Significance
Discovery of novel metastasis suppressor genes in breast cancer using genomic efforts has been limited, potentially due to overlooking their regulation by epigenetic mechanisms. We report the discovery of SDPR as a novel metastasis suppressor gene localized to 2q32-33, a region associated with significant loss of heterozygosity in breast cancer, using comparative gene expression analysis of a breast cancer progression model system in conjunction with in silico metaanalysis of publicly available datasets. SDPR is silenced epigenetically by promoter DNA methylation and its loss of expression correlates with significantly reduced distant-metastasis–free and relapse-free survival of breast cancer patients. Overexpression of SDPR reduces cell migration and intravasation/extravasation potential, favors cell death, and suppresses experimental lung metastasis of breast cancer cells.
Keywords: metastasis, breast cancer, SDPR, epigenetics, metastasis suppressor
Abstract
Metastatic dissemination of breast cancer cells represents a significant clinical obstacle to curative therapy. The loss of function of metastasis suppressor genes is a major rate-limiting step in breast cancer progression that prevents the formation of new colonies at distal sites. However, the discovery of new metastasis suppressor genes in breast cancer using genomic efforts has been slow, potentially due to their primary regulation by epigenetic mechanisms. Here, we report the use of model cell lines with the same genetic lineage for the identification of a novel metastasis suppressor gene, serum deprivation response (SDPR), localized to 2q32-33, a region reported to be associated with significant loss of heterozygosity in breast cancer. In silico metaanalysis of publicly available gene expression datasets suggests that the loss of expression of SDPR correlates with significantly reduced distant-metastasis–free and relapse-free survival of breast cancer patients who underwent therapy. Furthermore, we found that stable SDPR overexpression in highly metastatic breast cancer model cell lines inhibited prosurvival pathways, shifted the balance of Bcl-2 family proteins in favor of apoptosis, and decreased migration and intravasation/extravasation potential, with a corresponding drastic suppression of metastatic nodule formation in the lungs of NOD/SCID mice. Moreover, SDPR expression is silenced by promoter DNA methylation, and as such it exemplifies epigenetic regulation of metastatic breast cancer progression. These observations highlight SDPR as a potential prognostic biomarker and a target for future therapeutic applications.
The metastatic progression of breast cancer accounts for the majority of disease-related mortality. A major rate-limiting step in metastasis is the loss of function of the metastasis suppressor genes, which block a cascade of crucial steps including the loss of adhesion of primary tumor cells, intravasation into the blood and lymphatics with subsequent extravasation at distant sites, and the formation of new colonies. Despite the identification of the first metastasis suppressor gene, nonmetastatic 23 (NM23), nearly two decades ago (1), only a handful of new metastasis suppressors have been identified in recent years using candidate gene approaches (2, 3). It is likely that the current catalog of metastasis suppressor genes remains incomplete despite the vast sequencing efforts due to the possibility that a subset of genes regulated by epigenetic mechanisms may have eluded traditional discovery procedures (4–6). To identify these elusive metastasis suppressor genes, which are functionally compromised in late-stage disease (7–9), we took advantage of a well-established breast cancer progression cell line model system sharing the same genetic linage (Fig. 1A) (10). This model system consists of five cell lines that represent the various stages of breast cancer progression based on the MCF10A cell line: MCF10AneoT (NeoT), MCF10AT1Kcl2 (MII), MCF10CA1h (MIII), and MCF10CA1a (MIV). NeoT cells were generated by overexpression of HRAS in MCF10A cells and rarely exhibit growth following injection into nude mice. MII cells were generated by single xenograft passaging of NeoT cells. When injected subcutaneously (s.c.) into nude mice, MII cells generally form benign tumors that progress to carcinoma one out of four times; hence they mimic the early stage, carcinoma in situ. MIII and MIV cells were isolated from tumors formed by MII cells. MIII cells represent carcinoma, as in general they metastasize at a very low frequency, which requires a prolonged incubation period. On the other hand, MIV cells have the potential to readily seed lung metastases and represent the final stages of a breast cancer, metastatic carcinoma. We compared the gene expression profiles of these latter three model cell lines and leveraged large amounts of publically available breast tumor gene expression profiling data (11–13) by applying multiple bioinformatics filters to identify candidate metastasis suppressor genes.
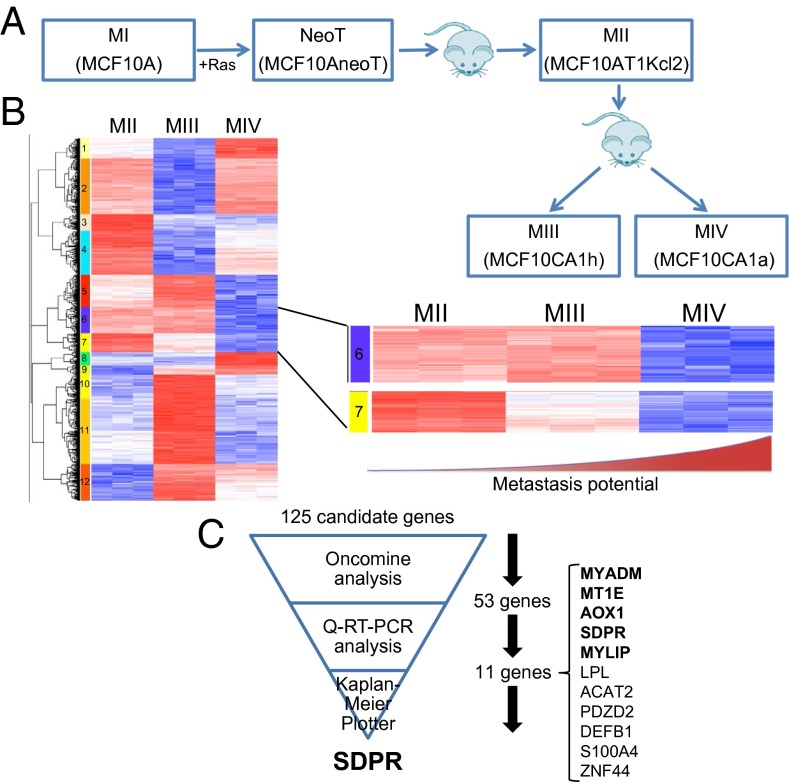
Fig. 1.
Identification of SDPR as a candidate metastasis suppressor gene. (A) Schematic depiction of the generation of breast cancer progression cell line model system. The model consists of five cell lines representing different stages of breast cancer progression. MI, normal breast epithelial cells; NeoT and MII, carcinoma in situ; MIII, carcinoma; and MIV, metastatic carcinoma. (B) Hierarchical clustering of gene expression profiles from MII, MIII, and MIV cells for the genes whose expression differ at least twofold between each cell line. Two clusters, cluster 6 and 7, are magnified because expressions of the genes in these two clusters are significantly suppressed in metastatic MIV cells compared with nonmetastatic MII and MIII. (C) The schematic summary of our strategy for the selection of SDPR as the top candidate metastasis suppressor.
Here, we report the discovery of the phosphatidylserine-interacting protein, serum deprivation response (SDPR) (also known as cavin-2), as a bona fide metastasis suppressor. Thus far, studies on SDPR function have been limited to its role as a regulator of caveolae formation (14), and its potential direct involvement in cancer has not been previously described. However, it has been reported that SDPR expression is down-regulated significantly in not only breast cancer but also in cancers of kidney and prostate (15). Moreover, SDPR protein down-regulation was observed in serum from patients with malignant kidney tumors, and hence it was suggested as a possible diagnostic marker to discriminate malignant tumors from benign formations (16). Interestingly, SDPR is localized to 2q32-33, a region with a significant level of loss of heterozygosity that is associated with a high degree of recurrence in breast cancer (17, 18). Our results indicate that SDPR is capable of specifically inhibiting the metastatic growth of breast cancer cells.
Results
SDPR Is Significantly Down-Regulated During Breast Cancer Progression.
To identify potential metastasis suppressor genes, we examined the gene expression profiles of MII, MIII, and MIV model cell lines (Fig. 1A) and focused on the genes down-regulated in metastatic MIV cells, relative to nonmetastatic MII and MIII cells (Fig. 1B and Dataset S1). Hierarchical clustering across these three cell lines revealed two clusters, clusters 6 (70 genes) and 7 (55 genes) in which the genes were specifically repressed in the metastatic MIV cells (Fig. 1B). Although, in cluster 6, gene expression was at a comparable level in MII and MIII and repressed in metastatic MIV, in cluster 7, the gene expression levels were high in MII, moderate in MIII, and low in MIV. Overall, the expression pattern of these 125 genes was inversely correlated with the metastatic potential of these model cell lines. Therefore, we hypothesized that these clusters consist of metastasis suppressor genes.
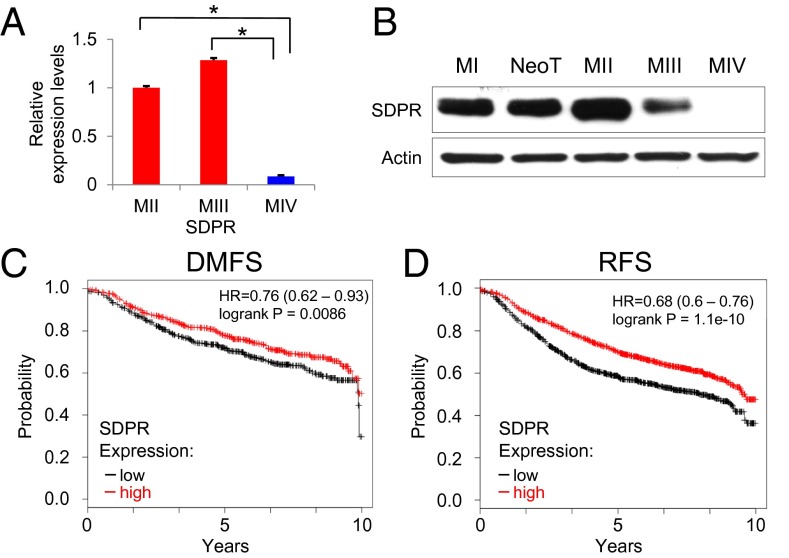
Because the two clusters of interest contained a total of 125 genes, we used a filtering strategy to select the most promising metastasis suppressors (Fig. 1C). First, we interrogated each gene by accessing the Oncomine database, which revealed that 53 out of the 125 genes were down-regulated in cancers compared with control tissues (SI Appendix, Table S1) (13). Because Oncomine analysis incorporates independent gene expression studies that used clinical samples from patients, it gave us the confidence that the results we obtained from hierarchical clustering of gene expression profiles of model cell lines is likely to be representative of the different stages of human breast cancer progression. Next, we confirmed our microarray results by quantitative RT-PCR. The expected expression pattern, the loss of expression in metastasis, was observed for 23 out of 53 genes (SI Appendix, Fig. S1). We further eliminated 12 of these 23 genes by setting a stringent criterion of at least a threefold change in gene expression between each model cell line. This resulted with 11 candidates (Fig. 1C and SI Appendix, Fig. S2). Finally, we used in silico Kaplan–Meier analysis to generate relapse-free survival curves based on the expression level of each gene (SI Appendix, Fig. S3) (11). At this point, SDPR started to emerge as a promising candidate metastasis suppressor gene, significantly associated with low level of expression in tumors based on Oncomine analyses (SI Appendix, Fig. S4) (13). SDPR was clearly suppressed in the metastatic MIV cell line at both transcript and protein levels (Fig. 2 A and B). Importantly, Kaplan–Meier plotter analysis also revealed that the degrees of distant-metastasis–free survival (DMFS) and relapse-free survival (RFS) were significantly decreased in patients with lower levels of SDPR expression (Fig. 2 C and D). Taken together, these data enabled us to hypothesize that SDPR is likely to be a metastasis suppressor gene in breast cancer.
Fig. 2.
Expression analysis of SDPR in clinical samples and model cell lines. (A) SDPR mRNA levels in metastatic MIV cells compared with nonmetastatic MII (P = 0.00047) and MIII (P = 0.0005) cells. (B) Endogenous SDPR protein levels in the model cell lines were assessed by Western blot. (C) In silico Kaplan–Meier analysis depicting the association between SDPR expression and distant-metastasis–free survival (DMFS). The analysis was run on a cohort with 1,211 breast cancer patients, P = 0.0086. (D) In silico Kaplan–Meier analysis depicting the association between SDPR expression and relapse-free survival (RFS). The analysis was run on a cohort with 2,785 breast cancer patients, P = 1.1e-10. *P < 0.05.
SDPR Suppresses Metastatic Potential of Breast Cancer Cells.
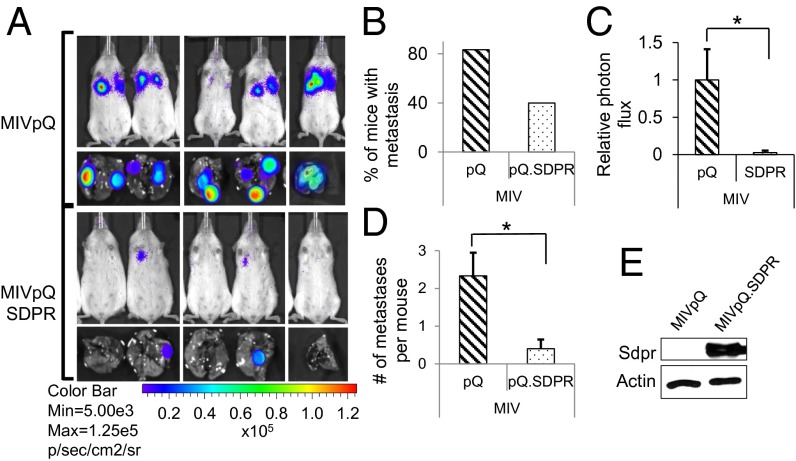
To test whether SDPR could function as a metastasis suppressor, we generated MIV cells with stable expression of SDPR (SI Appendix, Fig. S5). Following tail vein injections of MIV cells, we observed that SDPR overexpression caused a 52% reduction in the number of mice exhibiting lung metastases (Fig. 3 A and B). The significant decrease in metastatic burden on the mice injected with MIVpQ.SDPR cells was also clearly evident in the relative photon flux measurements (Fig. 3C). In addition, the number of macrometastatic nodules per mouse decreased from 2.3 to 0.4 (Fig. 3D and SI Appendix, Fig. S6). This effect was apparently specific to the metastatic potential of MIV cells because SDPR did not significantly affect the growth of these cells as primary tumors following s.c. injections (SI Appendix, Fig. S7). To determine whether SDPR can exert a similar effect as a metastasis suppressor in a different metastatic breast cancer cell line, we overexpressed SDPR in MDA-MB-231LM2 (LM2) cells (SI Appendix, Fig. S8) (19). We found that SDPR overexpression in LM2 cells caused a 60% reduction in the number of mice exhibiting lung metastases, with corresponding significant decreases in the relative photon flux as well as the number of macrometastatic nodules per mouse, from 2.8 to 0.8 (SI Appendix, Fig. S9 A–D). Overall, these observations were consistent with the function of SDPR as a metastasis suppressor in breast cancer.
Fig. 3.
SDPR suppresses lung colonization of breast cancer. (A) Bioluminescent imaging of animals 77 d after tail vein injections with 5 × 105 control, MIVpQ, or MIVpQ.SDPR cells. (B) The percentage of animals that developed lung metastases following tail vein injections is shown. (C) Quantification of metastases burden on mice was estimated by photon flux measurement, P = 0.026. (D) The average number of lung macrometastases observed per animal, P = 0.012. (E) SDPR overexpression was assessed by Western blotting. *P < 0.05.
SDPR Expression Leads to Decreased Migration and Increased Apoptosis.
To elucidate the mechanism of SDPR action, we examined the effects of SDPR on the critical regulators of various cellular functions including proliferation, epithelial-to-mesenchymal transition (EMT), migration, and apoptosis. SDPR expression did not alter the overall cell proliferation rate of MIV cells (SI Appendix, Fig. S10). The effect of SDPR overexpression on the levels of a known metastasis suppressor, NME1, matrix metalloproteinases (MMPs), and tissue inhibitors of metalloproteinases (TIMPs) were variable (SI Appendix, Fig. S11A). Interestingly, SDPR overexpression promoted epithelial features based on the changes in EMT and tight junction protein markers, but it did not alter expression of known EMT transcription factors like SNAIL in a consistent manner (SI Appendix, Fig. S11 A and B). Because these results hinted at the potential of SDPR to block migration and intravasation/extravasation of metastatic cancer cells, we carried out various migration assays. First, we found that SDPR expression inhibited the rate of wound closure in a scratch wound-healing assay (SI Appendix, Fig. S12A) as well as inhibited cell migration through a membrane in the Boyden chamber assay (SI Appendix, Fig. S12B). In addition, we observed that the number of MIVpQ.SDPR cells that migrated through an endothelial cell layer decreased markedly, by 38.1%, compared with control cells when we performed transendothelial migration assays mimicking in vitro the conditions of intravasation/extravasation (Fig. 4A). Similarly, the number of LM2 cells that migrated through an endothelial cell layer was even more significantly reduced (by 92.8%) (Fig. 4B). Overall, these observations suggest that SDPR expression hinders the migration and intravasation/extravasation potentials of metastatic breast cancer cells.
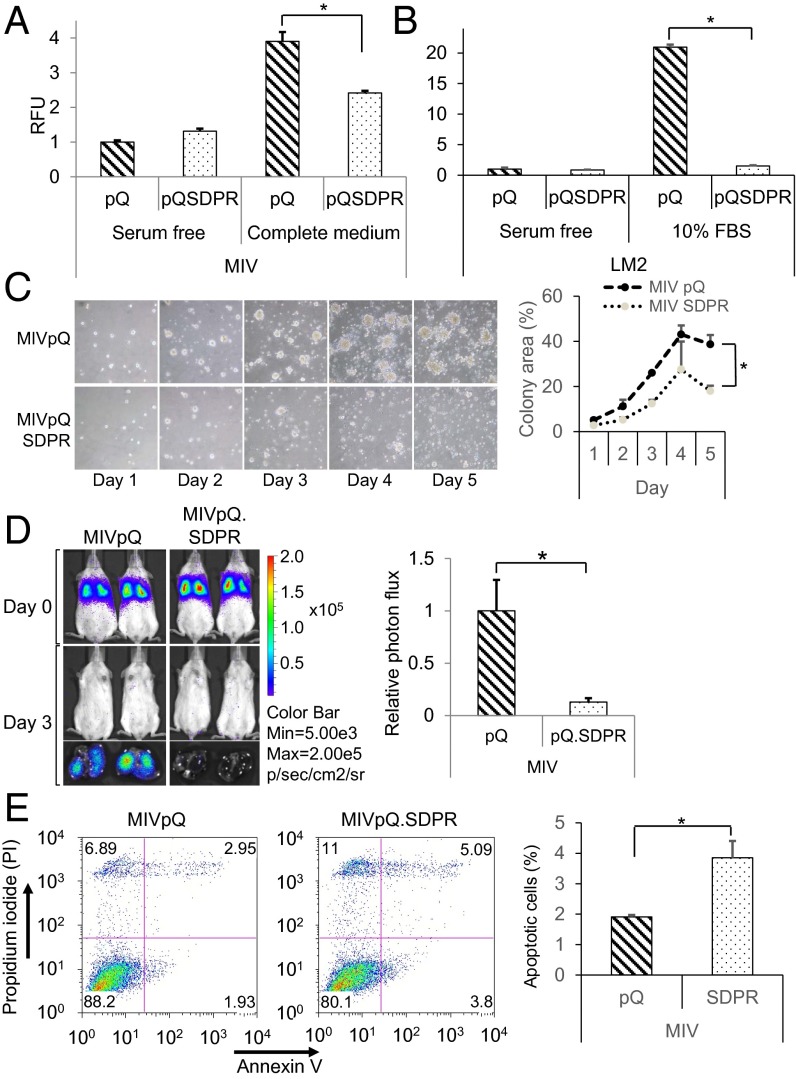
Fig. 4.
SDPR primes MIV cells for apoptosis and inhibits extravasation. (A) Transendothelial cell migration potential of the control and MIVpQ.SDPR cells toward serum-free or complete media was assessed 48 h after the seeding by calcein staining, P = 0.0374. RFU, relative florescence unit. (B) Effect of SDPR on the extravasation potential of LM2 cells was quantified, P = 7.87479E-07. (C) Growth of control and MIVpQ.SDPR cells were monitored over time in 3D cell culture and quantified, on the Right, by measuring colony area, P = 0.01. (D) Effect of SDPR overexpression on survivability of MIV cells was monitored 72 h after the tail vein injections by Caliper IVIS Spectrum. Whole-animal imaging is presented on the Upper Left, and extracted lungs are shown on Lower Left. Quantification of cell survivability was assessed on the Right, based on photon flux, P = 0.0014, npQ = 5, npQSDPR = 8. (E) Annexin V and PI staining were used to assess the basal level of apoptosis in control and MIVpQ.SDPR cells. Quantification of three independent Annexin V experiments is shown, P = 0.04. *P < 0.05.
We also investigated the effect of SDPR overexpression in 3D cell culture, as a possible indicator of survival potential as well as ability to form colonies at distant sites during metastasis (7, 20, 21). When MIV cells were grown in 3D cell culture, SDPR expression caused a significant decrease in the size of the colonies growing in aggregates (Fig. 4C). Consistent with these observations in MIV cells, SDPR overexpression also rendered LM2 cells with decreased ability to grow in aggregates in 3D culture (SI Appendix, Fig. S13). We infer that these observations suggest that SDPR overexpression mediates a decrease in the ability of MIV and LM2 cells to seed and proliferate at distal sites, blocking lung colonization.
We hypothesized that the significant decrease in metastatic potential of MIVpQ.SDPR cells could be explained by a possible decline in survivability of these cells in the lung microenvironment. To test this hypothesis, we performed tail vein injections and assessed cell survival after 72 h. We found that there was a significant decrease in the number of surviving MIVpQ.SDPR cells compared with control cells (Fig. 4D and SI Appendix, Fig. S14). This observation suggested that SDPR overexpression rendered breast cancer cells with a significantly decreased adaptability to survive in the lung microenvironment, potentially due to promotion of apoptosis. These observations were consistent with a significant increase (∼96%) in the basal level of apoptosis (from 1.93% to 3.8%) as assessed by Annexin/propidium iodide (PI) staining upon overexpression of SDPR in MIV cells (Fig. 4E).
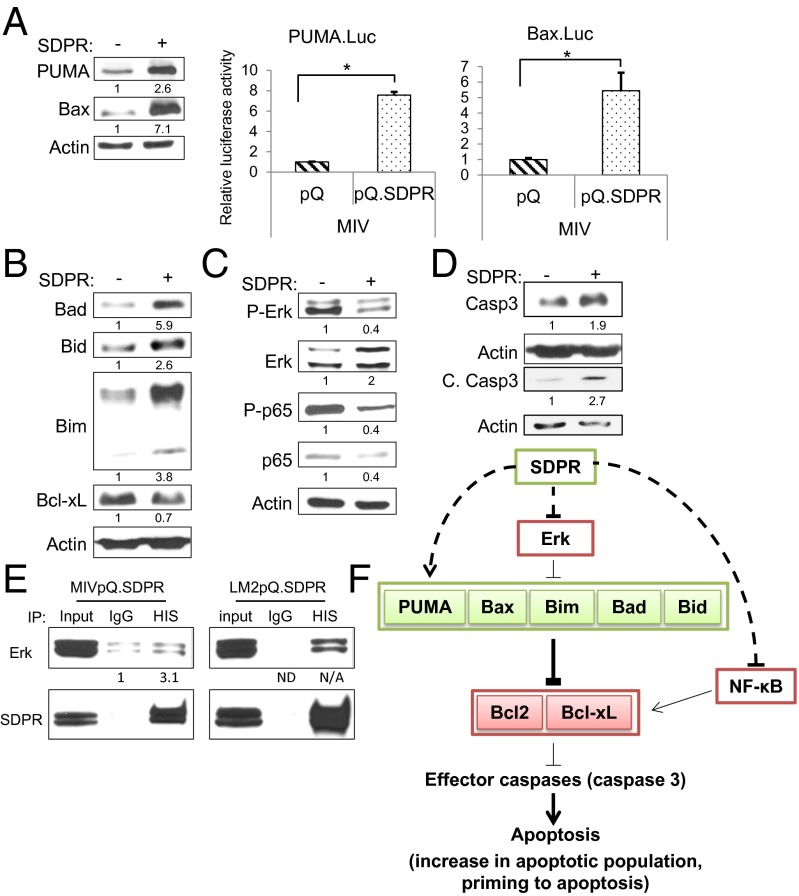
To determine the molecular basis for the increase in apoptosis, we examined the B-cell lymphoma-2 (BCL-2) family of proteins that play crucial roles in the apoptotic pathway. Consistent with the increase in apoptosis, we found that proapoptotic, PUMA and Bax expressions were induced in MIVpQ.SDPR cells (Fig. 5A). We also tested the promoter activity of these genes using luciferase reporters and observed that they were significantly induced in MIVpQ.SDPR cells (Fig. 5A). Interestingly, although the proapoptotic Bcl-2 family proteins Bad, Bid, and Bim (22) were also expressed at higher levels in MIVpQ.SDPR cells (Fig. 5B), there was a decrease in the antiapoptotic Bcl-xL protein. The ERK and NF-κB pathways are major regulators of Bcl2 family members, controlling the activities of Bim (23) and Bcl-xL (24), respectively. In MIVpQ.SDPR cells, we observed a decrease in ERK and p65 phosphorylation (Fig. 5C), suggesting that SDPR can restrain the activity of these antiapoptotic pathways. In conjunction with these observations, the levels of caspase 3, an effector caspase, and its cleaved product were also increased in MIVpQ.SDPR cells, indicating the activation of caspase-3 (Fig. 5D). Collectively, these observations pointed to a shift in the balance of apoptotic regulators that favor cell death in the presence of SDPR.
Fig. 5.
SDPR function and apoptosis. (A) The effect of SDPR overexpression on the proapoptotic PUMA and Bax expression in MIV cells was evaluated by Western blotting and luciferase reporter assays, pPUMA = 0.000003, pBax = 0.03 for all n = 3. (B) Protein levels of proapoptotic Bcl2 family members, Bad, Bid, and Bim, and antiapoptotic Bcl-xL were measured by Western blotting in control and MIpQ.SDPR cells. (C) The effect of SDPR overexpression on the activity of ERK and NF-κB pathways was evaluated by Western blotting against phosphorylated Erk and p65 protein levels, respectively. (D) Total and cleaved caspase-3 protein levels in control and MIVpQ.SDPR cells were measured by Western blotting. (E) Co-IP was carried out in MIV and LM2 cells using HIS (mouse) antibody to precipitate SDPR and mouse IgG as control. Western blot was used to assess the levels of precipitated SDPR and coprecipitated Erk. (F) SDPR overexpression, directly or indirectly, causes increases in proapoptotic BCL-2 family proteins. Additionally, the levels of the antiapoptotic protein Bcl-xL and the activity of prosurvival Erk and NF-κB signaling pathways were decreased due to SDPR overexpression. Overall, SDPR can alter the balance of regulatory proteins in the apoptotic pathway to favor cell death. *P < 0.05.
The proapoptotic effect mediated by SDPR was not limited to MIV cells. SDPR overexpression significantly increased the basal level of apoptosis in LM2 cells (from ∼0.6% to 14.5%) as well (SI Appendix, Fig. S15). Furthermore, SDPR overexpression also caused a dramatic increase in proapoptotic Bcl-2 family proteins, accompanied by a decrease in antiapoptotic Bcl-xL (SI Appendix, Fig. S16 A and B). Moreover, perturbations of ERK and NF-κB signaling, together with increased caspase-3 levels, were also observed in LM2pQ.SDPR cells (SI Appendix, Fig. S16 C and D). Additionally, following loss of adhesion, there was also a corresponding drastic increase in cleaved PARP levels, further supporting apoptotic cell death in LM2pQ.SDPR cells (SI Appendix, Fig. S17).
To investigate whether SDPR could affect these proapoptotic and antiapoptotic regulators by directly interacting with them, we performed coimmunoprecipitation (Co-IP) assays in MIV and LM2 cells. We found that Erk interacted with SDPR in both MIV and LM2 cell lines (Fig. 5E). These data suggest that SDPR can interact with Erk and possibly inhibit its activation, thereby affecting downstream targets and ultimately the cell fate during the metastatic cascade (Fig. 5F).
It is reported that SDPR can bind to phosphatidylserine (PS) (17). To investigate whether the proapoptotic role of SDPR was related to this interaction, we performed affinity assays between SDPR and PS. Both MIVpQSDPR and LM2pQSDPR cells showed increased levels of cleaved PARP protein when they were grown as tumorspheres compared with adherent cells, indicating an increase in apoptosis (SI Appendix, Fig. S18 A and B). Therefore, we compared SDPR–PS interaction levels between adherent and tumorsphere cells by using PS beads. In MIVpQSDPR cells, the PS–SDPR interaction was not significantly different between adherent and tumorsphere cells (SI Appendix, Fig. S18C). However, with LM2pQSDPR cells, when cells were undergoing apoptosis, the PS–SDPR interaction was markedly enhanced (SI Appendix, Fig. S18D). This result may indicate that, in LM2 cells, SDPR can shift its cellular localization and regulate other proteins in the vicinity during apoptosis.
Next, we examined whether loss of SDPR was sufficient to induce prosurvival signaling by performing loss-of-function experiments in NeoT cells, the nonmetastatic precursor of the metastatic MIV cells. SDPR knockdown in NeoT cells (SI Appendix, Fig. S19) caused a significant increase in surviving cell population (Annexin V−/PI−) from 83% to 88% (SI Appendix, Fig. S20A). This was consistent with the observed decreased levels of PUMA and Bax proteins and their corresponding transcriptional activation (SI Appendix, Fig. S20B). Although the proapoptotic Bcl-2 family members, Bad and Bim, as well as caspase-3 levels were all decreased following SDPR knockdown, ERK phosphorylation was increased (SI Appendix, Fig. S21 A–C). Furthermore, NeoTshSDPR cells exhibited enhanced cell growth potential in aggregates in 3D cell culture (SI Appendix, Fig. S21D). Thus, these observations support the notion that loss of SDPR expression is sufficient to alter critical proapoptotic and antiapoptotic regulators.
Overall, our studies found that SDPR acts as a crucial regulator that blocks metastasis in breast cancer, not only through the inhibition of tumor cell migration, intravasation/extravasation, and self-renewal, but also by promoting apoptosis.
SDPR Expression Is Lost in a Wide Variety of Cancers.
We found that SDPR loss was not limited to breast cancer, as tumor samples from bladder, colorectal, lung, pancreatic, and ovarian cancers as well as sarcomas also exhibited loss of SDPR expression from Oncomine analysis (SI Appendix, Table S2) (13). Furthermore, SDPR expression was significantly reduced in metastatic prostate cancer (SI Appendix, Fig. S22) (25). Moreover, higher SDPR transcript levels were significantly associated with increased chance of overall survival in lung cancer patients (SI Appendix, Fig. S23) (26). In summary, these observations suggest that the role of SDPR as a metastasis suppressor may have broader clinical relevance beyond breast cancer.
SDPR Is Epigenetically Silenced During Metastatic Cancer Progression.
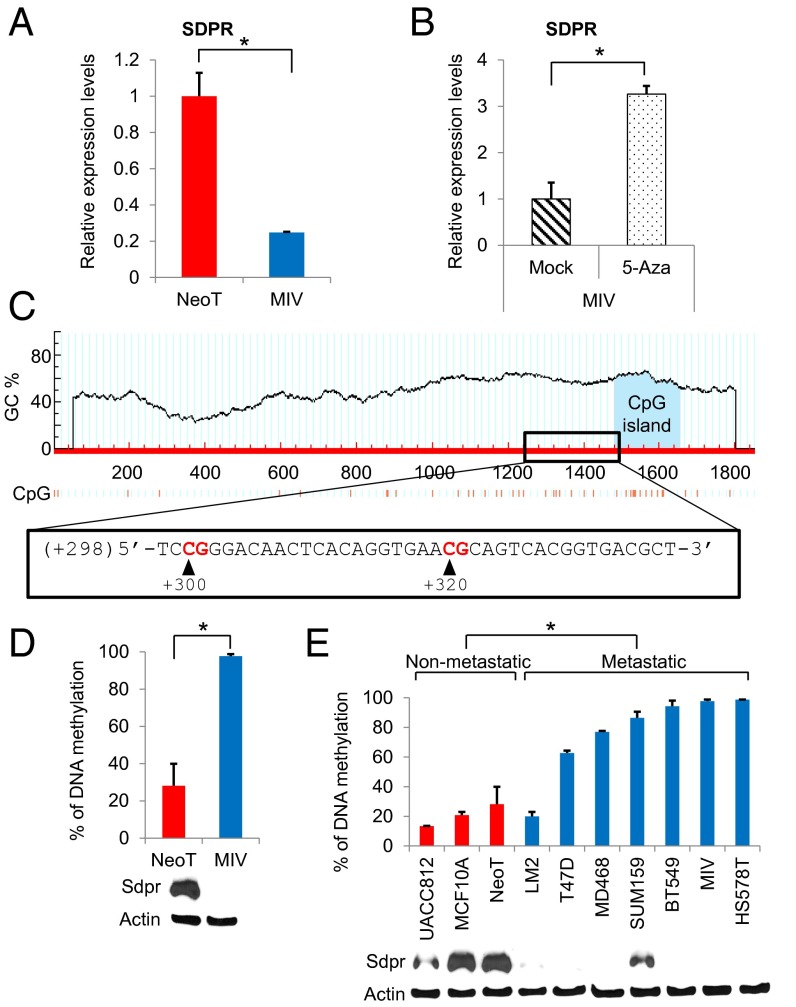
The fact that SDPR failed to emerge as a frequent target for mutational inactivation in the recent high-throughput next-generation sequencing efforts suggested that it is likely to be inactivated by epigenetic mechanisms (4–6). Therefore, we investigated the effect of 5-aza-2′-deoxycytidine (5-aza) treatment on SDPR expression in MIV cells. Indeed, the exposure to 5-aza caused a significant increase in the transcript level of SDPR and restored it to a comparable level to what was observed in nonmetastatic NeoT cells (Fig. 6 A and B). We followed up these observations by analyzing the promoter region of SDPR using the MethPrimer software to predict the likely location of CpG sites targeted for methylation (27). A GC percentage graph plotted by MethPrimer was used to design methylation-specific primers targeting the CpG sites at +300 and +320 positions of the CpG island shore (Fig. 6C). Quantitative methylation-specific PCR analysis revealed that the SDPR promoter region is significantly hypermethylated in metastatic MIV cells compared with the nonmetastatic NeoT cells (Fig. 6D). The significant hypermethylation of CpG sites at +300 and +320, along with suppression of SDPR expression, was observed in the majority of the metastatic breast cancer cell lines tested (Fig. 6E). As expected, the degree of DNA methylation was inversely correlated with the SDPR protein expression. Furthermore, loss of SDPR expression at the level of protein was observed in breast cancers as reported in a comprehensive antibody-based proteomics study of human tumors in the Human Protein Atlas (SI Appendix, Fig. S24) (12, 15, 28). When we treated metastatic breast cancer cell lines with 5-aza, we observed a significant growth inhibition in five out of six cell lines (SI Appendix, Fig. S25). Overall, these findings suggest that, similar to other metastasis suppressors (29–32), SDPR is epigenetically silenced due to DNA hypermethylation in metastatic breast cancer cells.
Fig. 6.
Epigenetic regulation of SDPR expression. (A) The relative expression levels of SDPR in NeoT and MIV cells was measured by quantitative RT-PCR, P = 0.0127, n = 3. (B) Effect of 5-Aza treatment on SDPR mRNA levels in MIV cells, P = 0.02, n = 3. (C) Analysis of −1000 to +1000 region of SDPR transcription start site for CpG sites using the MethPrimer online tool. (D) Methylation-specific quantitative PCR was used to assess the percentage of DNA methylation at the SDPR promoter region in NeoT and MIV cell lines, P = 0.0116, n = 3. SDPR protein levels are depicted below the graph. (E) The percentage of DNA methylation at the SDPR promoter region across nonmetastatic and metastatic breast cancer cell lines, P = 0.0007. SDPR protein levels are depicted below the graph. *P < 0.05.
Discussion
There are only a few metastasis suppressors that are specifically believed to have roles in breast cancer, and their contributions to the metastatic process are still being worked out (2, 3, 33, 34). Despite the advent of advanced technologies for mutational analysis, success in revealing major differences between metastatic lesions and primary tumors has been limited (4, 35). Thus, it is crucial to understand how cancer cells adapt to new microenvironments, a context in which epigenetic rather than genetic mechanisms of gene regulation may play a major role in acquiring metastatic properties.
Here, we report the successful exploitation of a breast cancer progression model system to identify a novel metastasis suppressor gene, SDPR. The strength of this model lies on its development from a single immortalized parental cell line, MCF10A, and the existence of derivatives representing premalignant, malignant, and metastatic carcinoma stages of breast cancer. Our findings ascribe that SDPR could play a previously unrecognized significant role in breast cancer progression as a bona fide metastasis suppressor gene, based on its loss of function aiding in the removal of major barriers to the metastatic cascade by promoting the loss of adhesion of primary tumor cells, intravasation into the blood and lymphatics, and subsequent extravasation and colonization at distal sites. Additionally, we want to note that, although our focus was on the metastasis suppressors, specifically SDPR in this report, the gene expression profiling data generated in this study also uncovered other potentially critical genes that could function as prometastatic oncogenes [clusters 8 and 9 in Fig. 1B (36)].
Upon arrival in the lung microenvironment, even 3 days after tail vein injections, the survival advantage provided by the loss of SDPR expression in the metastasizing breast cancer cells was significant compared with the cells overexpressing SDPR. These observations prompted us to examine the molecular basis underlying the function of SDPR as a metastasis suppressor. We found that there was increased expression of multidomain proapoptotic proteins such as Bax as well as BH3-only proteins such as PUMA, Bad, Bid, and Bim, with a corresponding decrease in prosurvival proteins such as Bcl-xL as well as an increase in cleaved caspase-3 levels indicating activation of Casp3 (Fig. 5). Additionally, the fact that SDPR interacts with Erk and inhibits prosurvival ERK and NF-κB signaling pathways is also consistent with promotion of apoptosis (Fig. 5F) (23, 24).
We found that SDPR was suppressed not only in breast cancer but also in other types of cancers, suggesting the exciting possibility that the functional role of SDPR as a metastasis suppressor is not likely to be limited to breast cancer (25, 26). Furthermore, our studies also suggested that silencing of SDPR expression due to DNA hypermethylation could be a key mechanism for its loss of function during metastatic breast cancer progression. Previous studies also found that the metastasis suppressors, CDH1 and CASP-8, are silenced due to promoter DNA hypermethylation (29, 30). Moreover, the expression of the metastasis suppressors DRG1 and NME1 were also found to dramatically increase upon 5-aza treatment (31, 32). These findings suggest that DNA methylation is an important mechanism for the regulation of metastasis suppressor genes. However, interestingly, although LM2 cells exhibited low levels of SDPR promoter DNA methylation but reduced expression, the SUM159 cells harbored a high degree of methylation accompanied with relatively strong SDPR protein expression. These observations indicate that, although methylation of the promoter seems to be the predominant mechanism for the majority of the cell lines we tested, it may not be the only epigenetic mechanism for SDPR regulation in cancers. Additionally, one cannot exclude other modes of regulations such as those at the level of translation, protein stability, or requirement for/inhibition by cofactors.
In conclusion, our observations support the hypothesis that SDPR is a metastasis suppressor, which elicits its effect by inhibiting EMT, migration, and intravasation/extravasation accompanied with promotion of apoptosis to halt the metastatic progression of breast and potentially other cancers. During breast cancer progression, loss of function of SDPR is likely to be primarily mediated by promoter DNA hypermethylation. Future studies should be focused on deciphering the regulation of SDPR in more detail and on generating a complete understanding of the pathways regulated by it to help with the identification of effective therapeutic targets.
Methods
In Vivo Metastasis and Tumorigenicity Assays.
Six-week-old female NOD.CB17-Prkdcscid/J mice were used for all in vivo metastasis and tumorigenicity assays. Bioluminescence imaging was performed with the Caliper IVIS Spectrum Imaging System (PerkinElmer). On necroscopy, lungs were extracted and imaged to count the number of macrometastases in each lung.
Annexin V Staining.
To quantify the apoptotic population, we used the Annexin V Apoptosis Detection Kit FITC from eBioscience and followed the provided protocol. We analyzed the samples using a FACSCalibur run by CellQuest Pro, version 5.2, software.
Three-Dimensional Cell Culture.
Ninety-six-well plates were coated with 100 µL of Matrigel, and 5,000 cells were seeded into the each well suspended in 100 µL of 2% (vol/vol) Matrigel/complete medium solution. Cell growth was monitored daily for 5 d by light microscopy. Quantification was done by using ImageJ and the plugin, ColonyArea (37).
Methylation-Specific Quantitative PCR.
Genomic DNA was isolated by Qiagen DNeasy Blood and Tissue Kit following the manufacturer’s protocol. Bisulfite conversion reactions were carried out using EpiTect Bisulfite Kits from Qiagen following the manufacturer’s protocol. Methylation-specific primer sets were designed by using MethPrimer (27).
Gene Expression Profiling.
Total RNA was isolated from MII, MIII, and MIV cell lines in triplicate using TRIzol, and RNA samples were cleaned with Qiagen RNeasy Kit following the manufacturer’s protocol. Following RNA quality control, samples were hybridized to GeneChip Human Genome U133 Plus 2.0 Arrays from Affymetrix (GEO accession no. GSE49156).
For additional methods, please refer to SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by grants from Susan G. Komen for the Cure (KG081435) and the NIH (CA165707) (to S.T.), Research Promotion Foundation of Cyprus (DIDAKTOR 0609/24) (to P.P.), and Department of Defense, Breast Cancer Research Program (W81XWH-11-1-0060) (to A.W.L.). We also acknowledge a seed grant from the Boston University Genome Science Institute, and support from the IVIS/Metabolic Phenotyping Core/Medicine, BU Flow Cytometry Core, and the core facilities at Boston University Clinical and Translational Science Institute (NIH CTSA Award UL1-TR000157). We thank Drs. Bert Vogelstein and Jian Yu (Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins), Steven Santner (Karmanos Cancer Institute), Joan Massague (Memorial Sloan Kettering Cancer Center), and Ramon Parsons (Icahn School of Medicine at Mount Sinai) for generously providing reagents and cell lines. We also thank Drs. David C. Seldin, Gerald V. Denis, and M. Isabel Dominguez for their valuable suggestions for the research project. Our thanks also go to Dr. Deniz Civril (Georgetown University) for her help with data analysis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE49156).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514663113/-/DCSupplemental.
References
- 1.Leone A, Flatow U, VanHoutte K, Steeg PS. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: Effects on tumor metastatic potential, colonization and enzymatic activity. Oncogene. 1993;8(9):2325–2333. [PubMed] [Google Scholar]
- 2.Montagner M, et al. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487(7407):380–384. doi: 10.1038/nature11207. [DOI] [PubMed] [Google Scholar]
- 3.Chen D, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med. 2012;18(10):1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephens PJ, et al. Oslo Breast Cancer Consortium (OSBREAC) The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steeg PS. Tumor metastasis: Mechanistic insights and clinical challenges. Nat Med. 2006;12(8):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 9.Stafford LJ, Vaidya KS, Welch DR. Metastasis suppressors genes in cancer. Int J Biochem Cell Biol. 2008;40(5):874–891. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Strickland LB, Dawson PJ, Santner SJ, Miller FR. Progression of premalignant MCF10AT generates heterogeneous malignant variants with characteristic histologic types and immunohistochemical markers. Breast Cancer Res Treat. 2000;64(3):235–240. doi: 10.1023/a:1026562720218. [DOI] [PubMed] [Google Scholar]
- 11.Györffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 12.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 13. Thermo Fisher Scientific (2013) Oncomine (Thermo Fisher Scientific, Ann Arbor, MI). Available at https://www.oncomine.org/. Accessed December 1, 2014.
- 14.Hansen CG, Bright NA, Howard G, Nichols BJ. SDPR induces membrane curvature and functions in the formation of caveolae. Nat Cell Biol. 2009;11(7):807–814. doi: 10.1038/ncb1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, et al. Coordinate suppression of Sdpr and Fhl1 expression in tumors of the breast, kidney, and prostate. Cancer Sci. 2008;99(7):1326–1333. doi: 10.1111/j.1349-7006.2008.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gianazza E, et al. Alterations of the serum peptidome in renal cell carcinoma discriminating benign and malignant kidney tumors. J Proteomics. 2012;76(Spec No):125–140. doi: 10.1016/j.jprot.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 17.Gustincich S, et al. The human serum deprivation response gene (SDPR) maps to 2q32-q33 and codes for a phosphatidylserine-binding protein. Genomics. 1999;57(1):120–129. doi: 10.1006/geno.1998.5733. [DOI] [PubMed] [Google Scholar]
- 18.Friedrich K, et al. Chromosomal genotype in breast cancer progression: Comparison of primary and secondary manifestations. Cell Oncol. 2008;30(1):39–50. doi: 10.1155/2008/209142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2(8):563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 21.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7(11):834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Youle RJ, Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9(1):47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 23.Weston CR, et al. Activation of ERK1/2 by deltaRaf-1:ER* represses Bim expression independently of the JNK or PI3K pathways. Oncogene. 2003;22(9):1281–1293. doi: 10.1038/sj.onc.1206261. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Edelstein LC, Gélinas C. The Rel/NF-kappaB family directly activates expression of the apoptosis inhibitor Bcl-xL. Mol Cell Biol. 2000;20(8):2687–2695. doi: 10.1128/mcb.20.8.2687-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 28.Uhlén M, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4(12):1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 30.Martinez R, et al. CpG island promoter hypermethylation of the pro-apoptotic gene caspase-8 is a common hallmark of relapsed glioblastoma multiforme. Carcinogenesis. 2007;28(6):1264–1268. doi: 10.1093/carcin/bgm014. [DOI] [PubMed] [Google Scholar]
- 31.Bandyopadhyay S, et al. Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene. 2004;23(33):5675–5681. doi: 10.1038/sj.onc.1207734. [DOI] [PubMed] [Google Scholar]
- 32.Hartsough MT, et al. Elevation of breast carcinoma Nm23-H1 metastasis suppressor gene expression and reduced motility by DNA methylation inhibition. Cancer Res. 2001;61(5):2320–2327. [PubMed] [Google Scholar]
- 33.Ozturk S, Lambert AW, Wong CK, Thiagalingam S. Cancer metastasis. In: Thiagalingam S, editor. Systems Biology of Cancer. Cambridge Univ Press; Cambridge, UK: 2015. pp. 282–294. [Google Scholar]
- 34.Smith SC, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer. 2009;9(4):253–264. doi: 10.1038/nrc2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanton C, Burrell RA, Futreal PA. Breast cancer genome heterogeneity: A challenge to personalised medicine? Breast Cancer Res. 2011;13(1):104. doi: 10.1186/bcr2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papageorgis P, et al. Targeting IL13Ralpha2 activates STAT6-TP63 pathway to suppress breast cancer lung metastasis. Breast Cancer Res. 2015;17(1):98. doi: 10.1186/s13058-015-0607-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guzmán C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: An ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 2014;9(3):e92444. doi: 10.1371/journal.pone.0092444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.