Significance
We developed a small protein tag enabling fluorescent labeling of proteins in living cells and in multicellular organisms through the specific binding and activation of a cell-permeant and nontoxic fluorogenic ligand. This tag, called Yellow Fluorescence-Activating and absorption-Shifting Tag (Y-FAST), was engineered by directed evolution from the Photoactive Yellow Protein. Y-FAST distinguishes itself from other labeling methods because the fluorogen binding is highly dynamic and fully reversible. Apart from providing new opportunities in superresolution imaging and biosensor design, this feature enables rapid switching on and off of the fluorescence of a fusion protein by addition or withdrawing of the fluorogenic ligand, opening exciting ways to perform sequential multiplexing imaging.
Keywords: fluorescence imaging, fluorogenic ligand, directed evolution
Abstract
This paper presents Yellow Fluorescence-Activating and absorption-Shifting Tag (Y-FAST), a small monomeric protein tag, half as large as the green fluorescent protein, enabling fluorescent labeling of proteins in a reversible and specific manner through the reversible binding and activation of a cell-permeant and nontoxic fluorogenic ligand (a so-called fluorogen). A unique fluorogen activation mechanism based on two spectroscopic changes, increase of fluorescence quantum yield and absorption red shift, provides high labeling selectivity. Y-FAST was engineered from the 14-kDa photoactive yellow protein by directed evolution using yeast display and fluorescence-activated cell sorting. Y-FAST is as bright as common fluorescent proteins, exhibits good photostability, and allows the efficient labeling of proteins in various organelles and hosts. Upon fluorogen binding, fluorescence appears instantaneously, allowing monitoring of rapid processes in near real time. Y-FAST distinguishes itself from other tagging systems because the fluorogen binding is highly dynamic and fully reversible, which enables rapid labeling and unlabeling of proteins by addition and withdrawal of the fluorogen, opening new exciting prospects for the development of multiplexing imaging protocols based on sequential labeling.
Deciphering the complex mechanisms controlling cells and organisms requires effective imaging systems and fluorescent probes to observe biomolecules in real time with high spatiotemporal resolution. Ideal fluorescent probes should be highly specific for their target, bright, photostable, nontoxic, and as small as possible to avoid perturbing the function of their target. They should also exhibit instantaneous and robust fluorescence, and offer the possibility of tuning at will the fluorescence of the system for sophisticated imaging protocols. GFP-like fluorescent proteins have revolutionized cell biology, providing an easy way to fluorescently tag any protein of interest with absolute specificity through genetic fusion (1–3). However, an increasing number of studies indicate that they are not always optimal probes, as (i) their size and tendency to oligomerize can lead to dysfunctional fusion proteins (4); (ii) their oxygen-dependent fluorescence precludes their use for anaerobic biology (5); (iii) their long maturation (up to 1 h) prevents real-time monitoring of rapid processes (6); and (iv) they display confounding photophysics like photoswitching, kindling, or dark-state conversion, which can complicate the interpretation of some experiments (6, 7).
The importance of fluorescent proteins for bioimaging has motivated biologists and chemists to develop alternative strategies to fluorescently label proteins by taking advantage of the unique behavior of fluorogenic chromophores (8). In these approaches, a protein of interest is fused to a protein tag that binds a fluorogenic ligand (so-called fluorogen) and activates its fluorescence. Because the fluorogenic ligand is nonfluorescent by its own and becomes strongly fluorescent only upon binding its cognate tag, unspecific fluorescence background in cells remains minimal even in the presence of an excess of fluorogen, thus ensuring high imaging contrast. Flavin-based fluorescent proteins such as FbFPs (9), iLOV (10), and mini-SOG (11) or bilirubin-binding UnaG (12) have been recently proposed as alternatives to GFP because of their small size and oxygen-independent fluorescence. Biliverdin-based fluorescent proteins [IFP1.4 (13), iRFP (14)] have opened new possibilities for imaging protein in deep tissue and in vivo using infrared excitation. Other interesting developments include labeling strategies relying on protein tags, such as SNAP-tag (15), PYP-tag (16), CRABPII (17), or FAPs (18–21), binding (covalently or noncovalently) an exogenously applied fluorogen. These systems present two main advantages: First, the photophysical properties of exogenous fluorogens can be tailored by molecular engineering; second, their flexibility opens new opportunities for on-demand applications wherein fluorescence is desired only at a specific time or at a given density (22).
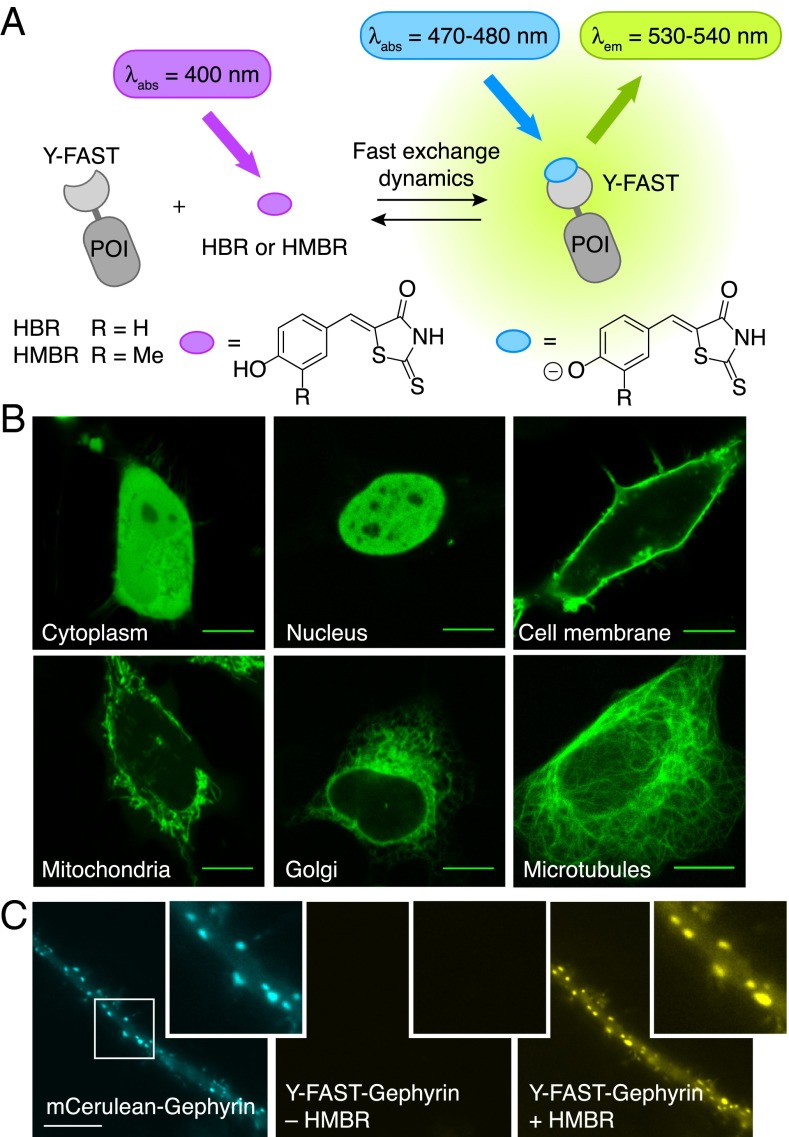
Herein we present the development of Yellow Fluorescence-Activating and absorption-Shifting Tag (Y-FAST), a small protein tag enabling fluorescent labeling of proteins in living cells and in multicellular organisms. Y-FAST is an engineered variant of the monomeric 14-kDa Photoactive Yellow Protein (PYP) [a blue-light photoreceptor from Halorhodospira halophila (23–25)] that we evolved to reversibly bind 4-hydroxybenzylidene-rhodanine (HBR) or 4-hydroxy-3-methylbenzylidene-rhodanine (HMBR), two fluorogens identified in the course of this study (Fig. 1A). HBR and HMBR are nonfluorescent by themselves, but they fluoresce yellow light upon blue-light excitation when bound to Y-FAST. Y-FAST distinguishes itself from existing labeling systems because the binding is not only specific and instantaneous but also highly dynamic and fully reversible. Fluorescence can thus be rapidly switched on and off simply by addition or withdrawal of the fluorogen, providing an additional degree of control. The fast binding dynamics might moreover decrease the apparent photobleaching rate by continuous renewal of the fluorogen, as suggested in previous reports (26). Designing a fluorogen-based reporter characterized by a reversible binding that is both specific and highly dynamic was, however, challenging, as the high off-rate necessary for a fast exchange dynamics tends to decrease the affinity required for high selectivity. Thus, to maintain high selectivity, we relied on two spectroscopic changes for fluorogen activation: first, binding of H(M)BR to Y-FAST results in a significant increase of fluorescence quantum yield, and, second, it induces large absorption red shift. Because Y-FAST is the only species promoting these two spectroscopic changes, free or nonspecifically bound fluorogen does not contribute to the fluorescence signal, ensuring high imaging contrast.
Fig. 1.
Y-FAST enables specific labeling of fusion proteins in living cells. (A) Y-FAST binds the fluorogenic HBR and HMBR and activates their fluorescence (POI, protein of interest). Binding induces two spectroscopic changes: an increase of the fluorescence quantum yield and an absorption red shift (due to ionization). (B) Confocal micrographs of live HeLa cells expressing various Y-FAST fusions labeled with 5 μM HMBR (Ex/Em 488/493–797 nm). Cytoplasm: Y-FAST; Nucleus: H2B-Y-FAST; Cell membrane: Lyn11-Y-FAST; Mitochondria: Mito-FAST (Mito = Mitochondrial targeting sequence from subunit VIII of human cytochrome c oxidase); Golgi: Golgi-Y-FAST (Golgi = N-terminal 81 amino acids of the human beta 1,4-galactosyltransferase); Microtubules: Ensconsin-Y-FAST. (C) Epifluorescence micrographs of a dendritic segment of a spinal cord neuron cotransfected with mCerulean−Gephyrin that accumulates at inhibitory synapses (Ex/Em 427/472 ± 15 nm; Left) and a Y-FAST-tagged Gephyrin construct (Ex/Em 504/542 ± 14 nm; Center). After 10 s of incubation with 10 μM HMBR, the fluorescence of Y-FAST was detected in the yellow emission range (Ex/Em 504/542 ± 14 nm; Right). (Scale bars, 10 μm.)
Results
Fluorogen Design.
HBR is easily obtained in one step by in-water condensation of the rhodanine to the parahydroxybenzaldehyde. It is composed of an electron-donating phenol conjugated to an electron-withdrawing rhodanine (Fig. 1A). This push−pull structure is analogous to the GFP chromophore 4-hydroxybenzylidene-5-imidazolinone (5), known to deexcite nonradiatively in solution but to relax to the ground state radiatively in the rigid barrel of GFP (27). HBR drew our attention as putative fluorogen for the design of Y-FAST for several reasons. First, HBR is almost fully protonated at physiological pH (its pKA is 8.4 ± 0.1), and it undergoes a 50-nm absorption red shift upon deprotonation as a result of the stronger electron donation of the phenolate (Table 1 and SI Appendix, Fig. S1 A and B). We therefore anticipated that a protein tag stabilizing deprotonated HBR would exhibit a red-shifted absorption with respect to free HBR in pH 7.4 solutions, enabling discrimination of the free and bound states by their absorption properties. Secondly, HBR fluorescence is highly environment-sensitive. In water, the protonated and deprotonated states of HBR emit at 470 nm and 545 nm with fluorescence quantum yields of 0.02% and 0.3% (Table 1 and SI Appendix, Fig. S1C), whereas, in viscous solutions (containing 40% glycerol), they exhibit sixfold and threefold higher brightness, respectively. Taken together these spectroscopic properties allowed us to anticipate that binding of HBR to a well-designed protein tag could provide a unique fluorogenic effect based on two spectroscopic changes: an absorption red shift through binding-induced deprotonation and a fluorescence quantum yield increase via fluorogen immobilization.
Table 1.
Physicochemical properties of the fluorogens and their complex with Y-FAST
| Compound | λabs, nm | λem, nm | ε, M–1⋅cm–1 | ϕ, % | KD, μM | 10−7 × kON, M−1⋅s−1 | kOFF, s−1 |
| HBR (pH 6.8)* | 397 | 470 | 33,000 | 0.02 | |||
| HBR (pH 10.1)* | 449 | 545 | 34,500 | 0.3 | |||
| Y-FAST:HBR (pH 7.4) | 467 | 527 | 44,000 | 9 | 0.62 ± 0.05 | 3† (2.9 ± 0.4‡) | 17† (8.5 ± 1.2‡) |
| HMBR (pH 5.8)§ | 401 | 480 | 29,500 | 0.04 | |||
| HMBR (pH 10.5)§ | 461 | 561 | 33,500 | 0.2 | |||
| Y-FAST:HMBR (pH 7.4) | 481 | 540 | 45,000 | 33 | 0.13 ± 0.01 | 6.3 ± 0.9 | 6.3 ± 0.7 |
Abbreviations are as follows: λabs, wavelength of maximal absorption; λem, wavelength of maximal emission; ε, molar absorption coefficient at λabs; ϕ, fluorescence quantum yield; KD, dissociation constant; kON, on-rate kinetic constant; kOFF, off-rate kinetic constant. Thermodynamic and kinetic constants are given as ± the SE of the fits (n = 3). Temperature is 25 °C.
The pKA of HBR is 8.4 ± 0.1.
Kinetic constants at 25 °C were extrapolated from the kinetic parameters shown in SI Appendix, Table S2.
Kinetic constants were determined experimentally at 20 °C.
The pKA of HMBR is 8.7 ± 0.1.
Y-FAST Is a Variant of PYP Engineered by Directed Evolution.
PYP was chosen as scaffold for the design of Y-FAST for several reasons. First, its parahydroxycinnamoyl (HC) chromophore—covalently tethered to Cys69 and responsible for its blue-light photosensing properties (23–25)—shares structural features with HBR, suggesting that the binding site of PYP could be engineered to bind HBR selectively and reversibly. Moreover, the binding pocket of PYP accommodates HC in its phenolate deprotonated form (25), providing a platform for designing variants able to stabilize deprotonated HBR and thus obtain absorption red shift upon binding. Finally, wild-type PYP has a proven ability as a recombinant protein tag (16, 28) and is a small protein (14 kDa) compared with GFP-like fluorescent proteins (26−30 kDa).
To engineer the binding cavity of PYP, we randomized loops and residues in close proximity with the chromophore pocket by saturation mutagenesis (SI Appendix, Fig. S2). By screening yeast surface-displayed libraries by fluorescence-activating cell sorting (29) in the presence of HBR, we successfully identified 47 clones specifically activating HBR fluorescence (SI Appendix, Fig. S3). The selected clones all belonged to the library constructed by randomizing the loop 94-101 that gates the entrance of the binding pocket. The emergence of the consensus sequence WxIPTxxx confirmed convergence of the selection process.
Physicochemical Characterization.
The most promising variants were expressed in Escherichia coli and purified by affinity chromatography for in vitro characterization. Titration experiments relying on fluorescence increase to assay complex formation established that all selected variants bound HBR with 0.5- to 1-μM affinities (SI Appendix, Table S1 and Fig. S4; see SI Appendix, Text S1 for the thermodynamic analysis). When bound to Y-FAST, the best variant of our selection, HBR fluoresces at ∼530 nm three orders of magnitude more than in solution, and absorbed maximally at ∼470 nm instead of ∼400 nm in pH 7.4 solutions, in accordance with HBR being deprotonated when bound (Table 1 and SI Appendix, Fig. S5).
Y-FAST:HBR complex was shown to form extremely rapidly. Addition of HBR to Y-FAST solutions instantaneously produced yellow fluorescence (Movie S1). The on- and off-rate kinetic constants were further determined by stopped-flow experiments (Table 1; see SI Appendix, Text S1, Fig. S6, and Tables S2 and S3 for the kinetic analysis). This kinetic analysis indicated that, when [HBR] = KD, the relaxation time of binding at 25 °C was 30 ms. The analysis demonstrated, moreover, that the binding was not only rapid but also highly dynamic, because the residence time (reciprocal of the off-rate constant) of HBR in the bound state was only 60 ms at 25 °C.
Finally, Y-FAST was proved to exist as a monomer in solution up to millimolar concentrations by determining its apparent size by analytical size exclusion chromatography (SI Appendix, Fig. S7) and by measuring its global tumbling correlation time and translational diffusion coefficient using NMR spectroscopy (see SI Appendix, Text S2 and Table S4). These latter NMR experiments showed, in particular, that the diffusion coefficient of Y-FAST at 20 °C was 1.1 × 10−10 m2⋅s−1, in perfect agreement with values reported for monomeric wild-type PYP (30), which corresponds to a hydrodynamic radius of 1.9 nm (see SI Appendix, Text S2 and Table S4).
Brightness Optimization.
The complex between HBR and Y-FAST displays a fluorescence quantum yield of 9% and a brightness of 4,000 M–1⋅cm–1 (Table 1). To improve its brightness properties, we screened HBR analogs for enhanced fluorescence performance (SI Appendix, Fig. S8). We found that HMBR, an analog bearing an additional methyl group on the aromatic ring (Fig. 1a), formed a complex with Y-FAST fivefold tighter (Table 1 and SI Appendix, Fig. S9) and fourfold brighter than HBR (Table 1 and SI Appendix, Fig. S10 A and B). The complex Y-FAST:HMBR (i) exhibits a fluorescence quantum yield of 33% and an absorption coefficient of 45,000 M–1⋅cm–1 (Table 1), attaining the fluorescence performance of common fluorescent proteins (1), and (ii) still displays a red-shifted absorption (Table 1 and SI Appendix, Fig. S10C), in agreement with HMBR being deprotonated in the complex (pKA 8.7 ± 0.1; see SI Appendix, Fig. S11). Kinetic analysis revealed that, despite the gain in affinity, the binding remained both fast (the relaxation time is 70 ms at 25 °C when [HMBR] = KD) and highly dynamic (the residence time of HMBR is 160 ms at 25 °C) (Table 1; see also SI Appendix, Text S1, Fig. S6, and Tables S2 and S3).
Specific and Efficient Labeling of Fusion Proteins in Various Cellular Systems.
The labeling selectivity and efficiency was studied in various hosts (bacteria, yeast, and mammalian cells) by flow cytometry and confocal microscopy (SI Appendix, Fig. S12 and Table S5). The analysis concluded that HBR and HMBR (i) are cell-permeant, (ii) generate no or negligible fluorescence background, and (iii) do not bind PYP, illustrating the high selectivity of Y-FAST labeling in living cells. The analysis also confirmed that HMBR outperformed HBR for fluorescent labeling of Y-FAST in living cells. Moreover, when labeled with HMBR, Y-FAST reached brightness and photostability performance comparable with the green fluorescent proteins EGFP (31) and UnaG (12) (SI Appendix, Fig. S13). Finally, no toxic or adverse effects were observed at the fluorogen concentrations typically used for imaging (SI Appendix, Fig. S14), suggesting that Y-FAST should enable long-term imaging in mammalian cells.
Specific Labeling of Fusion Proteins in Various Subcellular Locations.
Y-FAST labeling was shown to be robust in the biologically relevant pH range 5.5–8 (SI Appendix, Text S3 and Fig. S15), thus allowing anticipation of efficient labeling in most organelles. Labeling of HeLa cells expressing various Y-FAST fusions enabled visualization of proteins in various subcellular locations such as the cytoplasm, the nucleus, the cell membrane, the mitochondria, the Golgi apparatus, and the cytoskeleton (Fig. 1B). The general applicability of Y-FAST was further demonstrated by visualizing postsynaptic clusters of Gephyrin at inhibitory synapses in dissociated spinal cord neurons, illustrating the ability of Y-FAST to label proteins in more delicate cells and confined cellular compartments (Fig. 1C).
Specific Labeling of Fusion Proteins in Multicellular Organisms.
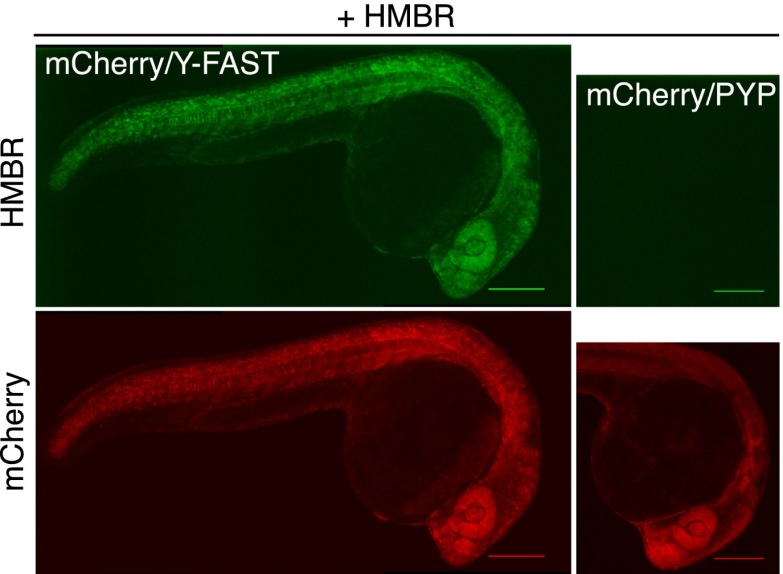
The labeling of Y-FAST was then validated in zebrafish embryo, as a model of a multicellular organism. Labeling of zebrafish embryos coexpressing mCherry and Y-FAST at different stages during embryogenesis revealed an expression pattern for Y-FAST indistinguishable from that of mCherry (Fig. 2 and SI Appendix, Fig. S16A), indicating that Y-FAST labeling was highly specific in vivo. Evaluation of embryo fitness after prolonged exposure (19 h) to HMBR revealed no mortality or developmental anomalies (SI Appendix, Fig. S16B), proving that HMBR was nontoxic for zebrafish embryos and suggesting therefore that Y-FAST could enable long-term imaging during embryogenesis.
Fig. 2.
Specific labeling of fusion proteins in vivo. Spinning-disk confocal micrographs of live zebrafish embryos coexpressing mCherry/Y-FAST or mCherry/PYP labeled with 5 μM HMBR at 24 h postfertilization (HMBR channel: Ex/Em 491/525–539 nm; mCherry channel: Ex/Em 561/605–664 nm). (Scale bars, 200 μm.) Side-by-side images were recorded using the same settings.
Real-Time Monitoring of Rapid Processes.
Most GFP-like fluorescent proteins fail to report on protein synthesis in real time because the rate of appearance of their fluorescence depends not only on the protein synthesis itself but also on the posttranslational formation of their chromophore. To show that the rapid labeling of Y-FAST could be an advantage in this context, we followed the cell-free expression of a fusion between mCherry and Y-FAST by monitoring their fluorescence emissions simultaneously (SI Appendix, Fig. S17). Even though a single protein was synthesized, we observed different rates of appearance for Y-FAST and mCherry fluorescence: Although Y-FAST could already be detected as soon as 10 min after the initiation of the protein synthesis, reaching saturation within 90 min, the mCherry signal only started to appear after 50 min and took over 4 h to reach saturation as a result of the slow maturation of its chromophore (32, 33). In addition, we compared the expression of mCherry−Y-FAST with that of EGFP and Venus (34), reported to mature within 10 min and 40 min in vitro, respectively (35), the bilirubin-inducible UnaG, and the Firefly luciferase, the latter often used as a reporter of protein synthesis. Although the expression of the different proteins was controlled by the same T7 promoter and should therefore occur at the same rate, we observed various rates of luminescence appearance (SI Appendix, Fig. S17). Our experiments revealed that Y-FAST clearly outperforms Venus and mCherry to report on protein synthesis in near real time, and provides kinetic information comparable with Firefly luciferase, EGFP, and UnaG.
The Labeling of Y-FAST is Highly Tunable.
Given that Y-FAST fluorescence directly depends on HMBR concentration, Y-FAST labeling can be controlled on demand. This feature was used to control the density of emitters independently of the protein expression level by tuning the fluorogen concentration (SI Appendix, Fig. S18). Titration experiments in cells were in good agreement with the in vitro results, indicating that the concentration of HMBR in the milieu reflected its intracellular level.
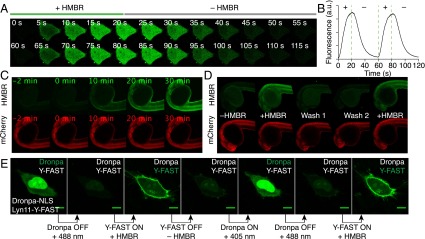
Because of its rapid, reversible, and highly dynamic labeling, Y-FAST can be rapidly switched on and off by addition and removal of HMBR. Labeling was shown to occur in cells within about 10 s, in accordance with high cell permeability of HMBR and immediate complex formation (Fig. 3 A and B). Rapid replacement of the medium with HMBR-free medium led to protein unlabeling on a similar timescale (Fig. 3 A and B), in accordance with the short residence time of HMBR. Using a microfluidic device and a multifunctional fluidic controller to switch repeatedly between the labeling and washing solutions, 10 cycles of labeling and unlabeling could be performed (Fig. 3 A and B and Movie S2).
Fig. 3.
On/off fluorescence switching by iterative labeling/unlabeling. (A and B) HeLa cells expressing mCherry-Y-FAST were grown in a microfluidic channel and repeatedly incubated with HMBR-containing culture medium for 20 s and HMBR-free culture medium for 40 s. A multifunctional fluidic controller enabled several cycles of labeling/unlabeling. HMBR concentration was 5 μM. (A) Confocal time lapse showing two cycles of labeling/unlabeling (Ex/Em 488/493–575 nm). Movie S2 shows 10 cycles of labeling/unlabeling. (B) Temporal evolution of the cell fluorescence upon addition (+) and removal (–) of HMBR. (C) Confocal time lapse showing the labeling kinetics in a zebrafish embryo expressing Y-FAST and mCherry (HMBR channel: Ex/Em 491/525–539 nm; mCherry channel: Ex/Em 561/605–664 nm). HMBR concentration was 10 μM. See also Movie S3. (D) A zebrafish embryo expressing Y-FAST and mCherry was imaged before addition of HMBR (–HMBR), 20 min after incubation with 10 μM HMBR (+HMBR), after two washings of 20 min (Wash 1 and 2), and after reincubation with 10 μM HMBR (+HMBR). (E) Confocal micrographs of live HeLa cells expressing Dronpa−NLS (nucleus) and lyn11−Y-FAST (membrane) showing sequential imaging of nuclear Dronpa and membrane-anchored Y-FAST through sequential on/off labeling of Y-FAST intercalated with on/off photoswitching of Dronpa (Ex/Em 488/493–797 nm). HMBR concentration was 5 μM. (Scale bars, 10 μm.)
The ability to rapidly switch Y-FAST on and off was further evaluated in zebrafish embryo. Significant staining was obtained within 20–30 min of incubation with HMBR (Fig. 3C and Movie S3). Dimensional analysis revealed that this timescale was in good agreement with the cell experiments (SI Appendix, Text S4), demonstrating that HMBR was also highly permeant in zebrafish embryo. This latter feature enabled reversing of the labeling, by washing away HMBR, and repeating of the labeling (Fig. 3D).
Y-FAST Opens New Opportunities for Multiplexing Imaging.
The ability to reverse the labeling allows the observation of spectrally indistinguishable targets using sequential rounds of fluorogenic labeling, imaging, and fluorogen removal. To validate this strategy, we expressed Y-FAST (fused to a membrane anchoring sequence) and the photoswitchable fluorescent protein Dronpa (36) (fused to a nuclear localization signal) in mammalian cells. Iterative labeling/unlabeling of Y-FAST combined with on/off photoswitching of Dronpa enabled imaging of the two proteins sequentially (Fig. 3E).
Discussion
Y-FAST is a PYP variant engineered to specifically activate the fluorogenic HMBR through two specific spectroscopic alterations—increase of fluorescence quantum yield and absorption red shift—providing a unique spectroscopic signature that ensures high imaging contrast. Y-FAST is comparable to common fluorescent proteins in terms of brightness and photostability, and is well suited for imaging proteins in various organelles and in a large variety of systems, from mammalian cells (including neurons) to microorganisms (e.g., E. coli, Saccharomyces cerevisiae) and zebrafish. Interestingly, Y-FAST is (i) half as large as GFP-like fluorescent proteins, which should ensure minimal functional perturbation within fusion proteins, and (ii) fully monomeric up to millimolar concentrations. Note that being monomeric up to millimolar concentrations corresponds to exhibiting no driving force for self-association up to an average intermolecular distance of about 10 nm. This latter feature should permit avoidance of unexpected oligomerization as encountered in certain cases with GFP-like proteins expressed in dense and compact environments (e.g., membranes) or within multimodule fusions (e.g., biosensors). All together, these properties make Y-FAST a good alternative to GFP-like fluorescent proteins.
Unlike GFP-like fluorescent proteins, which are fully fluorescent only after posttranslational formation of their chromophore, Y-FAST is fluorescent as soon as it is folded, provided that the fluorogen is present. This feature enabled following protein synthesis in near-real-time and opens new opportunities for (i) reporting on fast processes such as early promoter activation, (ii) labeling proteins with short lifetimes, or (iii) monitoring single translation or folding events in near real time.
The ability to control the fluorescence level of Y-FAST independently of its expression level (by choosing the concentration of fluorogen) could find interesting applications in quantitative biology (22). One challenge in this field is to detect changes in protein copy numbers in single cells or within specific compartments to understand their phenotypic state. However, protein copy numbers can vary by six orders of magnitude in mammalian cells (37), surpassing the dynamic range of most fluorescence detectors (typically three orders of magnitude). Systems like Y-FAST should allow the quantitative comparison of populations of cells displaying disparate protein copy numbers, just by adjusting the concentration of fluorogen.
Y-FAST distinguishes itself from other labeling systems because labeling is fully reversible. This feature provides high tunability and opens new exciting prospects to obtain multiplexed images for a large number of distinct target species. Indeed, the number of molecular species that can be imaged simultaneously is often limited by the spectral overlap between labels. Even though spectral deconvolution can be used, the number of molecular readouts that can be simultaneously measured in single cells remains limited, in most cases, to a handful of species. Several strategies have been proposed for multiplexing in fixed cells relying on iterative staining and removal (38–41): A first target is labeled with a first stain and imaged; the label is removed by physical or chemical means, after which a second orthogonal stain can be applied to label a second target, and so on. Even though these multiplexing approaches open very interesting prospects for imaging several tens of targets in a single cell, they remain limited to the study of fixed, permeabilized cells in which targets are labeled with oligonucleotide- or antibody-based probes. A collection of systems such as Y-FAST could extend this strategy to live cells, particularly when combined with recently developed targeted genome editing techniques (e.g., CRISPR-Cas9 system) (42).
The possibility to control the fluorescence on demand should also facilitate the implementation of FRET measurements. Estimate of FRET signal requires extensive controls to determine the extent of cross talk between donor and acceptor (43). The ability to perform multiple experiments on the same sample in the absence or presence of the fluorogen (and therefore with and without the contribution of Y-FAST) could improve FRET imaging protocols. Y-FAST could play the acceptor in a pair with CFP or the donor in a pair with mCherry (see SI Appendix, Text S5 and Fig. S19 for FRET characterization between Y-FAST and mCherry). The use of Y-FAST as acceptor could, in particular, permit easy determination of FRET efficiency by measuring the quenching of the donor fluorescence upon addition of the fluorogen, or, conversely, the decrease in fluorescence quenching by rapid washing of the fluorogen, thus competing with donor recovery after acceptor photobleaching techniques, but with the additional advantage of reversibility. The small size of Y-FAST is also an advantage for FRET, as it enhances the energy transfer efficiency by enabling a priori shorter Förster distances than GFP-like fluorescent proteins. FRET detection could further benefit from the ability to control the labeling density of Y-FAST independently of its expression level to set the donor:acceptor stoichiometry within the range of 1:10–10:1 to ensure detectable FRET signals.
Finally, the fast exchange dynamics of Y-FAST could be advantageously exploited for superresolution imaging in live cells. As Y-FAST interconverts spontaneously and rapidly at the single-molecule level between a dark (unbound) state and a bright (bound) state, it should behave as a blinking fluorophore (44). Fine-tuning of the exchange dynamics could give access to blinking rates adequate for Single-molecule Localization Microscopies (41, 45, 46) or Superresolution Optical Fluctuation Imaging (44, 47).
In conclusion, the strategy developed in this work is generic and may open new routes for the design of smart probes and biosensors. In particular, HMBR belongs to a series of conjugated donor−acceptor compounds exhibiting various photophysical/photochemical behaviors (48) that could facilitate the design of a collection of FASTs covering the whole visible spectrum for various applications in multiplexed bioimaging and biosensing.
Materials and Methods
Mammalian Cell Culture.
HEK293 and HeLa cells were cultured in DMEM supplemented with phenol red, Glutamax I, 10% (vol/vol) fetal calf serum (FCS), and 1% penicillin−streptomycin at 37 °C within a 5% CO2 atmosphere. For microscopic imaging, cells were seeded in μDish IBIDI (Biovalley) coated with poly-l-lysine. Cells were transiently transfected using Genejuice (Merck) according to the manufacturer’s protocol. Before imaging, cells were washed with PBS and incubated in DMEM (without phenol red) complemented with HBR or HMBR at the indicated concentration.
Neuron Cultures.
Cultures of dissociated spinal cord neurons were prepared from Sprague−Dawley rats (at embryonic day 14) as described previously (49). Neurons were maintained in neurobasal medium containing B27, 2 mM glutamax, 5 U/mL penicillin, and 5 µg/mL streptomycin at 36 °C and 5% CO2, cotransfected at day in vitro (DIV) 15 with Y-FAST−Gephyrin and mCerulean−Gephyrin plasmid DNA using Lipofectamine 2000 (Invitrogen), and used for experiments on DIV17. Neurons were imaged at 35 °C in MEM medium without phenol red, containing 33 mM glucose, 20 mM Hepes, 2 mM glutamax, 1 mM sodium pyruvate, and B27. HMBR was added by bath application at a final concentration of 10 µM in imaging buffer.
Zebrafish Experiments.
Zebrafish were maintained and staged according to Westerfield (50). Experiments were performed using the standard Ab wild type strain. The embryos were incubated at 28 °C. The animal facility obtained a French agreement from the ministry of agriculture for all of the experiments performed in this manuscript (Agreement C 75-05-12). The mRNA synthesis was performed using the mMESSAGE mMACHINE Transcription Kit (Ambion, Inc.). Equivalent volume of 100 ng/mL mRNA was injected into one-cell stage embryos. Embryos were allowed to grow in Volvic mineral water until imaging. To evaluate the effect of HMBR on embryogenesis, groups of about 50 embryos were incubated with HMBR solution at the indicated concentrations from 50% epiboly to 24 hours postfertilization.
Fluorescence Analysis.
Flow cytometry analyses were performed on an Accuri C6 cytometer (BD Biosciences). Confocal micrographs were acquired on a Zeiss LSM 710 Laser Scanning Microscope equipped with a Plan Apochromat 63×/1.4 NA oil immersion objective. ZEN software was used to collect the data. Images were analyzed with Image J. Spinning-disk confocal micrographs were acquired on a Nikon Eclipse Ti microscope equipped with a 4×/0.15 N.A objective and a coolSnap HQ2/CDDcamera (Princeton Instrument). Metamorph premier 7.6 software (Molecular Devices) was used to collect the data. Live epifluorescence imaging was performed on an inverted Nikon Eclipse Ti microscope with a 100× oil immersion objective (N.A. 1.49), a 1.5× magnifying lens, and a mercury lamp. Images were acquired with an Andor iXon EMCCD camera (image pixel size 107 nm).
SI Appendix, Materials and Methods contains Chemical Synthesis; Plasmid Constructions; Yeast Display; Protein Expression, Purification and Characterization; NMR Spectroscopy Experiments; Cell-Free Expression; Viability Cellular Assay; and Microfluidics.
Supplementary Material
Acknowledgments
We thank K. D. Wittrup, for providing us with the pCTCON2 vector and the EBY100 yeast strain for the yeast display selection, and A. Miyawaki, for providing us with plasmids for the expression of UnaG in bacteria and in mammalian cells. We also thank the Flow Cytometry Facility (IFR83) of the University Pierre and Marie Curie, and, more particularly, Annie Munier for her assistance. This work was supported by the Agence National de la Recherche (ANR-09-MNPS-013-01, ANR-11-BSV8-021-01, and ANR-14-CE09-0002-01), the Region Ile-de-France in the framework of C'Nano IdF (C'Nano IdF is the nanoscience competence of Paris Region, supported by CNRS, Commissariat à l'énergie atomique et aux énergies alternatives (CEA), Ministère de l'Enseignement et de la Recherche, and Region Ile-de-France), PSL Research University (project IMRESOV), France BioImaging, the Equipex Morphoscope 2, the ERC advanced research grant “PlasltInhib,” and program “Investissements d’Avenir” (ANR-10-LABX-54 MEMO LIFE and ANR-11-IDEX-0001-02 PSL Research University).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.M.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513094113/-/DCSupplemental.
References
- 1.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2(12):905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 2.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90(3):1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 3.Snapp EL. Fluorescent proteins: A cell biologist’s user guide. Trends Cell Biol. 2009;19(11):649–655. doi: 10.1016/j.tcb.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiedenmann J, Oswald F, Nienhaus GU. Fluorescent proteins for live cell imaging: Opportunities, limitations, and challenges. IUBMB Life. 2009;61(11):1029–1042. doi: 10.1002/iub.256. [DOI] [PubMed] [Google Scholar]
- 5.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67(1):509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 6.Remington SJ. Fluorescent proteins: Maturation, photochemistry and photophysics. Curr Opin Struct Biol. 2006;16(6):714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Zimmer M. Green fluorescent protein (GFP): Applications, structure, and related photophysical behavior. Chem Rev. 2002;102(3):759–781. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 8.Jullien L, Gautier A. Fluorogen-based reporters for fluorescence imaging: A review. Methods Appl Fluoresc. 2015;3(4):042007. doi: 10.1088/2050-6120/3/4/042007. [DOI] [PubMed] [Google Scholar]
- 9.Drepper T, et al. Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol. 2007;25(4):443–445. doi: 10.1038/nbt1293. [DOI] [PubMed] [Google Scholar]
- 10.Chapman S, et al. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc Natl Acad Sci USA. 2008;105(50):20038–20043. doi: 10.1073/pnas.0807551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu X, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9(4):e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumagai A, et al. A bilirubin-inducible fluorescent protein from eel muscle. Cell. 2013;153(7):1602–1611. doi: 10.1016/j.cell.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 13.Shu X, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324(5928):804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29(8):757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukinavičius G, et al. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat Chem. 2013;5(2):132–139. doi: 10.1038/nchem.1546. [DOI] [PubMed] [Google Scholar]
- 16.Hori Y, et al. Development of fluorogenic probes for quick no-wash live-cell imaging of intracellular proteins. J Am Chem Soc. 2013;135(33):12360–12365. doi: 10.1021/ja405745v. [DOI] [PubMed] [Google Scholar]
- 17.Yapici I, et al. “Turn-on” protein fluorescence: In situ formation of cyanine dyes. J Am Chem Soc. 2015;137(3):1073–1080. doi: 10.1021/ja506376j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szent-Gyorgyi C, et al. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat Biotechnol. 2008;26(2):235–240. doi: 10.1038/nbt1368. [DOI] [PubMed] [Google Scholar]
- 19.Shank NI, Zanotti KJ, Lanni F, Berget PB, Armitage BA. Enhanced photostability of genetically encodable fluoromodules based on fluorogenic cyanine dyes and a promiscuous protein partner. J Am Chem Soc. 2009;131(36):12960–12969. doi: 10.1021/ja9016864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozhalici-Unal H, et al. A rainbow of fluoromodules: A promiscuous scFv protein binds to and activates a diverse set of fluorogenic cyanine dyes. J Am Chem Soc. 2008;130(38):12620–12621. doi: 10.1021/ja805042p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Telmer CA, et al. Rapid, specific, no-wash, far-red fluorogen activation in subcellular compartments by targeted fluorogen activating proteins. ACS Chem Biol. 2015;10(5):1239–1246. doi: 10.1021/cb500957k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz SL, et al. Fluorogen-activating proteins provide tunable labeling densities for tracking FcεRI independent of IgE. ACS Chem Biol. 2015;10(2):539–546. doi: 10.1021/cb5005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McRee DE, et al. Crystallographic structure of a photoreceptor protein at 2.4 Å resolution. Proc Natl Acad Sci USA. 1989;86(17):6533–6537. doi: 10.1073/pnas.86.17.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baca M, et al. Complete chemical structure of photoactive yellow protein: Novel thioester-linked 4-hydroxycinnamyl chromophore and photocycle chemistry. Biochemistry. 1994;33(48):14369–14377. doi: 10.1021/bi00252a001. [DOI] [PubMed] [Google Scholar]
- 25.Borgstahl GE, Williams DR, Getzoff ED. 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: Unusual fold, active site, and chromophore. Biochemistry. 1995;34(19):6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 26.Dean KM, Palmer AE. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat Chem Biol. 2014;10(7):512–523. doi: 10.1038/nchembio.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber W, Helms V, McCammon JA, Langhoff PW. Shedding light on the dark and weakly fluorescent states of green fluorescent proteins. Proc Natl Acad Sci USA. 1999;96(11):6177–6182. doi: 10.1073/pnas.96.11.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hori Y, Ueno H, Mizukami S, Kikuchi K. Photoactive yellow protein-based protein labeling system with turn-on fluorescence intensity. J Am Chem Soc. 2009;131(46):16610–16611. doi: 10.1021/ja904800k. [DOI] [PubMed] [Google Scholar]
- 29.Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol. 2007;17(4):467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan JS, Imamoto Y, Harigai M, Kataoka M, Terazima M. Conformational changes of PYP monitored by diffusion coefficient: Effect of N-terminal α-helices. Biophys J. 2006;90(10):3686–3693. doi: 10.1529/biophysj.105.078196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373(6516):663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 32.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 33.Macdonald PJ, Chen Y, Mueller JD. Chromophore maturation and fluorescence fluctuation spectroscopy of fluorescent proteins in a cell-free expression system. Anal Biochem. 2012;421(1):291–298. doi: 10.1016/j.ab.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20(1):87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 35.Iizuka R, Yamagishi-Shirasaki M, Funatsu T. Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal Biochem. 2011;414(2):173–178. doi: 10.1016/j.ab.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 36.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306(5700):1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 37.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 38.Schubert W, et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol. 2006;24(10):1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 39.Zrazhevskiy P, True LD, Gao X. Multicolor multicycle molecular profiling with quantum dots for single-cell analysis. Nat Protoc. 2013;8(10):1852–1869. doi: 10.1038/nprot.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerdes MJ, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci USA. 2013;110(29):11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jungmann R, et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods. 2014;11(3):313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piston DW, Kremers G-J. Fluorescent protein FRET: The good, the bad and the ugly. Trends Biochem Sci. 2007;32(9):407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 44.Xu J, et al. Labeling cytosolic targets in live cells with blinking probes. J Phys Chem Lett. 2013;4(13):2138–2146. doi: 10.1021/jz400682m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uno S-N, et al. A spontaneously blinking fluorophore based on intramolecular spirocyclization for live-cell super-resolution imaging. Nat Chem. 2014;6(8):681–689. doi: 10.1038/nchem.2002. [DOI] [PubMed] [Google Scholar]
- 46.Kiuchi T, Higuchi M, Takamura A, Maruoka M, Watanabe N. Multitarget super-resolution microscopy with high-density labeling by exchangeable probes. Nat Methods. 2015;12(8):743–746. doi: 10.1038/nmeth.3466. [DOI] [PubMed] [Google Scholar]
- 47.Dertinger T, Colyer R, Iyer G, Weiss S, Enderlein J. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI) Proc Natl Acad Sci USA. 2009;106(52):22287–22292. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanchard-Desce M, et al. Large quadratic hyperpolarizabilities with donor−acceptor polyenes exhibiting optimum bond length alternation: Correlation between structure and hyperpolarizability. Chemistry. 1997;3(7):1091–1104. [Google Scholar]
- 49.Calamai M, et al. Gephyrin oligomerization controls GlyR mobility and synaptic clustering. J Neurosci. 2009;29(24):7639–7648. doi: 10.1523/JNEUROSCI.5711-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westerfield M. 1994. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish Danio (Brachydanio rerio) (Univ Oregon, Eugene, OR)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.