Significance
Efficient delivery of therapeutic molecules inside cells by nontransgenic approaches is key as gene editing/correction, directed differentiation, and in vivo cell modulation/tracking are translated for regenerative medicine applications. Here we describe a peptide-based system engineered to enhance the activity of cell-penetrating peptides to achieve exceptional intracellular transduction. Glycosaminoglycan-binding enhanced transduction (GET) uses peptides that interact with cell membrane heparan sulfates and promote cell-penetrating peptide-mediated endocytosis into cells. The system is not dependent on extensive positive charge and can be tailored to deliver peptides, recombinant proteins, nucleic acids, nanoparticles, and antibodies. Importantly, this approach does not affect cell proliferation and viability and can be used to deliver a plethora of functional cargoes.
Keywords: cell-penetrating peptides, transduction, human embryonic stem cells, differentiation, heparin-binding domain
Abstract
Protein transduction domains (PTDs) are powerful nongenetic tools that allow intracellular delivery of conjugated cargoes to modify cell behavior. Their use in biomedicine has been hampered by inefficient delivery to nuclear and cytoplasmic targets. Here we overcame this deficiency by developing a series of novel fusion proteins that couple a membrane-docking peptide to heparan sulfate glycosaminoglycans (GAGs) with a PTD. We showed that this GET (GAG-binding enhanced transduction) system could deliver enzymes (Cre, neomycin phosphotransferase), transcription factors (NANOG, MYOD), antibodies, native proteins (cytochrome C), magnetic nanoparticles (MNPs), and nucleic acids [plasmid (p)DNA, modified (mod)RNA, and small inhibitory RNA] at efficiencies of up to two orders of magnitude higher than previously reported in cell types considered hard to transduce, such as mouse embryonic stem cells (mESCs), human ESCs (hESCs), and induced pluripotent stem cells (hiPSCs). This technology represents an efficient strategy for controlling cell labeling and directing cell fate or behavior that has broad applicability for basic research, disease modeling, and clinical application.
Nongenetic tools that afford controllable stoichiometry in the absence of genome integration (1) are attractive for use in directing cell fate and unraveling protein function. In this regard, cationic protein transduction domains (PTDs) have been derived from the HIV (HIV-1) TAT protein basic domain (e.g., RKKRRQRRR) or engineered from poly-arginine or poly-lysine. PTDs have been used to deliver bioactive cargoes of proteins, nucleic acids, nanoparticles, and pharmacologic agents to cells in culture and to preclinical models in vivo (2, 3). As an alternative to unstructured cationic PTDs, there has been development of supercharged (sc) protein carriers that have been extracted from natural proteins or have been engineered [i.e., scGFP (+36 GFP) (4)]. These proteins [defined as >0.75 theoretical positive charge/kDa mass (5)] have exceptional delivery into mammalian cells, but because of their extensive positive charge, they can be cytotoxic.
Although the mechanism of PTD and sc-protein uptake is still poorly defined (1, 6–8), it is clear that their extracellular concentration dictates the efficiency of delivery. Recent studies have demonstrated that PTD-mediated protein delivery is mediated by endocytotic pathways such as lipid raft-dependant macropinocytosis (8). After internalization, cargos are contained within macropinocytomes and not free within cytosol, restricting correct localization and activity (9, 10). This means high (>200 nM) (11, 12) in vivo extracellular concentrations of PTD proteins are needed to drive translocation and permit biologically useful quantities to escape endosomes. Various techniques have been developed to stimulate the vesicular release of cargo, including ultrasound, cotreatment with endosome-disruptive peptides, and chemical treatments (1, 13). However, the efficiency of these processes still leaves the majority of protein trapped away from cytosol (14, 15).
To overcome the inefficiencies in delivery of biologically active PTD conjugates, we developed a multidomain protein comprising a glycosaminoglycan (GAG)-binding peptide to stimulate cell interaction and a PTD for high-efficiency membrane transduction. We provided proof of principle of the efficacy of this system by delivering fluorescent proteins, enzymes, transcription factors, antibodies, nucleic acids, and nanoparticles to mouse and human cells, including pluripotent stem cells. Thus, the development of synergistic cell-targeting and cell-penetrating peptides enables functional quantities of therapeutic cargoes to be delivered to pluripotent and other clinically relevant cell types.
Results
Isolation of P21, a HBD that Enhances PTD Function through GAG Interaction.
We first focused on improving the initial PTD/cell lipid–bilayer interaction and cellular uptake of cargo proteins. For this, we used monomeric red fluorescent protein (mRFP1; mR) as a self-reporting cargo that is readily expressed and purified in Escherichia coli (SI Appendix, Fig. S1A). We confirmed that although TAT- and 8R PTD-tagged mR interact with most cell types, levels of cell uptake were highest in somatic cells (NIH3t3, C2C12, mouse embryonic fibroblasts, ∼5–50-fold over control; P < 0.01; SI Appendix, Fig. S1) and poor in mouse embryonic stem cells (mESCs), human ESCs (hESCs), and induced pluripotent stem cells (hiPSCs) (∼one- to threefold over control; SI Appendix, Figs. S1 and S2B). To improve cellular uptake of cargo proteins into these poorly transduced cell types, we screened several short peptides (identified in the literature) that have been reported to interact with ubiquitous molecules on cell membranes, including integrins, CD markers, and GAGs. We fused peptides to the N-terminal mR with screening of variants, yielding the isolation of a short 21-residue peptide, termed P21 (KRKKKGKGLGKKRDPCLRKYK) (Fig. 1). P21 enhances the association of the mR reporter with both mouse and human pluripotent stem cells. Furthermore, P21 also demonstrates significant transduction activity itself, exhibiting punctate intracellular fluorescence indicative of endosomal localization (Fig. 1 B and C). P21 is the heparin-binding domain (HBD) from heparin-binding epidermal growth factor (HB-EGF) that shows a strong affinity to heparin specifically through P21 (16). Heparin binding is essential for its optimal binding to EGFR and for promoting its growth/migratory activity in vascular smooth muscle cells (17, 18). Interestingly HB-EGF has recently been described as a natural “supercharged” protein with a theoretical net charge:molecular weight ratio >0.75 (5). The fragment of HB-EGF (Pfam: Q99075, aa72–aa147 with extra N-terminal residues, 79 aa) used in that study contained the P21 HBD sequence, and when compared with our P21 fragment (aa93–aa113), that study’s truncated HB-EGF fragment yields similar transduction activity (5). We hypothesized that cell uptake activity intrinsic to P21 is a result of its supercharged nature; however, not all positively charged sequences can be considered HBDs. For confirmation, we demonstrated the direct binding of P21 to heparin by showing that P21-tagged mR was efficiently and reversibly sequestered on heparin-Sepharose (96.2 ± 5.3%; P < 0.01) (SI Appendix, Fig. S3).
Fig. 1.
P21 improves PTD-mediated cellular uptake. (A) Schematic of the proteins created after screening domains that improve efficiency of protein delivery to cells. mR and mR-8R are described in Fig. S1. P21-mR is mRFP with an N-terminal fusion of the P21 domain of HB-EGF. P21-mR-8R is mRFP with N-terminal fusion of P21 and C-terminal fusion of 8R. (B) Fusion of P21 to mR-8R significantly improves uptake into NIH3t3 cells. Fluorescence microscopy images of NIH3t3 cells treated with proteins (20 µg/mL) for 12 h in standard media conditions. (Scale bar, 100 µm.) (C) P21-mR-8R is efficiently taken into hESCs and mESCs (HUES7 and CGR-8, respectively) and hiPSCs (IPS2) and mouse cardiomyocyte cell line HL1. Flow cytometry analyses of the mR-8R inefficiently delivered cell lines treated with proteins mR-8R (20 µg/mL) for 12 h. (D) P21-mR-8R initially strongly interacts with cell membranes and progressively is taken up and localized perinuclearly. Fluorescence (Top) and confocal laser scanning microscopy (Bottom) images of NIH3t3 cells treated with P21-mR-8R (20 µg/mL) for 1 h, 1 h with washes and a further 5 h incubation (in serum-free media), or 6 h treatment. Cells were preincubated for 1 h in serum-free media and transduced for the desired time in serum-free media. (Scale bars, top, 50 µm; bottom, 10 µm.) (E) Enhancement of cellular uptake mediated by P21 and 8R are affected by Trypsin proteolysis. Flow cytometry analyses NIH3t3 cells treated with proteins (20 µg/mL) for 1 h and a further 5 h incubation (in serum-free media), with or without 10 min predigestion with Trypsin or treatment with nonproteolytic cell dissociation solution (CDS). Cells were preincubated for 1 h in serum-free media, treated with Trypsin, and transduced for 1 h in serum-free media. (F) Cell surface interaction of P21-containing proteins is disrupted by Tritonx100 treatment. Flow cytometry analyses of NIH3t3 cells treated with proteins (20 µg/mL) for 1 h and a further 5 h incubation (in serum-free media) with 10 min pretreatment of PBS or PBS containing 0.1% (vol/vol) Tritonx100 (Tx100). Cells were preincubated for 1 h in serum-free media, treated with PBS or PBS with Tx100, and transduced for 1 h in serum-free media. Error bars indicate SD. n = 6.
We hypothesized that enhanced cell binding coupled with a cell-penetrating peptide would significantly enhance cell uptake. This would also indicate that further charging of P21-fused proteins via a PTD would enhance uptake if functioning primarily through heparan sulfate binding. To test this, we combined both moieties in one molecule (Fig. 1A). The inclusion of both P21 and 8R synergized to enhance uptake of all cell lines tested (Fig. 1B). Importantly, mouse and human pluripotent stem cells (CGR-8, HUES7, and IPS2) and cardiomyocytes (HL1) only possessed efficient uptake with the inclusion of P21. P21-mediated activity was more than two orders of magnitude more efficient than PTD-mediated uptake alone (Fig. 1C). We tested these motifs in tandem at both the N terminus and C terminus of mRFP or switched their termini. All variants demonstrated similar synergistic behavior (SI Appendix, Fig. S4), even when 8R was swapped for alternative cationic PTDs [TAT, 8K, and 8RQ (2); SI Appendix, Fig. S5]. We describe this synergistic delivery mechanism as GAG-binding enhanced transduction (GET).
We attempted to address the issue of cationic charge and its influence on P21 activity more thoroughly. Addition of the 8R PTD adds 8+ and P21 adds 11+ to the mRFP reporter (SI Appendix, Table S1), so to discount any primary cationic effect of P21 and to confirm that P21 genuinely targeted membrane-bound heparin sulfates, we constructed variants with scrambled P21 sequence (P21*) and also with all but the positive residues (K & R) deleted (termed P21 KR only) (SI Appendix, Fig. S6). Neither of these variants with similar charge could be taken up with wild-type P21 efficiency; however, they did endocytose, confirming positive charge is a prerequisite for cell delivery. These data importantly demonstrate that P21 is not a simple unstructured cationic peptide.
For completeness, we also compared uptake of mRFP with more extensive positive charging by attaching longer poly-arginines (4R, 8R as before, 16R, 24R, 32R, and 48R). mRFP-24R has a net positive charge of +19.5 versus P21-mRFP-8R with +14.4 at pH 7.0 but did not deliver to the same magnitude; nor did longer poly-arginine versions (SI Appendix, Fig. S6). Interestingly, if longer poly-arginine stretches were included with P21, there was greater synergy (until a saturation point: ∼+25), demonstrating P21 does not elicit its activity through simple cationic charging.
GET Requires the Presence of Trypsin-Sensitive and Detergent-Soluble Cell Membrane Molecules.
To evaluate the mechanism of GET interaction and uptake by cells, we performed a series of experiments that have been previously used to assess PTD activity. We showed enzymatic digestion of cell membrane proteins with trypsin reduced GET by 8.4-fold (P < 0.05) (Fig. 1E). In contrast, it was unaffected by cell dissociation solution, a nonenzymatic method for cell dispersal. Furthermore, we depleted detergent-soluble cell membrane molecules with Triton X-100 and observed a ∼2.2-fold decrease in GET (P < 0.05) without a decrease in viability. Therefore, both protein and detergent-soluble moieties on the cell membrane affect the efficacy of P21 and PTD synergy in GET.
Synergy Between HDBs and PTDs Is a General Phenomenon.
To determine whether P21 had unique activity or whether other natural HBDs elicit the same synergy with PTDs, we probed the literature and constructed a series of other mRFP-GET proteins with HBDs taken from different growth-factor families and extracellular matrix proteins (SI Appendix, Table S1 and Fig. S7). Importantly, some of these were essentially neutrally or negatively charged, which would allow us to highlight the importance of cationic charge for enhanced transduction through HBDs. We were able to show that all HBDs analyzed promoted PTD-mediated delivery of mRFP into cells. Without a PTD, some HBDs promoted transduction themselves, but this was not a result of simple cationic charge, as some had a high positive charge (FGF4 and 7 B domains; SI Appendix, Table S1). Also, with the addition of 8R PTD, there was no link between the overall charge of the molecule and its transduction activity (SI Appendix, Fig. S7 C and D). We tested a selection of these HBDs (with and without the addition of 8R) in other cell types to demonstrate any cell-type specific activity. We could confirm that a number of HBDs enhance transduction more strongly in certain cell-types (i.e., FGF2B, PDGF; SI Appendix, Fig. S7C), whereas many produced similar lower levels of enhancement in all of the three cell lines tested (NIH3t3, CGR8, and HUES7). In conclusion, the intrinsic transduction activity and enhancing function of HBDs on PTDs appears independent on its own positive charge, and this activity can be more potent in particular cell types.
GET Enhances Cre-Mediated Genome Modification.
It was imperative to assess the proportion of delivered protein that escaped endosomes and therefore could be considered as functional by GET. Previous studies have the avoided issues associated with direct measurement of fluorescent-tagged proteins (such as being unable to distinguish membrane, vesicle, or functional cytosolic/nuclear protein) by assaying for the successful nuclear activity of Cre recombinase (8). We also take advantage of this system and measured Cre-mediated recombination of a loxP-STOP-loxP (LSL) enhanced green fluorescent protein (eGFP) reporter gene in live NIH3t3 mouse fibroblast cells (NIH3t3: LSL-eGFP cells) (Fig. 2A). This system can be considered a measure of functional transduction as well as cellular uptake, as activation of green fluorescence requires exogenous Cre protein to enter the cell, undergo nuclear translocation, and excise the LSL fragment of the transgene.
Fig. 2.
GET of Cre recombinase. (A) Schematic of the construct created to mark Cre activity in cells. Cre-mediated excision of a transcriptional STOP region flanked by loxP sites induces the constitutive expression of eGFP. Pr, promoter; βGal, β-galactosidase; Neo, Neomycin phosphotransferase. The NIH3t3 LSL-eGFP cell line was created by transfection and selection of NIH3t3 cells. (B) eGFP expression in untreated NIH3t3 LSP-eGFP cells or those transduced with SIN Cre lentivirus. (Left) Fluorescence microscopy. (Right) Flow cytometry histogram of eGFP expression. (Scale bar, 50 µm.) (C) Scheme of testing transduction of Cre activity in NIH3t3 LSL-eGFP cells. Cells were transduced with Cre proteins for 1 h and washed and cultured for 2 d before analyses. (D and E) P21-mR-Cre-8R is efficiently transduced and recombines DNA. (D) Fluorescence microscopy images Cre-transduced NIH3t3 LSL-eGFP with the variety of dosages. (Scale bar, 50 µm.) (E) Flow cytometry analyses of NIH3t3 LSL-eGFP cells transduced for 1 h with mR-Cre, mR-Cre-8R, and P21-mR-Cre-8R at a variety of dosages (0, 1, 10, 100, and 500 µg/mL), washed and cultured for 2 d. Graph shows percentage recombination (i.e., percentage of eGFP positive from total cell population). Error bars indicate SD. n = 6.
Transduction of NIH3t3:LSL-eGFP cells with SIN Cre lentiviruses to overexpress Cre transgenically led to near-complete (92 ± 6%; P < 0.001) activation of eGFP expression in all cells, confirming the utility of this system (Fig. 2B). Reporter activation requires only one functional Cre recombinase complex (four molecules) to be delivered, which does not allow the determination of the precise amount of cargo delivered. To overcome this issue, we delivered Cre proteins at limiting dilutions for a short exposure time (1 h) and determined the minimum dose required to activate green fluorescence after 48 h (Fig. 2C).
Treatment of NIH3t3:LSL-eGFP cells with mR-Cre (mRFP fused to Cre) resulted in recombination and eGFP activation (22.1 ± 6.7%; P < 0.05) at the highest doses (500 µg/mL; Fig. 2D). eGFP activation was inhibited at 4 °C and negatively and concentration-dependently affected by serum. mR-Cre-8R demonstrated that the 8R PTD enhanced functional delivery of Cre (∼22-fold; P < 0.01). GET-Cre (P21-mR-Cre-8R) required as little as 1 min incubation with cells at a low dose (1 µg/mL; ∼30 nM) to elicit recombination (4.3 ± 2.5%; P < 0.05), confirming that binding and internalization is an efficient and rapid process. For a moderate dose (10 µg/mL; ∼300 nM), GET achieved a complete functional delivery and recombined all NIH3t3; this is ∼15-fold (P < 0.01) above PTD-only levels and ∼340-fold higher than mR-Cre (P < 0.001) (Fig. 2 D and E). We repeated heparinase III, free heparin, and serum-free experiments and confirmed that heparinase III pretreatment reduced recombination to basal levels and that media serum plays a role in replenishing cell membrane GAGs depleted by heparinization (SI Appendix, Figs. S8–S11). Overall, these data correlate with the fluorescence delivery conclusions and show synergy between P21 and PTD moieties to achieve significant increases in functional transduction of protein cargo.
GET of NANOG Promotes Self-Renewal of Pluripotency.
If GET technology is to be adopted for clinical applications, demonstration of its use to alter cell fate is crucial. An important application would be in the driving of reprogramming, self-renewal, and differentiation of stem cells. iPSC technology has been swiftly developed to allow genome nonintegrating DNA-based (19), RNA-based (20), and protein-based (11, 12) technologies to supersede the original retroviral protocols (21) (Fig. 3).
Fig. 3.
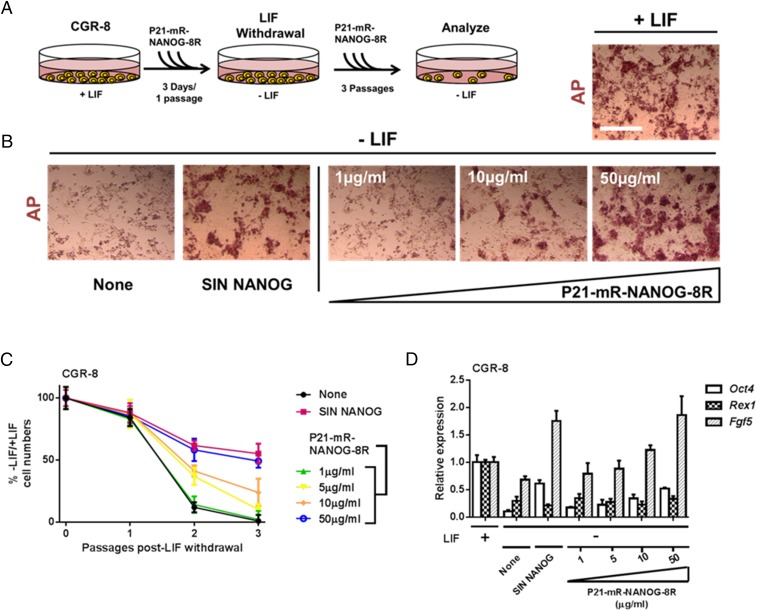
GET of NANOG promotes the self-renewal of mouse embryonic stem cells. (A) Scheme of testing activity of transduced NANOG in CGR-8 cells. Cells were transduced with P21-mR-NANOG-8R proteins (0, 1, 10, and 50 µg/mL) for 3 consecutive days (1 passage, 1:3 split), passaged 1:3, and plated into growth media with P21-mR-NANOG-8R but lacking LIF (−LIF). Cells were fed daily with –LIF media containing P21-mR-NANOG-8R and passaged 1:3 every 3 d for 2 passages (a total of 3 passages –LIF). (B) P21-mR-NANOG-8R rescues self-renewal of mESCs lacking LIF dose-dependently. AP staining of CGR-8 cells treated with P21-mR-NANOG-8R proteins and LIF withdrawal. AP activity and colony morphology is retained in CGR-8 cells cultured in LIF or without LIF but supplemented with SIN NANOG (to overexpress NANOG) or transduced with P21-mR-NANOG-8R. (Scale bar, 100 µm.) (C) P21-mR-NANOG-8R maintains the proliferation of mESCs lacking LIF dose-dependently. Percentage of the number of CGR-8 cells cultured without LIF versus those with LIF (percentage −LIF/+LIF) at passaging. In LIF-deficient CGR-8 cultures, proliferation is promoted when supplemented with SIN NANOG (to overexpress NANOG) or transduced with P21-mR-NANOG-8R. Error bars indicate SD. (D) NANOG-dependent rescue in LIF-deficient cultures generates a more epiblast-like gene expression profile. Relative gene expression analyses of LIF-deficient CGR-8 cultures using quantitative PCR. Cultures supplemented with SIN NANOG (to overexpress NANOG) or transduced with P21-mR-NANOG-8R have increased Fgf5 expression, reduced Rex1 expression, and retain Oct4 expression. Error bars indicate SE. n = 6.
We used CGR-8 mESCs to determine whether GET-mediated delivery can sustain their pluripotent self-renewing phenotype with the withdrawal of leukemia inhibitory factor (LIF). We delivered GET NANOG-cargo (P21-mR-NANOG-8R) in an assay (22) similar to that used to initially isolate the role of Nanog in mESCs (23). P21-mR-NANOG-8R rescued pluripotency-associated alkaline phosphatase (AP) activity in significant numbers of CGR-8Z, even with relatively low doses (10 µg/mL) (Fig. 3B). AP activity in high-dose P21-mR-NANOG-8R samples was similar to that achieved by the SIN NANOG lentiviral transgenic. Transgenics and high-dose P21-mR-NANOG-8R-transduced CGR-8 cells proliferated to a similar level in LIF-deficient cultures (∼87.6-fold more; P < 0.001) (Fig. 3C) and also retained Oct4 expression to a similar level (albeit lower than in LIF-containing cultures), indicative of retention of pluripotency (both P < 0.05) (Fig. 3D). As observed previously, rescued cells by both transgenic and protein methods up-regulated Fgf5 and down-regulated Rex1 expression, indicative of an inner cell mass-to-epiblast transition phenotype (22). A cell-penetrating peptide (CPP) version (mR-NANOG-8R) of this protein did not confer LIF independence to cells (SI Appendix, Fig. S12).
GET of MYOD Drives Myogenesis.
Given the high efficiency of functional PTD-mediated cargo delivery by P21 enhancement, we reasoned that GET technology might also be used to redirect pluripotent cells toward differentiated cell fates, using the transduction to deliver recombinant transcription factors (24–26).
For this, we used the delivery of the efficacious MYOD myogenic factor (27) to drive skeletal muscle specification (Fig. 4). We devised an in vitro differentiation protocol in which HUES7 cells were transduced with a GET MYOD cargo, P21-mR-MYOD-8R (Fig. 4A). We show that a high percentage of large multinucleated MYOGENIN-positive myotubes (62.1 ± 8.9%; P < 0.01) (Fig. 4 B–F) that had elevated MYOD and ACTA1 expression (P < 0.01 and < 0.05, respectively) (Fig. 4C) could be generated by GET-MYOD. This was comparable to the differentiation observed in the SIN MYOD lentiviral transgenic when P21-mR-MYOD-8R was delivered at higher doses. A CPP version (mR-MYOD-8R) of this protein did not promote myogenesis at these concentrations (SI Appendix, Fig. S13). Taken together, these experiments provide proof of principle that GET-delivered protein can direct the fate of pluripotent cells to a terminally differentiated somatic cell type.
Fig. 4.
GET of MYOD promotes myogenic differentiation of hESCs. (A) Scheme of testing the differentiation activity of transduced MYOD in HUES7 cells. HUES7 cells were plated onto gelatinized plastic and cultured in DMEM containing 10% (vol/vol) FCS. Cells were fed daily with DMEM containing 10% (vol/vol) FCS and P21-mR-MYOD-8R (0, 1, 5, 10, or 50 µg/mL) for 7 d. Media was then changed to DMEM containing 2% (vol/vol) horse serum (HS), human recombinant insulin, and P21-mR-MYOD-8R and fed daily for 3 d. (B–F) P21-mR-MYOD-8R drives myogenic differentiation of HUES7 cells to multinucleated Myotubes. (B) Light microscopy of HUES7 cells cultured under the myogenic regime supplemented with SIN MYOD (to overexpress MYOD) or transduced with P21-mR-MYOD-8R. Elongated fused Myotubes and single myocytes are generated with SIN-MYOD or high doses of P21-mR-MYOD-8R. (Scale bar, 100 µm.) (C) MYOD-dependent myogenic differentiation of hESCs. Relative gene expression analyses of HUES7 cultures using quantitative PCR. Cultures supplemented with SIN MYOD (to overexpress MYOD) or transduced with P21-mR-MYOD-8R have increased endogenous MYOD expression and skeletal muscle-specific ACTA1 expression. Error bars indicate SE. (D and E) P21-mR-MYOD-8R differentiated cells are multinucleated. (D) Quantitation of mean nuclei number per cell using PI staining. Error bars indicate SD. (E) Fluorescence microscopy images of HUES7 cells differentiated with P21-mR-MYOD-8R (50 µg/mL) and stained with nuclear dye DAPI. (Scale bar, 50 µm.) (F) P21-mR-MYOD-8R differentiated cells are MYOGENIN-positive. Quantitation of the percentage of MYOGENIN-positive cells using immunolabeling. Error bars indicate SD. n = 6.
GET Can Be Coupled to a Variety of Clinically Useful Cargoes.
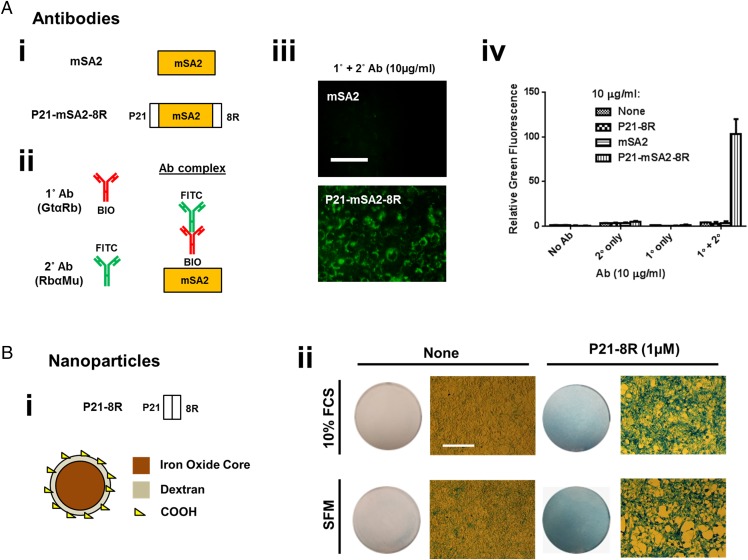
Because GET can effectively deliver functional recombinant proteins, we assessed whether the P21 and 8R peptide moieties can be linked to a variety of other cargoes to enhance intracellular delivery. We initially tested other protein cargoes (Fig. 5 and SI Appendix, Figs. S14 and S15). The first strategy was to produce a cargo-interacting P21-8R variant by cloning and recombinantly expressing monomeric streptavidin (mSA2) fused between P21-8R to effectively target biotinylated cargoes (Fig. 5 A and I and SI Appendix, Fig. S16), or by using staphylococcal protein A B domains, which have strong affinity for IgG antibodies (SI Appendix, Fig. S25). By simple mixing GET-mSA2 (P21-mSA2-8R; Fig. 5A), we show a significant enhancement in transduction: a naturally extracted (bovine heart) proapoptotic factor, cytochrome C (Cyt-C), which we biotinylated in vitro (to produce BIO-Cyt-C) to allow GET-mSA2 interaction. Only with the GET system, and specifically with biotinylated Cyt-C (BIO-Cyt-C), along with endosomal escape (using chloroquine), did cells respond to Cyt-C delivery and lose viability (SI Appendix, Fig. S14). Addition of GET-mSA2 to biotinylated primary antibodies (which in turn bind to complementary secondary antibodies) yielded efficient delivery of the complex (measured by secondary antibody fluorescence intracellularly; 26-fold over primary and secondary antibodies alone) (Fig. 5 B and C). Using GET-staphylococcal protein A B protein, IgG antibodies could be directly delivered to cell without requirement of biotinylation (∼786-fold over antibody alone), demonstrating the utility of GET as a biotinylated-cargo or IgG antibody transduction reagent (SI Appendix, Fig. S15).
Fig. 5.
GET of antibodies and nanoparticles. (A) GET of biotinylated cargoes using monomeric streptavidin (mSA2). (A, i) Schematic of the mSA2 proteins engineered to bind to and enable the uptake of biotinylated cargoes. We used P21-8R as a noninteracting control, mSA2 as a nontransducing control, and P21-mSA2-8R as the test protein. (A, ii) Schematic of the antibody (Ab) complexes of a biotinylated primary (1°) antibody (goat anti-rabbit; GtαRb) bound to an FITC-conjugated secondary (2°) antibody (rabbit anti-mouse; Rb αMu) used to test activity. (A, iii) GET delivery of Ab complexes was visible by fluorescence microscopy. (Scale bar, 50 µm.) With coincubation of P21-mSA2-8R (Bottom, 10 µg/mL), Ab complexes were efficiency delivered to cells. (A, iv) Flow cytometry demonstrating that 1°/2° Ab complexes (1 µg/mL) are taken into NIH3t3 cells poorly by direct incubation or when coincubated with mSA2 only. (B) GET of magnetic nanoparticles. (B, i) Schematic of the P21-8R peptide synthesized and test MNPs. We tested 250-nm Nanomag-D dextran shell/iron oxide core MNPs and conjugated P21-8R peptide to surface COOH groups. (B, ii) MNPs are taken into NIH3t3 cells most efficiently in serum-free media (SFM; Left). Light microscopy images of Prussian blue iron-stained NIH3t3 cells treated with MNPs (50 µg/mL) for 12 h in standard media conditions [10% (wt/vol) FCS] or SFM. Conjugation of P21-8R to MNPs significantly increases cellular uptake in both 10% (wt/vol) FCS and SFM conditions (circular image is of entire well). (Scale bar, 100 µm.) n = 6.
We hypothesized the same approach could be used for nucleic acid delivery. We used the pan-nucleic acid interaction sequence LK15 and synthesized GET-LK15 peptides (Fig. 6). After charge ratio optimization for each test nucleic acid, we were able to demonstrate significant transfection activity for P21-LK15-8R for plasmid DNA (pDNA; transfecting SIN-GFP), modified nucleotide mRNA [transfecting GFP modRNA (20)], and small inhibitory RNAs (siRNAs; FAM-labeled GAPDH siRNA). The transfection efficiencies of optimized protocols were similar to Lipofectamine 2000 (LIPO2000; Invitrogen), and GET-transfection retained activity in serum-containing transfections in which LIPO2000 was significantly inhibited. Colloidal stability of GET peptide/nucleic acid particles remained with addition of the serum, demonstrating that there was no loss of stability (by aggregation), and no efficiency was lost as a result of the serum-rich environment.
Fig. 6.
GET of nucleic acids. (A) Schematic of the LK15 proteins engineered to bind to and transduce nucleic acids. (B) Transfection of human mesenchymal stem cells using GET-LK15. Initially we assessed binding capacity of LK15 peptides for plasmid (p)DNA (SIN GFP, to express GFP on transfection), modified synthetic messenger RNA (modRNA) (Miltenyi Biotech; to express GFP on transfection), and small inhibitory (si)RNAs (labeled with FAM fluorophore to detect delivery). After optimizing ratios, we transfected human mesenchymal stem cells with P21-LK15-8R and pDNA (10 µg), modRNA (10 µg), or siRNA (1 µg) and visualized transfection by fluorescence microscopy. (Scale bar, 100 µm.) (C) Quantification of GET-LK15 transfection of human mesenchymal stem cells by flow cytometry (percentage transfection efficiency or relative fluorescence for siRNA) compared with LIPO2000 as a commercial standard. Error bars indicate SD.
To extend our demonstration of the GET system to large cargoes (the largest demonstrated here being ∼150 kDa IgG), we attempted to enhance delivery of magnetic nanoparticles (MNPs) by covalently coupling P21-8R peptide to commercially available MNPs (coupled to Nanomag-D 250-nm MNPs through N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDAC)/n-hydroxysuccinimide (NHS) reaction with MNP COOH functional group; estimated 1.5–2.0 μg peptide/mg MNPs; ∼400 peptide molecules/particle; Fig. 5B). Conjugation of P21-8R to MNP increased their size (from 255.4 ± 1.6 nm to 372.9 ± 2.3 nm) and charge (from −33.7 ± 0.5 to +19.3 ± 0.4). In both serum- and serum-free media conditions, MNP uptake by cells was significantly enhanced by P21-8R conjugation (assessed by Prussian blue staining), and particles retained colloidal stability. These data indicate that GET may be used for any choice of cargo, dependent on providing an interaction or conjugation of P21-8R to the target.
Discussion
Using the synergistic combination of a PTD with a cell membrane binding peptide, we aimed to improve intracellular targeting of proteins and other cargoes. With this approach, we have developed a technology that enables highly efficient delivery of functionally relevant proteins to direct a variety of cell behaviors even in hard-to-transduce cell types (see SI Appendix, Figs. S17–S19 for enhancement of delivery with GET, viability with high protein concentrations and repeated incubation, and viability after cell uptake of a variety of clinically relevant cell types). High concentrations of GET peptide are not required to deliver functional amounts of cargo into cells; this is unlike cationic CPP technologies, which are primarily driven to transduce by high extracellular concentrations. We demonstrated that the GET system can be harnessed to promote survival or self-renewal or direct the differentiation of pluripotent cells toward a desired lineage. Furthermore, other proteins can be delivered by coupling to GET peptides (such as antibodies), and this can be extended to other chemically distinct targets such as nucleic acids or MNPs. This system is not technically complex, as it is for modified RNA systems (20), and offers several key advantages over established techniques to deliver the exogenous function of a gene or protein or delivering other reagents. Fundamentally, because GET is protein-based, it completely eliminates the risk of genomic integration and insertional mutagenesis inherent to all DNA-based methodologies (1). Moreover, if endosomal escape can be improved, our approach will allow protein stoichiometry to be tightly regulated within cells. This will avoid stochastic variation in expression typical of integrating vectors, as well as the uncontrollable effects of viral silencing. Importantly, the GET peptides do elicit some endosomal escape, which could be a result of the net positive charge and a mechanism similar to that of previously described CPPs. It is also likely that the GET peptides will be degraded both before and after endosomal escape, as for other CPPs.
Given the significant function of recombinant proteins delivered with our methodology, it is possible that GET technology may also be directly applied to reprogramming and programming approaches. As there are stepwise phenotypic changes observed during pluripotency induction (28, 29) and in directed-differentiation protocols (30), it seems likely that individual transcription factors play distinct, stage-specific roles. The unprecedented potential for temporal control over individual factor function afforded by GET technology should enable these variables to be tested to improve efficiency and kinetics of cell fate control. Furthermore, the delivery of DNA, RNA, siRNA, and MNPs by GET enables its use for genome editing, gene knockdown, and physical manipulation or imaging of cells, respectively.
The fact that we isolated heparan sulfate binding domains/proteins as PTD enhancers is an important finding. Although GAGs have long been considered important in the process of PTD-mediated transduction, their exact role is presently a point of contention, with the most recent hypothesis being that GAG is merely important by directly binding to PTDs, thereby enhancing lipid–bilayer translocation, but not being necessary for its occurrence (8). We also demonstrated that HBDs can elicit this function without themselves intrinsically being positively charged; however, an overall positive charge is required, provided by the PTD, to mediate the transduction. This makes GET distinct from simple cationic CPP-based approaches. Using noncationic CPPs with HBDs may completely negate the requirement for an overall positive charge for transduction. Importantly, the effect of HBDs on PTDs and its cargo is not simple “supercharging,” and therefore can be considered distinct from other attempts to use such technologies (4, 5).
Furthermore, the demonstration that a variety of HBDs can target different cell types more specifically (SI Appendix, Fig. S7) adds a further advantage to GET. At this time, there are >100 detailed GAG-binding proteins (31), and these natural or synthetic peptides screened to specifically bind heparan sulfate molecules presented on particular cells will allow GET to be adapted to tightly and efficiently transduce target cells.
Our discovery that promoting heparan sulfate interaction significantly and synergistically improves the efficiency of PTD-tagged biologically active, macromolecular cargo will open up new avenues for the treatment, imaging, and experimental investigation of disease. We believe that our improvement of the long-established PTD technologies has the potential to become a major enabling technology for cell-based therapies and regenerative medicine.
Materials and Methods
Recombinant Protein Expression, Cell Culture, and Transduction of Cargoes by GET.
Recombinant proteins were expressed as GST-tagged proteins and cleaved using PreScission Protease (GE Healthcare) during purification. Cell culture techniques have been previously described (32) and further detailed in SI Appendix, Materials and Methods. Proteins were transduced into cells by addition to culture media for the specific cell line. Cells were harvested by trypsinization unless otherwise stated and analyzed for fluorescence intensity by flow cytometry or for gene expression by real-time PCR. Transfection of nucleic acids or transduction of antibody complexes was achieved by complexation of the peptide with the cargo in OptiMEM (Invitrogen). Peptides were covalently attached to MNPs presenting COOH groups (Nanogmag-D, MicroMod) with EDAC/NHS chemistry.
Statistical Analysis.
Statistical comparisons were carried out using the GraphPad Prism software package. Comparisons were made using one-way Tukey-Kramer analysis of variance. Results were considered significant if P < 0.05. Experiments were completed six times (n = 6), and data depict mean values (six replicates of duplicates) with SD or for quantitative PCR with SEM.
Supplementary Material
Acknowledgments
We thank Dr. Andrew D. Johnson (University of Nottingham) and Dr. Catherine Merry (University of Manchester) for helpful discussions. The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant Agreement 227845. J.E.D. and K.M.S. acknowledge the support of the Medical Research Council, the Engineering and Physical Sciences Research Council, and the Biotechnology and Biological Sciences Research Council UK Regenerative Medicine Platform Hub “Acellular Approaches for Therapeutic Delivery” (MR/K026682/1). C.D. is supported by British Heart Foundation (04BX14CDLG, PG/14/59/31000, RG/14/1/30588, P47352); Medical Research Council (MR/M017354/1); National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC/K000225/1, 35911-259146); and Heart Research UK (TRP01/12).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518634113/-/DCSupplemental.
References
- 1.Gump JM, Dowdy SF. TAT transduction: The molecular mechanism and therapeutic prospects. Trends Mol Med. 2007;13(10):443–448. doi: 10.1016/j.molmed.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 2.El-Andaloussi S, Holm T, Langel U. Cell-penetrating peptides: Mechanisms and applications. Curr Pharm Des. 2005;11(28):3597–3611. doi: 10.2174/138161205774580796. [DOI] [PubMed] [Google Scholar]
- 3.Meade BR, Dowdy SF. Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv Drug Deliv Rev. 2007;59(2-3):134–140. doi: 10.1016/j.addr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Thompson DB, Cronican JJ, Liu DR. Engineering and identifying supercharged proteins for macromolecule delivery into mammalian cells. Methods Enzymol. 2012;503:293–319. doi: 10.1016/B978-0-12-396962-0.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cronican JJ, et al. A class of human proteins that deliver functional proteins into mammalian cells in vitro and in vivo. Chem Biol. 2011;18(7):833–838. doi: 10.1016/j.chembiol.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakase I, Takeuchi T, Tanaka G, Futaki S. Methodological and cellular aspects that govern the internalization mechanisms of arginine-rich cell-penetrating peptides. Adv Drug Deliv Rev. 2008;60(4-5):598–607. doi: 10.1016/j.addr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: From molecular mechanisms to therapeutics. Br J Pharmacol. 2009;157(2):195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gump JM, June RK, Dowdy SF. Revised role of glycosaminoglycans in TAT protein transduction domain-mediated cellular transduction. J Biol Chem. 2010;285(2):1500–1507. doi: 10.1074/jbc.M109.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meier O, et al. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J Cell Biol. 2002;158(6):1119–1131. doi: 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422(6927):37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 11.Kim D, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4(6):472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou HY, et al. Generation of Induced Pluripotent Stem Cells Using Recombinant Proteins. Cell Stem Cell. 2009;4(5):381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10(3):310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 14.Skehel JJ, Cross K, Steinhauer D, Wiley DC. Influenza fusion peptides. Biochem Soc Trans. 2001;29(Pt 4):623–626. doi: 10.1042/bst0290623. [DOI] [PubMed] [Google Scholar]
- 15.Han X, Bushweller JH, Cafiso DS, Tamm LK. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat Struct Biol. 2001;8(8):715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 16.Sakuma T, Higashiyama S, Hosoe S, Hayashi S, Taniguchi N. CD9 antigen interacts with heparin-binding EGF-like growth factor through its heparin-binding domain. J Biochem. 1997;122(2):474–480. doi: 10.1093/oxfordjournals.jbchem.a021776. [DOI] [PubMed] [Google Scholar]
- 17.Higashiyama S, Abraham JA, Klagsbrun M. Heparin-binding EGF-like growth factor stimulation of smooth muscle cell migration: Dependence on interactions with cell surface heparan sulfate. J Cell Biol. 1993;122(4):933–940. doi: 10.1083/jcb.122.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson SA, et al. Characterization of sequences within heparin-binding EGF-like growth factor that mediate interaction with heparin. J Biol Chem. 1994;269(4):2541–2549. [PubMed] [Google Scholar]
- 19.Yu J, Chau KF, Vodyanik MA, Jiang J, Jiang Y. Efficient feeder-free episomal reprogramming with small molecules. PLoS One. 2011;6(3):e17557. doi: 10.1371/journal.pone.0017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 22.Dixon JE, et al. Axolotl Nanog activity in mouse embryonic stem cells demonstrates that ground state pluripotency is conserved from urodele amphibians to mammals. Development. 2010;137(18):2973–2980. doi: 10.1242/dev.049262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 24.Kwon YD, et al. Cellular manipulation of human embryonic stem cells by TAT-PDX1 protein transduction. Mol Ther. 2005;12(1):28–32. doi: 10.1016/j.ymthe.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Hidema S, Tonomura Y, Date S, Nishimori K. Effects of protein transduction with intact myogenic transcription factors tagged with HIV-1 Tat-PTD (T-PTD) on myogenic differentiation of mouse primary cells. J Biosci Bioeng. 2012;113(1):5–11. doi: 10.1016/j.jbiosc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 26.Liang QL, Mo Z, Li XF, Wang XX, Li RM. Pdx1 protein induces human embryonic stem cells into the pancreatic endocrine lineage. Cell Biol Int. 2013;37(1):2–10. doi: 10.1002/cbin.10001. [DOI] [PubMed] [Google Scholar]
- 27.Bichsel C, et al. Direct reprogramming of fibroblasts to myocytes via bacterial injection of MyoD protein. Cell Reprogram. 2013;15(2):117–125. doi: 10.1089/cell.2012.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27(11):1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 29.Smith KP, Luong MX, Stein GS. Pluripotency: Toward a gold standard for human ES and iPS cells. J Cell Physiol. 2009;220(1):21–29. doi: 10.1002/jcp.21681. [DOI] [PubMed] [Google Scholar]
- 30.Burridge PW, et al. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6(4):e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esko JD, Linhardt RJ. Proteins that bind sulfated glycosaminoglycans. In: Varki A, et al., editors. Essentials of Glycobiology. 2nd Ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2009. [PubMed] [Google Scholar]
- 32.Dixon JE, et al. Combined hydrogels that switch human pluripotent stem cells from self-renewal to differentiation. Proc Natl Acad Sci USA. 2014;111(15):5580–5585. doi: 10.1073/pnas.1319685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.