Significance
Ras activation has been detected in many types of human cancers, and its activity can be regulated by its association with GTPase-activating proteins (GAPs). However, the underlying mechanism of Ras regulation by GAP is incompletely understood. Here we show that FAM129B phosphorylation by EGFR reduced the association of p120-RasGAP with H-Ras, thereby enhancing H-Ras activation, the Warburg effect, and tumorigenesis. Given that FAM129B is overexpressed in human cancer and that approaches to directly target Ras have not been successful, disruption of Ras activation by interfering with FAM129B function can be an alternative approach for treating human cancers with activated Ras.
Keywords: EGFR, FAM129B, Ras, p120-RasGAP, Warburg effect
Abstract
Ras GTPase-activating proteins (GAPs) are important regulators for Ras activation, which is instrumental in tumor development. However, the mechanism underlying this regulation remains elusive. We demonstrate here that activated EGFR phosphorylates the Y593 residue of the protein known as family with sequence similarity 129, member B (FAM129B), which is overexpressed in many types of human cancer. FAM129B phosphorylation increased the interaction between FAM129B and Ras, resulting in reduced binding of p120-RasGAP to Ras. FAM129B phosphorylation promoted Ras activation, increasing ERK1/2- and PKM2-dependent β-catenin transactivation and leading to the enhanced glycolytic gene expression and the Warburg effect; promoting tumor cell proliferation and invasion; and supporting brain tumorigenesis. Our studies unearthed a novel and important mechanism underlying EGFR-mediated Ras activation in tumor development.
Overexpression or mutation of EGFR has been detected in many types of human tumors and correlates with poor clinical prognosis (1, 2). EGFR activation initiates an array of signaling events, including activation of Ras family proteins (3–5). Three members of the RAS family—H-Ras, N-Ras, and K-Ras—are activated and play direct roles in cancer development (6). Ras proteins are active in a GTP-bound form and inactive when bound to GDP (7, 8). Guanine exchange factors (GEF), such as Son of Sevenless (SOS), enhance the rate of GDP dissociation for Ras activation, whereas Ras GTPase-activating proteins (GAPs), such as p120-RasGAP and neurofibromin 1 (NF1), accelerate the GTPase activity of Ras to promote Ras inactivation (9, 10). Activated Ras activates Ras effectors, including Raf, phosphatidylinositol 3-kinase (PI3K), and RAL-GDS, a GDP-GTP exchange factor for RAL proteins (9, 11). Ras regulation has been intensively studied. However, the mechanism underlying the alleviation of RasGAP inhibition on Ras remains elusive.
Like EGFR, family with sequence similarity 129, member B (FAM129B), also known as MINERVA, is up-regulated in many types of cancer, including breast, kidney, large intestine, lung, and endometrial cancers as well as hematopoietic and central nervous system tumors (12, 13). FAM129B contains a pleckstrin homology (PH) domain near the amine terminus and a proline-rich domain near the carboxyl end. FAM129B has been found to promote tumor cell invasion via extracellular signal-regulated protein kinases 1 and 2 (ERK1/2)-dependent phosphorylation (14). In addition, FAM129B depletion inhibits Wnt3A/β-catenin target gene AXIN2 expression and promotes TNFα-induced apoptosis (15, 16). However, the mechanism by which FAM129B is regulated, thereby promoting tumor development, is largely unknown.
In this report, we show that EGFR phosphorylated FAM129B at Y593, which promoted the interaction between FAM129B and Ras and subsequently reduced binding of p120-RasGAP to H-Ras. Enhanced H-Ras activity resulted in increased β-catenin transactivation, leading to an enhanced Warburg effect and tumorigenesis.
Results
FAM129B Promotes EGF-Induced Ras Activation.
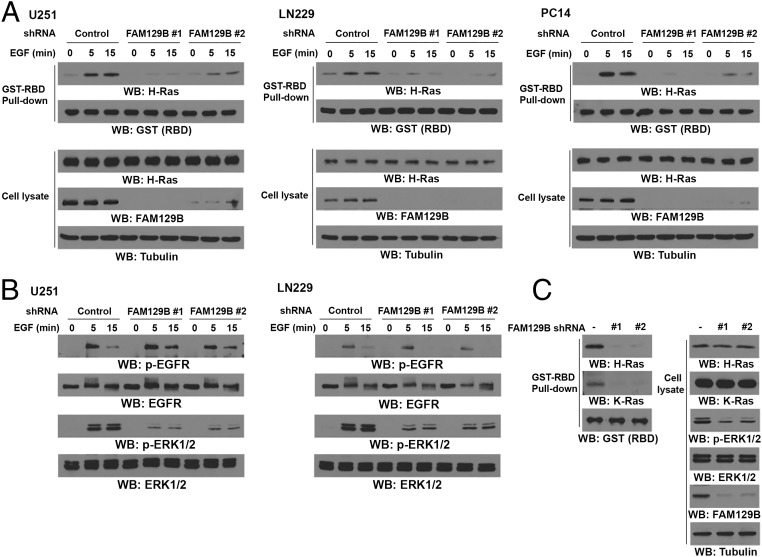
H-Ras, a major regulator in response to EGFR activation, is highly activated in glioblastoma (GBM) and is required for GBM cell proliferation (8, 17). To determine whether FAM129B is involved in regulation of EGFR-activated H-Ras, we used two FAM129B shRNAs targeting different FAM129B regions and depleted FAM129B in U251 and LN229 GBM cells and PC14 nonsmall cell lung cancer cells and treated these cells with EGF for different periods of time. We then examined the H-Ras activity in these cells by using GST-tagged Ras-binding domain of Raf-1 (GST-Raf-RBD) to pull down GTP–H-Ras from cell lysates. As expected, EGF stimulation enhanced H-Ras activity (Fig. 1A). Intriguingly, depletion of FAM129B by expressing its shRNA largely reduced the basal and EGF-enhanced H-Ras activity. In line with this finding, EGF-induced Ras-downstream ERK1/2 activation was inhibited by FAM129B depletion (Fig. 1B). In addition, depletion of FAM129B greatly reduced the activities of H-Ras and ERK1/2 in U87 cells expressing the constitutively activated EGFRvIII mutant (Fig. 1C), which lacks 267 amino acids from its extracellular domain and is found frequently in GBM (18, 19); Notably, K-Ras activity was also significantly reduced in U87/EGFRvIII cells with FAM129B depletion. These results indicate that FAM129B regulates EGF-induced activation of H-Ras and K-Ras.
Fig. 1.
FAM129B promotes EGF-induced H-Ras activation. (A) U251, LN229, and PC14 cells with or without stable expression of the indicated FAM129B shRNA or a control shRNA were treated with EGF (100 ng/mL) for the indicated time intervals. H-Ras activity was determined by using GST-Raf-RBD to pulldown GTP–H-Ras from cell lysates. Immunoblotting analyses were performed with the indicated antibodies. (B) U251 and LN229 cells with or without stable expression of FAM129B shRNA or a control shRNA were treated with EGF (100 ng/mL) for the indicated time intervals. Immunoblotting analyses were performed with the indicated antibodies. (C) The activities of H-Ras and K-Ras were determined in U87/EGFRvIII cells with or without expression of FAM129B shRNA or a control shRNA. Immunoblotting analyses were performed with the indicated antibodies.
EGFR Phosphorylates FAM129B at Y593 and Promotes the Interaction Between FAM129B and H-Ras.
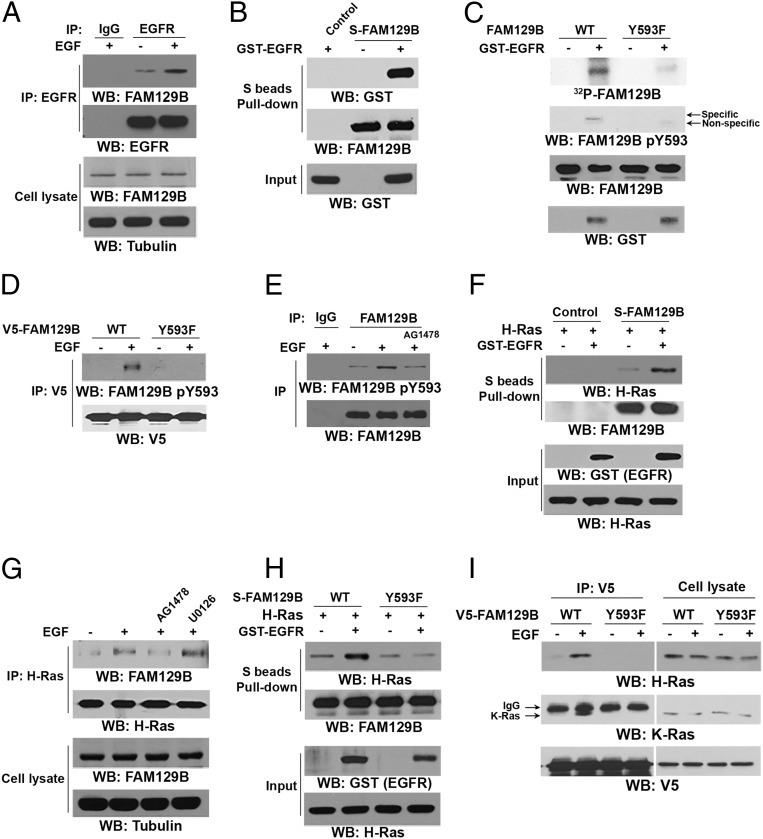
To determine the mechanism of FAM129B-regulated and EGFR activation–dependent H-Ras activation, we first investigated whether EGFR directly regulates FAM129B. Immunoprecipitation with an anti-EGFR antibody followed by immunoblotting with an anti-FAM129B antibody showed that EGF treatment induced an interaction between EGFR and S-tagged FAM129B (Fig. 2A). Incubation of purified GST-tagged intracellular domain of EGFR (GST-EGFR) with bacterially purified FAM129B proteins showed that these two proteins directly interacted with each other (Fig. 2B).
Fig. 2.
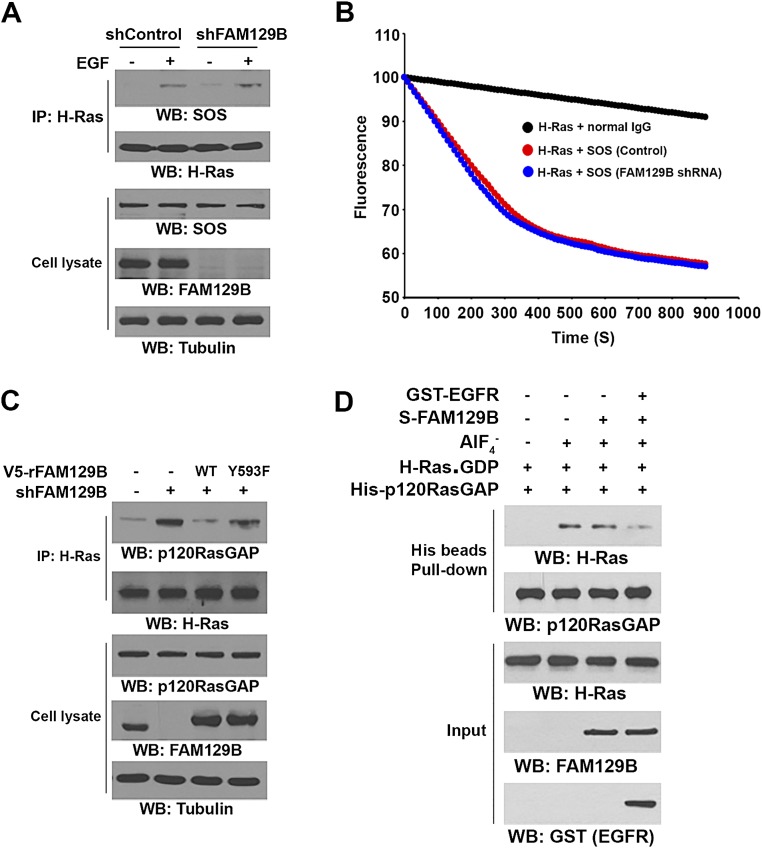
EGFR phosphorylates FAM129B at Y593 and promotes the interaction between FAM129B and H-Ras. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. (A) U251 cells were treated with EGF (100 ng/mL) for 15 min. (B) Purified bacterially expressed S-tagged FAM129B protein was incubated with purified active GST-EGFR (intracellular domain). A pulldown assay was performed. (C) In vitro kinase assays were performed with purified bacterially expressed WT FAM129B or FAM129B Y593F with or without GST-EGFR. (D) U251 cells expressing the indicated V5-tagged FAM129B were treated with or without EGF (100 ng/mL) for 15 min. (E) U251 cells were treated or not treated with AG1478 (1 μM) for 30 min before stimulation with or without EGF (100 ng/mL) for 15 min. (F and H) In vitro kinase assays were performed by mixing purified bacterially expressed S-tagged WT FAM129B (F and H) or FAM129B Y593F (H) with or without active GST-EGFR, which was followed by incubation with purified GST–H-Ras for a S-tag FAM129B pulldown assay. (G) U251 cells were treated with EGFR inhibitor AG1478 (1 μM) or MEK/ERK inhibitor U0126 (20 μM) for 30 min before stimulation with EGF (100 ng/mL) for 15 min. (I) U251 cells overexpressing the indicated V5-tagged FAM129B were treated with or without EGF (100 ng/mL) for 15 min.
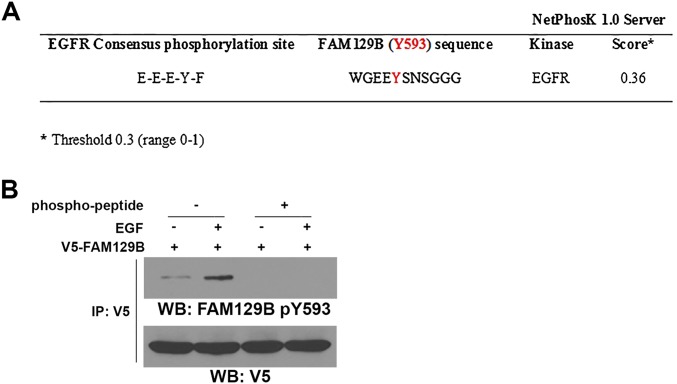
To examine whether EGFR phosphorylates FAM129B, we performed an in vitro phosphorylation assay by mixing GST-EGFR and S-tagged FAM129B in the presence of [γ-32P]ATP. Fig. 2C shows that EGFR phosphorylated FAM129B. Analysis of the FAM129B protein sequence by NetPhosK-1.0 (www.cbs.dtu.dk/services/NetPhosK/) revealed that Y593 is a potential site of phosphorylation by EGFR (Fig. S1A). Mutation of Y593 to phenylalanine (F) largely reduced EGFR-mediated FAM129B phosphorylation, which was confirmed by a phospho-FAM129B Y593 antibody. The specificity of this FAM129B pY593 antibody was validated by using a specific phosphorylation-blocking peptide, which blocked basal and EGF-induced FAM129B Y593 phosphorylation (Fig. S1B). In addition, EGF stimulation of U251 cells resulted in enhanced phosphorylation of V5-tagged wild-type (WT) FAM129B but not of the FAM129B Y593F mutant (Fig. 2D). Pretreatment with EGFR inhibitor AG1478 blocked EGF-induced phosphorylation of endogenous FAM129B Y593 (Fig. 2E). These results indicate that EGFR phosphorylates FAM129B at Y593 in vitro and in vivo.
Fig. S1.
Analysis of the FAM129B protein sequence by NetPhosK-1.0 and validation of the specificity of an anti-FAM129B pY593 antibody. (A) The FAM129B protein sequence was analyzed by NetPhosK-1.0. FAM129B Y593 is a potential site of phosphorylation by EGFR. (B) pCep4–EGFR was cotransfected with V5-tagged FAM129B into 293T cells. These cells were treated with EGF (100 ng/mL) for 15 min. Immunoprecipitation of V5 was followed by immunoblotting with an anti-FAM129B pY593 antibody in the presence or absence of specific competing phosphopeptides.
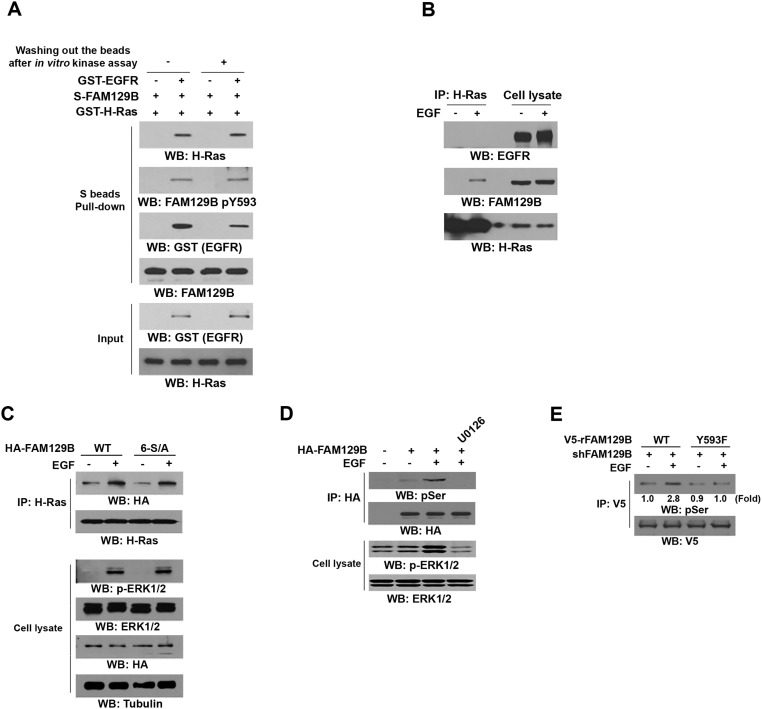
To determine the effect of FAM129B Y593 phosphorylation on H-Ras activation, we examined the potential interaction between phosphorylated FAM129B and H-Ras. Incubation of purified S-tagged WT FAM129B with purified GST–H-Ras in the presence or absence of purified activated GST-EGFR showed that binding of WT FAM129B to H-Ras was limited, and this binding was dramatically increased in the presence of activated EGFR (Fig. 2F). The interaction between EGFR-phosphorylated FAM129B and H-Ras was mostly not affected by removal of GST-EGFR from the reaction buffer, suggesting that EGFR is not an adaptor for this interaction and that phosphorylation of FAM129B by EGFR promotes the binding of FAM129B to H-Ras (Fig. S2A). In line with this finding, EGF stimulation, which did not result in obvious association of EGFR with H-Ras (Fig. S2B), increased the binding of H-Ras to WT FAM129B, which was blocked by AG1478 (Fig. 2G). In addition, activated EGFR failed to enhance the association of purified FAM129B Y593F with H-Ras (Fig. 2H), further supporting the finding that FAM129B Y593 phosphorylation is required for FAM129B binding to H-Ras.
Fig. S2.
FAM129B Y593 phosphorylation is required for ERK1/2-dependent FAM129B phosphorylation at Ser residues. (A) Purified immobilized S-FAM129B on agarose beads was incubated with or without purified active GST–EGFR for an in vitro phosphosphorylation reaction, which was followed with or without washing with PBS before incubation with GST–H-Ras. Immunoblotting analyses were performed with the indicated antibodies. (B) U251 cells were stimulated with EGF (100 ng/mL) for 15 min. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. (C) U251 cells expressing HA-tagged WT FAM129B or FAM129B 6-S/A mutant were treated with EGF (100 ng/mL) for 15 min. Immunoblotting analyses were performed with the indicated antibodies. (D) U251 cells expressing or not expressing HA-tagged WT FAM129B were pretreated or not pretreated with MEK/ERK inhibitor U0126 (20 μM) for 30 min before stimulation with EGF (100 ng/mL) for 15 min. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. (E) U251 cells depleted of FAM129B and with reconstituted expression of V5-tagged WT rFAM129B or rFAM129B Y593F mutant were treated with EGF (100 ng/mL) for 15 min. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies.
It was previously reported that ERK1/2 may phosphorylate FAM129B at 6 Ser residues (S641, S646, S665, S681, S692, and S696) (14). Expression of FAM129B 6-S/A mutant (Fig. S2C, Top) or treatment of U251 cells with MEK/ERK inhibitor U0126 (Fig. 2G), which inhibited Ser phosphorylation of HA-FAM129B (Fig. S2D), did not affect EGF-induced interaction between FAM129B and H-Ras. In addition, FAM129B 6-S/A expression did not affect EGFR-regulated ERK1/2 activation (Fig. S2C, Bottom). These results strongly suggest that ERK1/2-dependent FAM129B phosphorylation does not regulate the binding of FAM129B to H-Ras and subsequent ERK1/2 activation (Fig. 2G). In contrast, EGFR phosphorylation-resistant FAM129B Y593F mutant largely abrogated EGF-enhanced binding of FAM129B to both H-Ras and K-Ras (Fig. 2I). Notably, FAM129B Y593F expression reduced EGF-induced and ERK1/2-dependent Ser phosphorylation of FAM129B (Fig. S2E) (14). These results indicate that EGFR-mediated FAM129B Y593 phosphorylation promoted the binding of FAM129B to H-Ras and K-Ras, which in turn promoted downstream ERK1/2-dependent FAM129B phosphorylation in a feedback manner.
FAM129B Y593 Phosphorylation Reduces Binding of p120RasGAP to H-Ras and Enhances Ras Activity and Subsequent β-Catenin Transactivation.
H-Ras activity is mediated by Ras-GEF and Ras-GAP proteins. FAM129B depletion did not affect EGF-induced SOS binding to H-Ras in U251 cells as determined by coimmunoprecipitation analyses (Fig. S3A). In addition, immunoprecipitated SOS activity was not obviously altered in the presence or absence of depletion of FAM129B (Fig. S3B), suggesting that FAM129B does not directly regulate EGF-induced SOS binding to H-Ras or SOS activity.
Fig. S3.
FAM129B Y593 phosphorylation reduces binding of p120-RasGAP to H-Ras. (A) U251 cells depleted or not depleted of FAM129B were treated with EGF (100 ng/mL) for 15 min. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. (B) U251 cells were treated with EGF (100 ng/mL) for 15 min. SOS was immunoprecipitated and its activity was measured. Data represent the mean ± SD of three independent experiments. (C) U87/EGFRvIII cells depleted or not depleted of FAM129B and with reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant were analyzed by immunoprecipitation and immunoblotting analyses with the indicated antibodies. (D) Purified His-p120-RasGAP and GDP-bound GST–H-Ras in the presence or absence of tetrafluoroaluminate (AlF4−; 10 mM NaF and 30 μM AlCl3) were incubated with or without GST–EGFR-phosphorylated or nonphosphorylated S-FAM129B. A His-pulldown assay was performed.
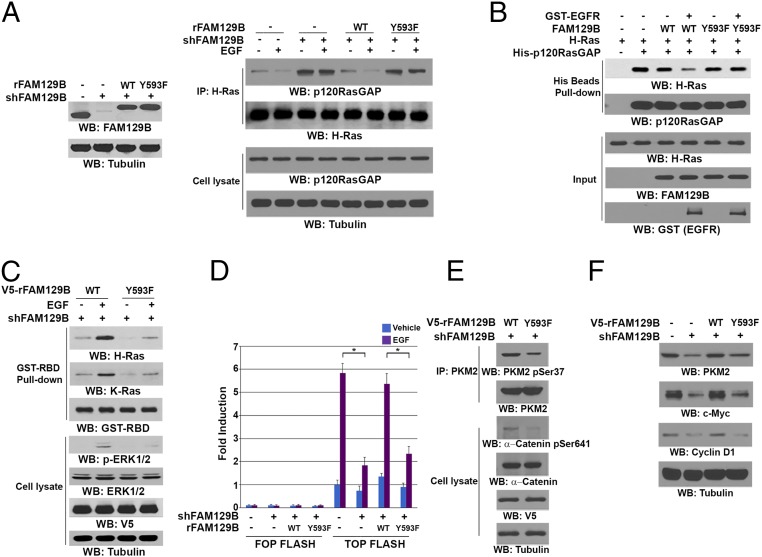
In contrast to the increased binding of SOS to H-Ras, p120-RasGAP was dissociated from H-Ras upon EGF stimulation (Fig. 3A, Right). Notably, FAM129B depletion (Fig. 3A, Left) increased the basal level of the interaction between p120-RasGAP and H-Ras and largely abrogated EGF-induced dissociation of p120-RasGAP from H-Ras (Fig. 3A, Right). The FAM129B depletion-increased association of p120-RasGAP with H-Ras was alleviated by reconstituted expression of WT FAM129B but not of FAM129B Y593F in U251 cells (Fig. 3A). Similar results were observed in U87/EGFRvIII cells, showing that FAM129B depletion increased binding of p120-RasGAP to H-Ras, which was abrogated by reconstituted expression of WT FAM129B but not of FAM129B Y593F (Fig. S3C). In addition, an in vitro GST pulldown assay showed that purified His-p120-RasGAP protein interacted with purified GST–H-Ras, and this interaction was modestly reduced by inclusion of WT FAM129B (Fig. 3B), which had limited binding to Ras (Fig. 2F). In contrast, inclusion of EGFR-phosphorylated WT FAM129B significantly abrogated the interaction between p120-RasGAP and H-Ras. As expected, FAM129B Y593F abrogated the activated EGFR-enhanced dissociation of p120-RasGAP from H-Ras. In line with this finding, His-p120-RasGAP, which did not associate with GDP-bound GST–H-Ras, interacted with GST–H-Ras in the presence of GDP and tetrafluoroaluminate (AlF4−), which occupies the γ-phosphate–binding site and mimics the transition state of Ras (GTP) during the GTP hydrolysis reaction (20); however, this interaction was largely reduced by inclusion of EGFR-phosphorylated FAM129B, but not nonphosphorylated FAM129B, in the pulldown assay (Fig. S3D). These results indicate that FAM129B Y593 phosphorylation promotes the dissociation of p120-RasGAP from H-Ras.
Fig. 3.
FAM129B Y593 phosphorylation reduces binding of p120-RasGAP to H-Ras and enhances Ras activity and subsequent β-catenin transactivation. Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. (A) U251 cells with or without expressing FAM129B shRNA and with or without reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant (Left) were treated or not treated with EGF (100 ng/mL) for 15 min (Right). (B) In vitro kinase assays were performed by mixing purified indicated S-tagged FAM129B protein with or without active GST-EGFR, which was followed by incubation with purified GST–H-Ras and His-tagged p120-RasGAP for a His-pulldown assay. (C) The activates of H-Ras and K-Ras of FAM129B-depleted U251 cells with or without reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant were determined by a pulldown assay using GST-Raf-RBD protein. (D) TOP-FLASH or FOP-FLASH plasmids were transfected into U251 cells expressing FAM129B shRNA with or without reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant, which were treated or not treated with EGF (100 ng/mL) for 8 h. Luciferase activity was measured. The relative levels of luciferase activity were normalized to the levels of untreated cells and to the levels of luciferase activity in the Renilla control plasmid. Data represent mean ± SD of three independent experiments. *P < 0.01, Student’s t test. (E) U87/EGFRvIII cells expressing FAM129B shRNA and reconstituted expression of WT rFAM129B or rFAM129B Y593F were analyzed by immunoblotting assays. (F) U87/EGFRvIII cells expressing or not expressing FAM129B shRNA and with reconstituted expression of WT rFAM129B or rFAM129B Y593F were analyzed by immunoblotting assays.
FAM129B Y593 phosphorylation-reduced binding of p120-RasGAP to Ras likely affects Ras activity. As expected, FAM129B depletion-dependent inhibition of EGF-induced H-Ras and K-Ras activity and ERK1/2 phosphorylation was rescued by reconstituted expression of WT FAM129B but not of FAM129B Y593F (Fig. 3C). These results indicate an essential role for FAM129B Y593 phosphorylation in full activation of H-Ras and K-Ras and its downstream ERK1/2.
ERK1/2 regulates many signaling events, including β-catenin transactivation (21, 22). ERK1/2 phosphorylates and activates CK2α, leading to CK2α-dependent α-catenin S641 phosphorylation and subsequent α-catenin dissociation from β-catenin in adherence junctions for enhanced nuclear translocation and transactivation of β-catenin (22, 23). In addition, ERK1/2 phosphorylates PKM2 at S37, leading to the nuclear translocation of PKM2 and subsequent binding of PKM2 to β-catenin in the nucleus for β-catenin transactivation (19, 21). To examine whether FAM129B Y593 phosphorylation plays a role in β-catenin regulation, we performed T-cell factor/lymphoid enhancer factor-1 (TCF/LEF-1) luciferase reporter analyses by expressing TCF-responsive promoter reporter (TOP-FLASH) and non-responsive control reporter (FOP-FLASH) plasmids, showing that FAM129B depletion significantly inhibited EGF-induced β-catenin transactivation (Fig. 3D). This inhibition was abrogated by reconstituted expression of WT FAM129B but not of FAM129B Y593F. Notably, FAM129B Y593F reduced phosphorylation of PKM2 S37 and α-catenin S641 in U87/EGFRvIII cells (Fig. 3E). Consistent with this finding, β-catenin transactivation-dependent expression of c-Myc, cyclin D1, and PKM2 was reduced by FAM129B depletion, and the reduced protein expression was restored by reconstituted expression of WT FAM129B but not of FAM129B Y593F (Fig. 3F). These results indicate that FAM129B Y593 phosphorylation promotes H-Ras activation and subsequent ERK1/2-dependent β-catenin transactivation.
FAM129B Y593 Phosphorylation Promotes the Warburg Effect, Tumor Cell Proliferation and Invasion, and Brain Tumorigenesis.
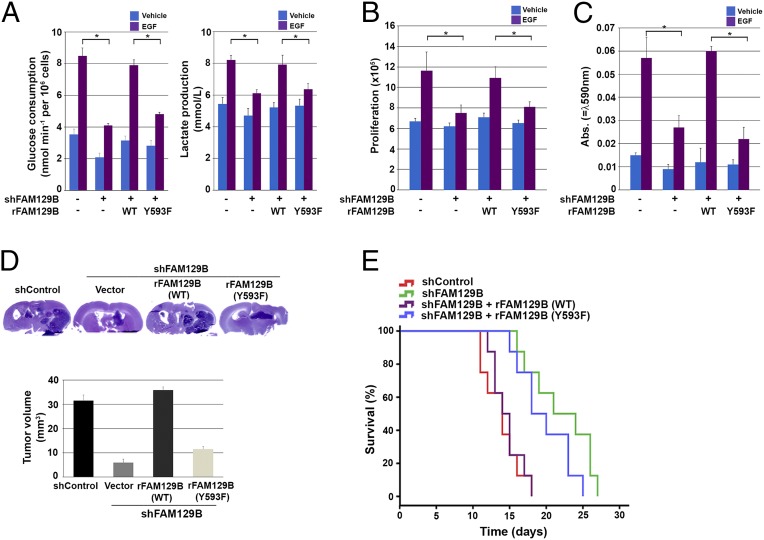
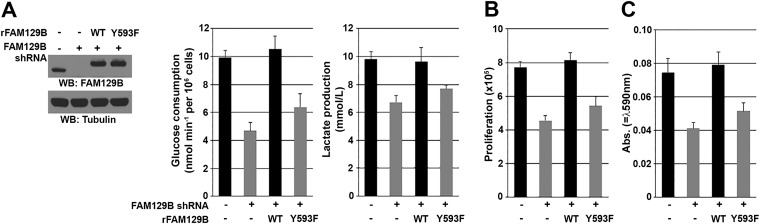
β-Catenin transactivation-dependent expression of c-Myc and PKM2 promotes glycolysis, whereas β-catenin–enhanced expression of cyclin D1 accelerates G1–S transition (24–26). Therefore, we next investigated the role of FAM129B Y593 phosphorylation in EGFR-regulated glycolysis of both U251 cells and U87/EGFRvIII. FAM129B depletion significantly reduced EGFR activation-promoted glucose consumption and lactate production, which were rescued by reconstituted expression of WT FAM129B but not of FAM129B Y593F (Fig. 4A and Fig. S4A). Similarly, cell counting (Fig. 4B and Fig. S4B) and Transwell invasion (Fig. 4C and Fig. S4C) assays showed that EGFR-promoted cell proliferation (Fig. 4B and Fig. S4B) and invasion (Fig. 4C and Fig. S4C) were inhibited by FAM129B depletion, and in both cases the inhibition was abrogated by reconstituted expression of WT FAM129B but not of FAM129B Y593F. These results indicate that FAM129B Y593 phosphorylation promotes the Warburg effect and tumor cell proliferation and invasion.
Fig. 4.
FAM129B Y593 phosphorylation promotes the Warburg effect, tumor cell proliferation and invasion, and brain tumorigenesis. (A) U251 cells depleted or not depleted of FAM129B and with reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant were cultured in no-serum DMEM with or without EGF (100 ng/mL) for 18 h. The media were collected for analysis of glucose consumption (Left) or lactate production (Right). Data represent the means ± SD of three independent experiments. *P < 0.01, Student’s t test. (B) U251 cells depleted or not depleted of FAM129B and with reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant were cultured with or without EGF (100 ng/mL) for 5 d and harvested for cell counting. Data represent the mean ± SD of three independent experiments. *P < 0.01, Student’s t test. (C) U251 cells depleted or not depleted of FAM129B and with reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant were plated on the surface of Matrigel coating the upper chamber of Transwell chambers. After 16 h, cells that migrated to the opposite side of the insert were stained with crystal violet. The membranes with invaded cells were dissolved in 4% (wt/vol) deoxycholic acid and read colorimetrically at 590 nm for quantification of invasion. Data represent the mean ± SD of three independent experiments. *P < 0.01, Student’s t test. (D) U87/EGFRvIII cells depleted or not depleted of FAM129B and with reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant were intracranially injected into athymic nude mice. After 2 wk, mice were killed and tumor growth was examined. Hematoxylin and eosin-stained coronal brain sections show representative tumor xenografts. (E) U87/EGFRvIII cells depleted or not depleted of FAM129B and with reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant were intracranially injected into athymic nude mice. The mouse survival times were recorded. Data represent the means ± SD of eight mice.
Fig. S4.
FAM129B Y593 phosphorylation promotes the Warburg effect and tumor cell proliferation and invasion. (A) U87/EGFRvIII cells depleted or not depleted of FAM129B and with reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant (Left) were cultured in no-serum DMEM for 18 h. The media were collected for analysis of glucose consumption (Middle) or lactate production (Right). Data represent the means ± SD of three independent experiments. Immunoblotting analyses were performed with the indicated antibodies. (B) U87/EGFRvIII cells depleted or not depleted of FAM129B and with reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant were cultured for 5 d and harvested for cell counting. Data represent the mean ± SD of three independent experiments. (C) U87/EGFRvIII cells depleted or not depleted of FAM129B and with reconstituted expression of WT rFAM129B or rFAM129B Y593F mutant were plated on the surface of Matrigel coating the upper chamber of Transwell chambers. After 16 h, cells that migrated to the opposite side of the insert were stained with crystal violet. The membranes with invaded cells were dissolved in 4% (wt/vol) deoxycholic acid and read colorimetrically at 590 nm for quantification of invasion. Data represent the mean ± SD of three independent experiments.
To investigate the role of FAM129B Y593 phosphorylation in brain tumorigenesis, we injected U87/EGFRvIII, U87/EGFRvIII–FAM129B shRNA, or U87/EGFRvIII–FAM129B shRNA cells with reconstituted expression of WT FAM129B or FAM129B Y593F intracranially into athymic nude mice. Depletion of FAM129B significantly reduced the growth of brain tumors (Fig. 4D) and prolonged the survival time of mice (Fig. 4E); these effects were reversed by reconstituted expression of WT FAM129B but not of FAM129B Y593F. These results highlight the significance of EGFR-dependent FAM129B Y593 phosphorylation in brain tumor development.
Discussion
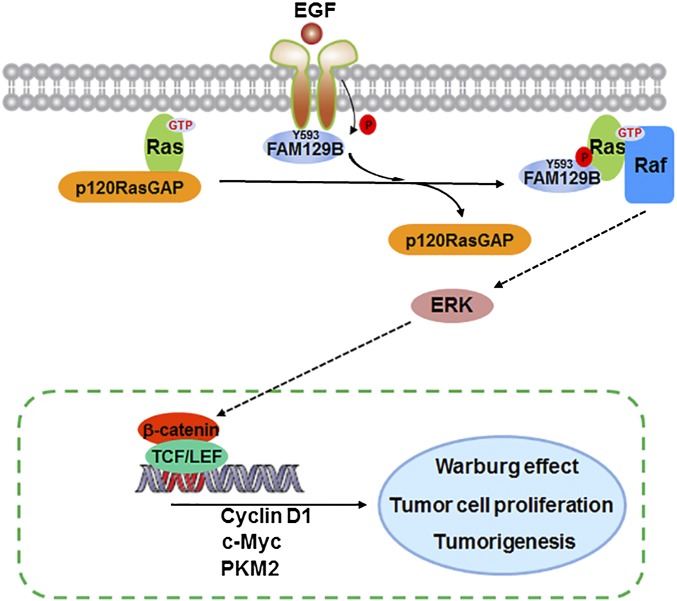
Ras activation has been detected in many types of human cancers, and its activity can be regulated by its association with GTP or GDP, facilitated by GEF or GAP proteins, respectively (6). We demonstrated that activation of EGFR by EGF stimulation or expression of active EGFRvIII mutant results in an association of EGFR and FAM129B, leading to EGFR-dependent FAM129B phosphorylation at Y593 and subsequent binding of phosphorylated FAM129B to H-Ras and K-Ras. The interaction between FAM129B and Ras reduced the association of p120-RasGAP with Ras, thereby enhancing Ras activation for ERK1/2-dependent β-catenin transactivation. The activated β-catenin/TCF/LEF transcriptional complex promoted the expression of cyclin D1, c-Myc, and PKM2, which in turn promoted the Warburg effect, tumor cell proliferation, and brain tumorigenesis (Fig. 5).
Fig. 5.
A mechanism of FAM129B-promoted H-Ras activation by dissociation of p120-RasGAP from H-Ras. EGFR phosphorylates FAM129B, resulting in binding of phosphorylated FAM129B to H-Ras and reduced the association of p120-RasGAP with H-Ras, thereby enhancing H-Ras activation for ERK1/2-dependent β-catenin transactivation for enhanced Warburg effect, tumor cell proliferation, and brain tumorigenesis.
Because FAM129B structure has not been revealed, it is unclear whether FAM129B and p120-RasGAP have some structural similarities that would allow them to compete for activated Ras. However, the evidence that FAM129B binds to Ras in a FAM129B phosphorylation-dependent manner suggests that FAM129B and p120-RasGAP do not share the same mechanism of binding to Ras. Because Ras has no reported phosphotyrosine binding domain, FAM129B Y593 phosphorylation may alter the FAM129B structure to promote the binding of FAM129B to Ras, which in turn results in structural changes in Ras that facilitate subsequent dissociation of p120-RasGAP from Ras.
A previous report showed that ERK1/2 phosphorylated FAM129B at six Ser residues and mutation of these residues reduced melanoma cell invasion via an unknown mechanism (14). We found that expression of FAM129B 6-S/A mutant or inhibition of ERK1/2 did not affect the binding of FAM129B to H-Ras. In addition, FAM129B 6-S/A expression did not affect EGFR-regulated ERK1/2 activation. In contrast, FAM129B Y593 phosphorylation is required for H-Ras and K-Ras activation and subsequent ERK1/2 activation and ERK1/2-dependent FAM129B Ser phosphorylation. These results indicate that EGFR-mediated FAM129B Y593 phosphorylation is a molecular event upstream to ERK1/2-dependent FAM129B phosphorylation and provides regulatory feedback for the functions of FAM129B.
FAM129B depletion inhibits WNT3A-mediated activation of β-catenin transactivation in melanoma cells via undefined mechanisms (16). We previously showed that ERK1/2 phosphorylates PKM2 at S37, leading to the binding of PKM2 to β-catenin in the nucleus for β-catenin transactivation (19, 21), and that ERK1/2 phosphorylates CK2α, which activates CK2α and results in CK2α-mediated α-catenin S641 phosphorylation, promoting the dissociation of inhibitory α-catenin from β-catenin in adherens junctions and β-catenin transactivation (22, 23). Here we show that expression of FAM129B Y593F mutant largely reduced Ras and downstream ERK1/2 activation and subsequent phosphorylation of PKM2 S37 and α-catenin S641 and β-catenin transactivation. Thus, our findings elucidate a novel mechanism underlying FAM129B regulation of β-catenin transactivation and β-catenin–promoted glycolysis and tumor cell proliferation and invasion upon EGFR activation.
p120-RasGAP plays an important role in regulation of Ras activation. However, the mechanism underlying regulation of p120-RasGAP and thus enhancement of Ras activation is unclear. Here we present a previously unknown mechanism showing that EGFR-mediated FAM129B Y593 phosphorylation results in binding of FAM129B to Ras, leading to reduced association of p120-RasGAP with Ras. Given that FAM129B is overexpressed in human cancer and that Ras has been targeted for human cancer treatment (6, 12, 13), disruption of Ras activation by interfering with FAM129B function can be an alternative approach for treating human cancers that overexpress FAM129B and activated Ras.
Materials and Methods
Phosphorylation of FAM129B Y593 was detected in Western blots using validated antibody from Signalway Antibody. H-Ras GTP charging reactions were performed by incubation with 1 mM GTP in a buffer solution containing 50 mM Tris⋅HCl (pH 7.5), 2 mM EDTA, 1 mM MgCl2, 100 μg/mL BSA, and 2 mM DTT for 1 h at 4 °C. The reaction was terminated by addition of 20 mM MgCl2. For nucleotide exchange assays, the rate of nucleotide dissociation was determined using H-Ras preloaded with BODIPY-GDP (Life Technologies), and reactions were performed with addition of excess GTP in the presence or absence of immunoprecipitated SOS from tested cells. Transcriptional activities of TCF/LEF-1 were measured as described previously (22). Statistical significance was calculated by using the two-tailed Student’s t test. P < 0.05 was considered significant. All mouse experiments were approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center.
Further details of materials and methods are included in SI Materials and Methods.
SI Materials and Methods
Cells and Cell Culture Conditions.
U251, LN229, PC14, and U87/EGFRvIII GBM cells and 293T cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% (vol/vol) bovine calf serum (HyClone). Cell cultures were made quiescent by growing them to confluence and then replacing the medium with fresh medium containing 0.5% serum for 1 d.
Materials.
Rabbit polyclonal antibodies recognizing phospho-FAM129B Y593, FAM129B (for immunoblotting analyses), and PKM2 were obtained from Signalway Antibody. Polyclonal antibodies for ERK, H-Ras, GST, and p120-RasGAP and monoclonal antibody for phospho-ERK1/2 were purchased from Santa Cruz Biotechnology. Polyclonal antibodies for K-Ras was purchased from Proteintech. EGF and mouse monoclonal antibodies for FLAG, His, tubulin, sodium fluoride (NaF), and aluminum chloride (A1C13) were purchased from Sigma. Mouse monoclonal antibodies for H-Ras, EGFR, p120-RasGAP, SOS1, and antiphosphorylated serine antibody were purchased from BD Biosciences. Rabbit polyclonal antibody recognizing FAM129B for immunoprecipitation analyses was purchased from Cell Signaling Technology. The mouse monoclonal antibody for V5 tag was obtained from Abcam. The mouse monoclonal antibody for c-Myc was obtained from EMD Millipore. Hygromycin, puromycin, MEK/ERK inhibitor U0126, and EGFR inhibitor AG1478 were purchased from EMD Biosciences. Blasticidin S was purchased from Life Technologies. Active GST-EGFR (695-end) was obtained from SignalChem. HyFect transfection reagents were obtained from Denville Scientific.
DNA Constructs and Mutagenesis.
PCR-amplified human FAM129B was cloned into pcDNA6-V5-His vector and pCold-S tag vector between HindIII and KpnI and into pET32a vector and pGEX-4T1 vector between EcoRI and XhoI. PCR-amplified human H-Ras was cloned into pCDH-SFB vector between NheI and BamHI. PCR-amplified human p120-RasGAP was cloned into pCold-S tag vector (TaKaRa) between BamHI and SalI.
FAM129B Y593F, WT rFAM129B, and Y593F mutant in pcDNA6-V5-His vector were made by using the QuikChange site-directed mutagenesis kit (Stratagene). pCDNA 3.1 rFAM129B contained these mutations: G81A, C84T, and C85T.
pGIPZ control was generated with control oligonucleotide GCTTCTAACACCGGAGGTCTT. The following FAM129B shRNAs was used: shFAM129B no. 1 (TACGACTACGACAGCAG) and shFAM129B no. 2 (GACATGAACCTGAACGTCA).
Purification of Recombinant Proteins.
GST-tagged Ras-binding domain of Raf-1 (GST-Raf-RBD) and the WT and mutants of His–S–FAM129B, GST–FAM129B, WT GST–H-Ras, and WT His–S–p120-RasGAP were expressed in bacteria and purified as described previously (22).
The purified H-Ras was incubated with 1 mM GTP or 1 mM GDP in a buffer solution containing 50 mM Tris⋅HCl (pH 7.5), 2 mM EDTA, 1 mM MgCl2, 100 μg/mL BSA, and 2 mM DTT for 1 h at 4 °C. The reaction was terminated before pulldown assays by addition of 20 mM MgCl2, as described previously (27, 28).
In Vitro Kinase Assays.
The kinase reactions were performed as described previously (22).
Pulldown Assay.
GST pulldown assays were performed as described previously (29).
In Vitro Invasion Assay.
Cell invasion was assessed by quantifying invasion of cells through Matrigel-coated Transwell inserts (Greiner Bio-One North America) during a 16-h time frame, as described previously (22). The membranes with invaded cells were dissolved in 4% (wt/vol) deoxycholic acid and the solutions read colorimetrically at 590 nm for quantification of invasion. Each experiment was repeated at least three times.
Immunoblotting Analysis.
Extraction of proteins from cultured cells using a modified buffer was followed by immunoblotting with corresponding antibodies, as described previously (30). Each experiment was repeated at least three times.
Luciferase Reporter Gene Assay.
Transcriptional activities of TCF/LEF-1 in tested cells were measured as described previously (22). EGF (100 ng/mL) was added 8 h before harvesting. Each experiment was performed three times.
Guanine Nucleotide Exchange Assay.
Fluorescein-based nucleotide exchange assays were conducted by using Ras loaded with BODIPY-GDP (Life Technologies) in a buffer containing 25 mM Tris (pH 7.5), 50 mM NaCl, and 1 mM MgCl2 in a black 96-well plate. Baseline fluorescence was recorded, which was followed by addition of excess GTP in the presence or absence of immunoprecipitated SOS from U251 cells. Nucleotide exchange was monitored as a decrease in fluorescence with time. Final reactions contained 1 μM BODIPY-GDP–loaded H-Ras, 200 μM unlabeled GTP, and immunoprecipitated SOS from U251 cells. Changes in fluorescence were monitored using a Hamamatsu FDSS 6000 with readings conducted every 3 s for 15 min.
Measurements of Glucose Consumption and Lactate Production.
The levels of glucose and lactate in cells were determined as described previously (21). Glucose levels were determined using a glucose (GO) assay kit (Sigma). Glucose consumption was the difference in glucose concentration compared with DMEM. Lactate levels were determined by using a lactate assay kit (Eton Bioscience).
Intracranial Implantation of GBM Cells in Mice.
We injected 5 × 105 U87/EGFRvIII cells (in 5 μL of DMEM per mouse), with or without regulation of FAM129B expression, intracranially into 4-wk-old male athymic nude mice, as described previously (19). The animals were killed 2 wk after GBM cell injection. The brain of each mouse was harvested, fixed in 4% (vol/vol) formaldehyde, and embedded in paraffin. Tumor formation and phenotype were determined by histological analysis of hematoxylin and eosin-stained sections.
Acknowledgments
We thank Natalie Ahn (University of Colorado) for the FAM129B plasmids, Hans Clevers (Netherlands Institute for Developmental Biology, Hubrecht Laboratory) for the pTOP-FLASH and pFOP-FLASH plasmids, Johanna Ivaska (University of Turku) for the p120-RasGAP plasmids, and Kathryn Hale for her critical reading of this manuscript. This work was supported by National Cancer Institute Grants 2R01 CA109035 and 1R0 CA169603 (to Z.L.), National Institute of Neurological Disorders and Stroke Grant 1R01 NS089754 (to Z.L.), MD Anderson Support Grant CA016672 and the James S. McDonnell Foundation 21st Century Science Initiative in Brain Cancer Research Award 220020318 (to Z.L.), 2P50 CA127001 (Brain Cancer SPORE), a Sister Institution Network Fund from MD Anderson (to Z.L.), the Jeffrey Lee Cousins Fellowship at MD Anderson Cancer Center (to H.J.), and the Opening Project of Zhejiang Provincial Top Key Discipline of Clinical Medicine (to J.L. and Z.L.). Z.L. is a Ruby E. Rutherford Distinguished Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517112113/-/DCSupplemental.
References
- 1.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: Decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12(2):104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 2.Lu Z, Hunter T. Degradation of activated protein kinases by ubiquitination. Annu Rev Biochem. 2009;78:435–475. doi: 10.1146/annurev.biochem.013008.092711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 5.Karnoub AE, Weinberg RA. Ras oncogenes: Split personalities. Nat Rev Mol Cell Biol. 2008;9(7):517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3(1):112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- 7.Bernards A, Settleman J. GAP control: Regulating the regulators of small GTPases. Trends Cell Biol. 2004;14(7):377–385. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7(4):295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 9.Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2(3):344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 2007;129(5):865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 11.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: Post-translational modification of RAS. Nat Rev Mol Cell Biol. 2012;13(1):39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes SA, et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bamford S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91(2):355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Old WM, et al. Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Mol Cell. 2009;34(1):115–131. doi: 10.1016/j.molcel.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, Evans HG, Evans DR. FAM129B/MINERVA, a novel adherens junction-associated protein, suppresses apoptosis in HeLa cells. J Biol Chem. 2011;286(12):10201–10209. doi: 10.1074/jbc.M110.175273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conrad W, et al. FAM129B is a novel regulator of Wnt/β-catenin signal transduction in melanoma cells. F1000 Res. 2013;2:134. doi: 10.12688/f1000research.2-134.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guha A, Feldkamp MM, Lau N, Boss G, Pawson A. Proliferation of human malignant astrocytomas is dependent on Ras activation. Oncogene. 1997;15(23):2755–2765. doi: 10.1038/sj.onc.1201455. [DOI] [PubMed] [Google Scholar]
- 18.Kuan CT, Wikstrand CJ, Bigner DD. EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr Relat Cancer. 2001;8(2):83–96. doi: 10.1677/erc.0.0080083. [DOI] [PubMed] [Google Scholar]
- 19.Yang W, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mittal R, Ahmadian MR, Goody RS, Wittinghofer A. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science. 1996;273(5271):115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- 21.Yang W, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji H, et al. EGF-induced ERK activation promotes CK2-mediated disassociation of alpha-Catenin from beta-Catenin and transactivation of beta-Catenin. Mol Cell. 2009;36(4):547–559. doi: 10.1016/j.molcel.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji H, Wang J, Fang B, Fang X, Lu Z. α-Catenin inhibits glioma cell migration, invasion, and proliferation by suppression of β-catenin transactivation. J Neurooncol. 2011;103(3):445–451. doi: 10.1007/s11060-010-0413-4. [DOI] [PubMed] [Google Scholar]
- 24.Yang W, Lu Z. Nuclear PKM2 regulates the Warburg effect. Cell Cycle. 2013;12(19):3154–3158. doi: 10.4161/cc.26182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Z. Nonmetabolic functions of pyruvate kinase isoform M2 in controlling cell cycle progression and tumorigenesis. Chin J Cancer. 2012;31(1):5–7. doi: 10.5732/cjc.011.10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Lu Z. Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett. 2013;339(2):153–158. doi: 10.1016/j.canlet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chuang E, et al. Critical binding and regulatory interactions between Ras and Raf occur through a small, stable N-terminal domain of Raf and specific Ras effector residues. Mol Cell Biol. 1994;14(8):5318–5325. doi: 10.1128/mcb.14.8.5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korlach J, et al. Spontaneous nucleotide exchange in low molecular weight GTPases by fluorescently labeled gamma-phosphate-linked GTP analogs. Proc Natl Acad Sci USA. 2004;101(9):2800–2805. doi: 10.1073/pnas.0308579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji H, et al. AKT-dependent phosphorylation of Niban regulates nucleophosmin- and MDM2-mediated p53 stability and cell apoptosis. EMBO Rep. 2012;13(6):554–560. doi: 10.1038/embor.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Z, et al. Activation of protein kinase C triggers its ubiquitination and degradation. Mol Cell Biol. 1998;18(2):839–845. doi: 10.1128/mcb.18.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]