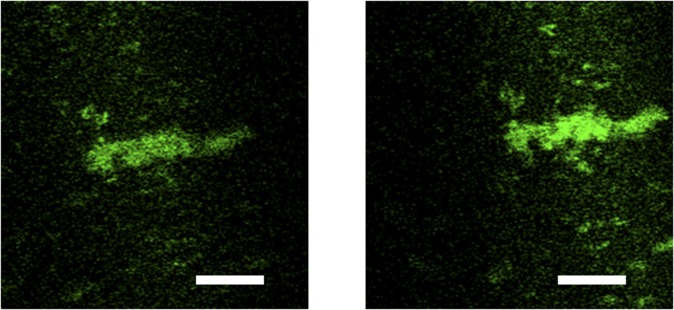
To show that FasL and TRAIL on the drop’s surface are functional, we incubate FasL- and TRAIL-coated drops with Jurkat cells at 37 °C for 90 min (Fig. 3C). For controls, we incubate cells with GFP-TM–coated drops and in vitro-synthesized TRAIL without drops. In all experiments, the total number of cells is kept constant. To ensure that all in vitro-synthesized TRAIL is available, we added the entire mixture, including the cell-free synthesis solution, in the cell-based assays. The fraction of apoptotic Jurkat cells in the presence of drops mixed with the cell-free system absent DNA was within error bars of zero. Similarly, the fraction of apoptotic Jurkat cells in the presence of GFP-TM drops was within error bars of zero. These control experiments indicate that the cell-free solution and oil drops alone or oil drops coated with GFP-TM do not induce significant apoptosis of Jurkat cells. Remarkably, FasL- and TRAIL-coated drops induce apoptosis in ∼30% of cells (Fig. 3D), indicating that surface-anchored FasL and TRAIL are functional. It seems that oil drops are critical for the functionality, because TRAIL synthesized in the absence of oil drops, which were immediately aggregated, induces apoptosis only in ∼2% cells (Fig. 3D). It seems that cells must contact TRAIL-coated drops to undergo apoptosis and that TRAIL that comes off the drops (probably as aggregates) does not induce a high level of apoptosis (Fig. S7). Taken together, drop-anchored FasL and TRAIL exhibit high apoptosis-inducing activity in Jurkat cells and seem to be more effective than a similar amount of the soluble ectodomain. We speculate that the oil drops not only solubilize in vitro-synthesized FasL and TRAIL but also, provide a fluidic surface on which these cell surface ligands can orient properly and move laterally (Fig. S8). Such functionalized oil surface could create a high local concentration of the ligands and lead to more efficient interactions with the receptors on the cell surface (Fig. 3D) (26), similar to the high apoptosis rate induced by TRAIL-coated leukocytes in the work by Mitchell et al. (27).
Fig. S7.
Percentage of apoptotic Jurkat cells during a 90-min incubation period while exposed to oil drops coated with TRAIL.
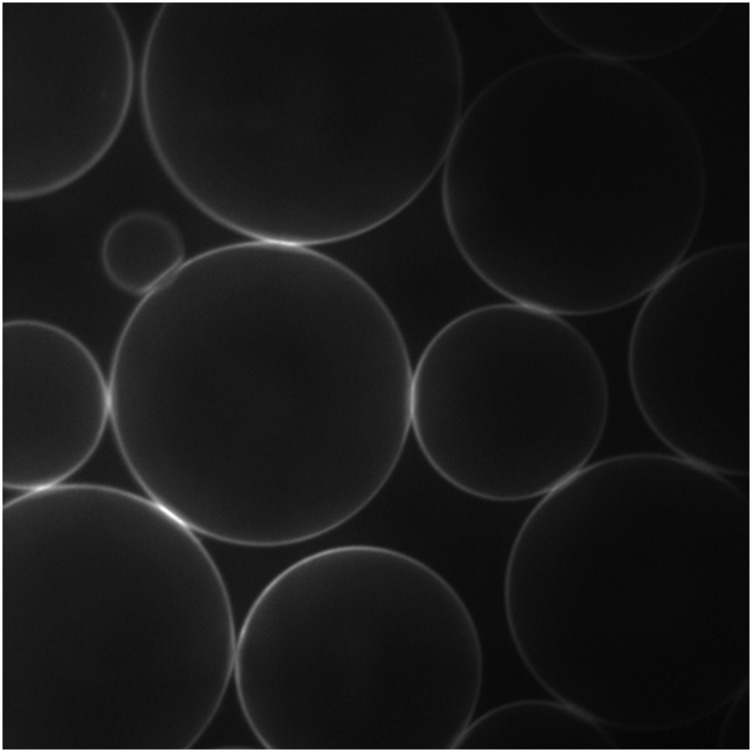
Fig. S8.
Images of GFP-TM on the oil–water interface showing that these proteins remain mobile after adsorption. The image in Right was taken ∼1 s after the image in Left. (Scale bar: 10 μm.)
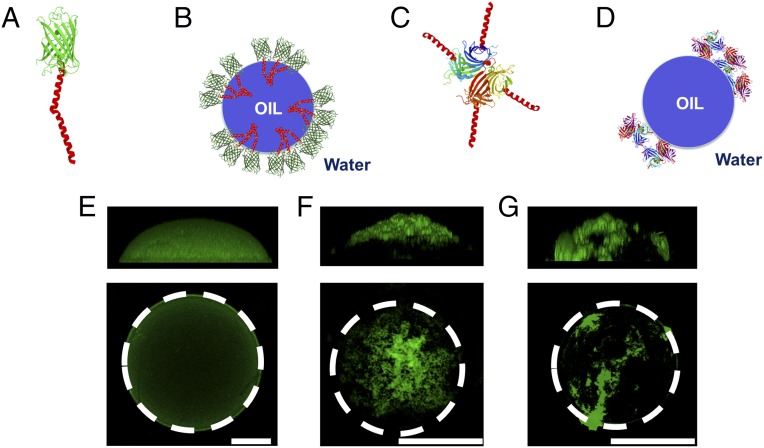
In the next set of experiments, we manipulate the assembly of synthetic ssMPs to form clusters on the oil drop surface. We first exploit the hydrophobic interaction of the TM helix by fusing a second TM to GFP-TM, creating a GFP with an artificially elongated TM helix (GFP-2xTM) (Fig. 4A). GFP-2xTM appears to cluster on the oil drop surface, forming a sparse network with regions densely packed with GFP-2xTM as well as regions that appear void of the protein (Fig. 4 B, scheme and F). In comparison, GFP-TM was more uniformly distributed across the drop surface (Fig. 4E).
Fig. 4.
(A) Cartoon depiction of GFP-2xTM. (B) Cartoon depiction of possible distribution of GFP-2xTM on an oil drop. (C) Cartoon depiction of SA-TM. (D) Cartoon depiction of possible distribution of SA-TM on an oil drop. (E–G) Experimental images of (Upper) side views and (Lower) z projections of (E) a GFP-TM–coated oil drop, (F) a GFP-2xTM–coated oil drop, and (G) an SA-TM–coated oil drop. (Scale bar: 100 μm.)
We next create another synthetic ssMP by fusing a TM to streptavidin (SA), which naturally forms a tetramer (SA-TM) (Fig. 4C). We synthesized SA-TM in vitro in the presence of oil drops; in contrast to GFP-TM but similar to GFP-2xTM, SA-TM forms dense clusters on the drop surface (Fig. 4 D, scheme and G). Based on the synthesis yield, we calculate SA-TM coats drops with a coverage density measured to be 4.0 ± 0.5 × 1012 active SA proteins per 1 cm2 (SI Materials and Methods and Fig. S9). This coverage density is ∼20 times higher than that maximum packing oil drop surface allows (28). These data suggest that SA-TM forms 3D structures on the drop surface, which can be visualized by confocal fluorescence microscopy (Fig. 4G). Because each SA-TM tetramer has four TM helices, we suspect that some TM helices participate in high-order hydrophobic interactions with each other rather than oil drops, thus forming dense clusters before and during localization onto the drop surface. Here, cotranslation of SA-TM with oil drops allows assembly of 3D structures at the water–oil interface.
Fig. S9.
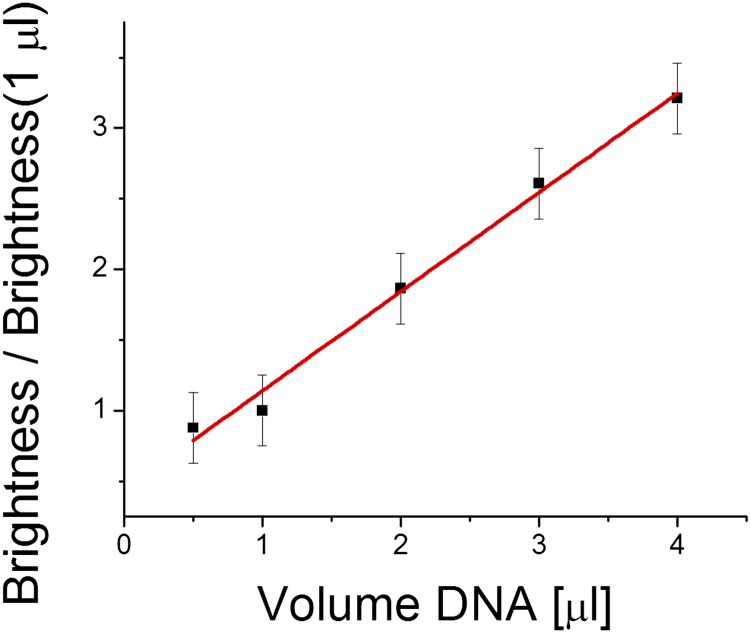
Fluorescent signal of biotinylated FITC attached to the SA-TM on oil–water interface as a function of the volume of DNA (at a fixed concentration) used during in vitro synthesis. The amount of SA-TM and thus, biotinylated FITC increases linearly.
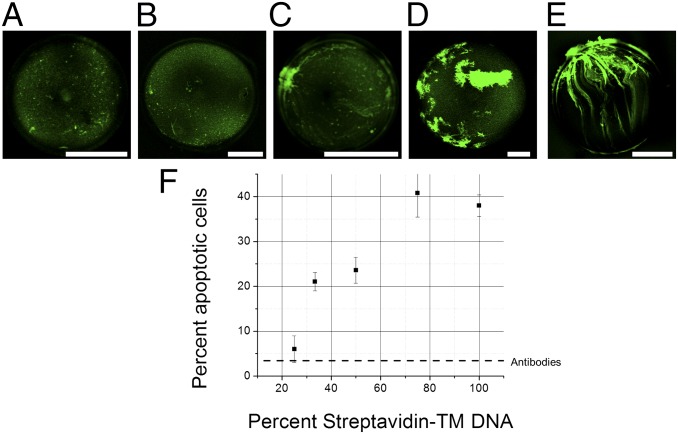
To examine whether the number of TM helices per SA tetramer is correlated to the clustering of SA-TM, we coexpress SA and SA-TM in the cell-free system in the presence of oil drops. By using increasing amounts of SA-TM DNA relative to SA DNA, we intend to synthesize hybrid tetramers with one to four TM helices per tetramer (Fig. 5 A–E). Based on the SDS/PAGE analyses, roughly equal amounts of the SA proteins/domains are synthesized in all cases and anchored on oil drops (Fig. S6). When only 0.25 of the DNA is for SA-TM, the tetramers, on average, only have a single TM helix, and like GFP-TM, this single-helix tetramer disperses evenly over the oil–water interface (Fig. 5A). As we increase the fraction of SA-TM DNA, the extent of SA clustering, indicated by the typical cluster size, increases as well (Fig. 5 A–E). Preclustering protein ligands could enhance ligand–receptor communications between cells (29). To show such application, oil drops with different extents of SA clustering are used to conveniently anchor and precluster biotinylated antibodies that recognize cell surface receptors. In this case, we use biotinylated anti-human CD95 mAb (biotin anti-CD95) capable of inducing apoptosis by cross-linking the receptor CD95. Biotin anti-CD95 alone in solution induces apoptosis in 8% of Jurkat cells. Remarkably, as we anchor biotin anti-CD95 on SA-coated oil drops and increase the extent of SA clustering, the fractions of apoptotic cells also increase up to 35%, where the maximal clustering of SA tetramer is achieved (Fig. 5). The data suggest that preclustering of the receptor ligands (in this case, biotin anti-CD95) on oil drops is directly correlated to the effectiveness of inducing apoptosis through the CD95 receptor. This result is consistent with the known effect of the CD95 receptor clustering on apoptosis (30–33).
Fig. 5.
Experimental z projections of oil drops coated with SA tetramers produced from (A) 25%, (B) 33%, (C) 50%, (D) 75%, and (E) 100% SA-TM DNA, with the remainder being WT SA DNA. (Scale bar: 100 μm.) (F) Percentage of apoptotic Jurkat cells during a 90-min incubation period while exposed to oil drops coated with SA and biotinylated anti-CD95 as a function of the percentage of SA-TM DNA. The dashed line corresponds to the percentage of apoptotic cells induced by freely suspended antibodies.
In summary, we describe a one-pot method to synthesize and display natural or synthetic ssMPs on oil drop surfaces. Our approach applies to a truncated form of the full-length natural ssMP, consisting of the ectodomain and the TM helix and without the intracellular domains. Many natural ssMPs have small intracelluar domains, some of which comprise only a few amino acid residues. The functionality of ssMPs is largely exhibited by the interactions of their membrane-anchored ectodomains with cell surface receptors of other cells. Therefore, our method overcomes one of the bottlenecks in functional studies of ssMPs, which are, in general, difficult to deal with experimentally because of the presence of TM helices. For instance, we can rapidly produce a large number of ssMPs by cell-free synthesis and use ssMP-coated oil drops to mimic cell surface conditions for direct cell-based functional assays. Additionally, this method permits us to exploit hydrophobicity to modulate protein interactions at the water–oil interface, thereby showing that hydrophobic interactions may be used to facilitate, rather than limit, the studies of TM proteins.
Detergents and membrane mimetics have been used to solubilize membrane proteins synthesized in cell-free systems. However, subsequent processes of purifying functional membrane proteins can be difficult and lengthy. In comparison, our approach immobilizes ssMPs on oil drops; therefore, by simply washing oil drops, functional ssMPs may be separated from most of the aggregated and nonfunctional ssMPs and components of cell-free systems. We find that oil drops still bind many components of the cell-free system, even after repeated washing (Figs. S4, S5, and S6). This nonspecific binding to oil surface is likely caused by hydrophobic areas on the surfaces of soluble proteins and could be further minimized by using more stringent washing conditions, blocking agents, and different surfactants.
ssMP-coated oil drops represent a platform to communicate with cells that has advantages over current approaches. For instance, soluble protein ligands, such as TRAIL, or antibodies can bind cell surface receptors but cannot do so in the context in which membrane-bound ligands normally function and therefore, may not have the same activity and effectiveness. Conjugation of antibodies and ectodomains on nanoparticles (34) and liposomes through chemical reactions (35) or affinity tags (27) intends to mimic the cell surface, but the orientation and position of chemically conjugated protein ligands can be difficult to control. It is not possible to form extensive clusters on nanoparticles and liposomes that can potentially induce strong biological responses (32, 33). In contrast, ssMPs are more likely to orient properly on oil drops, capable of being spatially clustered, and in a fluidic environment that facilitates the communication with receptors on cell surfaces.
SI Materials and Methods
Stability of GFP-TM on Oil Drops.
For these drops to be useful for future in vivo or in vitro experiments and applications, the amphiphilic proteins coating the drops must remain on the drops’ surfaces for long periods of time. Thus, we performed a series of practical tests to show that amphiphilic proteins remain attached to drop surfaces over time and while processing these drops. Drops coated with GFP-TM were stored at 4 °C and 37 °C. The fluorescent intensity (Fl) at the edge of these drops was measured with a confocal microscope and averaged over many drops. This measurement was repeated every ∼24 h, and the surrounding water was then replaced. The process was repeated for 28 d to accurately quantify the presence of GFP on drop surfaces over time (Fig. S1A); the decay of the fluorescent signal is proportional to the rate at which individual GFP-TM proteins escape from the surface of the oil drop. At 4 °C, GFP-TM is very stable anchored to the drop surface; it takes ∼28 d for the signal from GFP-TM to completely disappear. At 37 °C, GFP-TM detaches from the drop surface at a higher rate; however, it still takes ∼8 d for the signal from GFP-TM to completely disappear. Thus, for in vivo experiments that last ∼4 d or fewer, membrane proteins are sufficiently anchored to the drop surface.
Next, we tested if the amphiphilic proteins are easily sheared off by flowing water. Drops coated with GFP-TM were inserted in a microfluidic chamber that is 1,700 μm wide, 50 μm tall, and ∼2 cm long. Some drops get stuck behind notches that partially block the chamber exit. Water was then flowed through the chamber at a rate of ∼50 μL/s, replacing the water surrounding the GFP-TM drops ∼4,000 times over the course of ∼4 h. Fl was measured with a confocal microscope (Fig. S1B). Fl did not significantly decrease during these experiments. These experiments show that, on a practical level, these amphiphilic TM proteins are stably anchored to the oil drop surface.
TRAIL-Coated Oil Drops Kill Jurkat Cells by Contact.
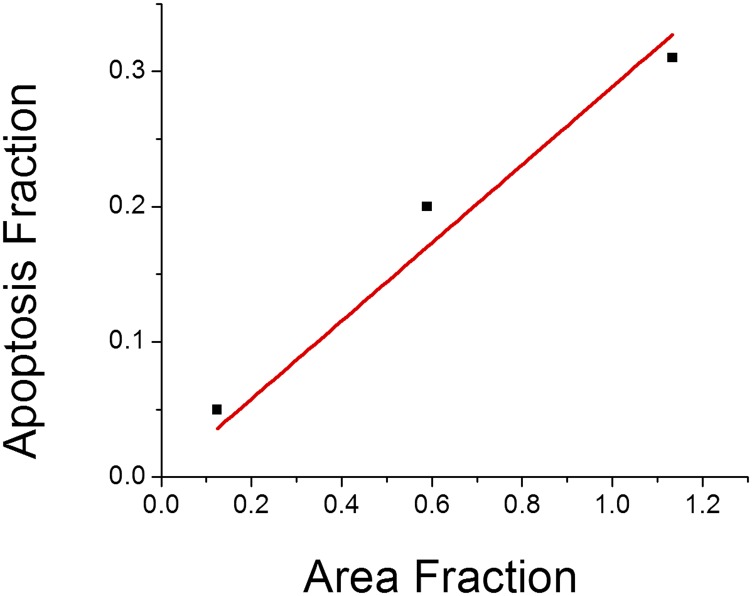
To test if cell-to-drop contact is necessary for apoptosis to occur, we varied the size of the container in which Jurkat cells and TRAIL-coated drops were incubated. The oil drops sediment quickly, because they are denser than water and have large diameters, and then, they cover some fraction of the well plate bottom. Jurkat cells are smaller and less dense, and therefore, they sediment at a slower rate. Thus, Jurkat cells are less likely to contact drops in larger wells than in smaller wells. In fact, the fraction of apoptotic cells increases linearly with the fraction of area covered by oil drops (Fig. S7). The best linear fit has an intercept of 2.9 ± 2.5 and R2 = 0.96. Thus, these data support the picture that cells must contact protein-coated drops to undergo apoptosis, and they do not support the idea that the proteins may leave the surface of the drop and then, induce apoptosis.
Washing Drops in the Presence of GFP and GFP-TM.
To show the ability of oil drops to capture amphiphilic proteins at a higher rate than more hydrophilic proteins, we performed a series of washing experiments; 100 μL cell-free synthesis solution was used to make GFP in one experiment and GFP-TM in another experiment. Before washing, both the GFP and the GFP-TM synthesized produce large fluorescent signals. After the oil drops sedimented (after ∼10 s), 90 μL solution was removed and replaced with 500 μL water, constituting the first wash. This procedure was repeated, removing all but ∼10 μL solution and replacing it with 500 μL water multiple times (Fig. 2). After three to four washes, the fluorescent signal from GFP was decreased to nearly nothing, whereas the GFP-TM still produced a large signal on the drop surfaces, indicating that GFP-TM was captured more efficiently than GFP.
Mobility of Proteins on Oil Drop Surfaces.
The motion of GFP-TM proteins on the oil–water interface is evident in zoomed-in confocal images (Fig. S8). A very large oil drop, ∼1 mm in diameter, was prepared, and GFP-TM was synthesized around it following the same procedure described above. Then, fluorescent images were record on the drop surface, showing that these proteins remain mobile after adsorption.
In Vitro Synthesis of TRAIL and SDS/PAGE Analyses.
SDS/PAGE analyses of in vitro synthesis of FasL, TRAIL, and mixtures of SA and SA-TM in the absence or presence of oil drops (Figs. S4–S6).
Different Oil and TM Tails.
To test the generality of this approach, we repeated our GFP-TM experiments with five total TM tails (human PD1, p23, p24, CD40, and CD45) as well as three total oils (HFE-7500, Silicon 1000, and Miglyol 812). Although different viscosities and surface tensions resulted in different drop size dispersions, in all 15 cases, the GFP-TM coated the oil drops (Fig. S3).
Localization of Aggregated GFP-TM on Bare Oil Drops.
We synthesized GFP-TM–absent oil drops, thus allowing it to aggregate. Then, we added oil drops at room temperature. After 4 h, the drops were coated with GFP-TM (Fig. S2).
Materials and Methods
Reagents.
Fluorescein (FITC)-conjugated mouse anti-human Fas ligand mAb (clone SB93a), soluble TRAIL recombinant human protein (PHC1634), and apoptosis inducing anti-human CD95 mAb (clone NOK-1) were purchased from Life Technologies. Phycoeerythrin-conjugated mouse anti-human TRAIL mAb (clone 2E5) was purchased from Abcam. In vitro protein synthesis reagents (IVT) are from New England Biolabs (PURExpress Kit). Perfluorinated oil (Novec-7500) was purchased from 3M.
DNA Template Preparation.
The genes for human TM helices of human PD1 (PD1-TM; residues 171–191), TM emp23 domain-containing protein 10 (p23-TM; residues 186–206), TM emp24 domain-containing protein 2 (p24-TM; residues 169–189), TNF receptor superfamily member 5 (CD40-TM; residues 194–215), and receptor type tyrosine-protein phosphatase C (CD45-TM; residues 576–597) were synthesized by Integrated DNA Technologies. The amino acid sequences of these TM helices are shown below:
PD1 TM VVGVVGGLLGSLVLLVWVLAV,
p23 TM VLYFSIFSMFCLIGLATWQVF,
p24 TM VVLWSFFEALVLVAMTLGQIY,
CD40 TM ALVVIPIIFGILFAILLVLVFI, and
CD45 TM ALIAFLAFLIIVTSIALLVVLY.
The genes for human Fas ligand (23 kDa; residues 80–281, omitting the cytoplasmic domain), TRAIL (31 kDa; residues 18–281, omitting the cytoplasmic domain), and SA were synthesized by Integrated DNA Technologies and cloned into pUCA105T7 vector, a pUC19 derivative with a T7 promoter (36).
The genes for GFP and SA were amplified by PCR and cloned into pUCA105T7 to create the GFP and SA DNA templates. The gene for TM or 2xTM was amplified by PCR and cloned into the C terminus of GFP or SA to create the DNA templates: GFP-TM, GFP-2xTM, or SA-TM.
In Vivo Expression of GFP and GFP-TM in E. coli Cells.
DNA templates for GFP and GFP-TM were transformed into an E. coli expression strain: T7 Express lysY/Iq (New England Biolabs). The cells were grown in LB at 37 °C to the log phase. Isopropyl-beta-D-thiogalactopyranoside (0.1 mM) was then added to induce protein expression. After incubation at 37 °C for another 2 h, cells were examined under fluorescent confocal microscope.
In Vitro Protein Synthesis in the Presence of Oil Drops.
To synthesize proteins in vitro, the IVT solution was mixed with DNA templates (10 ng/μL) and incubated at 37 °C for 4 h [for some qualitative microscopy-only experiments (i.e., experiments that do not involve apoptosis), incubation time was 3 h]; the resulting protein solution was stored at 4 °C or on ice.
To measure the amounts of TRAIL and FasL proteins produced by IVT, we ran the in vitro synthesis reactions (2.5 μL) on SDS/PAGE gels. The gels were stained by SimplyBlue Safe Stain (Life Technologies) and scanned by the ODYSSEY Infrared Imaging System (LI-COR Biosciences). We quantified newly synthesized proteins using ODYSSEY quantification software and an internal standard protein with a known quantity on the same gels. We estimated that 100 μL IVT produces ∼10 μg FasL and 20 μg TRAIL (Figs. S4 and S5).
To synthesize proteins in vitro in the presence of oil drops, the IVT solution (100 μL) containing DNA templates (10 ng/μL) was mixed with 20 μL oil in a flat-bottomed vial. The vial was then vortexed for ∼10 s to directly generate oil drops in the IVT solution. In the case of monodisperse drops, they were first made with microfluidic devices (37) and then, mixed with the IVT solution. The IVT and oil drop mixture was then incubated at 37 °C for 3–4 h to allow protein synthesis in the presence of oil drops.
To determine the amounts of TRAIL stably anchored on oil drops, we washed oil drops three times after protein synthesis and extracted proteins remaining on oil drops by adding 5 μL 10% (mass/mass) Nonidet P-40 and 10 μL 3× SDS Sample Buffer (NEB). The solutions were heated at 95 °C for 5 min and then, directly loaded on SDS gels. Based on the analyses of the scanned gels, we estimated that, from 100 μL IVT reaction, 130 ng FasL [1.3% (mass/mass) total synthesized protein] and 600 ng TRAIL [3% (mass/mass) total synthesized protein] were anchored on the oil drops. When these oil drops were mixed with 1 mL cell suspension, the amounts of FasL and TRAIL corresponded to 5.6 and 19 nM, respectively.
Biotinylated FITC (Sigma-Aldrich; at concentrations of ∼50 nM) was used to visualize SA-coated drops. Anti-Fas ligand antibodies conjugated to FITC (Life Sciences) were used (at concentrations of ∼50 nM) to image Fas ligand-coated oil drops. Anti-TRAIL antibodies conjugated to phycoeerythrin (Abcam) were used (at concentrations of ∼50 nM) to image TRAIL-coated oil drops.
Interaction of Protein-Coated Oil Drops with Natural Cells.
Jurkat cells are purchased from ATCC and cultured in DMEM with 2% (vol/vol) FBS and 0.1% PenStrep at 37 °C and 5% (vol/vol) CO2 (culture media from Life Technologies). Jurkat cells, suspended at ∼106 cells per 1 mL, were placed in 48-well plates (Corning) and returned to the incubator for 2–4 h. The cells were then mixed with ∼105 oil drops and/or the cell-free protein synthesis solutions. These materials were simply pipetted into the wells, where they were then free to diffuse or sediment. To induce apoptosis with soluble TRAIL, recombinant human soluble TRAIL (20 kDa; 50 μg; Life Technologies) was resuspended in 250 μL water. We added 2 μL soluble TRAIL solution (0.2 mg/mL) in 1 mL cell suspension, resulting in a final soluble TRAIL concentration of 20 nM. To induce apoptosis with antibody, biotinylated anti-human CD95 mAb (100 μg; Sigma-Aldrich) was resuspended in 1 mL water. We mixed 10 μL anti-CD95 (0.1 mg/mL) with SA-TM oil drops and added to 1 mL cell suspension, which corresponds to the final concentration of 7 nM anti-CD95.
These mixtures were then returned to the incubator at 37 °C and 5% (vol/vol) CO2 for 90 min to induce apoptosis. After 90 min, the well plates were removed from the incubator, and the remaining steps proceeded at room temperature (∼22 °C). To identify apoptotic (and necrotic) cells, an apoptosis assay (Sigma-Aldrich) was used.
Annexin V suspension (5 μL; Sigma-Aldrich) per 500 μL cell suspension was added to the cell suspension to fluorescently label cells undergoing apoptosis (38). A calcium binding buffer was also needed for Annexin V to bind properly; this binding buffer was added at a ratio of ∼50 μL per 500 μL. However, Annexin V can also attach to necrotic cells; propidium iodide will only attach to necrotic cells, and therefore, 10 μL propidium iodide per 500 μL cell suspension was also added. Cells were identified as undergoing apoptosis if they were labeled with Annexin V but not if they were labeled with propidium iodide.
Apoptotic cells were counted by fluorescent image analysis. Well plates were placed on a confocal microscope (Leica SP5). Images were taken in bright-field mode to count the total number of cells. Fluorescent micrographs were also taken to determine the levels of Annexin V and propidium iodide in each cell. Computational particle tracking analyses counted the number of cells as well as their total green and red fluorescent levels. The analysis technique was validated by counting manually as well.
Footnotes
Conflict of interest statement: A patent application by P.J.Y., D.A.W., and S.C. may be affected by the publication of this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504992113/-/DCSupplemental.
References
- 1.Hubert P, et al. Single-spanning transmembrane domains in cell growth and cell-cell interactions: More than meets the eye? Cell Adhes Migr. 2010;4(2):313–324. doi: 10.4161/cam.4.2.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons K, Helenius A, Leonard K, Sarvas M, Gething MJ. Formation of protein micelles from amphiphilic membrane proteins. Proc Natl Acad Sci USA. 1978;75(11):5306–5310. doi: 10.1073/pnas.75.11.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katzen F, et al. Insertion of membrane proteins into discoidal membranes using a cell-free protein expression approach. J Proteome Res. 2008;7(8):3535–3542. doi: 10.1021/pr800265f. [DOI] [PubMed] [Google Scholar]
- 4.Katzen F, Peterson TC, Kudlicki W. Membrane protein expression: No cells required. Trends Biotechnol. 2009;27(8):455–460. doi: 10.1016/j.tibtech.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Shadiac N, Nagarajan Y, Waters S, Hrmova M. Close allies in membrane protein research: Cell-free synthesis and nanotechnology. Mol Membr Biol. 2013;30(3):229–245. doi: 10.3109/09687688.2012.762125. [DOI] [PubMed] [Google Scholar]
- 6.Junge F, et al. Large-scale production of functional membrane proteins. Cell Mol Life Sci. 2008;65(11):1729–1755. doi: 10.1007/s00018-008-8067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cappuccio JA, et al. Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Mol Cell Proteomics. 2008;7(11):2246–2253. doi: 10.1074/mcp.M800191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang X, et al. Interfacial assembly of protein-polymer nano-conjugates into stimulus-responsive biomimetic protocells. Nat Commun. 2013;4:2239. doi: 10.1038/ncomms3239. [DOI] [PubMed] [Google Scholar]
- 9.Feng L, Pontani L-L, Dreyfus R, Chaikin P, Brujic J. Specificity, flexibility and valence of DNA bonds guide emulsion architecture. Soft Matter. 2013;9:9816–9823. [Google Scholar]
- 10.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19(8):751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 11.Chong S. Overview of cell-free protein synthesis: Historic landmarks, commercial systems, and expanding applications. Curr Protoc Mol Biol. 2014;108(8):16.30.1–16.30.11. doi: 10.1002/0471142727.mb1630s108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourouina N, Husson J, Hivroz C, Henry N. Biomimetic droplets for artificial engagement of living cell surface receptors: The specific case of the T-cell. Langmuir. 2012;28(14):6106–6113. doi: 10.1021/la300398a. [DOI] [PubMed] [Google Scholar]
- 13.Ben M’Barek K, et al. Phagocytosis of immunoglobulin-coated emulsion droplets. Biomaterials. 2015;51:270–277. doi: 10.1016/j.biomaterials.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 14.Pontani L-L, Jorjadze I, Viasnoff V, Brujic J. Biomimetic emulsions reveal the effect of mechanical forces on cell-cell adhesion. Proc Natl Acad Sci USA. 2012;109(25):9839–9844. doi: 10.1073/pnas.1201499109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fattaccioli J, Baudry J, Henry N, Brochard-Wyart F, Bibette J. Specific wetting probed with biomimetic emulsion droplets. Soft Matter. 2008;4:2434–2440. [Google Scholar]
- 16.Bottier C, et al. Active transport of oil droplets along oriented microtubules by kinesin molecular motors. Lab Chip. 2009;9(12):1694–1700. doi: 10.1039/b822519b. [DOI] [PubMed] [Google Scholar]
- 17.Pontani L-L, Haase MF, Raczkowska I, Brujic J. Immiscible lipids control the morphology of patchy emulsions. Soft Matter. 2013;9:7150–7157. [Google Scholar]
- 18.Ranji A, Wu JC, Bundy BC, Jewett MC. Transforming Synthetic Biology with Cell-Free Systems. Elsevier; Amsterdam: 2013. pp. 277–301. [Google Scholar]
- 19.Laaksonen P, et al. Genetic engineering of biomimetic nanocomposites: Diblock proteins, graphene, and nanofibrillated cellulose. Angew Chem Int Ed Engl. 2011;50(37):8688–8691. doi: 10.1002/anie.201102973. [DOI] [PubMed] [Google Scholar]
- 20.Hudalla GA, et al. Gradated assembly of multiple proteins into supramolecular nanomaterials. Nat Mater. 2014;13(8):829–836. doi: 10.1038/nmat3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finger LR, et al. The human PD-1 gene: Complete cDNA, genomic organization, and developmentally regulated expression in B cell progenitors. Gene. 1997;197(1-2):177–187. doi: 10.1016/s0378-1119(97)00260-6. [DOI] [PubMed] [Google Scholar]
- 22.Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281(5381):1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Lostao L, Marzo I, Anel A, Naval J. Targeting the Apo2L/TRAIL system for the therapy of autoimmune diseases and cancer. Biochem Pharmacol. 2012;83(11):1475–1483. doi: 10.1016/j.bcp.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 24.van Dijk M, Halpin-McCormick A, Sessler T, Samali A, Szegezdi E. Resistance to TRAIL in non-transformed cells is due to multiple redundant pathways. Cell Death Dis. 2013;4(2013):e702. doi: 10.1038/cddis.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werner AB, de Vries E, Tait SWG, Bontjer I, Borst J. TRAIL receptor and CD95 signal to mitochondria via FADD, caspase-8/10, Bid, and Bax but differentially regulate events downstream from truncated Bid. J Biol Chem. 2002;277(43):40760–40767. doi: 10.1074/jbc.M204351200. [DOI] [PubMed] [Google Scholar]
- 26.Nair PM, et al. Enhancing the antitumor efficacy of a cell-surface death ligand by covalent membrane display. Proc Natl Acad Sci USA. 2015;112(18):5679–5684. doi: 10.1073/pnas.1418962112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell MJ, Wayne E, Rana K, Schaffer CB, King MR. TRAIL-coated leukocytes that kill cancer cells in the circulation. Proc Natl Acad Sci USA. 2014;111(3):930–935. doi: 10.1073/pnas.1316312111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson KE, et al. Surface characterization of mixed self-assembled monolayers designed for streptavidin immobilization. Langmuir. 2001;17(9):2807–2816. [Google Scholar]
- 29.Cochran JR, Aivazian D, Cameron TO, Stern LJ. Receptor clustering and transmembrane signaling in T cells. Trends Biochem Sci. 2001;26(5):304–310. doi: 10.1016/s0968-0004(01)01815-1. [DOI] [PubMed] [Google Scholar]
- 30.Gulbins E, Grassmé H. Ceramide and cell death receptor clustering. Biochim Biophys Acta. 2002;1585(2-3):139–145. doi: 10.1016/s1388-1981(02)00334-7. [DOI] [PubMed] [Google Scholar]
- 31.Grassmé H, Cremesti A, Kolesnick R, Gulbins E. Ceramide-mediated clustering is required for CD95-DISC formation. Oncogene. 2003;22(35):5457–5470. doi: 10.1038/sj.onc.1206540. [DOI] [PubMed] [Google Scholar]
- 32.Caré BR, Soula HA. Impact of receptor clustering on ligand binding. BMC Syst Biol. 2011;5(2011):48. doi: 10.1186/1752-0509-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grochmal A, Ferrero E, Milanesi L, Tomas S. Modulation of in-membrane receptor clustering upon binding of multivalent ligands. J Am Chem Soc. 2013;135(27):10172–10177. doi: 10.1021/ja404428u. [DOI] [PubMed] [Google Scholar]
- 34.Cho K, Wang X, Nie S, Chen ZG, Shin DM. Therapeutic nanoparticles for drug delivery in cancer. Clin Cancer Res. 2008;14(5):1310–1316. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- 35.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12(4):278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 36.Asahara H, Chong S. In vitro genetic reconstruction of bacterial transcription initiation by coupled synthesis and detection of RNA polymerase holoenzyme. Nucleic Acids Res. 2010;38(13):e141. doi: 10.1093/nar/gkq377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 38.van Engeland M, Nieland LJ, Ramaekers FC, Schutte B, Reutelingsperger CP. Annexin V-affinity assay: A review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry. 1998;31(1):1–9. doi: 10.1002/(sici)1097-0320(19980101)31:1<1::aid-cyto1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]