Significance
Necrotizing fasciitis (NF) is a rapidly progressing fatal skin and muscle tissue lesion. We studied a human case of NF and found that the infection was caused by multiple strains of A. hydrophila (NF1–NF4). The latter three strains constitute a clonal group, whereas NF1 is phylogenetically distinct. We tested these strains individually in a mouse intramuscular model of infection and observed NF1 to be less virulent than NF2. However, when NF1 and NF2 were mixed, NF1 exhibited more virulence and it decreased NF2 virulence. The cross-talk between NF1 and NF2 was due to the presence of ExoA toxin in NF2, ability of NF1 and NF2 to differentially modulate innate immune mechanism(s), and direct killing of NF2 by NF1.
Keywords: Aeromonas hydrophila, necrotizing fasciitis, mixed infections, intramuscular mouse model, metagenomics
Abstract
Necrotizing fasciitis (NF) caused by flesh-eating bacteria is associated with high case fatality. In an earlier study, we reported infection of an immunocompetent individual with multiple strains of Aeromonas hydrophila (NF1–NF4), the latter three constituted a clonal group whereas NF1 was phylogenetically distinct. To understand the complex interactions of these strains in NF pathophysiology, a mouse model was used, whereby either single or mixed A. hydrophila strains were injected intramuscularly. NF2, which harbors exotoxin A (exoA) gene, was highly virulent when injected alone, but its virulence was attenuated in the presence of NF1 (exoA-minus). NF1 alone, although not lethal to animals, became highly virulent when combined with NF2, its virulence augmented by cis-exoA expression when injected alone in mice. Based on metagenomics and microbiological analyses, it was found that, in mixed infection, NF1 selectively disseminated to mouse peripheral organs, whereas the other strains (NF2, NF3, and NF4) were confined to the injection site and eventually cleared. In vitro studies showed NF2 to be more effectively phagocytized and killed by macrophages than NF1. NF1 inhibited growth of NF2 on solid media, but ExoA of NF2 augmented virulence of NF1 and the presence of NF1 facilitated clearance of NF2 from animals either by enhanced priming of host immune system or direct killing via a contact-dependent mechanism.
Necrotizing fasciitis (NF) is a deadly necrotic inflammation of skin, s.c. tissues, and muscle bundles, most frequently caused by Streptococcus species, notably Streptococcus pyogenes, either alone or in combination with Streptococcus hemolyticus, Staphylococcus aureus, or both (1, 2). NF is classified as polymicrobial (type-I) or monomicrobial (type-II), and type-I is more prevalent and frequently linked to immune status of the patient (2, 3).
In recent years, type-I and type-II–associated NF caused by Aeromonas hydrophila have been reported at an increasing rate (4–6). These infections progress to septicemia via hematogenous access despite aggressive antibiotic treatment (6). Recently, we described an infection with A. hydrophila of wounds and bloodstream of a young immunocompetent NF patient. The patient, as a result of the infection, had to undergo several lifesaving surgical procedures, including amputations of limbs (7). Although this case of NF may be considered monomicrobial, because only a single Aeromonas species was involved, genomic analysis indicated mixed infection due to four strains representing two paraphyletic lineages of A. hydrophila. Three of the four strains, NF2, NF3, and NF4, exhibited minimal difference in genome sequence [12 high-quality single nucleotide polymorphisms (SNPs)] and identical genome content and synteny (7), thus can be considered a clonal group and are assumed to share similar pathodynamics of infection. The fourth strain, NF1, was phylogenetically distinct and, accordingly, differed in virulence from the other NF strains (7). It was speculated that the presence of multiple strains of A. hydrophila influenced disease progression and outcome significantly than if the individual strains had been involved alone. Plausibly, necrotic lesions were caused by A. hydrophila strains producing a variety of toxins (8, 9) and secreted toxins of one of the NF strains may have had an influential role in pathogenesis, in concert with the other strains during infection. Indeed, a notable difference in the genomes of the strains was presence of a gene, exoA, encoding exotoxin A (ExoA), a homolog of Pseudomonas aeruginosa ExoA, in the genomes of NF2, NF3, and NF4, but not NF1 (7, 10). ExoA has ADP ribosylating activity for eukaryotic elongation factor-2 (eEF-2), leading to inhibition of protein synthesis and host cell death (11).
In the present study, we provide evidence that during mixed infection with NF1 and NF2, NF1 benefited by greater dissemination induced by ExoA secreted from NF2. However, the presence of NF1 in the infection mixture either directly and/or via host innate immune mechanisms antagonized virulence of NF2 by preventing its dissemination and aiding in clearing of NF2 from the injection site.
Results
Virulence of A. hydrophila NF2.
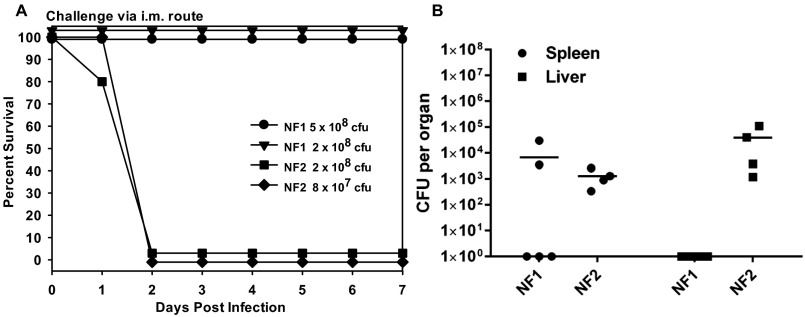
To understand virulence attributes of NF1 and NF2, an i.m. (intramuscular) model of mouse infection was used with these strains separately. A. hydrophila NF2 exhibited greater virulence compared with NF1, measured by animal mortality and bacterial dissemination to peripheral organs (Fig. S1 A and B). On day 2 p.i. (postinfection), NF2 at infection dose of 8 × 107 cfu (colony-forming units) caused 100% animal mortality, in contrast to an approximately sixfold higher dose of NF1 with no mortality at 7 d (Fig. S1 A and B). All animals infected with NF2 at an infection dose of 2 × 108 cfu showed bacterial dissemination to both spleen and liver, at a level of 103 to 105 cfu per organ. However, only two of five mice inoculated with NF1 yielded bacterial counts in the spleen and no detectable bacterial growth was observed in the liver.
Fig. S1.
Virulence characteristics of A. hydrophila strains NF1 and NF2 in a mouse model of infection. (A) Swiss–Webster mice (n = 5) were infected with the indicated infection doses of the individual NF strains by i.m., and mortality rate was recorded at 7 d. (B) A group of four or five animals was infected with strain NF1 or NF2 at a dose of 2 × 108 cfu per animal i.m. and after 48 h, the entire spleen and liver from each of the surviving animal were excised, and the bacterial counts determined. The horizontal lines represent the arithmetic means of the bacterial counts.
Attenuation of Virulence of A. hydrophila NF2 in the Presence of NF1.
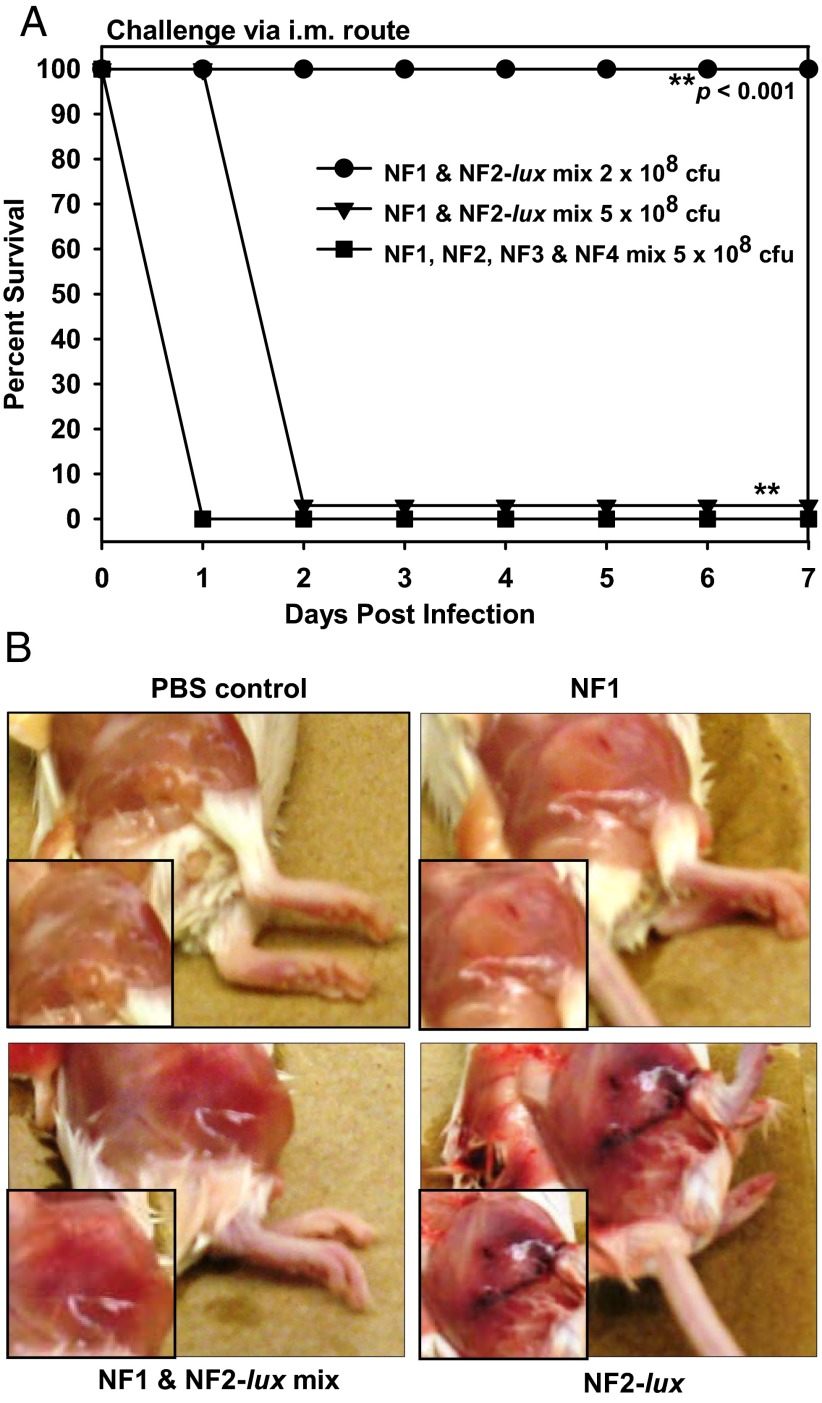
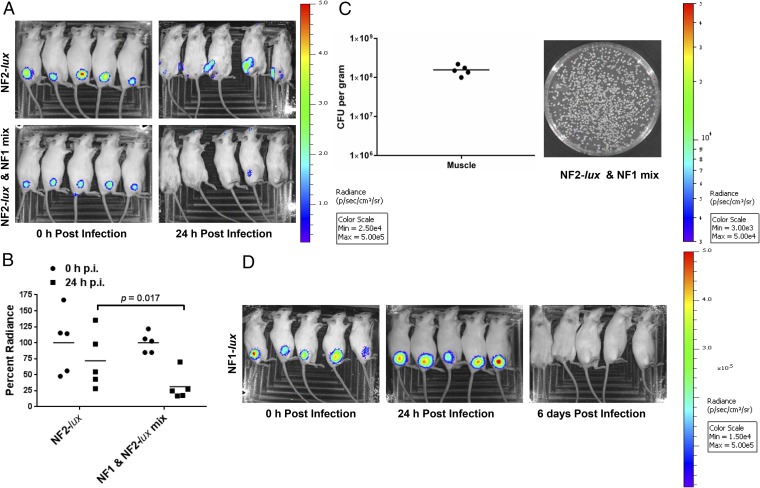
An infection dose of 5 × 108 cfu (7) with all four strains (NF1, NF2, NF3, and NF4; 1.25 × 108 cfu per strain) caused 100% mortality within 24 h p.i. after i.m. injection (Fig. 1A). The same dose (5 × 108 cfu, 2.5 × 108 cfu per strain) of NF1 and NF2 combined also resulted in 100% mortality within 48 h p.i. (Fig. 1A). A mixed infection of NF1 and NF2 at a dose of 2 × 108 cfu (1 × 108 cfu per strain) resulted in 100% survivability of mice, whereas an approximately threefold lower dose caused 100% mortality when only NF2 was injected (Fig. S1A). These data can be interpreted as NF1 modulating the virulence of NF2 in mixed culture. These results were substantiated because after 24 h p.i., gross necrotic lesions around the site of the i.m. injection in mice, when qualitatively scored, were mild for NF1 infection (1+), moderate for NF1 and NF2 mixed infection (2+), and severe for NF2 infection alone (3+) (Fig. 1B).
Fig. 1.
Virulence features of mono and mixed infections with A. hydrophila NF strains. (A) A group of five mice was infected with the noted dose of NF1 and NF2-lux or NF1 to NF4 mixture and animal mortality was observed over 7 d. Asterisks denote statistical significance among the indicated groups. (B) Three to five mice were infected with NF1, NF1 and NF2-lux mixture, or NF2-lux at 2 × 108 cfu per animal i.m. Control animals were injected with sterile PBS. After 24 h p.i., severity of inflammatory swelling and necrotic lesions were examined and scored. Based on the severity of necrosis, the scale used was as follows: 0 (no necrosis, e.g., for the PBS group), 1+ (mild necrosis, for the NF1-infected group of mice), 2+ (moderate necrosis, for the NF1 and NF2-infected group of mice), 3+ (severe necrosis, for the NF2-infected group of mice). Representative images are shown.
Selective Dissemination of A. hydrophila NF1 to Peripheral Organs in Mixed Infection and Role of exoA Gene.
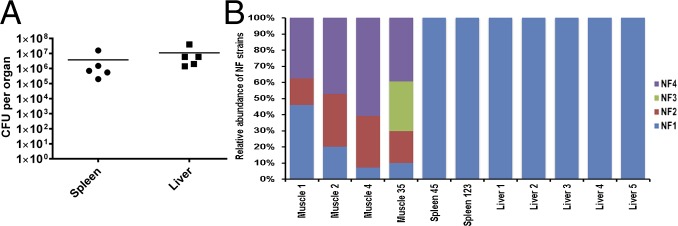
A large number of strain-specific patterns associated with NF1 is indicative of its distinct phylogenetic lineage, compared with other three isolates, NF2, NF3, and NF4, which form a clonal group (7). Mixed infection with all four strains injected i.m., yielded counts of ∼105 to 107 cfu in the spleen and liver after 24 h p.i. (Fig. 2A). Although the four NF strains were mixed in equal ratios, it was not known whether all four strains would disseminate similarly or differentially to peripheral organs of mice. Therefore, we used state-of-the-art unbiased whole genome shotgun metagenomics and GENIUS software package (CosmosID) to detect and quantify NF strains in muscle, spleen, and liver tissue of infected animals.
Fig. 2.
Dissemination characteristics of A. hydrophila NF strains during mixed infection in a mouse model. (A) Mice (n = 5) were injected with a mixture of NF1 to NF4 at an infection dose of 5 × 108 cfu (1.25 × 108 cfu per strain) per animal via i.m. route. After 24 h, whole spleen and liver from each animal were homogenized and an aliquot from each sample was subjected to bacterial colony count. The horizontal lines represent the arithmetic means of the bacterial counts (B). The remaining portion of the homogenates was processed for total DNA isolation. The isolated DNA for some organs representing different animals was combined because of the low yield and was labeled as muscle35, spleen45, and/or spleen123, respectively. The isolated DNA was subjected to deep sequencing and metagenomic analysis to identify the NF strains. Relative distribution of four A. hydrophila strains NF1, NF2, NF3, and NF4 in different metagenomic datasets derived from muscle, spleen, and liver samples is shown.
A key feature of GENIUS is the ability to incorporate new genome sequences into the database, and use the modified database to probe metagenome datasets. The newly sequenced NF genomes (NF1, NF2, NF3, and NF4) were incorporated into the existing bacterial database, which placed these four NF strains into the Aeromonas clade along with other A. hydrophila strains and identified unique patterns (biomarkers) associated with each of the four NF strains (n = 41,483, 147, 47, and 63 for NF1, NF2, NF3, and NF4, respectively). Indeed, GENIUS analysis of metagenomic datasets derived from muscle, spleen, and liver was able to identify and differentially detect individual NF strains (Table S1).
Table S1.
GENIUS identification and quantification of NF strains in muscle, liver, and spleen samples
| Metagenomes | Strain name | Weighted score | Percent unique hits | Percent added hits | Relative abundance (RA) | Confidence interval of RA |
| Mixed Culture (NF1-4): Muscle-1 | A. hydrophila NF1 | 2,341.94 | 98.34 | 98.41 | 87.31 | 86.64–87.94 |
| A. hydrophila NF2 | 113.27 | 24.49 | 94.18 | 4.22 | 3.834–4.63 | |
| A. hydrophila NF4 | 227.03 | 31.75 | 97.64 | 8.46 | 7.93–9.02 | |
| Mixed Culture (NF1-4): Muscle-2 | A. hydrophila NF1 | 514.81 | 85.4 | 85.97 | 69.68 | 68.77–70.57 |
| A. hydrophila NF2 | 87.16 | 27.21 | 93.92 | 11.80 | 11.18–12.44 | |
| A. hydrophila NF4 | 136.81 | 36.51 | 97.31 | 18.52 | 17.77–19.29 | |
| Mixed Culture (NF1-4): Muscle-35 | A. hydrophila NF1 | 1,945.27 | 98.67 | 98.73 | 40.46 | 39.50–41.43 |
| A. hydrophila NF2 | 753.60 | 41.5 | 95.49 | 15.68 | 14.98–16.41 | |
| A. hydrophila NF3 | 460.26 | 21.28 | 97.95 | 9.57 | 9.01–10.16 | |
| A. hydrophila NF4 | 1,643.28 | 47.62 | 98.19 | 34.18 | 33.26–35.12 | |
| A. hydrophila subsp hydrophila ATCC 7966 | 5.03 | 1.22 | 5.51 | 0.10 | 0.05–0.18 | |
| Mixed Culture (NF1-4): Muscle-4 | A. hydrophila NF1 | 976.69 | 82.45 | 83.16 | 41.96 | 40.99–42.93 |
| A. hydrophila NF2 | 388.78 | 28.57 | 94.5 | 16.70 | 15.98–17.44 | |
| A. hydrophila NF4 | 962.40 | 41.27 | 97.97 | 41.34 | 40.38–42.31 | |
| Mixed Culture (NF1-4): Spleen-123 | A. hydrophila NF1 | 0.79 | 0.45 | 0.46 | 100.00 | 99.96–100 |
| Mixed Culture (NF1-4): Spleen-45 | A. hydrophila NF1 | 14.36 | 5.76 | 5.92 | 100.00 | 99.96–100 |
| Mixed Culture (NF1-4): Liver-1 | A. hydrophila NF1 | 2.89 | 1.68 | 1.72 | 100.00 | 99.96–100 |
| Mixed Culture (NF1-4): Liver-2 | A. hydrophila NF1 | 15.59 | 7.42 | 7.59 | 91.84 | 91.29–92.36 |
| Escherichia_coli E1167 | 0.97 | 1.12 | 0.92 | 5.71 | 5.27–8.18 | |
| Enterococcus 71 Node | 0.42 | 0.3 | 0.3 | 2.45 | 2.16–2.77 | |
| Mixed Culture (NF1-4): Liver-3 | A. hydrophila NF1 | 2.92 | 1.3 | 1.35 | 100.00 | 99.96–100 |
| Mixed Culture (NF1-4): Liver-4 | A. hydrophila NF1 | 156.75 | 63.43 | 64.06 | 99.18 | 98.98–99.34 |
| Lactobacillus johnsonii NCC 533 | 0.98 | 1.06 | 1.76 | 0.62 | 0.48–0.79 | |
| L. johnsonii pf01 | 0.18 | 0.31 | 1.51 | 0.11 | 0.06–0.20 | |
| L. johnsonii DPC 6026 | 0.14 | 0.25 | 1.48 | 0.09 | 0.05–0.17 | |
| Mixed Culture (NF1-4): Liver-5 | A. hydrophila NF1 | 2.84 | 1.43 | 1.48 | 100.00 | 99.96–100 |
| NF2-lux + NF1-exoA: Muscle-1 | A. hydrophila NF1 | 5,519.63 | 98.72 | 98.77 | 97.58 | 97.26–97.86 |
| A. hydrophila NF2 | 134.07 | 29.25 | 94.55 | 2.37 | 2.09–2.69 | |
| Enterococcus face 17 Node | 2.72 | 1.08 | 1.22 | 0.05 | 0.02–0.12 | |
| NF2-lux + NF1-exoA: Muscle-2 | A. hydrophila NF1 | 1,988.79 | 96.41 | 96.56 | 85.34 | 84.63–86.02 |
| A. hydrophila NF2 | 341.55 | 31.97 | 94.76 | 14.66 | 13.98–15.37 | |
| NF2-lux + NF1-exoA: Muscle-3 | A. hydrophila NF1 | 3,781.49 | 98.67 | 98.72 | 96.67 | 96.30–97.00 |
| A. hydrophila NF2 | 118.42 | 27.21 | 94.39 | 3.03 | 2.71–3.38 | |
| Bacillus 79 Node | 12.00 | 3.64 | 3.48 | 0.31 | 0.22–0.44 | |
| NF2-lux + NF1-exoA: Liver-1* | A. hydrophila NF1 | 0.71 | 0.31 | 0.32 | 52.48 | 51.50–53.46 |
| Clostridiales bacterium 1–7-47FAA | 0.64 | 0.04 | 0.04 | 47.52 | 46.54–48.50 | |
| NF2-lux + NF1-exoA: Liver-3 | A. hydrophila NF1 | 13.15 | 3.75 | 3.73 | 94.10 | 93.62–84.55 |
| C. bacterium 1–7 -47FAA | 0.82 | 0.06 | 0.06 | 5.90 | 5.45–6.38 | |
| NF2-lux + NF1-exoA: Spleen-1* | A. hydrophila NF1 | 0.01 | 0.01 | 0.01 | 0.72 | 0.57–0.91 |
| C. bacterium 1–7-47FAA | 0.83 | 0.06 | 0.06 | 99.28 | 99.09–99.43 | |
| NF2-lux + NF1-exoA: Spleen-2* | A. hydrophila NF1 | 0.01 | 0.01 | 0.01 | 1.20 | 1.00–1.43 |
| C. bacterium 1–7-47FAA | 0.49 | 0.04 | 0.04 | 98.80 | 98.57–99.00 |
No calls identified above the threshold. Weighted score represents a weighted average of percent unique hits and unique hit frequency. Percent unique hits represent the percent of different unique kmers found in the sample of the total number of precalculated unique kmers available in the database. Similarly, percentage of added hits represents the percent of total (shared and unique) kmers for the organism of the total number of precalculated shared and unique kmers for the organism available in the database. Relative abundance is calculated by using a weighted average of percent unique hits and the frequency of unique hits in the sample and then normalized to represent the abundance of each organism. The confidence interval of the relative abundance estimated here is at the 95% confidence level.
Results showed that, at the site of injection in the thigh muscle of the animals and after 24 h p.i., all or the majority of the strains (NF1, NF2, NF3, and NF4) could be detected in varying proportions (Fig. 2B). Whereas strain-specific genomic biomarkers corresponding to strains NF1, NF2, and NF4 were detected in all muscle samples, NF3 was detected only in one of four muscle samples. Surprisingly, in the spleen and liver, only NF1-specific genomic signatures were detected, with no reads corresponding to NF2, NF3, or NF4, although a mixture of the four strains had been injected i.m. These data suggested that the bacteria in the spleen and liver of mice were only NF1 (Fig. 2A).
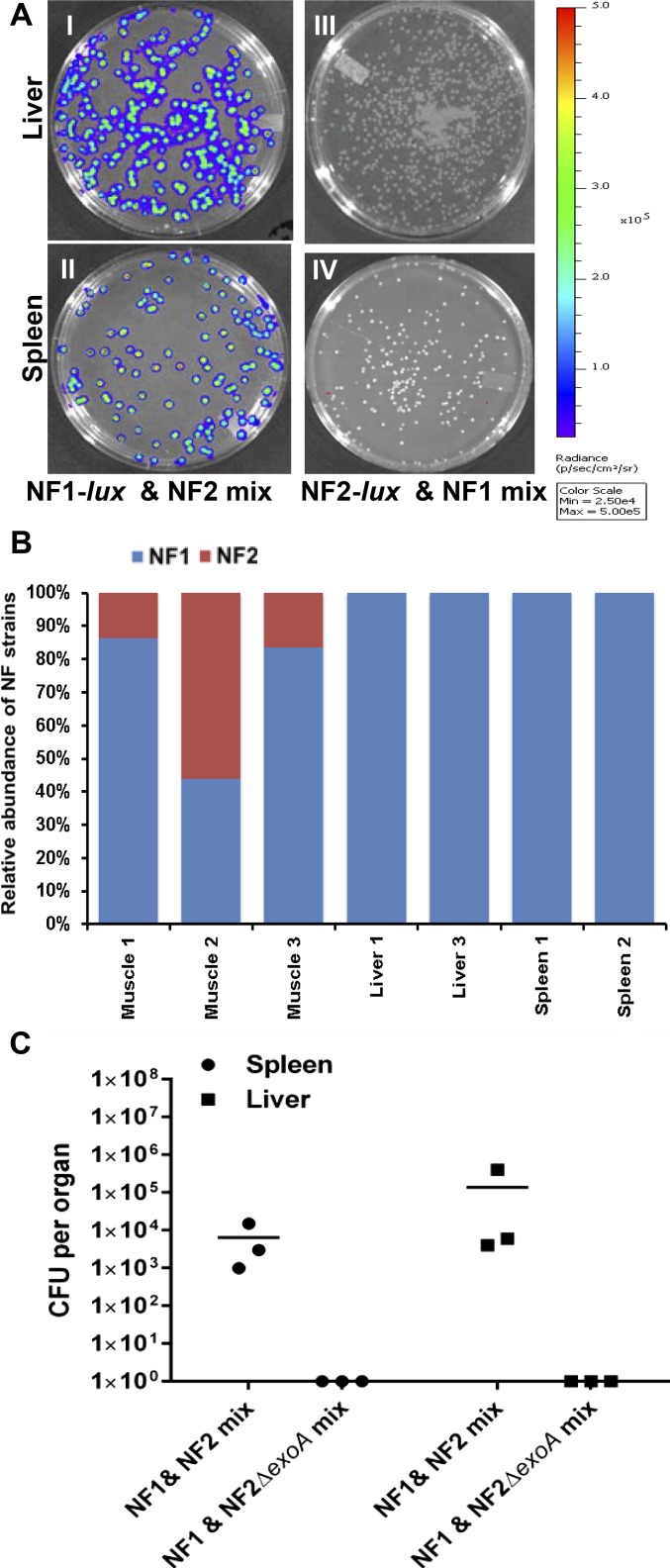
NF1 mixed with NF2, NF3, and NF4 or just with NF2 resulted in only NF1 migrating to the peripheral organs of the mice after i.m. injection. Antibiotic selection and expression of the luciferase gene (lux) in NF1-lux or NF2-lux confirmed the bacterial colonies recovered from the spleen and liver of mice injected in the mixed infection experiments were solely NF1 (Fig. S2 A and B), validating the finding of GENIUS metagenomic detection.
Fig. S2.
Selective dissemination of A. hydrophila strains in a mouse model. Bacterial dissemination to spleen and liver after 24 h of infection with NF1-lux and NF2, or NF1 and NF2-lux mixed cultures was observed by bioluminescence of colonies. (A) Representative images displayed confirmed that all of the disseminated bacterial colonies are NF1 in liver and spleen (I–IV). Further, DNA isolated from spleen and liver tissue samples from NF1 and NF2 mixed infections were subjected to sequencing and metagenomic analysis to determine identity of the strains (either NF1 or NF2) disseminated to these organs (B). The resulting analysis showed that the bacterial DNA recovered from the peripheral organs was specific to NF1. The quality and the concentration of DNA from liver 2 and spleen 3 samples were not high for sequencing. In addition, mice (n = 3) were infected with a mixture of NF1-lux and NF2, or NF1 and NF2∆exoA at a dose of 2 × 108 cfu (1 × 108 cfu per strain) per animal via i.m. route. After 24 h, entire spleen and liver from each animal were homogenized and bacterial counts were determined. The horizontal lines represent the arithmetic means of the bacterial counts (C). Samples collected from animals infected with NF1 and NF2∆exoA showed no dissemination of bacteria to peripheral organs.
Interestingly, when NF1 was mixed with the NF2∆exoA mutant, instead of the NF2 parental strain, and injected i.m., dissemination of NF1 to peripheral organs was not observed. The injected strains remained at the site of injection (Fig. S2C). Clearly, dissemination of NF1 after injection was influenced by the mixture of NF1 and NF2 and, most likely, due to ExoA secreted by the latter.
Elimination of NF2 from Site of Injection with NF1 and NF2.
When NF2-lux alone was injected in mice by the i.m. route at a dose of 2 or 5 × 108 cfu, bioluminescent signals were detected at 0 and 24 h p.i. at the site of injection in all animals (Fig. 3A). At 24 h p.i., bioluminescent intensity was comparatively less around the site of injection, likely due to the migration of NF2 from the site of infection to peripheral organs. Similarly, i.m. infection with the NF1-lux alone resulted in bacteria localized around the site of injection, between 24 and 72 h p.i. (Fig. 3D; 72 h data not shown). Eventually, the NF1-lux bioluminescent signal decreased and the animals no longer demonstrated localized bioluminescence at 6 d p.i., indicating clearing of the bacteria from the mice; all of the animals survived. However, mixed infection with NF1 and NF2-lux resulted in significant decrease in bioluminescent signal at 24 h p.i., compared with animals infected with NF2-lux alone (Fig. 3A). Furthermore, bacteria recovered from muscle tissue 24 h p.i. were bioluminescent-negative NF1 (Fig. 3C). Thus, these results confirmed clearing of NF2 within 24 h p.i. when injected with NF1.
Fig. 3.
Progression of infection from the local site of injection for NF strains in a mouse model. (A) A group of five mice was infected with NF2-lux or NF2-lux and NF1 in mixed culture (1:1 ratio) at 2 × 108 cfu per animal. Immediately after injection and at 24 h p.i., the animals were imaged for bioluminescence signal. (B) Total flux (p/s) was measured for each animal around the bioluminescent spot with the same shape and area across the images. The values between NF2-lux and the NF1 and NF2-lux mixed infection were then compared. (C) After 24 h p.i., the absence of bioluminescence from bacterial colonies recovered from muscle tissue of NF2-lux and NF1 mixed infection indicated elimination of NF2-lux from the site of injection. Bacterial load of the injected muscle tissue is also shown. The horizontal lines represent the arithmetic means of the bacterial counts. (D) Time course for progression of dissemination for NF1-lux strain was monitored for 6 d. Bioluminescence images are for 0 h, 24 h, and 6 d p.i.
Role of exoA in Murine Infection.
Genome sequencing and annotation revealed exoA gene, a major virulence factor in P. aeruginosa and related species, was present in A. hydrophila NF2, NF3, and NF4, but absent from NF1 (7). Similarly, diarrheal isolate SSU of A. dhakensis, serving as reference, carries the exoA gene, which is 97% homologous to exoA of NF2. Initially, the exoA gene was cloned with its cis-acting promoter from the SSU strain and transformed via mini-Tn7 transposition system to NF1; the resulting strain is referred to as NF1-exoA (Table S2). The exoA gene was inserted downstream of the glmS gene encoding glucosamine-6-phosphate synthase.
Table S2.
Bacterial strains and plasmids used in this study
| Strains or plasmids | Relevant characteristics | Source |
| A. hydrophila | ||
| NF1 | Isolated from NF wound site | 7 |
| NF1-lux | Transferred with Tn7-luciferase operon and spontaneous rifampicin resistance clone (RifR, KmR) | 7 |
| NF1-exoA | Transferred with Tn7-exoA and spontaneous rifampicin resistance clone (RifR, ApR) | This study |
| NF2, NF3, NF4 | Isolated from NF wound site following the amputation and surgical debridement | 7 |
| NF2-lux | Transferred with Tn7-luciferase operon and spontaneous rifampicin resistance clone (RifR, KmR) | This study |
| NF2ΔexoA | Isogenic deletion mutation for gene exoA in strain NF2 and spontaneous rifampicin resistance clone (RifR) | This study |
| A. dhakensis | ||
| SSU | Diarrheal isolate | 20 |
| SSUΔexoA | Isogenic deletion mutation for gene exoA in strain SSU and spontaneous rifampicin resistance clone (RifR) | This study |
| SSUΔexoA::exoA | SSUΔexoA complemented with Tn7-exoA and spontaneous rifampicin resistance clone (RifR, ApR) | This study |
| E.coli | ||
| SM10 λpir | The strain carrying Tn7-transposon helper plasmid, pTNS2 (ApR) | Laboratory stock |
| SM10 λpir | The strain carrying Tn7-transposon pUC18R6K-mini-Tn7T::Km-lux (KmR) or pUC18R6K-mini-Tn7T::Ap-exoA (ApR) | Laboratory stock |
| Plasmids | ||
| pTNS2 | A mobilizable helper plasmid encoding only the specific TnsABC+D transposition pathway (ApR) | 19 |
| pUC18R6K-mini-Tn7T::Ap-exoA (Tn7-exoA) | Tn7 based transposon system carries the gene exoA with its cis-acting promoter and ampicillin resistance selection marker (ApR) | This study |
| pUC18R6K-mini-Tn7T::Km-lux (Tn7-luciferase) | Tn7 based transposon system carries luciferase operon and kanamycin resistance selection marker (KmR) | This study |
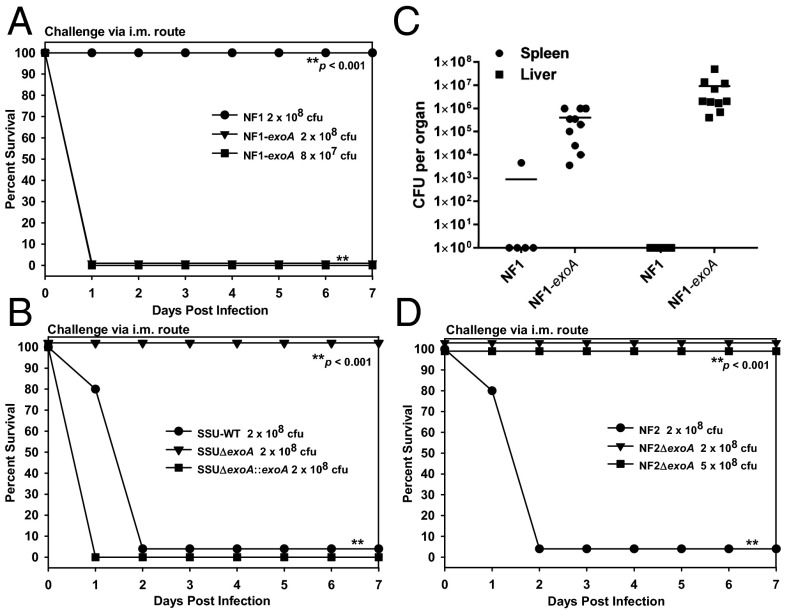
When NF1-exoA was injected i.m., all animals succumbed to infection by 24 h p.i. at an infection dose of 8 × 107 or 2 × 108 cfu (Fig. 4A). As a control, mice were infected with WT NF1 at a dose of 2 × 108 cfu, and these animals all survived. Similarly, all mice injected with WT SSU died by day 2, whereas animals injected with SSU∆exoA mutant at the same dose survived (Fig. 4B). In contrast, mice injected with SSU∆exoA::exoA at a dose of 2 × 108 cfu all died by 24 h p.i. (Fig. 4B), clearly indicating a role of ExoA in animal mortality and, by extrapolation, septic progression of NF.
Fig. 4.
Role of exoA gene on animal mortality and bacterial dissemination. Mice (n = 5) were infected with NF1 or NF1-exoA (A); WT SSU, SSUΔexoA, or SSUΔexoA::exoA (B); or NF2 or NF2∆exoA (D) at the indicated doses and observed for mortality. Asterisks denote statistical significance among the indicated groups. From mice (n = 5 or 10) infected with NF1 or NF1-exoA, bacterial dissemination was measured in spleen and liver tissues after 24 h p.i. The horizontal lines represent the arithmetic means of the bacterial counts (C).
At 24 h p.i., NF1-exoA is concluded to disseminate significantly more widely than when NF1 was injected. All animals injected with NF1-exoA yielded bacterial counts for both spleen and liver, i.e., 106 to 107 cfu per organ, respectively (Fig. 4C). In contrast, only one of five animals injected with NF1 yielded bacterial colony counts and then only ∼5 × 103 cfu per organ (spleen) (Fig. 4C). NF1 was not detected in the liver of any of the animals after 24 h p.i.
Subsequent construction of ∆exoA mutant of NF2 confirmed the role of this toxin in an i.m. murine model. As a control, all animals injected with NF2 parental strain at a dose of 2 × 108 cfu died by 48 h p.i. However, mice injected with NF2∆exoA at the same or 2.5-fold higher dose all survived (Fig. 4D).
Phagocytosis Efficiency and Elimination of NF2 by Macrophages.
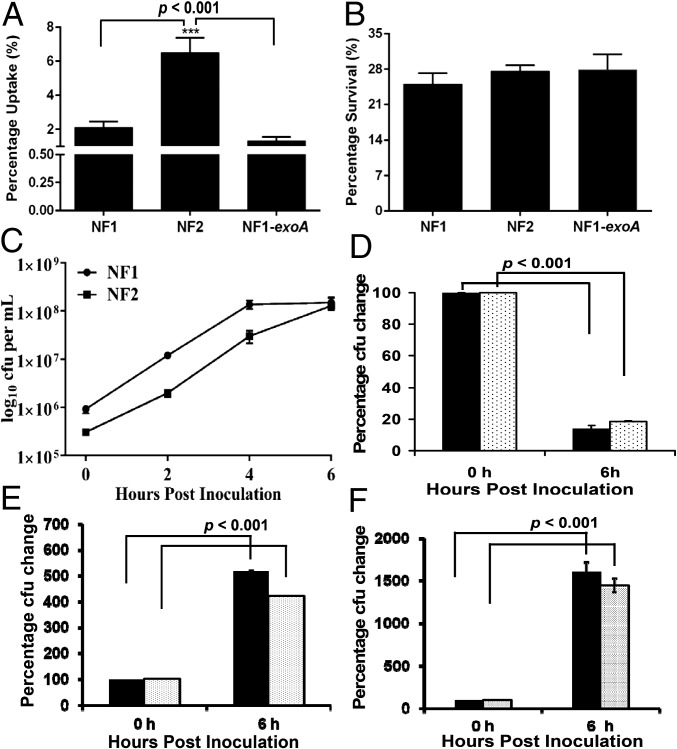
Murine macrophage RAW 264.7 infection in vitro with NF2 resulted in phagocytosis at a significantly higher rate compared with NF1 (Fig. 5A). Uptake of NF1 and NF1-exoA by macrophages was similar. Although NF2 was phagocytized to a greater extent, percent intracellular survival of NF1, NF2, and NF1-exoA was approximately the same, i.e., 25% (Fig. 5B).
Fig. 5.
Phagocytic elimination and in vitro growth dynamics of A. hydrophila strains. RAW 264.7 cells were infected with NF1, NF2, or NF1-exoA at a multiple of infection (moi) of 5, and percent bacterial uptake was calculated based on colony counts after gentamicin treatment (1.75 h p.i.) (A). At 2 h after gentamicin treatment, intracellular bacterial counts were determined to estimate percentage of intracellular bacterial survival (B). Furthermore, NF1 and NF2 were grown individually in LB medium at 37 °C for 6 h with 180 rpm shaking in an incubator (New Brunswick Scientific Co., Enfield, CT) and bacterial counts determined (C). At the same time, NF1 and NF2-lux were mixed at a ratio of 5:1 (solid bar) or 10:1 (dotted bar), respectively. The mixed cultures were either incubated at 37 °C on LB agar plates for 6 h (D) or in liquid LB medium with 180 rpm shaking for 6 h (E), and the growth of NF2-lux was measured. Similarly, when NF2 was mixed with NF1-lux at a ratio of 5:1 (solid bar) or 10:1 (dotted bar), respectively, and incubated at 37 °C on LB agar plates for 6 h, the number of NF1-lux was enumerated (F). Results were plotted with arithmetic means ± SD.
Suppression of Growth of NF2 by NF1.
When NF1 and NF2 were grown alone in vitro, both showed significant growth at 6 h (Fig. 5C). However, when NF1 and NF2-lux were mixed, 5:1 or 10:1, plated on agar, and incubated for 6 h, colony counts for NF2-lux were reduced approximately fivefold (Fig. 5D). In contrast, in a liquid medium, when NF1 and NF2-lux cultures were grown at 5:1 or 10:1 mixture, NF2-lux exhibited four- to fivefold growth at the 6 h time point (Fig. 5E). When NF2 was in excess compared with NF1-lux (5:1 or 10:1) in mixed culture, the growth of NF1-lux on solid agar plate was not inhibited by NF2 (Fig. 5F).
Influence of NF1 on Bacterial Motility via Expression of exoA.
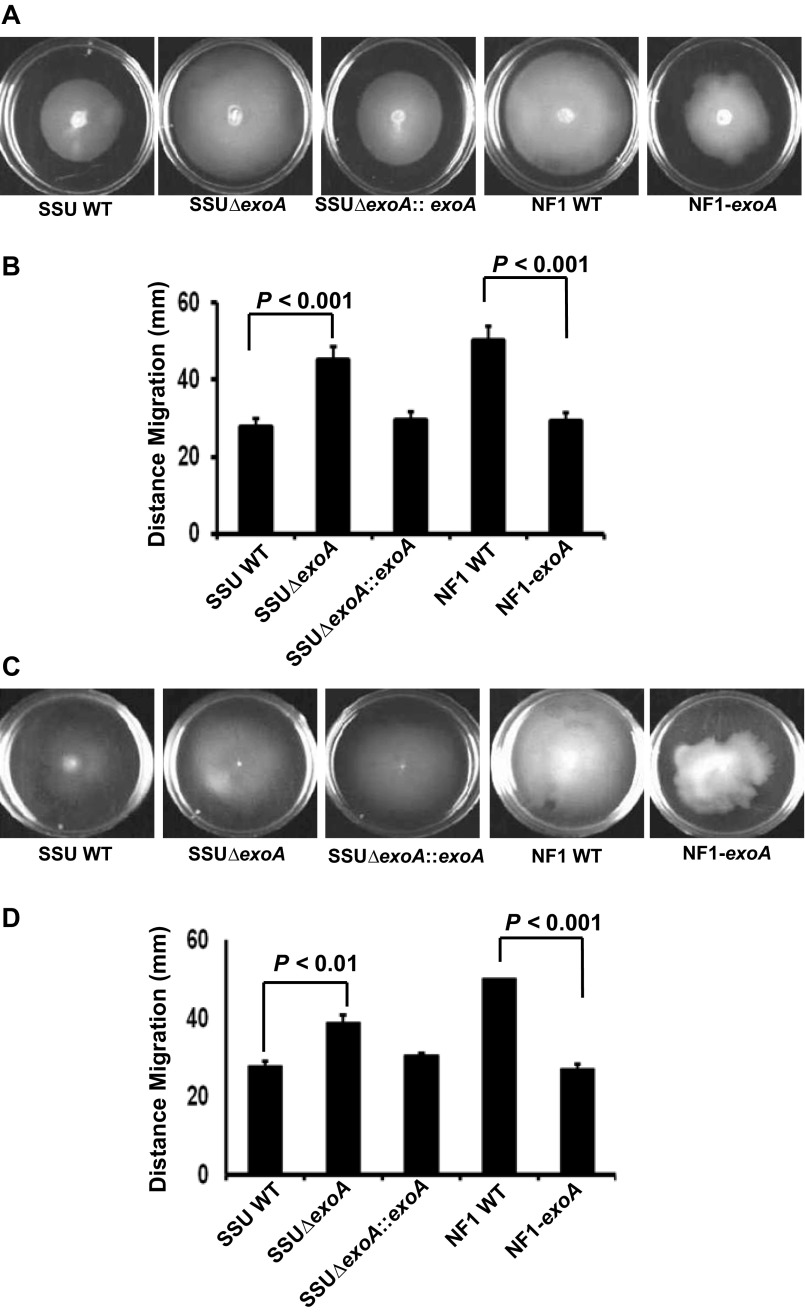
Deletion of exoA from SSU resulted in increased swimming and swarming motility, and motility returned to the level of A. hydrophila SSU WT when the mutant was complemented with exoA (Fig. S3). Equivalently, when exoA was expressed in NF1, swimming and swarming motility was significantly decreased compared with that of NF1 (Fig. S3). In accordance, the level of vfr gene expression, a negative regulator of motility, was inversely related to levels of fleQ transcript, a positive regulator of motility, in NF1, NF1-exoA, WT SSU, and SSUΔexoA. For example, WT SSU showed 2.29 ± 0.05-fold lower fleQ transcript level than that of SSUΔexoA mutant. Similarly, fleQ expression was down-regulated by 2.41 ± 0.029-fold in NF1-exoA compared with NF1. Conversely, the level of vfr gene expression in NF1-exoA was 1.40 ± 0.01-fold up-regulated compared with NF1. In SSUΔexoA, vfr gene expression was 1.45 ± 0.04-fold down-regulated compared with WT SSU.
Fig. S3.
Bacterial motility assays for A. hydrophila strains. Swimming (A and B) and swarming (C and D) motility of A. hydrophila SSU, SSUΔexoA, SSUΔexoA::exoA, NF1 and NF1-exoA were measured in 60-mm Petri plates. Three independent experiments were performed for each strain, and arithmetic means ± SD were plotted.
Discussion
A. hydrophila strains NF1, NF2, NF3, and NF4 were isolated from a patient with NF as a consequence of deep wound infection (7). Among the strains, NF2, NF3, and NF4 were characterized as a clonal group, with NF2 presenting as the dominant colony morphotype. Genomic analysis of NF1 demonstrated a phylogenetically distant relationship to the other three stains, indicating the patient, in reality, suffered from a mixed infection acquired from exposure to a natural water reservoir of A. hydrophila. Specifically, the presence of the exoA gene in NF2 led to increased virulence of strain NF2 when tested in a mouse model, compared with NF1 (Fig. S1) (7).
In contrast, NF1 was restricted to the local site of infection and eventually eliminated by the host innate immune system (Fig. 3D); NF1 rarely caused death of the infected animals. One of the striking differences between NF1 and NF2 was that the latter harbored the exoA gene, with its C-terminal ADP ribosylation domain sharing 77% homology to a similar domain of P. aeruginosa ExoA. ExoA is an eEF-2 inhibitor; the toxin is involved in catalytic transfer of ADP ribose to eEF-2 (12), resulting in inhibition of host cell protein synthesis.
ExoA causes necrosis and apoptosis of mouse liver in acute phase (10). Therefore, it is possible that ExoA secreted by NF2 destroys or liquefies muscle (hallmark of NF) at the site of injection (Fig. 1B), breaking the local barrier and allowing systemic spread of the organism. However, because NF1 does not natively produce ExoA, it persists at the injection site and eventually is cleared by host defense mechanisms (Fig. 3D).
Although studies of monomicrobial infection with only NF1 or NF2 help to understand A. hydrophila-associated NF, we can now offer the hypothesis that a mixture of NF1, NF2, NF3, and NF4 at a wound site significantly influences progression of infection. Interestingly, NF caused by group A β-hemolytic Streptococcus species is often polymicrobial (1). During such mixed infections, synergistic or antagonistic effects on virulence induced by the causative agents comprise a complex interplay leading to establishment and subsequent progression of disease (13, 14). For example, Mosser et al., recently reported enhanced virulence for several natural and experimentally paired Aeromonas strains in a Caenorhabditis elegans killing model (13). Interestingly, the synergistic effects they observed were limited to pairs that were composed of strains belonging to different species (13).
Synergistic virulence in wound infections has been reported for type-2 diabetic mouse models (14). In those animals, mixed infections with Escherichia coli, Bacteroides fragilis, and Clostridium perfringens led to a higher bacterial load of B. fragilis around the s.c. injection sites when E. coli was present. Similarly, C. perfringens and B. fragilis showed possible interplay enhancing their survival during infection. The results presented in this study of mixed infection with NF strains in a mouse model clearly indicate interactive processes of both synergistic augmentation and antagonistic attenuation of virulence of NF1 and NF2 (Fig. 6). What is noteworthy is that understanding this interplay among strains of the same species involved in a mixed infection is critical for determining molecular mechanisms involved in progression of A. hydrophila-associated NF.
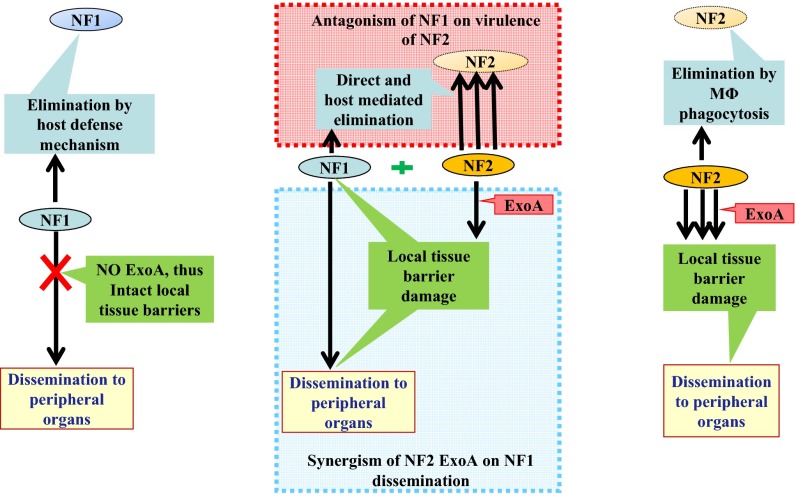
Fig. 6.
Schematic showing interaction of NF strains during mixed infections. Left indicates absence of ExoA in strain NF1 resulted in the bacterium being confined to the local site of infection and eventually eliminated by host defense mechanisms. In contrast, Right shows secretion of ExoA by strain NF2 allowed the bacterium to disseminate to peripheral organs, away from the site of infection, by destroying local tissue barriers. Middle reveals pathodynamics of mixed infection with NF1 and NF2. ExoA secreted by NF2 assists dissemination of NF1 to peripheral organs by destroying local tissue barriers. However, NF1 facilitates direct killing and/or host-mediated elimination of NF2 from the site of infection in muscle tissue. Triple arrow indicates overwhelming response compared with that indicated by the single arrow.
Reduction in mortality and elimination of NF2 from the site of injection in mice was observed when both NF1 and NF2 were injected (Figs. 1A and 3 A–C), indicating an antagonistic effect of NF1 on virulence of NF2. The results of in vitro growth experiments provided evidence that NF1 was lethal for NF2 when both were in close contact on a solid medium (Fig. 5D). These findings suggest the possible involvement of a secretion system capable of introducing toxic effector molecules from NF1 to NF2. One such secretion system reported for A. hydrophila is the type-VI secretion system (T6SS) (15). The genome profiles showed the presence of two clusters of T6SS synthesis genes and effectors [three copies of Hemolysin-coregulated protein (Hcp) and five copies of valine-glycine repeat G proteins (VgrGs)] in both NF1 and NF2 (7), raising the possibility that expression of these effector molecule-encoding genes may be differentially regulated in A. hydrophila NF1 and NF2. Alternatively, it is also plausible that NF1 but not NF2 may possess yet-unidentified specific bacterial toxic T6SS effectors. Likely, interaction of strain NF1 mutated for the T6SS locus with NF2 may provide a definitive answer under this experimental setting. Therefore, future studies to determine the role of T6SS in the interplay between NF1 and NF2 should be conducted to understand the survival and thriving strategies of these bacteria in a competitive environment, namely the aquatic ecosystem.
Macrophage phagocytosis and intracellular survival data for NF1 and NF2 (Fig. 5 A and B) suggest that there is a host intervening process in the elimination of NF2 when NF1 is also present at the site of infection. NF1 in mixed culture may favor recruitment and/or activation of macrophages, which would result in rapid elimination of NF2 before the bacterium enters systemic circulation (Fig. 6).
It can be noted that the motile version of P. aeruginosa strain is associated with enhanced macrophage recruitment to the site of infection and efficient activation via bacterial flagellar interaction with host pathogen recognition receptors (16–18). NF1 proved to have superior motility (Fig. S3) and, thus, can be expected to increase host innate immune surveillance at the local site of infection. Because NF2 was relatively readily phagocytized and subsequently killed by host macrophages (Fig. 5 A and B) in monomicrobial infection, ExoA-mediated local tissue damage would overwhelm the host macrophage-intervened defense process, allowing the bacterium to cause septicemic infection and acute mortality in animals.
After the antagonistic effect of NF1 on virulence of NF2 had been demonstrated, evidence was sought to delineate effects of NF2 on pathogenesis of NF1 in a mouse model of mixed infection. Results showed ExoA secreted by NF2 played an important role in progression of NF caused by NF1 during mixed infection. That is, ExoA causes tissue damage at the site of infection, thereby weakening host defense barriers. As a consequence, NF1 is less phagocytized and less likely to be killed by local macrophages, and can disseminate efficiently to peripheral organs (Fig. 6). When NF2ΔexoA mutant was mixed with NF1, dissemination of NF1 did not occur, confirming a role of ExoA in bacterial dissemination. Similarly, cis expression of the exoA gene in NF1 significantly increased bacterial dissemination to peripheral organs and increased animal mortality (Fig. 4 A and C).
In summary, when NF is caused by a mixed infection with strains of a single species, namely A. hydrophila, progression of NF follows a different course from that of a single strain of A. hydrophila. In this study, a murine NF model of infection and metagenomic analysis was used to show that ExoA of NF2 plays a significant role in both local necrotic inflammation and bacterial dissemination to peripheral organs. Therapeutic formulation that could inactivate ExoA should be considered when treating A. hydrophila-related NF. Future studies identifying T6SS effector molecule(s) produced by NF1 that blocks growth of NF2 might prove useful in designing antimicrobials against NF strains of A. hydrophila.
Materials and Methods
Bacterial Strains and Culture Conditions.
A. hydrophila NF strains are listed in Table S2 and were grown overnight at 37 °C in Luria–Bertani (LB) broth with 180 rpm shaking. The cultures were washed twice in sterile PBS and resuspended in the same buffer to prepare doses for injection in mice. All animal studies were performed under an approved Institutional Animal Care and Use Committee protocol at the University of Texas Medical Branch. Antibiotics, namely ampicillin (Ap) 250 μg/mL, kanamycin (Km) 100 μg/mL, and rifampicin (Rif) 200 μg/mL, were added as needed. LB agar plates were prepared with appropriate antibiotics when growth of the bacteria on solid medium was required.
Genetic Manipulation of the Bacterial Strains.
Construction of bioluminescent A. hydrophila NF2.
Luciferase enzyme-mediated bioluminescent strain of A. hydrophila NF2 was constructed as explained (7, 19). A detailed procedure is given in SI Materials and Methods.
Construction of A. hydrophila NF1-exoA strain.
The exoA gene from a diarrheal isolate SSU of A. dhakensis (20) was initially cloned, along with its native promoter, in a Tn7-based transposon system. Briefly, the strain was generated by triparental conjugation of Rifr NF1, E. coli SM10 λpir carrying the pTNS2 plasmid, and E. coli SM10 λpir harboring the pUC18R6K-mini-Tn7T::Ap-exoA plasmid. Insertion of the target gene at the correct location was confirmed by PCR, using a primer pair PTn7R:5′ ACAGCATAACTGGACTGATTTC 3′ and GlmSFwd: 5′ GCCAGTATCCCATTGCCATG 3′, followed by DNA sequencing.
Construction of A. hydrophila NF2ΔexoA mutant strain.
Upstream and downstream flanking regions corresponding to the exoA gene from NF2 strain were PCR amplified and the product cloned into pRE112 suicide vector to in-frame delete the target gene from the genome of NF2, as described (21). Subsequently, deletion of the exoA gene from the mutant was confirmed by PCR and genome sequencing.
Generation of A. dhakensis SSUΔexoA mutant strain and complementation of the mutant with the corresponding exoA gene.
In-frame deletion of the exoA gene from the genome of SSU was performed as described above for the NF2ΔexoA strain. To obtain the complemented strain, the SSUΔexoA mutant strain was transformed with the plasmid pBR322 carrying the native exoA gene with its 200-bp cis-operating promoter. We also transformed empty pBR322 vector into the WT SSU strain to serve as a negative control for some of the studies. Various primer sequences used during the study are listed in Table S3.
Table S3.
Oligonucleotides used and their PCR product sizes
| Primer | Oligonucleotide sequence | Use | PCR product size, bp |
| EflanF1up | 5′-TCATCGCGCTCGATCACCT-3′ | Gene exoA upstream fragment | 570 |
| EflanR1up | 5′-GTGATTTCCTTTGAGTAATGGCAA-3′ | ||
| EflanF2d | 5′-TTGCCATTACTCAAAGGAAATCACA | Gene exoA downstream fragment | 441 |
| TCGCGGGTCATCGATACAA-3′ | |||
| EPflanR2d | 5′-TCGACGGTGACGAGCTGAT-3′ | ||
| pPexoA322F | 5′-TCATCGCGCTCGATCACCT-3′ | Upstream-exoA- downstream fragment | 2,931 |
| pPexoA322R | 5′-TCGACGGTGACGAGCTGAT-3′ | ||
| fleQF | 5′-TGCTTCCTTGCTCATCCCGTA-3′ | fleQ fragment | 1,403 |
| fleQR | 5′-TGTTGATTGTTGACGCCGAT-3′ | ||
| vfrF | 5′-TCATTGGCAAACCGCAAA-3′ | vfr fragment | 621 |
| vfrR | 5′-ACACCACTATGGTCTTGCCGT-3′ |
In Vitro Characterization of the Bacterial Strains.
Bacterial swarming and swimming motilities were measured as described (7). A detailed method is provided in SI Materials and Methods. Furthermore, various in vitro assays, such as transcriptome measures for genes fleQ and vfr via RT-PCR, phagocytic assay and intracellular survival in macrophage cell culture, and growth dynamics of mixed NF strains on solid or in liquid medium, carried out for this study were elaborated in SI Materials and Methods.
Animal Infection and Mortality Pattern Analysis.
Healthy female Swiss–Webster mice (Taconic Farms) were infected via the i.m. route with infection doses of 8 × 107, 2 × 108, or 5 × 108 cfu per animal. For mixed infections, indicated doses represented equal numbers of cfu for each of the mixed strains. After infection, animals were observed for disease progression over a period of 7 d and the mortality rate was recorded daily.
Isolation of DNA from Murine Tissues Infected with A. hydrophila NF Strain Mixture.
Five animals were injected i.m. with a mixed culture of A. hydrophila, NF1, NF2, NF3, and NF4, at infection dose of 5 × 108 cfu per animal (1.25 × 108 cfu per strain). After 24 h p.i., ∼250 mg of muscle tissue around the injection sites were collected in sterile PBS. Similarly, the entire spleen and liver were excised from the animals and immersed in 1 and 2 mL, respectively, of sterile PBS. The tissues were thoroughly ground, and homogenates passed through 70-µm nylon filters. An aliquot (100 µL) of the filtrate from each sample was plated on sheep blood agar and colonies enumerated after incubation overnight at 37 °C. Remaining filtrates were centrifuged at 4,000 × g for 15 min to pellet animal tissue and bacteria. From the pellet fractions, total DNA was isolated by using the DNeasy blood and tissue kit (Qiagen).
Bacterial Dissemination Using a Murine Intramuscular Model.
Mice infected with the given NF strain or mixture of strains i.m. were killed at 24 or 48 h p.i. Approximately 250 mg of muscle tissue around the injection site and the entire liver and spleen were placed in sterile PBS. Samples were homogenized, serially diluted, and aliquots plated on LB agar plates supplemented with appropriate antibiotic(s). After incubation for 24 h at 37 °C, colonies were counted to calculate bacterial load per organ or gram of muscle tissue.
In Vitro Imaging of Animals Infected with A. hydrophila Strains and Bioluminescent Bacterial Colonies on Agar Plates.
Animals were infected with either A. hydrophila NF1-lux or NF2-lux i.m. Whole-body imaging was performed at indicated time points by using a IVIS 200 bioluminescence and fluorescence whole-body imaging workstation (Caliper), with auto-background and other appropriate default settings. Pixel intensity was adjusted to achieve uniform bioluminescence measurement for all experiments. Tissue homogenates were serially diluted and plated on LB agar and colony bioluminescence measured by using the IVIS system.
Metagenomic Sequencing and Analysis.
Bacterial community DNA extracted from muscle, spleen, and liver tissue was sequenced by using a MiSeq benchtop sequencer (Illumina). The DNA was quantified with a Qubit 2.0 flourometer (Life Technologies), diluted to appropriate concentration, and prepared for sequencing by using the Nextera sample preparation kit (Illumina). Paired-end DNA libraries were sequenced for a total of 500 cycles in a multiplex format (6 samples per run, on average). FastQ files were converted to FastA format and directly analyzed by using GENIUS software package (CosmosID) for rapid identification of bacterial species and strains with estimations of their relative abundance (22, 23). Work flow details of GENIUS software are given in SI Materials and Methods.
Statistical Analysis.
Animal survival rates were analyzed by Kaplan–Meier survival estimate with Bonferroni post hoc test with GraphPad Prism version 6.0 software. Other data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey post hoc test with GraphPad Prism version 6.0 software. In addition, unpaired t test was used to compare data from two groups when appropriate. Wherever applicable, P values are reported, and a P value of ≤0.05 is considered significant.
Materials and Methods
Construction of Bioluminescent A. hydrophila NF2.
Initially, a spontaneous Rifr mutant of A. hydrophila strain NF2 was harvested from LB agar plates containing 200 μg/mL of Rif after incubation for 48–72 h at 37 °C. The Rfr strain was transconjugated with a mixture of two Escherichia coli strains, SM10 λpir carrying the pTNS2 plasmid, and SM10 λpir harboring the pUC18R6K-mini-Tn7T::Km-lux plasmid (7, 19) (Table S2). The latter plasmid carries a lux (luciferase) luminescence operon with its own cis-operating promoter and Apr selection marker. The Tn7-based plasmid integrates downstream of the glmS gene encoding glucosamine-6-phosphate synthase on the bacterial chromosome, with the help of a transposase complex encoded by the pTNS2 plasmid (Table S2).
Subsequently, Rfr and Kmr colonies were screened for bioluminescence using the ImageQuant LAS4000 imaging workstation (GE Healthcare Sciences). Approximately, 10–15 colonies emitting strong bioluminescence signal were tested for oxidase reaction (Aeromonas species are oxidase positive and E. coli strains are oxidase negative). Integration of Tn7 at the correct location within the host bacterium was confirmed by PCR amplification of a portion of the Tn7 transposon. Virulence of the bioluminescent strain, compared with wild-type (WT) NF2, was assessed in a mouse model of infection to confirm integration of the lux gene in bacterial genome did not alter its animal mortality pattern.
In Vitro Characterization of the Bacterial Strains.
Bacterial motility.
LB medium with 0.35% Difco Bacto-agar (Difco Laboratories) was used to characterize swimming motility, whereas Difco nutrient broth with 0.5% Eiken agar (Eiken Chemical Co., Ltd.) was used to measure swarming motility. Briefly, the bacterial cultures were grown in LB medium at 37 °C for 18 h with shaking (180 rpm), adjusted to the same optical density representing similar colony forming units (cfu), and stabbed into 0.35% LB agar plates. After overnight incubation at 37 °C, swimming motility was monitored as described (7). Growth from the edge of the swimming zone was again inoculated to assess swarming motility. The swarming plates were inoculated at the center on the surface of the agar plates. Motility was evaluated after incubation overnight at 30 °C (7).
RT-PCR.
Total RNA was isolated from bacterial cultures grown overnight in LB medium, as recommended by the manufacturer (Ambion) and subjected to RT-PCR by using SuperScript-III Platinum (Invitrogen). Amplified products (fleQ and vfr) were identified by size specific separation on agarose gel and specificity of the amplicons confirmed by DNA sequencing. The relative levels of cDNAs from RT-PCR were determined by densitometric analysis using AlphaEasyFC software (AlphaInnotech) and 16S rRNA gene as reference.
Phagocytic assay and intracellular survival in macrophage cell culture.
Murine RAW 264.7 macrophages were infected with overnight-grown A. hydrophila (NF1, NF2, or NF1-exoA) at a multiplicity of infection (moi) of 5. Infected macrophages were incubated at 37 °C with 5% (vol/vol) CO2 for 45 min, followed by gentamicin treatment (50 µg/mL) for 1 h to kill extracellular bacteria. The number of bacteria inside the macrophages was enumerated immediately after gentamicin treatment (0 h time point) and at 2 h by serial dilution and plating. The percent of phagocytosis and intracellular survival was calculated as number of bacteria at 0 h over initial infection dose, and number of bacteria at 2 h over number at 0 h, respectively.
In vitro growth dynamics of mixed NF strains.
Various overnight grown NF1 and NF2 strains were diluted in fresh LB broth and allowed further growth for 2 h at 37 °C with 180 rpm shaking. NF1 and NF2-lux strains or NF2 and NF1-lux strains were mixed at a ratio of 5:1 or 10:1, respectively, and either spotted on nonselective LB agar plates or grown in the liquid LB medium for 6 h at 37 °C. The growth of NF2-lux in the mixture of NF1/NF2-lux or the number of NF1-lux in the mixture of NF2/NF1-lux was determined by serial dilution and plating on the kanamycin plates.
Metagenomic Sequence Analysis via GENIUS Software.
GENIUS uses a high performance data-mining algorithm that rapidly disambiguates hundreds of millions of short reads of a metagenomic sample into the discrete microorganisms engendering the particular sequences (22, 23). The pipeline has two separable comparators. The first consists of a precomputation phase and a per-sample computation. The input to the precomputation phase is a curated reference microbial database, and its output is a whole genome phylogeny tree, together with sets of fixed length k-mer fingerprints (biomarkers) that are uniquely identified with distinct nodes and leaves of the tree. The second per-sample, computational phase searches the hundreds of millions of short sequence reads against the fingerprint sets in minutes. The resulting statistics are analyzed to give fine-grain composition and relative abundance estimates at all nodes of the tree. Overall classification precision is maintained through aggregation statistics.
Acknowledgments
Financial support was provided to A.K.C. through Leon Bromberg and Robert E. Shope and John S. Dunn Distinguished Chair in Global Health endowments, University of Texas Medical Branch, and NIH Grant 2RO1A1039129 was awarded to R.R.C. D.P. was supported in part by the James W. McLaughlin Postdoctoral Fellowship. B.L.T. and J.A.A. were supported in part by the WHO Collaborating Center for Vaccine Development, UTMB. E.C.F. was supported in part by T32 Biodefense Training Grant AI060549.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523817113/-/DCSupplemental.
References
- 1.Roje Z, et al. Necrotizing fasciitis: Literature review of contemporary strategies for diagnosing and management with three case reports: Torso, abdominal wall, upper and lower limbs. World J Emerg Surg. 2011;6(1):46. doi: 10.1186/1749-7922-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarani B, Strong M, Pascual J, Schwab CW. Necrotizing fasciitis: Current concepts and review of the literature. J Am Coll Surg. 2009;208(2):279–288. doi: 10.1016/j.jamcollsurg.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 3.Liu SYW, Ng SSM, Lee JFY. Multi-limb necrotizing fasciitis in a patient with rectal cancer. World J Gastroenterol. 2006;12(32):5256–5258. doi: 10.3748/wjg.v12.i32.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minnaganti VR, Patel PJ, Iancu D, Schoch PE, Cunha BA. Necrotizing fasciitis caused by Aeromonas hydrophila. Heart Lung. 2000;29(4):306–308. doi: 10.1067/mhl.2000.106723. [DOI] [PubMed] [Google Scholar]
- 5.Gold WL, Salit IE. Aeromonas hydrophila infections of skin and soft tissue: Report of 11 cases and review. Clin Infect Dis. 1993;16(1):69–74. doi: 10.1093/clinids/16.1.69. [DOI] [PubMed] [Google Scholar]
- 6.Borger van der Burg BL, Bronkhorst MW, Pahlplatz PV. Aeromonas hydrophila necrotizing fasciitis. A case report. J Bone Joint Surg Am. 2006;88(6):1357–1360. doi: 10.2106/JBJS.C.00923. [DOI] [PubMed] [Google Scholar]
- 7.Grim CJ, et al. Functional genomic characterization of virulence factors from necrotizing fasciitis-causing strains of Aeromonas hydrophila. Appl Environ Microbiol. 2014;80(14):4162–4183. doi: 10.1128/AEM.00486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furusu A, et al. Aeromonas hydrophila necrotizing fasciitis and gas gangrene in a diabetic patient on haemodialysis. Nephrol Dial Transplant. 1997;12(8):1730–1734. doi: 10.1093/ndt/12.8.1730. [DOI] [PubMed] [Google Scholar]
- 9.Heckerling PS, Stine TM, Pottage JC, Jr, Levin S, Harris AA. Aeromonas hydrophila myonecrosis and gas gangrene in a nonimmunocompromised host. Arch Intern Med. 1983;143(10):2005–2007. [PubMed] [Google Scholar]
- 10.Schümann J, Angermüller S, Bang R, Lohoff M, Tiegs G. Acute hepatotoxicity of Pseudomonas aeruginosa exotoxin A in mice depends on T cells and TNF. J Immunol. 1998;161(10):5745–5754. [PubMed] [Google Scholar]
- 11.Zdanovsky AG, Chiron M, Pastan I, FitzGerald DJ. Mechanism of action of Pseudomonas exotoxin. Identification of a rate-limiting step. J Biol Chem. 1993;268(29):21791–21799. [PubMed] [Google Scholar]
- 12.Yates SP, et al. Structure-function analysis of water-soluble inhibitors of the catalytic domain of exotoxin A from Pseudomonas aeruginosa. Biochem J. 2005;385(Pt 3):667–675. doi: 10.1042/BJ20041480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosser T, et al. 2015. Exposure to pairs of Aeromonas strains enhances virulence in the Caenorhabditis elegans infection model. Front Microbiol 6(29):1218 10.3389/fmicb.2015.01218.
- 14.Mastropaolo MD, et al. Synergy in polymicrobial infections in a mouse model of type 2 diabetes. Infect Immun. 2005;73(9):6055–6063. doi: 10.1128/IAI.73.9.6055-6063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suarez G, et al. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb Pathog. 2008;44(4):344–361. doi: 10.1016/j.micpath.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutterwala FS, et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204(13):3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith KD, et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4(12):1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 18.Ramos HC, Rumbo M, Sirard JC. Bacterial flagellins: Mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 2004;12(11):509–517. doi: 10.1016/j.tim.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Choi K-H, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat Protoc. 2006;1(1):153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 20.Khajanchi BK, et al. Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: Suggestive evidence of water-to-human transmission. Appl Environ Microbiol. 2010;76(7):2313–2325. doi: 10.1128/AEM.02535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y, Xu T, Fossheim LE, Zhang XH. FliC, a flagellin protein, is essential for the growth and virulence of fish pathogen Edwardsiella tarda. PLoS One. 2012;7(9):e45070. doi: 10.1371/journal.pone.0045070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lax S, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345(6200):1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasan NA, et al. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS One. 2014;9(5):e97699. doi: 10.1371/journal.pone.0097699. [DOI] [PMC free article] [PubMed] [Google Scholar]