Significance
Drought and salinity are the major threats to crop productivity at a worldwide scale. A fundamental portion of the plant response to these environmental stresses occurs at the cell membrane, where the molecular machinery to preserve cell turgor and the appropriate balance of intracellular ions is found. The C2-domain ABA-related (CAR) family of proteins contributes to these processes by delivering the regulatory proteins controlling this machinery from other cell compartments to the cell membrane. Our analysis provides an explanation on how CAR proteins specifically reach a particular membrane place to develop their function and trigger the plant defense mechanism against stress.
Keywords: signaling, ion transport, membrane biology, abiotic stress
Abstract
Regulation of ion transport in plants is essential for cell function. Abiotic stress unbalances cell ion homeostasis, and plants tend to readjust it, regulating membrane transporters and channels. The plant hormone abscisic acid (ABA) and the second messenger Ca2+ are central in such processes, as they are involved in the regulation of protein kinases and phosphatases that control ion transport activity in response to environmental stimuli. The identification and characterization of the molecular mechanisms underlying the effect of ABA and Ca2+ signaling pathways on membrane function are central and could provide opportunities for crop improvement. The C2-domain ABA-related (CAR) family of small proteins is involved in the Ca2+-dependent recruitment of the pyrabactin resistance 1/PYR1-like (PYR/PYL) ABA receptors to the membrane. However, to fully understand CAR function, it is necessary to define a molecular mechanism that integrates Ca2+ sensing, membrane interaction, and the recognition of the PYR/PYL interacting partners. We present structural and biochemical data showing that CARs are peripheral membrane proteins that functionally cluster on the membrane and generate strong positive membrane curvature in a Ca2+-dependent manner. These features represent a mechanism for the generation, stabilization, and/or specific recognition of membrane discontinuities. Such structures may act as signaling platforms involved in the recruitment of PYR/PYL receptors and other signaling components involved in cell responses to stress.
Many of the plant-adaptive responses to environmental stresses occur at the cell membrane. In particular, those related to the regulation of plant ion transporters as stress unbalance cell ion homeostasis (1, 2). The phytohormone abcisic acid (ABA) and the second messenger Ca2+ have central roles in regulating plant stress tolerance through the control of the activity of various families of protein kinases and phosphatases that regulate the activation of different ion channels or transporters (3–7). Given the presence of the channel substrates at the cell membranes and the transient nature of their activation, the functioning of these systems relies on the regulated localization of different molecular entities in the vicinity of the channels via protein–protein (8, 9) and/or protein–membrane interactions (10, 11).
The pyrabactin resistance 1/PYR1-like (PYR/PYL)/regulatory components of ABA receptors (RCAR) receptors perceive intracellular ABA levels and, as a result, form ternary complexes with clade A protein phosphatases type 2C (PP2C), thereby inactivating them (12–15). This prevents the PP2C-mediated dephosphorylation of ABA-activated sucrose nonfermenting 1-related protein kinases (SnRKs) subfamily 2 (SnRK2s), which results in the activation of an SnRK2-dependent phosphorylation cascade affecting a high number of targets in the plant cell (16, 17). As a result, ABA-activated SnRK2s are key players in regulating transcriptional responses to ABA and processes at the plasma membrane such as the activation of the R- and S-type anion channels and the subsequent efflux of K+ and water that modulate, in turn, stomata aperture and plant transpiration (4, 18–20). Particularly, SnRK2.6/OST1 plays a crucial role in regulating the R-type QUAC1 and the S-type SLAC1 anion channels (20, 21). Thus, membrane-delimited events play a critical role in ABA signaling. Some members of the Calcineurin-B like (CBL) family of plant Ca2+ sensors are often localized at the plasma membrane (11, 22), where they mediate the Ca2+-dependent interaction and activation of the CBL-interacting protein kinases (CIPKs) (10) and the Ca2+-independent interaction of some ion channels such as AKT1 (23). CIPKs together with certain PP2Cs tune the phosphorylation state and the activity of the Na+/H+ SOS1 antiporter under salt stress (24–27) or the K+ transporter AKT1 under low K+ stress (8, 28). Interestingly, the OST1-regulated anion channel SLAC1 is also phosphorylated by certain CIPKs and Ca2+-dependent protein kinases (CPKs) (6, 29, 30). Conversely, the clade A PP2C ABI1 acts as a negative regulator not only for OST1 but also for the calcium-dependent CPK21/CPK23 and CIPK23 kinases (6, 29). Thus, the above examples illustrate a merging point between Ca2+ and ABA signaling at the cell membrane for the response to dehydration stress.
The family of C2-domain ABA-related (CAR) proteins in Arabidopsis thaliana consists of single C2 domains that mediate the Ca2+-dependent docking of PYR/PYL ABA receptors to the membrane. This process is relevant for ABA signaling, because car triple mutants display reduced sensitivity to ABA in vivo (31). Thus, CARs represent a signaling node that connects the ABA and Ca2+ signaling pathways to the membrane. The crystal structure of CAR4 (31) showed that CAR proteins display the canonical fold of C2 domains, consisting of a compact β-sandwich composed of two four-stranded β-sheets, plus an insertion of 43 amino acids that connects both β-sheets and folds as an α-helix followed by a β-hairpin. This motif represents a unique CAR-family signature that is absent in other known families of C2 domains but conserved in crop CAR orthologs (31). Biochemical data showed that this region is involved in the interaction and recruitment of the ABA receptors to the membrane (31). The analysis of the structure of CAR4 revealed that Ca2+-dependent membrane binding and PYR/PYL receptor binding could take place simultaneously as these interactions occur at opposite sides of the CAR molecule. However, available biochemical data on the CAR and PYR/PYL interaction showed no definite stoichiometry (31), suggesting a more complex mechanism of action for CAR proteins that merely act as molecular carriers of the PYR/PYL receptors to the membrane.
An additional function for some C2 domains consists of the ability to generate and sense membrane curvature by the concerted insertion of a protein segment from adjacent proteins into one leaflet of the membrane bilayer. In this situation, the C2 domain acts as a molecular wedge that produces membrane bending (32). This has been shown for the C2 domains of synaptotagmin that produce a strong positive curvature to orchestrate the formation of the SNARE complex and promote the fusion of the membrane (33). This process is relevant, as the regulated endocytosis of plasma membrane proteins and their subsequent recycling controls housekeeping cellular functions and different aspects of cellular homeostasis and response to environment (34). Indeed, it has been found that ABA triggers similar processes to control selectively the population of the Arabidopsis KAT1 K+ channel at the plasma membrane (35). Additionally, ABA receptors have been shown to be a substrate of the membrane-bound ubiquitin ligase RSL1 and to be targeted to degradation (36). Whether this ubiquitylation triggers membrane endocytosis and recycling remains to be investigated, but it has been suggested that CAR proteins might be involved in this process. Membrane curvature has also been shown to be essential for the generation of membrane raft-like liquid ordered phases and, in turn, for the localization of lipid-anchored proteins (37). Interestingly, CAR proteins, among other ABA signaling components, might be localized on membrane nanodomains (38). However, it is still unclear how these ABA nanodomains are generated and maintained. Thus, exploring the ability of CARs to form oligomers at the cell membrane and their effect on membrane structure may provide molecular insights on different membrane-delimited signaling events.
To investigate the molecular mechanism underlying the function of CAR proteins, we performed structural and biochemical studies and analyzed both Ca2+ and membrane-binding properties of CAR proteins. Our data show that CARs are peripheral membrane proteins that penetrate into the bilayer in a Ca2+-dependent manner and originate membrane curvature. Moreover, the crystallographic analysis of CAR1 and CAR4 provides a structural framework that integrates the membrane-related functions with PYR/PYL receptor binding. This model suggests that clusters of CAR proteins can act as a membrane assembly point for ABA receptors and other proteins participating in the regulation of ion homeostasis or those involved in the regulated degradation of the receptors.
Results and Discussion
The Structure of CAR Proteins Defines Two Lipid Binding Sites.
Despite the available biochemical and structural information, there are aspects of the mechanism of action of CAR proteins that are not understood. Little is known about the membrane binding properties of these proteins, especially those features related to the molecular mechanism underlying their lipid specificity. This relies on the structure and amino acid composition of the canonical Ca2+-dependent and the polybasic lipid binding sites (39–41). The binding of C2 domains through the latter determines the orientation of the domain with respect to the membrane. Thus, whether this lipid binding site is functional or not for CAR proteins is important, as it may determine their membrane localization and the topology of the binding for correct functioning (42). To gain molecular insight into these aspects, we determined the X-ray structures of (i) CAR1 in complex with one Ca2+ ion; (ii) CAR4 in complex with two Ca2+ ions and the phospholipid 1,2-dihexanoyl-sn-glycery-3-phospho-l-serine (PSF); (iii) CAR4 in complex with two Ca2+ ions, a Zn2+ ion, and two molecules of phosphorylcholine (POC); and (iv) CAR4 in complex with two Ca2+ ions and a Zn2+ ion (Fig. 1, Fig. S1, Table S1, and Materials and Methods). The overall structures of CAR1 and CAR4 are very similar [sequence identity, 68%; Cα backbone root-mean-square deviation (rmsd), 0.77 Å], with differences confined to the side chains of the residues implicated in Ca2+ binding and in the loops connecting α1 and βB, which are not conserved among the CAR family (Fig. S2). The POC and PSF molecules are bound to CAR4 at the concave side of the domain, defining a Ca2+-independent lipid binding site. The structures of the different CAR4 complexes are nearly identical (rmsd ranging from 0.14 to 0.30 Å).
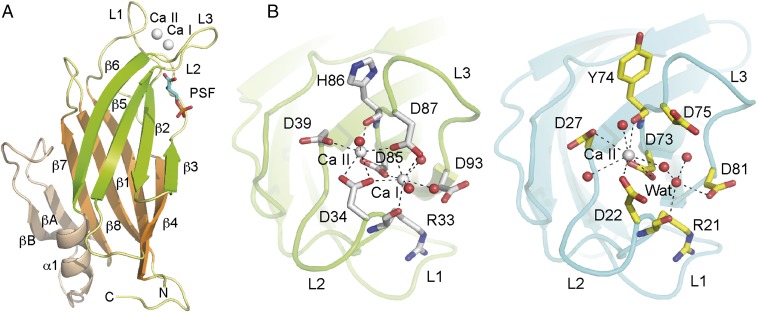
Fig. 1.
The crystal structures of CAR4 and CAR1. (A) A ribbon representation of CAR4 in complex with Ca2+ and PSF. (B) The ribbon representation of the Ca2+-dependent lipid binding sites of CAR4 (Left) and CAR1 (Right). The key structural features of the CAR fold are labeled and highlighted in different colors. The Ca2+ atoms are shown as gray spheres. Other nonprotein atoms and key residues at the Ca2+ binding sites are displayed in a ball-and-stick representation.
Fig. S1.
The lipid binding sites of CAR4. (A) The analysis of the anomalous difference density maps computed with data collected with crystals of CAR4 Ca2+ and Zn2+ at wavelength 1.7 (green) and 0.93 A (magenta) allows the unambiguous identification of the nature of the CAR4 metal centers, as Zn2+ and Ca2+ absorb radiation differentially at these wavelengths. The maps are contoured at 7 σ. (B) The polybasic binding site of CAR4 and three sections of the 2Fo-Fc simulated annealed omit map at the polybasic lipid binding site of CAR4 for the complex between CAR4 and PSF, the complex between CAR4 and POC, and the complex between CAR4, Zn, and POC.
Table S1.
Data collection and refinement statistics
| Crystallographic data | CAR4:Ca:PSF | CAR4:Ca:Zn:POC | CAR4:Ca:Zn | CAR1:Ca |
| Data collection | ||||
| Beam line ESRF | ID14-4 | ID14-1 | ID29 | ID23-1 |
| Wavelength | 0.94 | 0.93 | 1.71 | 0.98 |
| Resolution, Å | 46.7–1.06 (1.62–1.60) | 47.5–2.40 (2.5–2.40) | 46.90–2.20 (2.26–2.2) | 23.23–1.65 (1.75–1.65) |
| Space group | P212121 | P212121 | P212121 | C2 2 21 |
| a | 35.58 | 35.74 | 35.29 | 39.60 |
| b | 89.77 | 89.15 | 88.93 | 170.30 |
| c, Å | 109.64 | 112.47 | 110.57 | 53.21 |
| Rsym | 0.05 (0.38) | 0.09 (0.52) | 0.11 (0.86) | 0.06 (0.70) |
| Rpim | 0.02 (0.12) | 0.06 (0.33) | 0.03 (0.26) | — (—) |
| I/σ(I) | 32.1 (6.7) | 14.2 (3.3) | 17.2 (2.9) | 20.79 (3.07) |
| Completeness, % | 99.9 (99.0) | 99.2 (99.0) | 100 (100) | 99.4 (99.1) |
| Redundancy | 13.8 (11.2) | 6.3 (6.5) | 6.6 (6.2) | 5.9 (5.9) |
| Number of images | 360 | 161 | 360 | 200 |
| Δϕ, ° | 1 | 1 | 1 | 1 |
| Refinement | ||||
| No. reflections | 88,958 | 27,044 | 36,649 | 22,034 |
| Rwork/Rfree | 0.19/0.22 (0.24/0.27) | 0.21/0.26 (0.30/0.32) | 0.22/0.26 (0.32/0.35) | 0.21/0.24 (0.32/0.34) |
| No. atoms | ||||
| Protein | 2,631 | 2,634 | 2,630 | 1,215 |
| Water | 533 | 139 | 105 | 96 |
| Heteroatoms | 4 Ca | 4 Ca, 1 Zn | 4 Ca, 1 Zn | 1 Ca, 2 SO4, 1 GOL |
| Ligands | 1 PSF | 2 POC | ||
| rmsd | ||||
| Bond lengths, Å | 0.006 | 0.008 | 0.009 | 0.008 |
| Bond angles, ° | 1.075 | 1.210 | 1.297 | 1.247 |
The limits of the maximum resolution range and the corresponding statistics are indicated in parentheses.
Fig. S2.
The ribbon representation of the crystal structures of CAR4 (Left) and CAR1 (Right).
The Ca2+-dependent lipid binding site.
The structural basis of the Ca2+-dependent interaction of the C2 domains with membranes lies in the change of the electrostatic potential of the site upon Ca2+ binding. This change promotes the direct interaction of the phosphate moiety of lipids with the Ca2+ ion (43). The Ca2+ binding sites (Ca I and Ca II) of CAR4 and CAR1 are placed in a cavity formed by three loops (L1, L2, and L3) at one side of the β-sandwich. The side chains of five conserved aspartate residues and two main chain carbonyl oxygen atoms at loops L1 and L3 are responsible for Ca2+ binding (Fig. 1B). Three water molecules at site I and one at site II complete the coordination of the two observed CAR4 Ca2+ binding sites. Conversely, the CAR1 crystal structure shows that only site II binds Ca2+. Interestingly, Ca2+-containing buffers were not used during the purification and crystallization of CAR1. This suggests that Ca2+ site II displays a high affinity for Ca2+ and it is permanently occupied even at a low physiological Ca2+ concentration, whereas site I shows lower affinity for Ca2+. This observation suggests that Ca2+ site I in CAR proteins could be involved in the reported Ca2+-dependent phospholipid binding activity.
The polybasic lipid binding site.
Structural and biochemical studies on the C2 domains of PKC-α (39), synaptotagmin 1 (44), and rabphilin (40) demonstrated that a group of cationic and aromatic amino acids at the concave side of the C2 β-sandwich are responsible for the specific recognition of the polar head of anionic phospholipids such as phosphatidylinositol polyphosphate (40, 41, 44). The specificity of these sites determines the membrane localization and the functionality of the C2 domain. For instance, the C2B domain of synaptotagmin 1 bridges different membranes through the specific interaction of the polybasic region with phosphatidylinositol and the Ca2+-dependent interaction with phosphatidylserine (44). In addition, the occupancy of these sites determines the orientation of the C2 domain with respect to the membrane (42, 45) and membrane curvature (40).
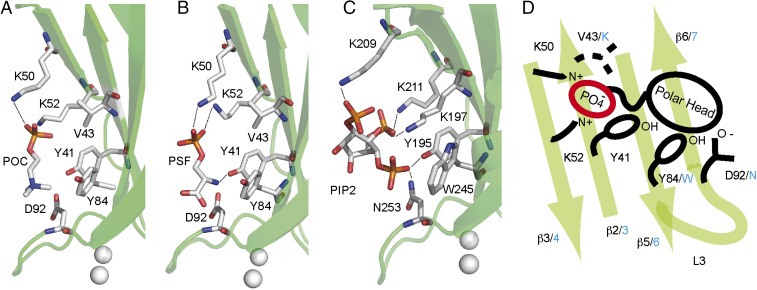
The electron density maps and the anomalous difference maps corresponding to the CAR4 complexes revealed the presence of molecules of POC, PSF, and one Zn2+ ion at the exposed cavity formed by the β strands β3, β2, β5, and β6 (Fig. 1A and Fig. S1). The analysis of these structures showed that Lys-50 and Lys-52 residues at β3 are involved in the stabilization of the phosphate groups of POC and PSF, whereas the positively charged trimethylamine moiety of POC and the primary amine form of PSF are interacting with the hydroxyl group of Tyr84 at β5, the aromatic ring of Tyr-41 at β2, and the Asp-92 side chain at the outer side of loop L3 (Fig. 2). This lipid binding site has been structurally characterized for the PKC-α in complex with the negatively charged phosphatidyl inositol biphosphate [PI(4,5)P2] (4, 5). The architecture of the pocket is remarkably conserved despite the different polarity of POC, PSF, and PI(4,5)P2. In all of the situations, the phosphoryl group of the polar lipid head is perpendicular to the strands forming the β-sheet and the polar head close to loop L3. Lys residues at β3 and the aromatic residues are conserved in the three structures. However, an additional Lys residue from the β-sheet and one Asn residue at L3 are stabilizing the negative charges of the PI(4,5)P2 in the C2 domain of PKC-α. In contrast, these residues correspond to Val-43 and Asp-92, respectively, in CAR4, thus precluding PI(4,5)P2 binding at this site. Interestingly, those residues are conserved among the members of the CAR family (31), thus suggesting that these lipid binding properties are a general feature of the family.
Fig. 2.
The polybasic lipid binding site of CAR4. (A) Structure of CAR4 in complex with POC and (B) with PSF. (C) Structure of the C2 domain of PKC-α in complex with PI(4,5)P2 is shown for comparison. The C2 domains are represented as green ribbons. Residues involved in ligand binding are represented as sticks. (D) Schematic representation of the polybasic lipid binding site highlighting the similarities and differences between CAR4 and PKC-α. Key CAR4 amino acids are labeled in black, and the corresponding amino acids in PKC-α are indicated in blue.
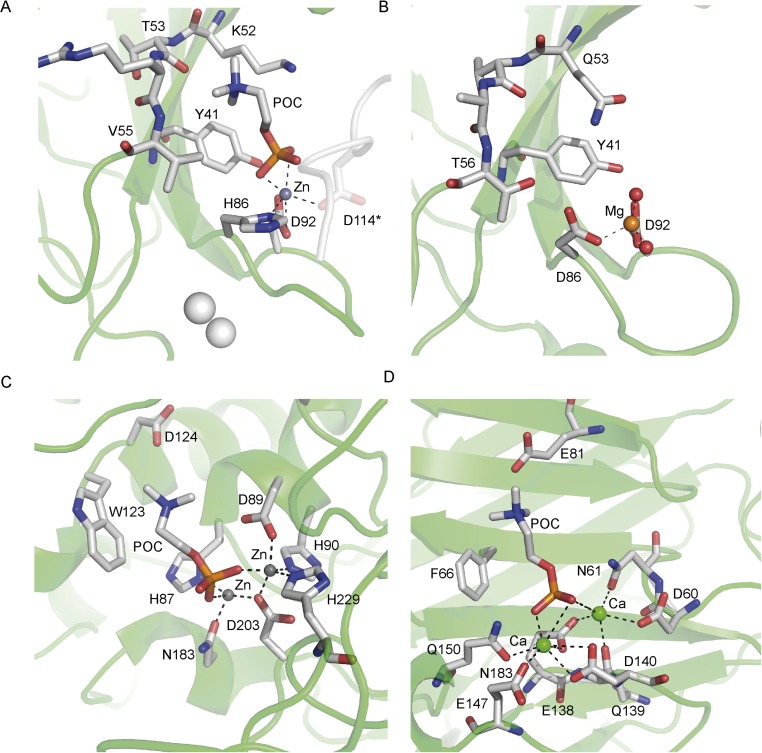
The crystals of CAR4 grown in the presence of Zn2+ coordinate this ion with Asp-92 and His-86 at loop L3 of one of the molecules of CAR4 in the asymmetric unit and with the carboxylate side chain of the symmetry-related Asp-114 (Fig. S3). The coordination sphere of this ion was completed alternatively with water molecules or, if POC is present in the crystallization conditions, with its phosphate group. In this situation, the positive charge of the POC trimethylamine is stabilized by the cation π interaction with the Tyr-41 side chain and with the Thr-52 main chain carboxylate from loop L2. Val52 and the hydrophophic moiety of the Lys-52 side chain further stabilize the POC moiety at this site. Interestingly, this pattern of interactions closely resembles those found for POC in the complexes with the pneumococcal phosphoryl choline esterase (46) or the C-reactive protein (47) where Zn2+ or Ca2+ ions, respectively, stabilize the phosphate moiety and an aromatic ring, together with a donor oxygen, interacts with the trimethyl amine group. In addition, a similar magnesium binding site has already been reported for the C2 from PKC-ε (48). Interestingly, this C2 domain has the ability to interact with membranes through this site in a Ca2+-independent manner.
Fig. S3.
Structural comparison of the metal-dependent POC binding sites of (A) CAR4, (B) the C2 domain of PKC-e (PDB ID code 1GMI), (C) phosphoryl choline esterase (PDB ID code 2BIB), and (D) C reactive protein (PDB ID code 1B09).
Altogether, the crystallographic data reveal that both the Ca2+-dependent and the polybasic lipid binding sites of CAR proteins are functional. However, the polybasic site does not display the characteristic PIP2 binding signature found in other C2 domains (41), which gives it the ability to recognize PIP2-enriched membranes. These features support a membrane binding model in which the unspecific binding of CAR proteins to the membrane via the polybasic site would reinforce the Ca2+-dependent binding. This would facilitate and restrict a closer contact of CAR proteins to the membrane.
CAR Proteins Are Calcium Sensors with a Basal Phospholipid Binding Activity.
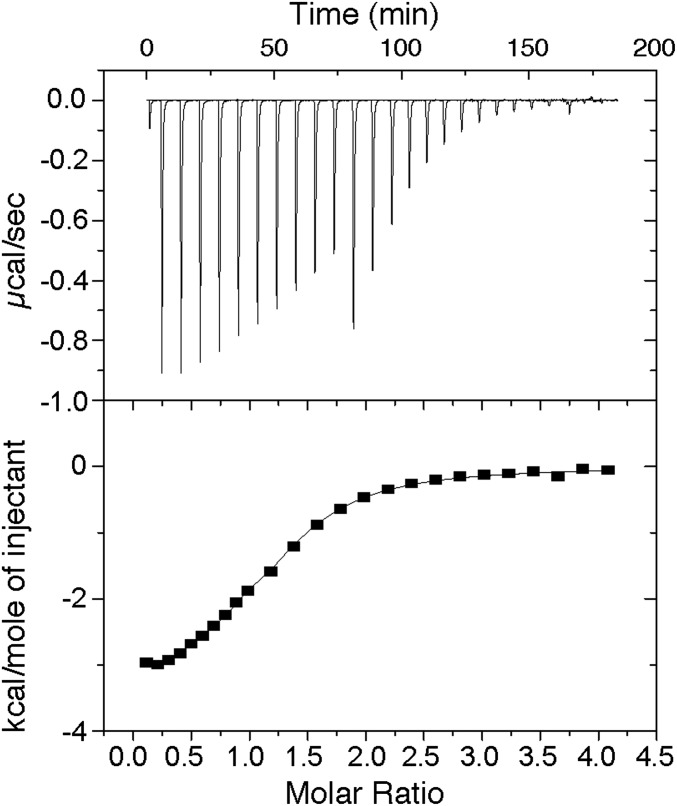
To quantify the Ca2+ binding properties of CAR proteins and to test if they display two different classes of Ca2+ binding sites—one to detect physiological μM variations of Ca2+ concentration and the other with a structural role—we used isothermal titration calorimetry (ITC). Our data show that the binding isotherm fits to the one-set-of-sites model in which CAR4 binds only one Ca2+ ion (n = 1.17) with a dissociation constant (Kd) of 10.0 μM (ΔH = –3.4 kcal∙mol−1) (Fig. 3). Identical results were obtained either including or not including Ca2+ during protein purification. These results indicate that only one of the two Ca2+ sites was depleted by exhaustive dialysis against a Ca2+-free buffer. Indeed, extensive dialysis of the CAR4 protein against the Ca2+ chelating agent EGTA induces precipitation of the protein. These thermodynamic data on CAR4 correlate well with the observed CAR structures and support the hypothesis that site I represents the lower affinity Ca2+ binding site that would be occupied depending on the physiological Ca2+ concentration whereas site II plays a structural function. These observations are consistent with those reported for other C2 domains (49, 50) and with those described for other plant Ca2+ sensors (51). However, in guard cells, ABA induces an increase in the Ca2+ cytosolic concentration from 0.07 to 1 μM (5–7). Taking into account that the observed CAR4:Ca Kd for site I is 10 μM, this might indicate that CAR4 is not a good Ca2+ sensor at physiological concentrations. However, in the presence of biological membranes, an enhancement of the Ca2+ affinity is expected, as Ca2+ bridges the protein with the membrane phospholipids. Therefore, the overall CAR4:Ca2+ Kd would be a combination of the Ca2+ affinity for the protein and the CAR4:Ca2+ affinity for the membrane. Moreover, the latter would depend on the lipid composition. This has been observed for the C2 domain of synaptotagmin 1 (50) and dysferlin (49), for which their Ca2+ affinity is markedly increased by the presence of synthetic liposomes. Indeed, the calculated CAR4 and CAR1 Kd for Ca2+ in the presence of liposomes are 7.4 μM and 1.4 μM, respectively (31), below the observed Kd measured in solution without lipids. Therefore, the ITC value for Ca2+ binding should be taken as an upper limit of the Ca2+ affinity in the in vivo membrane environment.
Fig. 3.
Calorimetric titration of CAR4 with Ca2+. (Upper) Representative thermogram obtained by the addition of 2 mM Ca2+ (1 × 1-μL, 10 × 5-μL, and 10 × 10-μL injections) to a solution of 64.4 μM CAR4 at 25 °C. (Lower) Dependence of the heat released per mol of Ca2+ injected as a function of the Ca2+:CAR4 molar ratio. The solid line corresponds to the best fit of the experimental data based on a one-set-of-sites model.
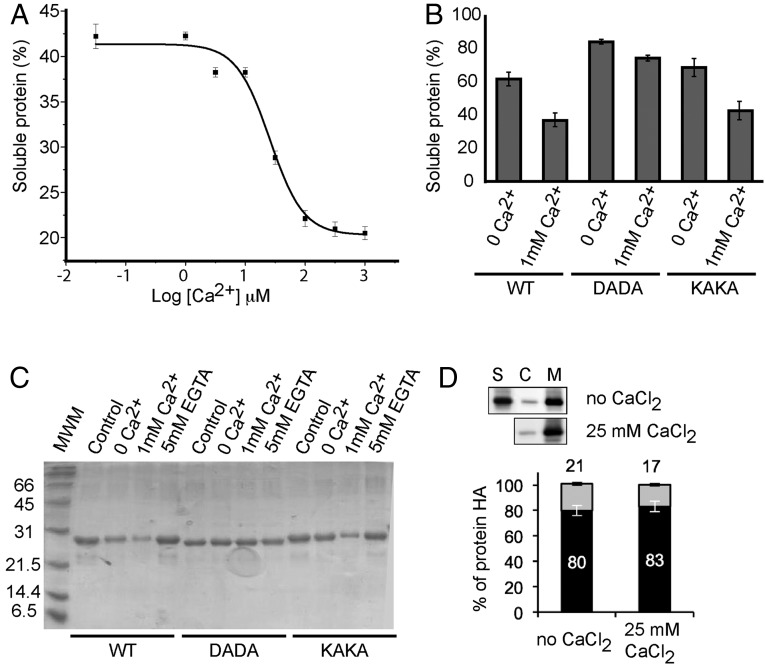
To further characterize the role of the two different Ca2+ sites, we tested whether CAR4 was able to bind negatively charged synthetic liposomes formed by 75% phosphatidyl choline and 25% phosphatidyl serine in the presence or absence of free Ca2+ (Fig. 4A) as previously reported for CAR1 (31). However, because our structural studies revealed that Ca2+ site II plays a structural role in CAR proteins, we performed the lipid cosedimentation assays without previously treating phospholipid vesicles with EGTA to avoid chelation of the Ca2+ present in the structural site. Our results show that CAR4 binds liposomes in the absence of exogenous Ca2+. This activity remains unaltered up to around 1 μM Ca2+ and increases at higher concentrations (Fig. 4A). This indicates that the basal liposome binding below 1 μM Ca2+ might depend on structural Ca2+ binding site II. However, it may also be possible that the polybasic lipid binding site would account for the basal membrane binding activity. To analyze this possibility, we generated a Lys50Ala Lys52Ala double mutant protein (CAR4-KAKA). These residues are directly involved in lipid binding at the polybasic binding site of CAR4 (Fig. 2), and equivalent mutations in the C2 domain of PKC-α abolish lipid binding to this site (40). Our results show a small reduction in the ability of this mutant to bind liposomes in the absence of free Ca2+ (Fig. 4B) that does not account for the observed basal activity—thus indicating that polybasic binding does not determine the basal membrane-binding properties of CAR4. To corroborate if the Ca2+ binding site accounts for this activity, we prepared the Asp85Ala Asp87Ala double mutant protein (CAR4-DADA), which is unable to stabilize lipids in the Ca2+-dependent lipid binding site (31). Our results show that CAR4-DADA does not retain the basal phospholipid binding (Fig. 4B). This shows that the canonical Ca2+-mediated lipid binding sites are mainly responsible for this activity and suggests that the structural Ca2+ binding site II is involved in the basal interaction with the liposomes. To further test this hypothesis, we incubated the CAR4-bound pelleted liposomes with EGTA and corroborated that CAR4 binding was reversed (Fig. 4C). All together, our data suggest that CAR proteins display site II-mediated basal membrane binding activity. In addition, the increase in the lipid binding activity of CAR4 observed above 1 μM Ca2+ would probably denote saturation of site I.
Fig. 4.
In vitro membrane binding and subcellular localization of CAR proteins. (A) Calcium-dependent lipid cosedimentation assays with CAR4. The protein was incubated with liposomes in the presence of increasing concentrations of Ca2+. (B) Comparative analyses of phospholipid binding of wild-type CAR4 and mutants at the Ca2+-dependent lipid binding site CAR4-DADA and the polybasic lipid binding site CAR4-KAKA. Protein quantifications of the soluble fraction after lipid pelleting on A and B were performed by measuring the optical density at 280 nm on a spectrophotometer. Lipid binding activity is expressed as the percentage of the unbound protein to lipids. The differences in the percentage of soluble protein in A and B are explained by the different protein concentration used in the experiments (Materials and Methods). Error bars indicate the SD calculated from three independent measurements. (C) A representative Coomassie blue-stained SDS/PAGE corresponding to the experiments shown in B. (D) Quantification of the subcellular location of CAR4 in the presence or absence of free Ca2+; S, C, and M represent the nonnuclear protein fraction, the cytosolic fraction, and the microsomal fraction, respectively. Immunoblot signals obtained in D were captured using the image analyzer LAS3000, and quantification of the protein signal was done using Image Guache version 4.0 software.
To explore if CAR proteins display a basal interaction with cell membranes, we examined the subcellular localization of the nonnuclear fraction of CAR4 protein by a fractionation technique (31) and analyzed the soluble cytosolic and the pelleted insoluble microsomal fractions in the absence or presence of exogenous Ca2+. Our analysis shows that at least 80% of CAR4 is localized in the microsomal fraction, thus supporting that CAR membrane localization is Ca2+-independent of the addition of exogenous Ca2+ (Fig. 4D). These experiments also show that 20% of CAR4 protein remains in the soluble cytosolic fraction. This is a general feature of other CAR proteins, the percentage of soluble fraction being variable from 8% to 20%. Hence, we cannot discard an uncharacterized cytosolic function of CAR proteins. However, it is worth noting that in vivo cell localization studies showed that both CAR proteins and CAR PYR/PYL complexes are localized at the cell membrane (31).
CAR Proteins Generate Membrane Curvature in a Ca2+-Dependent Manner.
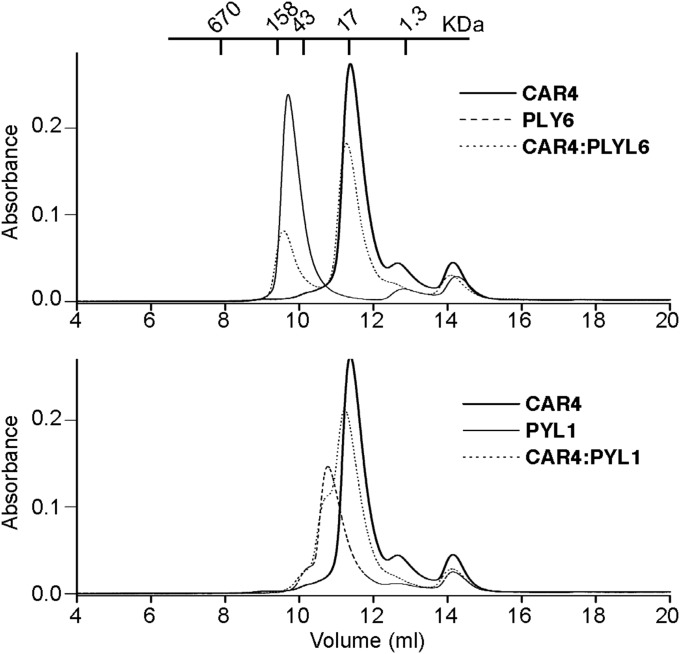
Previous studies showed that CAR proteins contain a protein–protein interaction module defined by the α1βAβB CAR signature, which is responsible for the membrane docking of PYR/PYL receptors to membranes (31). To analyze whether this interaction occurs in the absence of phospholipid membranes, we preformed in vitro binding assays in solution using the recombinant proteins. The analysis of the size and shape of protein samples by size exclusion chromatography indicates that there is no or very weak interaction upon mixing CAR4 and those PYR/PYL receptors reported to be recruited to membranes by CAR4 (31) (Fig. 5A). This supports that the CAR PYR/PYL interaction occurs at the cell membrane and suggests the notion that CAR binding to the plasma membrane stimulates the recruitment of the PYR/PYL receptors. This would imply that CAR proteins either undergo a structural rearrangement at the cell membrane and/or a change in their oligomerization state to promote PYR/PYL binding. Interestingly, the C2 domain B of synaptotagmin forms functional oligomers at the cell membrane to regulate membrane fusion. These oligomers are disrupted by physiological concentrations of Ca2+ to promote membrane fusion (52).
Fig. 5.
PYR/PYL ABA receptors do not interact with CAR4 in solution. Size exclusion chromatography profiles of CAR4, dimeric PYL1, monomeric PYL6, and their equimolecular mixtures as described in Materials and Methods.
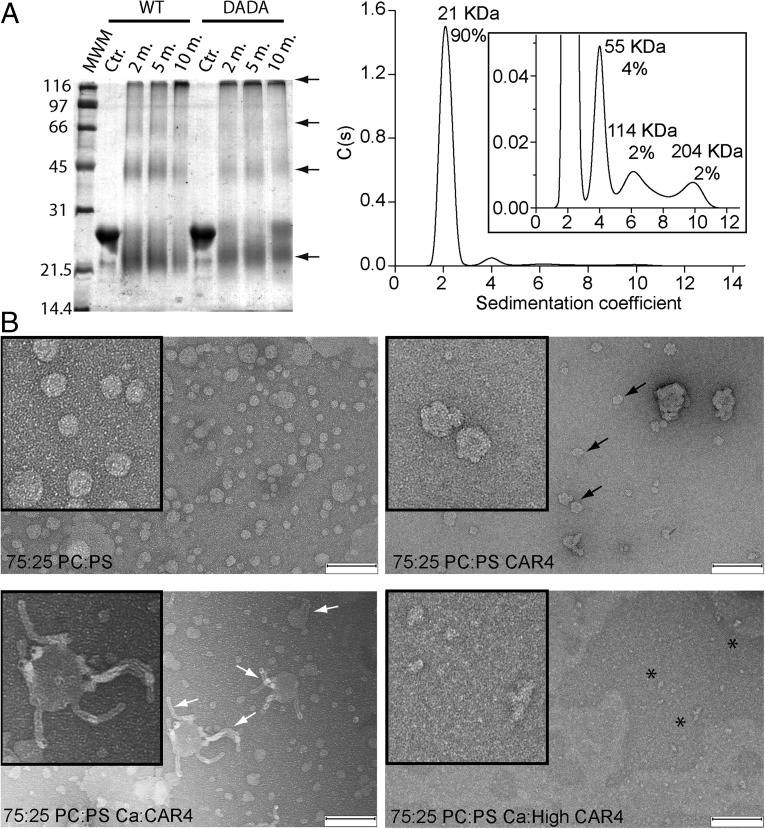
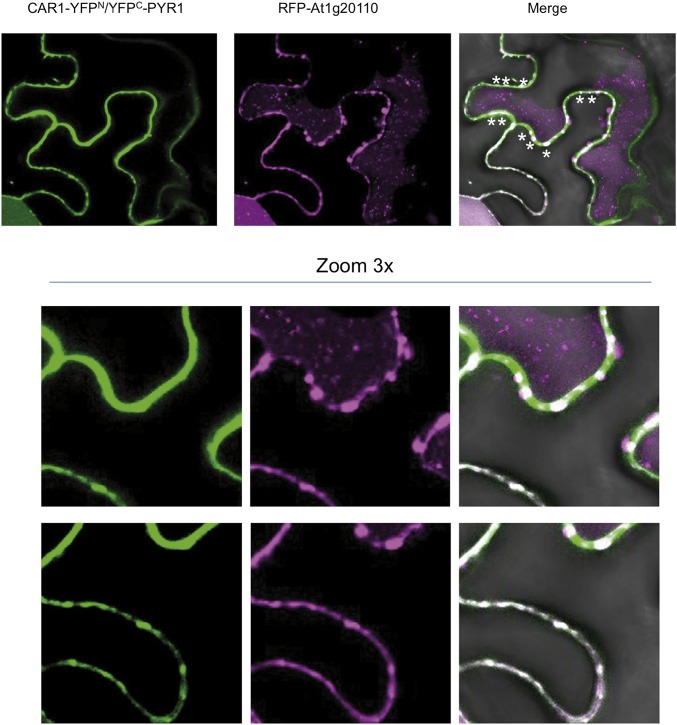
To investigate these possibilities, we first examined the formation of oligomeric structures of CAR4 and the Ca2+-insensitive mutant CAR4-DADA using glutaraldehyde as a chemical cross-linker agent followed by SDS/PAGE (53). Our results show that both proteins form dimers and higher order oligomers, revealing that CAR4 self-associates and that this process is Ca2+-independent. Additionally, we carried out analytical ultracentrifugation experiments in native conditions that further corroborate this observation (Fig. 6A). To examine whether these oligomeric structures are formed on the membrane, we used negative stain transmission electron microscopy (EM). This technique allows the direct identification of membrane-bound protein. This approach has been used to determine the molecular function of different proteins. EM was used to determine the scaffolding function of focal adhesion kinase by the visualization of protein clusters upon liposome binding (54) and to determine how the synagtotagmin C2 domain insertion into membranes induces curvature and promotes membrane fusion (33). Thus, we incubated liposomes with CAR4 in the absence or presence of free Ca2+ and analyzed these preparations using EM (Fig. 6B). Our results show that CAR4 produces roughness on the surface of the liposomes in the absence of free Ca2+. This can be explained as a consequence of the peripheral binding of CAR4 molecules to the membrane and further supports that CAR4 displays basal membrane binding activity. However, the addition of free Ca2+ to CAR4 liposome preparations produces strong tubulation of the membranes similar to the bilayer bending induced by the C2 domains of synagtotagmins (33). Remarkably, the average diameter of CAR4-induced tubules is around 17 nm, the size identical to that observed for the synagtotagmins. This indicates that CAR proteins and synagtotagmins share the capacity to induce and stabilize highly curved membrane. At the molecular level, it has been proposed that this phenomenon is a consequence of the coordinated shallow insertion of C2 domain clusters into one leaflet of the lipid bilayer, at the region occupied by the rigid lipid glycerol backbones. In this situation, the protein acts as a wedge that produces a local bending of the membrane, generating positive curvatures (32). Interestingly, EM experiments show that increasing the CAR4-to-liposome ratio that yields tubulation induces liposome disruption and the formation of CAR4 large-molecular-weight aggregates (Fig. 6B). Therefore, the joint analysis of the biochemical and the EM data suggest that CAR proteins self-associate and form clusters on the membrane. In addition, an increase in the Ca2+ concentration leads to membrane insertion of these oligomeric structures, producing membrane curvature. This necessarily implies that membrane insertion of CAR4 oligomers would be preferential in highly curved membranes with respect to flat membranes. Indeed, it has been suggested that CAR4 is found in membrane nanodomains (31), where the enrichment of bulkier lipid polar heads generates membrane curvature (38). Supporting these notions, we have been able to identify the complex between PYR1 and CAR1 on punctate and highly curved structures on the plant plasma membrane (Fig. S4). In other systems, this property contributes to the recruitment of peripherally bound proteins such as the lipidated proteins to nanodomains (37).
Fig. 6.
(A) CAR4 forms oligomeric structures in solution. (Right) Chemical cross-linking assays for self-association of CAR4 and CAR4-DADA. A Coomassie blue-stained SDS/PAGE showing the cross-linked species by 0.05% glutaraldehyde at different times in minutes. Cross-linked species as well as the monomer migrate aberrantly and are highlighted with arrows. (Left) Analytical ultracentrifugation analysis for CAR4 in native conditions and sedimentation coefficient c(s) distribution. The Inset corresponds to a zoomed area of the distribution. The position and the area under each peak reveal the molecular weight and the relative abundance of the CAR4 oligomers. (B) CAR4 binds to the periphery of liposomes and generates membrane tubules in a Ca2+-dependent manner. Negative-stain transmission electron micrographs of 12.5 μM liposomes (Upper Left) incubated with 8 μM CAR4 (Upper Right), 0.08 μM CAR4 plus 1 mM Ca2+ (Lower Left), and 8 μM CAR4 plus 1 mM Ca2+ (Lower Right). Peripherally membrane-bound CAR4 produces roughness on the surface of the liposomes and is indicated with black arrows, liposome tubules with white arrows, and protein aggregates as black asterisks. (Scale bar, 200 nm.)
Fig. S4.
Interaction of PYR1 and CAR1 generates punctate/globular structures in plasma membrane. Confocal images of transiently transformed tobacco epidermal cells coexpressing CAR1-YFPN/YFPC-PYR1 interacting proteins and a key cellular component for membrane trafficking. Asterisks indicate the presence of CAR1-PYR1 in membrane complexes decorated by At1g20110. To investigate the interaction of CAR1 and PYR1, we used the pSPYNE-35S and pYFPC43 vectors, respectively. At1g20110 was recombined by LR reaction into pH7WGR2 vector to coexpress RFP-At1g20110 together with CAR1-YFPN/YFPC-PYR1 using Agrobacterium-mediated transfection of tobacco leaves.
The Oligomerization of CAR Proteins at the Cell Membrane Affects ABA Signaling.
To investigate whether the formation of CAR oligomers at the cell membrane affects ABA signaling, we reasoned that proteins with oligomerization-dependent activity can be a target for dominant-negative mutations (55). That means that the presence in the oligomeric protein assembly of a nonfunctional mutant subunit, which is able to interact with wild-type subunits, can abrogate the function of the entire multimer. Thus, if CAR function depends on the establishment of oligomeric assembles, CAR mutants unable to bind Ca2+ might interfere with wild-type function and impair the formation or activity of the multimer. Therefore, we generated a mutant CAR1 version, CAR1 Asp22Ala Asp27Ala (CAR1-DADA), which contains a double Asp-to-Ala mutation in loop L1. As observed for CAR4-DADA, when assayed biochemically, CAR1-DADA showed a lack of Ca2+-dependent binding to phospholipid vesicles (31). We generated Arabidopsis transgenic lines expressing CAR1-DADA, and its expression in plant cells was verified by immunoblot analysis (Fig. 7A, Right). Next, we analyzed ABA sensitivity of CAR1-DADA overexpressing (OE) lines with respect to ABA-mediated inhibition of seedling establishment and early seedling growth (Fig. 7 A and B). Interestingly and in contrast to CAR1 OE lines (31), overexpression of CAR1-DADA led to reduced ABA sensitivity in those assays compared with wild type (Fig. 7B). Therefore, these results suggest that CAR1-DADA behaves as a dominant-negative mutation, which is able to antagonize the activity of the wild-type CAR1 protein or perhaps other CAR proteins from the 10-member family (31). As a control of reduced sensitivity to ABA, we used the HAB1 OE line, which displays enhanced expression of a negative regulator of ABA signaling (56). As expected, the HAB1 OE line shows higher ABA insensitivity than the CAR1-DADA line, as HAB1 is a PP2C acting as a global negative regulator of ABA signaling, whereas impairment of CAR function will affect mostly the fraction of ABA receptors acting at the membrane. Moreover, car triple mutants show a clear ABA insensitivity, albeit not so strong as triple pyr/pyl mutants (31). Conversely, transgenic lines that overexpress wild-type CAR1 (CAR1 OE) or lack the HAB1/ABI1 PP2Cs (hab1-1abi1-2 double mutant) showed enhanced ABA-mediated inhibition of seedling establishment and shoot growth (31). Thus, both loss-of-function and gain-of-function approaches have supported the positive role of CAR proteins in ABA signaling. Moreover, our data support the dependence of this role on the formation of functional CAR protein clusters.
Fig. 7.
Overexpression of CAR1-DADA leads to a dominant-negative phenotype with respect to ABA sensitivity. (A) CAR1-DADA OE lines show reduced sensitivity to ABA compared with Col WT (Upper Left). Expression of HA-tagged CAR1-DADA was verified by immunoblot analysis of the transgenic lines (Upper Right). Photographs are shown of representative seedlings from Col WT and two CAR1-DADA OE lines grown for 12 d on MS medium either lacking or supplemented with 0.5 μM ABA. Seedlings were rearranged on agar plates, and longitudinal or zenital photographs were taken to measure root length or the maximum rosette radius. The histograms show the quantification of root length (Lower Left) or the maximum rosette radius (Lower Right). (B) Quantification of ABA-mediated inhibition of seedling establishment of Col WT compared with hab1-1abi1-2 double mutant, CAR1 OE lines, and HAB1 OE and CAR1D22 D27A OE lines. Approximately 100 seeds of each genotype were sown on MS plates lacking or supplemented with either 0.5 or 1 mM ABA and scored for the presence of green expanded cotyledons 7 d later. *P < 0.05 (Student’s t test) when comparing data of each genetic background to Col WT plants in the same assay conditions. (C) The CAR1-DADA mutation abolishes the interaction of CAR1 with PYR1 in the plasma membrane. Confocal images of transiently transformed tobacco epidermal cells coexpressing the indicated constructs and increasing amounts of CAR1-DADA. The ratio of the relative concentration of agrobacteria in the different coinfiltrations is indicated by numbers (1×, 2×, or 4×).
At the molecular level, the dominant-negative effect of CAR1-DADA expression suggests the formation of CAR1:CAR1-DADA heterooligomers that prevent the CAR1 membrane association and the subsequent PYR/PYL interaction and, consequently, impair ABA signaling. To test this model, we used bimolecular fluorescence complementation (BiFC) assays to analyze whether CAR1 and CAR1-DADA compete for the PYR/PYL receptors in plant cells. Thus, using Agrobacterium-mediated transfection of tobacco plants, we coexpressed CAR1-YFPN and YFPC-PYR1 in the absence or the presence of increasing amounts of CAR1-DADA–YFPN (Fig. 7C). As previously reported (31), the fluorescence signal indicates that the CAR1–PYR1 interaction is solely observed at the plasma membrane or, less predominantly, in the nucleus, however as the amount of CAR1-DADA increases, the signal at the plasma membrane decreases, indicating that CAR1-DADA impedes proper CAR1 membrane localization and PYR1 binding. This result suggests that Ca+2-dependent oligomerization of CAR proteins in the plasma membrane is necessary for PYR/PYL docking (Fig. 8).
Fig. 8.
A schematic representation of the mechanism for CAR membrane interaction and PYR/PYL receptor recruitment. (1) Oligomeric CAR protein is recruited to the membrane. In the absence of a Ca2+ signal, the membrane interaction of CAR proteins is promoted but restricted both by a structural Ca2+ binding site (site I) and by the positively charged polybasic binding site. The dominant-negative CAR-DADA mutant precludes the anchoring of the oligomer to the membrane. (2) Abiotic stress induces an increase of the physiological Ca2+ concentration to the μM range. This triggers Ca2+ binding to site II and CAR membrane insertion. CAR molecules would act as molecular wedges into the membrane that have to overcome the lateral membrane energy barrier. (3) The bulkier side chain of lipids at membrane nanodomains generates membrane curvature. This facilitates CAR membrane insertion, as the membrane lateral pressure is relieved. The accumulation of CAR molecules favors the recruitment of PYR/PYL receptors. Additionally, this situation would help to recruit lipidated proteins for the control of ion homeostasis.
The Molecular Mechanism of the CAR PYR/PYL Recognition.
In plants, ABA and Ca2+-mediated signaling pathways at the cell membrane play a central role in regulating the adaptive response to environmental stress (3, 4). The discovery and characterization of the CAR family of small C2 proteins have provided a link between these pathways and the cell membrane, as they are involved in the Ca2+-dependent membrane localization of PYR/PYL ABA receptors. Previous structural and biochemical data showed that CAR proteins are small C2 proteins that additionally contain a characteristic α-helix β-hairpin insertion involved in the PYR/PYL interaction (31). This information suggested that the physiological Ca2+ fluctuations produced by different abiotic stresses would trigger the localization of CAR–PYR/PYL complexes at the cell membrane. However, signal transduction events at the cell membrane often rely on the formation of localized supramolecular complexes. These arrangements increase the signal specificity by the colocalization of substrates and products in response to a particular chemical signal. Scaffold proteins and membrane nanodomains are signaling platforms that mediate the formation of these complexes through the formation of specific protein–lipid and protein–protein interactions (57). The data presented here show that CAR proteins do not function as mere PYR/PYL carriers to the membrane. They rather provide the basis for a more complex model in which CAR proteins may be targeting PYR/PYL receptors to signaling platforms at the cell membrane for the control of the ABA-mediated stress response.
Our structural and biochemical data show that CAR oligomers are targeted to the membrane to develop their function. The topology of this binding is restricted by structural Ca2+ binding site II and through the polybasic binding site, which also contributes to CAR stabilization on the membrane. An increase in the Ca2+ concentration leads to the occupation of Ca2+ binding site I, and this in turn strengthens the CAR membrane interaction (Figs. 4 and 8). It has been reported for different C2 domains that this process involves the shallow insertion of the hydrophobic tip of loop L3, Met88, and Phe89 in CAR4 (Fig. 1). This is coupled with a change in the relative position of the protein with respect to the membrane due to the geometrical restraints imposed by the occupation of the different lipid binding sites (33, 42, 45). This suggests that the stress-induced specific Ca2+ signal triggers the coordinated rearrangement of CAR molecules within these oligomeric structures that cooperatively favor PYR/PYL docking (Fig. 8). This model explains that several CAR molecules are required to bind one PYR/PYL receptor (31). Such a process has biological relevance for ABA signaling, as introduction of a dominant-negative CAR1-DADA mutant in plant cells that can form oligomers in solution but is unable to bind the membrane in a Ca2+-dependent manner prevents the localization of PYR1 on the membrane (Figs. 7 and 8).
We have shown that CAR proteins are Ca2+ sensors that operate in the μM range of the Ca2+ concentration (Fig. 3). A correlated increase of the ABA and Ca2+ cytosolic concentrations is associated with stomata closure during dehydration stress (5, 7). At the molecular level, this relies on the ABA-dependent activation of SnRK2.6/OST1 and the Ca2+-dependent activation of CIPK23 and CPK6/CPK21/CPK23 kinases, which phosphorylate and induce the opening of the anion channel SLAC1 (6, 20, 29, 30). It has been shown that PP2C ABI1, which is inhibited by the ABA-dependent interaction with the PYR/PYL receptors, reverses this process. Full activation of SLAC1 likely requires an increase of the cytosolic Ca2+ concentration from the sub-μM at resting state to the μM range during stress, as for instance half-maximal kinase activity for CIPK23/CBL1 or CBL9 is 1.5 μM (6). Taken together, these data suggest that the targeting of PYR/PYL ABA receptors to the membrane occurs at the local μM Ca2+ concentration. In contrast, at resting Ca2+ concentrations, PYR/PYL receptors will not be delivered to the membrane, and consequently, ABI1 phosphatase would remain active, modulating SLAC1 anion transport. This information stresses the influence of Ca2+ signaling on different ABA-related pathways, such as those associated with stomata closure.
In this work, we show that CAR4 can bend membranes (Fig. 6) and provide in vivo data showing that CAR PYR/PYL complexes are localized in curved structures in the plasma membrane (Fig. S4). The ability to recognize membrane curvature is important in targeting proteins to specific membrane subdomains or nanodomains (58, 59). These structures provide a curved discontinuity at the cell membrane that functions as a platform for signaling proteins, as the presence of a bulkier lipid polar head leads to a reduction in the lateral energy barrier required for the insertion of hydrophobic protein segments and/or lipidated proteins (Fig. 8) (37). It is known that the membrane specificity of these proteins relies not only on the unambiguous binding to a particular lipid polar head but also on the recognition of intrinsically curved membrane architecture; furthermore, it has been demonstrated that protein membrane recognition will be more efficient if the membrane curvature is similar to that induced by the protein (33, 58). We have shown that CAR proteins do not display the characteristic phospho inositol phospholipid binding signature at the polybasic lipid binding site (41); rather, we provide crystallographic data and membrane binding assays showing that CAR proteins display a limited lipid specificity, as they interact with the widely distributed phosphatidylcholine and phosphatidylserine lipids. Thus, it may well be that CAR’s ability to form clusters and to bend membranes could be central to recognizing and/or stabilizing ABA-signaling nanodomains and to providing a platform for PYR/PYL receptors for ABA sensing. Indeed, the functional ABA signaling machinery, including PP2C phosphatases and Ca2+-dependent kinases, and the SLAH3 anion channel have been found in membrane nanodomains (38). In addition, it has been shown that CAR proteins are isolated from detergent-resistant Arabidopsis membranes (38), suggesting that they form part of the in vivo functional membrane domains (60). An additional function of CAR proteins in these domains may be to promote the concentration of other membrane-associated proteins involved in the regulation of ion transport and ion compartmentalization, such as the family of lipidated CBL Ca2+ sensors (10) or other proteins such as the membrane-bound ubiquitin ligase RSL1 (36) involved in PYR/PYL ubiquitylation.
Materials and Methods
Additional methods, including gene cloning, protein expression, and purification structure solution of CAR4 and CAR1 and CAR:PYR/PYL complex formation assays, are provided in SI Materials and Methods.
Chemical Cross-Linking Assays.
We incubated 50 μg of CAR4 and CAR4-DADA with 0.05% glutaraldehyde in a total volume of 10 μL at room temperature for 10, 20, and 30 min. The reaction was stopped by addition of SDS loading buffer and loaded and separated on a 12% (wt/vol) SDS/PAGE.
Analytical Ultracentrifugation.
Sedimentation velocity experiments were run using cells with double sector Epon-charcoal centerpieces. Raw sedimentation velocity profiles for this analysis were acquired using A280 at 148,288 × g, 20 °C, at different times. Differential sedimentation coefficients were calculated by least-squares boundary modeling of the experimental data with the program Sedfit and corrected to s20,w values (61). All measurements were performed in 50 mM Hepes, pH 7.5, 100 mM NaCl, 1 mM CaCl2 buffer at 20 °C using protein concentrations of ∼0.2 mg∙mL−1 in an Optima XL-A analytical ultracentrifuge (Beckman Coulter) with an AN50-Ti rotor.
Lipid Cosedimentation Assays.
Synthetic liposomes were prepared by mixing 75%:25% (wt:wt ratio) phosphatidylcholine and phosphatidylserine (Sigma Aldrich). Lipids were desiccated in a nonoxidizing atmosphere and subsequently hydrated in a 50 mM Hepes, pH 7.5, 100 mM NaCl buffer to a final concentration of 1 mg/mL. The sample was sonicated in a water bath for 5 min. Afterward, the liposomes were passed 11 times through 0.8-µm Nucleopore polycarbonate filters (Whatman) by syringe extrusion or 21 times through 0.1-µm filters for EM experiments.
The CAR4 Ca2+-dependent membrane binding activity was determined by incubating 7 µM CAR4 in 50 mM Hepes, pH 7.5, 100 mM NaCl with 1.6 mM liposomes in the presence of increasing concentrations of CaCl2. CAR4 protein samples were incubated for 30 min with the liposomes. The solution was then centrifuged at 65,000 rpm in a TLA100 rotor (Beckman) for 15 min. Supernatant and pellet fractions were separated, and the pellets were resuspended in equal volumes of reducing loading sample buffer. To determine the percentage of protein bound/unbound to liposomes, we measured the absorbance at 280 nm with a spectrophotometer Ultrospec 3100pro (Amersham Bioscience) of the soluble fraction after lipid pelleting. Data analysis was performed based on nonlinear least-squares fitting to Hill’s equation using Microcal Origin 6.0 software.
An identical protocol was used for the phospholipid binding experiments with CAR4 and the double mutant proteins CAR4-DADA and CAR4-KAKA. These experiments were performed at 2 µM CAR4 in the presence and absence of 1 mM CaCl2 or in the presence of 5 mM EGTA. Aliquots of the soluble fraction were run on 15% Bis–Tris SDS/PAGE gels. Protein and lipid concentration and phospholipid membrane composition were optimized to highlight the differences between CAR4 wild-type and mutant proteins.
EM.
Protein samples were prepared in the presence of 12.5-μM vesicles composed of a 75:25 PC:PS ratio incubated with 8 μM CAR4, 0.08 μM CAR4 and 1 mM Ca2+, and 8 μM CAR4 and 1 mM Ca2+. Samples were negatively stained on carbon-coated grids with uranyl acetate and imaged using a Tecnai G2 Spirit electron microscope (FEI) operated at 120 kV and a Slow Scan CCD (Gatan) or TemCam-F416 4,096 × 4,096-pixel camera (TVIPS GmbH).
Transgenic Plants.
To generate 35S:3HA-CAR1D22AD27A OE lines, the CAR1D22AD27A coding sequence (31) was cloned into pCR8/GW/TOPO entry vector (Invitrogen) and recombined using Gateway technology into pALLIGATOR2 vector (62). The resulting construct was transferred to Agrobacterium tumefaciens C58C1 (pGV2260) (63) by electroporation and used to transform Columbia wild-type plants by the floral dip method (64). T1 transgenic seeds were selected based on GFP visualization and sowed in soil to obtain the T2 generation. Homozygous T3 progeny was used for further studies, and expression of HA-tagged protein was verified by immunoblot analysis using anti–HA-peroxidase (Roche).
Seedling Establishment, Root Length, and Shoot Growth Assays.
After surface sterilization of the seeds, stratification was conducted in the dark at 4 °C for 3 d. Approximately 100 seeds of each genotype were sowed on MS plates supplemented with 0.5 or 1 μM ABA. Seedling establishment was scored as the percentage of seeds that developed green expanded cotyledons and the first pair of true leaves at 7 d. Root length and maximum rosette radius of seedlings grown for 12 d on MS medium supplemented with 0.5 μM ABA were scored to quantify ABA-mediated inhibition of growth.
Biochemical Fractionation, Protein Extraction, and Analysis.
Cytosolic and microsomal fractionation of HA-tagged proteins was performed as described previously (65). Briefly, 14-d-old seedlings of 35S:3HA-CAR4 plants grown in liquid MS medium were ground in liquid nitrogen and homogenized in an equal volume (1 g/mL) of extraction buffer (12% sucrose, 100 mM Tris∙HCl pH 7.5, 1 mM EDTA, 2 mM DTT, and 1× Complete Protease Inhibitor Mixture from Roche). The lysate was filtered through two layers of miracloth paper (Calbiochem) to remove insoluble plant debris, followed by centrifugation at 15,000 × g for 20 min at 4 °C. The nonnuclear soluble protein fraction was incubated at 4 °C in extraction buffer or buffer supplemented with 25 mM CaCl2 or 25 mM EGTA for 1 h. Then it was centrifuged at 100,000 × g for 45 min to pellet microsomal membranes and to obtain the cytosolic soluble fraction. HA-tagged protein was detected by immunoblot analysis using anti–HA-peroxidase (Roche). Antibody was used to a 1:10,000 dilution. Detection was performed using the ECL advance Western blotting chemiluminiscent detection kit (GE Healthcare). Image capture was done using the image analyzer LAS3000, and quantification of the protein signal was done using Image Guache V4.0 software.
Transient Protein Expression in Nicotiana Benthamiana and BiFC Assays.
To investigate the interaction of either CAR1 or CAR1-DADA and PYR1, we used the pSPYNE-35S and pYFPC43 vectors, respectively, as described in ref. 31. The different constructs were introduced into A. tumefaciens C58C1 (pGV2260) (63) by electroporation, and transformed cells were selected in LB plates supplemented with kanamycin (50 mg/L). Then, they were grown in liquid LB medium to late exponential phase, and cells were harvested by centrifugation and resuspended in 10 mM morpholinoethanesulphonic (Mes) acid–KOH pH 5.6 containing 10 mM MgCl2 and 150 mM acetosyringone to an OD at 600 nm of 1. These cells were mixed with an equal volume of Agrobacterium C58C1 (pCH32 35S:p19) expressing the silencing suppressor p19 of tomato bushy stunt virus so that the final density of Agrobacterium solution was about 1. Bacteria were incubated for 3 h at room temperature and then injected into young fully expanded leaves of 4-wk-old N. benthamiana plants. Leaves were examined 48–72 h after infiltration using confocal laser scanning microscopy.
SI Materials and Methods
Gene Cloning and Site-Directed Mutagenesis.
Cloning details of the pETM11:CAR4, pETM11:CAR1, pETM11:PYL6, and pETM11:PYL1 plasmids can be found in the literature (31). CAR1 D22A D27A and CAR4 D85A D87A mutants were generated using the PCR-overlap extension procedure and the oligonucleotides described in ref. 31. The CAR4K50A K52A mutant was generated using oligonucleotides FK50A K52A, ATGGGCAAACAGgcaTTGgcgACTCGTGTC, and RK50A K52A, GACACGAGTcgcCAAtgcCTGTTTGCCCAT. The sequence encoding the K50A K52A mutant residues was subcloned into pETM11-CAR4 by replacing the WT sequence using an NcoI-SalI double digestion.
Protein Expression and Purification.
The pETM11:CAR4, pETM11:PYL6, and pETM11:PYL1 plasmids were transformed into Escherichia coli strain BL21 (DE3) (Novagen) for protein expression using standard protocols. The same expression protocol was used for the three proteins. A total of 8 mL of overnight culture was subcultured into 800 mL of fresh 2TY broth (16 g Bacto tryptone, 10 g yeast extract, 5 g NaCl/L of solution) plus kanamicyn (50 μg∙mL−1) at 310 K. When the OD at 600 nm was 0.6–0.8, the overnight protein expression was induced with 0.3 mM isopropyl-β-d-thiogalactoside at 289 K. Cells were harvested by centrifugation (15 min, 4,500 × g). Pellets were resuspended in 20 mM Tris∙HCl pH 8.0, 200 mM NaCl, 50 mM imidazole, and 5 mM β-mercaptoethanol and disrupted by sonication. For CAR4 purification, 1 mM Ca2+ was also included in the lysis buffer. After centrifugation (40 min, 40,000 × g) at 277 K, the clear supernatant was filtered (pore diameter, 0.45 μm; Millipore Corporation). The His-tagged proteins were purified using nickel–nitrilotriacetic acid (Ni-NTA) Agarose (Qiagen) according to the manufacturer’s instructions. The filtered supernatant was mixed with the previously equilibrated beads. After incubation, a washing step with 10 volumes of 20 mM Tris∙HCl pH 8.0, 200 mM NaCl, 50 mM imidazole, and 5 mM β-mercaptoethanol buffer was performed. Protein was eluted from the Ni-NTA Agarose by washing with 500 mM imidazole buffer. Imidazole was removed using a PD-10 column (GE Healthcare), and the His-tag was cleaved using TEV protease. CAR4, PYL6, and PYL1 yielded in a SDS PAGE gel 23 kDa, 24 kDa, and 25 kDa products, respectively. A final polishing step was performed using a Superdex200 16/60 (Amersham Biosciences Limited) equilibrated in 20 mM Tris∙HCl pH 8.0, 200 mM NaCl, and 1 mM DTT.
E. coli BL21 (DE3) cells were transformed with the CAR1 constructs and grown in 2 L of LB medium containing 50 μg/mL kanamycin to an OD at 600 nm of 0.6–0.8. At this point, 1 mM IPTG was added, and the cells were harvested after overnight induction at 20 °C and stored at –80 °C before purification. The cells were resuspended in 180 mL lysis buffer (30 mM Tris pH 7.5, 500 mM NaCl, 15 mM imidazole, 1 mM β-mercaptoethanol, and protease mixture inhibitor) and lysed with a microfluidizer (Microfluidics). A cleared lysate was obtained after centrifugation at 40,000 × g for 45 min. The protein extract was incubated in 25 mL of Ni-NTA Agarose and washed with 125 mL of lysis buffer. The bound protein was eluted with a buffer containing 30 mM Tris pH 7.5, 300 mM NaCl, 250 mM imidazole, and 1 mM β-mercaptoethanol. The fractions containing CAR1 were pooled and dialyzed extensively against a buffer containing 30 mM Tris∙HCl pH 7.5, 150 mM NaCl, and 1 mM β-mercaptoethanol. The sample was then concentrated and purified by gel filtration in a Superdex 25 column (GE healthcare) equilibrated with 30 mM Tris∙HCl pH 7.5, 150 mM NaCl, and 1 mM β-mercaptoethanol. The size and purity of the recombinant protein was verified by SDS/PAGE. Purified CAR1 was concentrated to 20 mg/mL and either used directly or flash-frozen in liquid nitrogen for storage at –80 °C.
Protein Crystallization, Data Collection, and Structure Solution and Refinement.
Crystals of the CAR4:Ca2+ complex with POC, PSF, Zn2+, and Zn2+ plus POC were obtained in similar conditions as those described for the unbound form of the protein. To summarize, CAR4 was concentrated to 8 mg/mL and incubated with 0.1 mM CaCl2 and 1 mM PSF and 5 mM POC when appropriate. Prismatic colorless crystals were grown in a solution containing from 20% to 25% of polyethylene glycol 6K, 0.1 M Mes pH 6.0–6.5, and 0.1 M LiCl or ZnCl2. Crystals were observed in 3–4 d. Crystals were cryoprotected in mother liquor containing 20% (wt/vol) polyethylene glycol 400, mounted in fiber loops, and then flash-cooled in liquid nitrogen. X-ray diffraction data were collected at the ESRF.
The diffraction datasets were processed and scaled using XDS (66). Data were merged using AIMLESS from the CCP4 package (Collaborative Computational Project, Number 4, 1994) (67). A summary of the data collection statistics is given in Table S1. The X-ray structures of CAR4:Ca2+ in complex with POC, PSF, Zn2+, and Zn2+ plus POC were solved by molecular replacement with the program MolRep for CCP4 package (67) using the coordinates of CAR4:Ca2+ [Protein Data Bank (PDB) ID code 4V29]. The electron density map calculated using these phases was good enough to manually build the structure of the ligands. A cycle of restrained refinement with Phenix (68) and Refmac5 (67) and iterative model building with Coot (69) was carried out.
The CAR1 protein was assayed for crystallization by the vapor diffusion method at the High Throughput Crystallization Laboratory of the EMBL Grenoble Outstation (https://embl.fr/htxlab) (70). The CAR1 protein produced crystals by mixing one volume (100 nl) of protein sample with one volume of crystallization solution (0.5 M MgSO4, 0.5 M Hepes pH 7.0, and 1.6 M Lithium Sulfate) and equilibrating it against a reservoir containing 80 μL of precipitant solution. Crystals in the shape of cubes appeared within 2 d. Crystals were flash frozen in liquid nitrogen using 15% glycerol as cryo-protectant. X-ray diffraction data were collected at the ID14-1 beamline of the ESRF. Crystallographic data reduction and scaling were carried out with the software XDS (66). Initial phases were obtained using Phaser (68) by the molecular replacement method. Successive rounds of automatic refinement and manual building were carried out with REFMAC (67) and Coot (71).
The stereochemistry of the models was verified with MolProbity (68). Ribbon figures were produced using PyMOL (72). The refinement statistics are summarized in Table S1.
CAR:PYR/PYL Complex Formation Assays.
CAR4, PYL6, and PYL1 and their mixtures (CAR4:PYL6 and CAR:PYL1) were subjected to a size exclusion chromatography. Purified proteins were mixed in a 1:1 molar ratio when needed. The chromatographies were performed using a Bio-Silect SEC 250 column (BioRad) equilibrated in 20 mM Tris∙HCl pH 8.0, 200 mM NaCl, and 1 mM DTT.
ITC.
ITC measurements were performed at 25 °C using a MicroCal VP-ITC (GE-Healthcare). CAR4 was equilibrated in 50 mM Hepes and 200 mM NaCl pH 8.5, and Ca2+ solutions were prepared using the same buffer. All samples were thoroughly degassed before use. Titration was carried out by injecting consecutive aliquots of 2 mM Ca2+ (1 × 1 μL, 10 × 5 μL, and 10 × 10 μL) into the sample cell loaded with 64.4 μM CAR1. Heat that developed on Ca2+ dilution was found to be negligible. The thermodynamic parameters of binding were calculated by analyzing the binding isotherm with the MicroCal ITC Origin software.
Acknowledgments
A.A. and J.A.M. thank the European Syncrotron Radiation Facility and EMBL for access to the synchrotron radiation source. This work was funded by Ministerio de Economía y Competitividad (MINECO) Grants BFU2014-59796-R (to A.A.), BFU2011-28184-C02 (to M.J.S.-B.), and BIO2014-52537-R (to P.L.R.) and Comunidad de Madrid Grant S2010/BMD-2457 (to A.A and M.M.). M.J.S.-B. is supported by Ramón y Cajal Contract RYC-2008-03449 from MINECO and M.D. by a fellowship from Senacyt-Ifarhu. Access to the High Throughput Crystallization facility at European Molecular Biology Laboratory (EMBL) Grenoble was supported by the European Community’s Seventh Framework Programme through the Protein Production Platform project (P-CUBE) Grant 227764.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 5A4X , 5A50, and 5A51 (CAR4 complexes) and 5A52 (CAR1 complex).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512779113/-/DCSupplemental.
References
- 1.Serrano R, Rodriguez-Navarro A. Ion homeostasis during salt stress in plants. Curr Opin Cell Biol. 2001;13(4):399–404. doi: 10.1016/s0955-0674(00)00227-1. [DOI] [PubMed] [Google Scholar]
- 2.Bassil E, Blumwald E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H(+) transporters. Curr Opin Plant Biol. 2014;22:1–6. doi: 10.1016/j.pbi.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Batistič O, Kudla J. Analysis of calcium signaling pathways in plants. Biochim Biophys Acta. 2012;1820(8):1283–1293. doi: 10.1016/j.bbagen.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 5.McAinsh MR, Brownlee C, Hetherington AM. Abscisic acid-induced elevation of guard cell cytosolic Ca2+ precedes stomatal closure. Nature. 1990;343(6254):186–188. [Google Scholar]
- 6.Maierhofer T, et al. Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci Signal. 2014;7(342):ra86. doi: 10.1126/scisignal.2005703. [DOI] [PubMed] [Google Scholar]
- 7.Allen GJ, et al. Cameleon calcium indicator reports cytoplasmic calcium dynamics in Arabidopsis guard cells. Plant J. 1999;19(6):735–747. doi: 10.1046/j.1365-313x.1999.00574.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee SC, et al. A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA. 2007;104(40):15959–15964. doi: 10.1073/pnas.0707912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grefen C, et al. A vesicle-trafficking protein commandeers Kv channel voltage sensors for voltage-dependent secretion. Nature Plants. 2015;1(15108):1–11. doi: 10.1038/nplants.2015.108. [DOI] [PubMed] [Google Scholar]
- 10.Sánchez-Barrena MJ, Martínez-Ripoll M, Albert A. Structural biology of a major signaling network that regulates plant abiotic stress: The CBL-CIPK mediated pathway. Int J Mol Sci. 2013;14(3):5734–5749. doi: 10.3390/ijms14035734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quan R, et al. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 2007;19(4):1415–1431. doi: 10.1105/tpc.106.042291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 13.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santiago J, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60(4):575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura N, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61(2):290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, et al. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA. 2013;110(27):11205–11210. doi: 10.1073/pnas.1308974110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umezawa T, et al. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal. 2013;6(270):rs8. doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
- 18.Kollist H, Nuhkat M, Roelfsema MR. Closing gaps: Linking elements that control stomatal movement. New Phytol. 2014;203(1):44–62. doi: 10.1111/nph.12832. [DOI] [PubMed] [Google Scholar]
- 19.Lind C, et al. Stomatal guard cells co-opted an ancient ABA-dependent desiccation survival system to regulate stomatal closure. Curr Biol. 2015;25(7):928–935. doi: 10.1016/j.cub.2015.01.067. [DOI] [PubMed] [Google Scholar]
- 20.Geiger D, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA. 2009;106(50):21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imes D, et al. Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 2013;74(3):372–382. doi: 10.1111/tpj.12133. [DOI] [PubMed] [Google Scholar]
- 22.Ishitani M, et al. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 2000;12(9):1667–1678. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grefen C, Blatt MR. Do calcineurin B-like proteins interact independently of the serine threonine kinase CIPK23 with the K+ channel AKT1? Lessons learned from a ménage à trois. Plant Physiol. 2012;159(3):915–919. doi: 10.1104/pp.112.198051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA. 2002;99(12):8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintero FJ, et al. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Natl Acad Sci USA. 2011;108(6):2611–2616. doi: 10.1073/pnas.1018921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunez-Ramirez R, et al. Structural insights on the plant Salt-Overly-Sensitive 1 (SOS1) Na(+)/H(+) antiporter. J Mol Biol. 2012;424(5):283–294. doi: 10.1016/j.jmb.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Ohta M, Guo Y, Halfter U, Zhu JK. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci USA. 2003;100(20):11771–11776. doi: 10.1073/pnas.2034853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, et al. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125(7):1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Geiger D, et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA. 2010;107(17):8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandt B, et al. Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci USA. 2012;109(26):10593–10598. doi: 10.1073/pnas.1116590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez L, et al. C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in Arabidopsis. Plant Cell. 2014;26(12):4802–4820. doi: 10.1105/tpc.114.129973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martens S, McMahon HT. Mechanisms of membrane fusion: Disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9(7):543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 33.Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316(5828):1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 34.Jahn R, Lang T, Südhof TC. Membrane fusion. Cell. 2003;112(4):519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 35.Sutter JU, et al. Abscisic acid triggers the endocytosis of the arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Curr Biol. 2007;17(16):1396–1402. doi: 10.1016/j.cub.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 36.Bueso E, et al. The single subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2014;80(6):1057–1071. doi: 10.1111/tpj.12708. [DOI] [PubMed] [Google Scholar]
- 37.Larsen JB, et al. Membrane curvature enables N-Ras lipid anchor sorting to liquid-ordered membrane phases. Nat Chem Biol. 2015;11(3):192–194. doi: 10.1038/nchembio.1733. [DOI] [PubMed] [Google Scholar]
- 38.Demir F, et al. Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc Natl Acad Sci USA. 2013;110(20):8296–8301. doi: 10.1073/pnas.1211667110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerrero-Valero M, et al. Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2. Proc Natl Acad Sci USA. 2009;106(16):6603–6607. doi: 10.1073/pnas.0813099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guillén J, et al. Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1. Proc Natl Acad Sci USA. 2013;110(51):20503–20508. doi: 10.1073/pnas.1316179110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corbalan-Garcia S, Gómez-Fernández JC. Signaling through C2 domains: More than one lipid target. Biochim Biophys Acta. 2014;1838(6):1536–1547. doi: 10.1016/j.bbamem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta. 2006;1761(8):838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 43.Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gómez-Fernández JC. Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. EMBO J. 1999;18(22):6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honigmann A, et al. Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat Struct Mol Biol. 2013;20(6):679–686. doi: 10.1038/nsmb.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ausili A, Corbalán-García S, Gómez-Fernández JC, Marsh D. Membrane docking of the C2 domain from protein kinase Cα as seen by polarized ATR-IR. The role of PIP2. Biochim Biophys Acta. 2011;1808(3):684–695. doi: 10.1016/j.bbamem.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 46.Hermoso JA, et al. Insights into pneumococcal pathogenesis from the crystal structure of the modular teichoic acid phosphorylcholine esterase Pce. Nat Struct Mol Biol. 2005;12(6):533–538. doi: 10.1038/nsmb940. [DOI] [PubMed] [Google Scholar]
- 47.Thompson D, Pepys MB, Wood SP. The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure. 1999;7(2):169–177. doi: 10.1016/S0969-2126(99)80023-9. [DOI] [PubMed] [Google Scholar]
- 48.Ochoa WF, et al. Structure of the C2 domain from novel protein kinase Cepsilon. A membrane binding model for Ca(2+)-independent C2 domains. J Mol Biol. 2001;311(4):837–849. doi: 10.1006/jmbi.2001.4910. [DOI] [PubMed] [Google Scholar]
- 49.Fuson K, et al. Alternate splicing of dysferlin C2A confers Ca2+-dependent and Ca2+-independent binding for membrane repair. Structure. 2014;22(1):104–115. doi: 10.1016/j.str.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radhakrishnan A, Stein A, Jahn R, Fasshauer D. The Ca2+ affinity of synaptotagmin 1 is markedly increased by a specific interaction of its C2B domain with phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2009;284(38):25749–25760. doi: 10.1074/jbc.M109.042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schapire AL, et al. Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell. 2008;20(12):3374–3388. doi: 10.1105/tpc.108.063859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, et al. Calcium sensitive ring-like oligomers formed by synaptotagmin. Proc Natl Acad Sci USA. 2014;111(38):13966–13971. doi: 10.1073/pnas.1415849111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaenicke R, Rudolph R. Refolding and association of oligomeric proteins. Methods Enzymol. 1986;131:218–250. doi: 10.1016/0076-6879(86)31043-7. [DOI] [PubMed] [Google Scholar]
- 54.Goñi GM, et al. Phosphatidylinositol 4,5-bisphosphate triggers activation of focal adhesion kinase by inducing clustering and conformational changes. Proc Natl Acad Sci USA. 2014;111(31):E3177–E3186. doi: 10.1073/pnas.1317022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkie AO. The molecular basis of genetic dominance. J Med Genet. 1994;31(2):89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saez A, et al. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 2004;37(3):354–369. doi: 10.1046/j.1365-313x.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- 57.Simons K, Gerl MJ. Revitalizing membrane rafts: New tools and insights. Nat Rev Mol Cell Biol. 2010;11(10):688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 58.McMahon HT, Boucrot E. Membrane curvature at a glance. J Cell Sci. 2015;128(6):1065–1070. doi: 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tapken W, Murphy AS. Membrane nanodomains in plants: Capturing form, function, and movement. J Exp Bot. 2015;66(6):1573–1586. doi: 10.1093/jxb/erv054. [DOI] [PubMed] [Google Scholar]
- 60.Lingwood D, Simons K. Detergent resistance as a tool in membrane research. Nat Protoc. 2007;2(9):2159–2165. doi: 10.1038/nprot.2007.294. [DOI] [PubMed] [Google Scholar]
- 61.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bensmihen S, et al. Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 2004;561(1-3):127–131. doi: 10.1016/S0014-5793(04)00148-6. [DOI] [PubMed] [Google Scholar]
- 63.Deblaere R, et al. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 1985;13(13):4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 65.Hua J, Grisafi P, Cheng SH, Fink GR. Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 2001;15(17):2263–2272. doi: 10.1101/gad.918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 70.Dimasi N, Flot D, Dupeux F, Márquez JA. Expression, crystallization and X-ray data collection from microcrystals of the extracellular domain of the human inhibitory receptor expressed on myeloid cells IREM-1. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63(Pt 3):204–208. doi: 10.1107/S1744309107004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. DeLano (2002) The PyMOL Molecular Graphics System, 1.5.0.4 (DeLano Scientific, San Carlos, CA)