Abstract
Background:
Many studies have suggested that cigarette smoking and polymorphisms of resistin and glutathione peroxidase-1 (GPx-1) genes are closely correlated with the pathogenesis of nonalcoholic fatty liver disease (NAFLD). However, few reports have investigated these associations with respect to NAFLD susceptibility. We, therefore, examined the distribution of polymorphisms in GPx-1 and resistin genes in NAFLD patients and healthy controls and analyzed the relationship between these polymorphisms and smoking status.
Methods:
Nine hundred NAFLD patients and 900 healthy controls were selected, and the genetic polymorphisms of resistin gene promoter-420C/G and GPx-1 gene Pro198Leu were analyzed by polymorphism-polymerase chain reaction (PCR) in DNA extracted from peripheral blood leukocytes. Interactions between the two mutants and the gene–environment interaction with cigarette smoking were also analyzed.
Results:
Genotype frequencies of −420C/G (GG) and Pro198Leu (LL) were significantly higher in NAFLD cases (49.56% and 50.11%, respectively) compared with healthy controls (23.67% and 24.22%, respectively) (P = 0.0069; P = 0.0072). Moreover, the risk of NAFLD with −420C/G (GG) was significantly higher than in controls (odds ratio [OR] =3.1685, 95% confidence interval (CI) =1.9366–5.2073). Individuals carrying Pro198Leu (LL) had a high risk of NAFLD (OR = 3.1424, 95% CI = 1.7951–5.2367). Combined analysis of the polymorphisms showed that the −420C/G (GG)/Pro198Leu (LL) genotype was significantly more common in the NAFLD group than in the control group (39.44% vs. 12.78%, respectively, P = 0.0054), while individuals with −420C/G (GG)/Pro198Leu (LL) had a high risk of NAFLD (OR = 5.0357, 95% CI = 3.1852–7.8106). Moreover, the cigarette smoking rate in the NAFLD group was significantly higher than in the control group (OR = 1.8990, P = 0.0083 in the smoking index (SI) ≤400 subgroup; OR = 5.0937, P = 0.0051 in the SI >400 subgroup), and statistical analysis suggested a positive interaction between cigarette smoking and −420C/G (GG) (γ = 5.6018 in the SI ≤400 subgroup; γ = 4.4770 in the SI >400 subgroup) and Pro198Leu (LL) (γ = 5.7715 in the SI ≤400 subgroup; γ = 4.5985 in the SI >400 subgroup) in increasing the risk of NAFLD.
Conclusion:
NAFLD risk factors include -420C/G (GG), Pro198Leu (LL) and cigarette smoking, and these three factors have a significant additive effect on NAFLD risk.
Keywords: Cigarette Smoking, Glutathione Peroxidase-1 Gene, Nonalcoholic Fatty Liver Disease, Polymorphisms, Resistin Gene Promoter (420C/G)
INTRODUCTION
The disease mechanism of nonalcoholic fatty liver disease (NAFLD) is unclear, but the two-hit theory is more popular than other hypotheses. This theory states that the first hit in the pathogenesis of NAFLD is the accumulation of liver fat in hepatocytes caused by insulin resistance, which makes the liver more vulnerable to oxidative stress and subsequent lipid peroxidation. These factors constitute the second hit, which leads to the development of inflammation and fibrosis and eventually, NAFLD.[1,2] Cigarette combustion releases nicotine and carbon monoxide, which interfere with lipid metabolism while several ingredients of cigarette smoke stimulate and promote free radical lipid peroxidation, which participates in the development of NAFLD.[3]
Resistin is a peptide secreted mainly by white fat cells to combat the action of insulin, resulting in insulin resistance, and potentially inflammatory cytokine activity. This role of resistin is an important factor in promoting the progress of NAFLD.[4] The enzyme glutathione peroxidase-1 (GPx-1) scavenges free radicals and their derivatives while phospholipid hydroperoxide GPx constitutes an organic hydroperoxide reduction system together with catalase and glutathione S-transferase at different levels of substrate specificity. This system reduces the formation of lipid peroxides and enhances anti-oxidation, which is important in the progression of NAFLD.[5] The resistin gene and GPx-1 have a number of polymorphisms with multiple alleles encoding different protein activities. These polymorphisms can, therefore, affect the reaction of the body to the external environment (such as smoking), which is highly relevant in NAFLD susceptibility.
Studies into the correlation of resistin gene and GPx-1 polymorphisms and NAFLD susceptibility are increasing, but no reports have investigated the combined effect of cigarette smoking and gene polymorphisms on NAFLD susceptibility. This study, therefore, examined the distribution of resistin gene and GPx-1 polymorphisms in NAFLD patients and healthy controls and analyzed the relationship between the polymorphisms and smoking status to explore the effect of these interactions on NAFLD pathogenesis.
METHODS
Case and control selection
Between May 2009 and April 2013, 900 patients with confirmed NAFLD from the Department of Gastroenterology of The First Affiliated Hospital of Xinxiang Medical University were selected for this study. The diagnosis of NAFLD was based on guidelines for the diagnosis and treatment of NAFLD revised by the Fatty Liver and Alcoholic Liver Disease Study Group of the Chinese Liver Disease Association in 2010.[6] Inclusion criteria were: (1) conformed to NAFLD diagnostic criteria; (2) aged 17–69 years; (3) alanine transaminase or aspartate transaminase <80 U/L; and (4) voluntarily signed the informed consent form and passed the ethical evaluation. Exclusion criteria were: (1) a history of alcohol consumption >20 g/d; (2) co-existence of other liver diseases such as viral, drug-induced, auto-immune hepatitis; (3) suspected liver cirrhosis or liver cancer; (4) co-existence of other severe systematic disease or infectious disease such as malignant neoplasm, severe cardiopulmonary disease, neurological disorders, or HIV infection; (5) currently pregnant, breastfeeding, pregnancy anticipated during study, or planning to conceive; and (6) co-existence of mental disorders or severe neurosis, or unable to express symptoms subjectively and hindering connection and cooperation with researchers because of dysgnosia or aphasia.
The control group consisted of 900 healthy individuals and excluded those with type 2 diabetes mellitus, obesity, hypertension, hyperlipidemia, and other metabolic disorders, as well as those with a drinking history. NAFLD cases and healthy controls were divided into nonsmokers and smokers, the latter based on smoking at least one cigarette per day for more than 6 months. Smoking status was estimated by the smoking index (SI) = daily cigarette consumption (kg) × duration of cigarette smoking (years). Smokers were divided into SI S400 and SI >400 subgroups.
Sample collection
Three millilitres of blood was obtained by venipuncture and were centrifuged while still in the Monovette tube (Shanghai Qiwu Biological Technology Co., Ltd., Shanghai, China). Centrifugation speed was set at a value that corresponds to a 1500 × g centrifugal force. During centrifugation for at least 20 min, the samples were kept at room temperature (±20°C). From the Monovette tubes with anticoagulant, three blood fractions were obtained: Plasma, red blood cells and a buffy layer containing white blood cells. DNA was extracted from white blood cells using the QIAamp DNA extraction kit (Qiagen, Hilden, Germany) and stored at −30°C until required.
Analysis of the resistin gene promoter single nucleotide polymorphism −420C/G
Single nucleotide polymorphism (SNP) analysis was according to the method of Hossein-Nezhad et al.[7] It was based on polymerase chain reaction (PCR) amplification involving the primer pair 5′-GTTTGCATCAGCCACCCT-3′ (upstream primer) and 5′-GCACCGCAGCTCTTTCTT-3′ (downstream primer), both synthesized by Shanghai Sangon Biological Engineering Technology and Services Co., Ltd., (Shanghai, China). The PCR reaction included: 1-μl genomic DNA, 0.5 μl upstream and downstream primers, 25-μl Taq DNA polymerase mix (Takara Biotechnology Co., Ltd., [Dalian, China]). Reaction conditions were: Predegeneration at 94°C for 5 min, then 40 cycles of 94°C for 40 s, 55°C for 45 s, and 72°C for 30 s, followed by an extension at 72°C for 7 min. This amplified a PCR product of 286 bp, which was confirmed by agarose gel electrophoresis. Digestion with the restriction endonuclease Ear I for 4 h at 37°C resolved three different genotypes: Homozygous CC (179 bp and 107 bp bands), homozygous GG (286 bp band), and heterozygote CG (286 bp, 179 bp, and 107 bp bands). These were visualized by running 4-μl digestion products on agarose gel electrophoresis [Figure 1].
Figure 1.
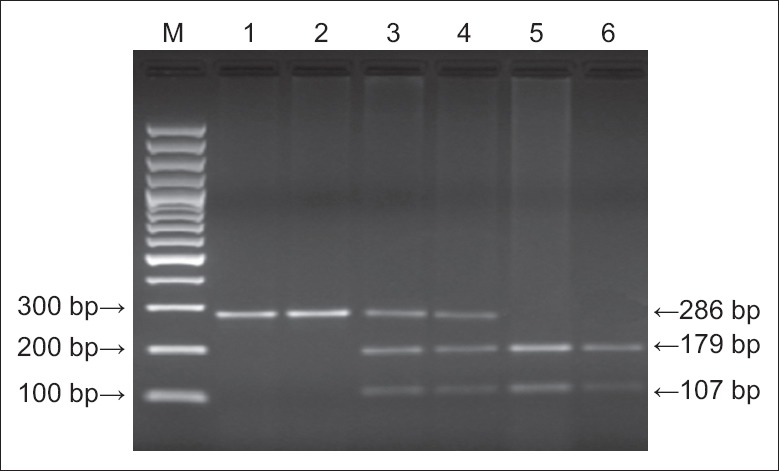
Electrophoretogram of polymerase chain reaction amplification products showing variants of the − 420C/G resistin gene promoter polymorphism. M: Marker; lanes 1, 2: G/G; lanes 3, 4: C/G; lanes 5, 6: C/C.
Analysis of the glutathione peroxidase-1 Pro198Leu polymorphism
This analysis was conducted according to the method of Suzen et al.[8] It also involved PCR amplification, using primers 5′-CCTACGCAGGTACAGCC-3′ (upstream primer) and 5′-CAACAGGACCAGCACCCATCTC-3′ (downstream primer) synthesized by Shanghai Sangon Biological Engineering Technology and Services Co., Ltd. The 25-μl PCR reaction used the Takara PCR Amplification Kit (Dalian Takara Biotechnology Co., Ltd.) and included: 1-μl template DNA, 2.5-μl × 10 PCR Buffer, 2-μl dNTP mixture, 0.625 U Takara Taq DNA polymerase, 15 pmol primers and 17.375-μl sterilized deionized water. PCR conditions were: Predegeneration at 93°C for 3 min, then 35 cycles of 93°C for 1 min, 66°C for 1 min, and 70°C for 1 min, followed by an extension at 70°C for 10 min. The 240 bp PCR products (12-μl) were incubated with the restriction endonuclease Dde I at 37°C, then analyzed using 3.5% agarose gel electrophoresis containing 0.5 μg/ml ethidium bromide. The electrophoretogram showed three different genotypes: Homozygous Pro198Leu (PP) (240 bp band), homozygous Pro198Leu (LL) (163 bp and 77 bp bands), and heterozygous Pro198Leu (PL) (240 bp, 163 bp, and 77 bp bands) [Figure 2].
Figure 2.
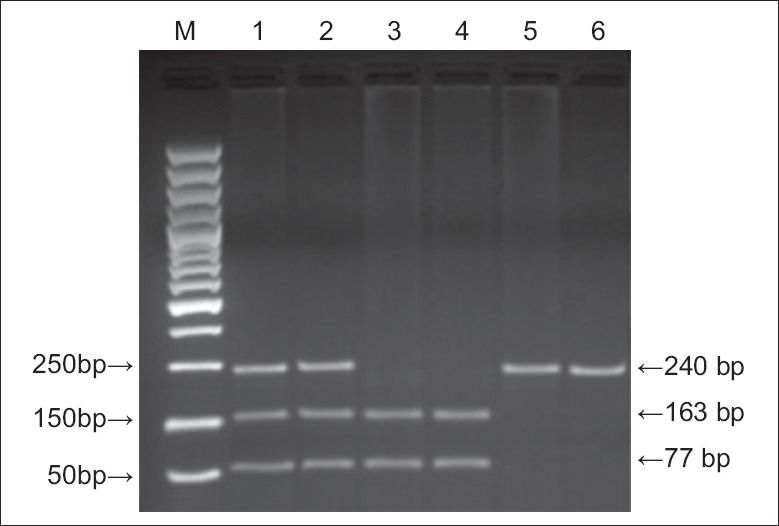
Electrophoretogram of polymerase chain reaction amplification products showing variants of the GPx-1 Pro198Leu polymorphism. M: Marker; lanes 1, 2: PL; lanes 3, 4: LL; lanes 5, 6: PP.
Analysis of the aldehyde dehydrogenase 2 gene polymorphism
Aldehyde dehydrogenase 2 gene (ALDH2) was PCR amplified as a control using primers 5′-CCCTTTGGTGGCTAGAA GA-TG-3′ (upstream primer) and 5′-CCACACTCACAGTTTTCTCTT-3′ (downstream primer). PCR was conducted in 25-μl reactions containing 1-μl DNA template, 1 μmol/L primers, 0.25 mmol/L × 4 dNTPs, 0.25 mmol/L MgCl2, 1 U Taq enzyme (Shanghai Sangon Biological Engineering Technology and Services Co., Ltd.), and 2.5-μl × 10 buffer. PCR conditions were: Predegeneration at 95°C for 10 min, then 35 cycles of 95°C for 1 min, 58°C for 2 min, and 72°C for 1 min, followed by an extension at 72°C for 5 min.
PCR products (3-μl) were digested with 5 U Mbo II restriction enzyme (Dalian Takara Biotechnology Co., Ltd.) in 10-μl × 10 buffer at 37°C for 2.5 h. Digestion products (8-μl) were electrophoresed on 15% polyacrylamide gels and stained with ethidium bromide. Three genotypes were visible: Homozygous GG (55 bp band), homozygous LL (65 bp band), and heterozygote GL (65 bp and 55 bp bands) [Figure 3].
Figure 3.
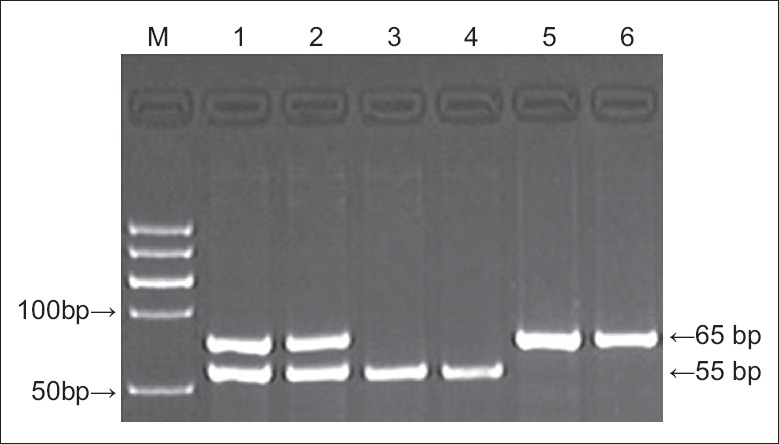
Electrophoretogram of polymerase chain reaction amplification products showing variants of the aldehyde dehydrogenase 2 polymorphism. M: Marker; lanes 1, 2: GL; lanes 3, 4: GG; lanes 5, 6: LL.
Statistical analysis
SPSS 11.0 for Windows (SPSS Inc., USA) was used for statistical analysis. Groups were tested to determine whether they were in Hardy–Weinberg equilibrium, with P > 0.05 indicating compliance with Hardy–Weinberg law. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were used to evaluate the relative risk between the NAFLD group and the control group, and gene and allele frequencies were tested by the Chi-square test. P < 0.05 was considered statistically significant.
Conditional logistic regression was applied to analyze interactions, using the following criteria according to the Khoury and Wagener model[9] of interaction and interaction coefficients (γ = βeg/βe) in determining gene–environment interactions. Judging criteria 1 were: γ >1 indicated that the effect of environmental exposure was amplified by genetic factors, that is., a positive interaction, while γ <1 indicated that the effect of environmental exposure was reduced by genetic factors, that is., negative interaction; γ = 1 indicated that gene and environmental exposure had no interaction. In this case–control study, γ equaled to lgReg divided by lgORe (lgOReg/lgORe). Judging criteria 2 were: OReg = ORe × ORg for the multiplicative model, OReg > ORe × ORg for the super-multiplicative model, OReg < ORe × ORg for the sub-multiplicative model, and OReg = ORe + ORg − 1 for the additive model.
RESULTS
Nonalcoholic fatty liver disease cases and controls
There was no significant difference between the two groups with respect to age, sex, ethnicity, and birthplace. The cigarette smoking rate in the NAFLD group was significantly higher than that in the control group (OR = 1.8990, P = 0.0083 in SI ≤400 subgroup; OR = 5.0937, P = 0.0051 in SI >400 subgroup) [Table 1].
Table 1.
Background information of NAFLD and control groups, n (%)
| Groups | Gender | Age | Smoking status | ||||
|---|---|---|---|---|---|---|---|
| Male | Female | ≤50 years | >50 years | Nonsmoking | SI ≤400 | SI >400 | |
| Control (n = 900) | 537 (59.67) | 363 (40.33) | 409 (45.44) | 491 (54.56) | 693 (77.00) | 116 (12.89) | 91 (10.11) |
| NAFLD (n = 900) | 542 (60.22) | 358 (39.78) | 406 (45.11) | 494 (54.89) | 453 (50.33) | 144 (16.00) | 303 (33.67) |
| OR* | 1.0000 | 1.8990 | 5.0937† | ||||
| 95% CI | 1.3728–2.5492 | 3.0195–8.3526 | |||||
| P | 0.1037 | 0.0974 | 0.0083 | 0.0051 | |||
*Adjusted for other factors; †P<0.05, compared with SI ≤400. NAFLD: Nonalcoholic fatty liver disease; CI: Confidence interval; OR: Odds ratio; SI: Smoking index.
Polymorphism distributions
The genotype distribution of the resistin − 420C/G polymorphism in the control group complied with HWE (P = 0.0865), indicating the representativeness of this group [Table 2]. We observed a significant difference between the two groups with respect to the CC + CG and GG genotype frequencies (P = 0.0069), and in the distribution of the G allele (P = 0.0085); the risk of NAFLD with the G allele was significantly higher than with the C allele. Under the recessive model, GG homozygotes were found at significantly higher frequencies in the NAFLD than the control group [P = 0.0069, [Table 2]. For the Pro198Leu genotype, the allele frequency and genetic correlation model were in accordance with the above rules [Table 3]. There was no significant difference between the two groups with respect to ALDH2 (GG + GL) and LL genotype frequencies (P = 0.1294), or in the distribution of the L allele (P = 0.0948) [Table 4].
Table 2.
Distribution of the resistin gene promoter −420C/G polymorphism genotypes, alleles, and analysis of the genetic model
| Groups | Genotype, n (%) | Allele, n (%) | Dominant model | Recessive model | ||
|---|---|---|---|---|---|---|
| CC+CG | GG | C | G | (CG+GG)/CC | GG/(CG+CC) | |
| Control (n = 900) | 687 (76.33) | 213 (23.67) | 966 (53.67) | 834 (46.33) | 621:279 | 213:687 |
| NAFLD (n = 900) | 454 (50.44) | 446 (49.56) | 701 (38.94) | 1099 (61.05) | 653:247 | 446:454 |
| OR* | 1.0000 | 3.1685 | 1.0000 | 1.8159 | 1.1878 | 3.1685 |
| 95% CI | 1.9366–5.2073 | 1.3047–2.8671 | 0.8387–1.7593 | 1.9366–5.2073 | ||
| P | 0.0069 | 0.0085 | 0.0857 | 0.0069 | ||
*Adjusted for sex, age, and smoking status. NAFLD: Nonalcoholic fatty liver disease; CI: Confidence interval; OR: Odds ratio.
Table 3.
Distribution of the GPx-1 Pro198Leu polymorphism genotypes, alleles, and analysis of the genetic model
| Groups | Genotype, n (%) | Allele, n (%) | Dominant model | Recessive model | ||
|---|---|---|---|---|---|---|
| PP+PL | LL | P | L | (PL+LL)/PP | LL/(PL+PP) | |
| Control (n = 900) | 682 (75.78) | 218 (24.22) | 959 (53.28) | 841 (46.72) | 623:277 | 218:682 |
| NAFLD (n = 900) | 449 (49.89) | 451 (50.11) | 700 (38.89) | 1100 (61.11) | 649:251 | 451:449 |
| OR* | 1.0000 | 3.1424 | 1.0000 | 1.7919 | 1.1496 | 3.1424 |
| 95% CI | 1.7951–5.2367 | 1.3583–4.0706 | 0.7095–1.9041 | 1.7951–5.2367 | ||
| P | 0.0072 | 0.0095 | 0.0894 | 0.0072 | ||
*Adjusted by sex, age, and smoking status. GPx-1: Glutathione peroxidase-1; NAFLD: Nonalcoholic fatty liver disease; CI: Confidence interval; OR: Odds ratio.
Table 4.
Distribution of the ALDH2 polymorphism genotypes, alleles, and analysis of the genetic model
| Groups | Genotype, n (%) | Allele, n (%) | Dominant model | Recessive model | ||
|---|---|---|---|---|---|---|
| GG + GL | LL | G | L | (GL+LL)/GG | LL/(GL+GG) | |
| Control (n = 900) | 719 (79.89) | 181 (20.11) | 1232 (68.44) | 568 (31.56) | 387:513 | 181:719 |
| NAFLD (n = 900) | 717 (79.67) | 183 (20.33) | 1226 (68.11) | 574 (31.89) | 391:509 | 183:717 |
| OR* | 1.0000 | 1.0138 | 1.0000 | 1.0155 | 1.0182 | 1.0138 |
| 95% CI | 0.4079–1.4825 | 0.5372–1.5285 | 0.5017–1.4720 | 0.4079–1.4825 | ||
| P | 0.1294 | 0.0948 | 0.0865 | 0.1294 | ||
*Adjusted by sex, age, and smoking status. ALDH2: Aldehyde dehydrogenase 2; NAFLD: Nonalcoholic fatty liver disease; CI: Confidence interval; OR: Odds ratio.
Interaction of the − 420C/G and Pro198Leu polymorphisms in nonalcoholic fatty liver disease
A total of 355 patients (39.44%) in the NAFLD group had both −420C/G (GG) and Pro198Leu (LL) genotypes, while only 115 (12.78%) in the control group had this combined genotype (P = 0.0054). Positive interactions were observed between −420C/G (GG) and Pro198Leu (LL) in the pathogenesis of NAFLD, with interaction coefficients γ1 = β1 × 2/β2 = 3.8919 and γ2 = β1 × 2/β1 = 3.8577 both >1. Moreover, OR1 × 2 > OR1 × OR2, revealing a super-multiplicative model [Table 5].
Table 5.
Interaction of polymorphisms −420C/G and Pro198Leu in NAFLD, n (%)
| Groups | Combined genotype of −420C/G and Pro198Leu | |||
|---|---|---|---|---|
| CC+CG/PP+PL | CC+CG/LL | GG/PP+PL | GG/LL | |
| Control (n = 900) | 584 (64.89) | 103 (11.44) | 98 (10.89) | 115 (12.78) |
| NAFLD (n = 900) | 358 (39.78) | 96 (10.67) | 91 (10.11) | 355 (39.44) |
| OR* | 1.0000 | 1.5204† (OR1) | 1.5148‡ (OR2) | 5.0357§ (OR1×2) |
| 95% CI | 0.9725–2.3982 | 0.9593–2.5481 | 3.1852–7.8106 | |
| β | 0.1820† (β1) | 0.1804‡ (β2) | 0.7021§ (β1×2) | |
| P | 0.0385 | 0.0269 | 0.0054 | |
*Adjusted for sex, age, and smoking status; †OR value by simple Pro198Leu homozygous mutant exposure (OR1 [β1]); ‡OR value by simple −420C/G homozygous mutant exposure (OR2 [β2]); §OR value by two interacting genes in homozygous mutant (OR1×2 [β1×2]). NAFLD: Nonalcoholic fatty liver disease; CI: Confidence interval; OR: Odds ratio.
Interaction of the −420C/G and Pro198Leu polymorphisms with cigarette smoking in nonalcoholic fatty liver disease
In the CC + CG/SI ≤400 subgroup, the ORe was 1.1942 (95% CI: 0.8721–1.6285), while this was 1.6754 (95% CI: 1.2470–2.1649) in the CC + CG/SI >400 subgroup. The ORg of − 420C/G (GG) alone was 1.3362 (95% CI: 0.9325–2.6457), but when smoking and −420C/G (GG) existed simultaneously, the interaction OReg was 2.7032 (95% CI: 1.9727–3.5193) in the GG/SI ≤400 subgroup and 10.0762 (95% CI: 7.0958–13.6125) in the GG/SI >400 subgroup. The interaction coefficient γ = βeg/βe = 5.6018 in the GG/SI ≤400 subgroup (or γ =4.4770 in the GG/SI >400 subgroup), and OReg > ORe × ORg, representing a super-multiplicative model [Table 6]. The interaction OReg with Pro198Leu (LL) and smoking was 2.6880 (95% CI: 1.9902–4.1749) in the LL/SI ≤400 subgroup and 9.9129 (95% CI: 6.5294–12.2581) in the LL/SI >400 subgroup, while the interaction coefficient γ = βeg/βe = 5.7715 in the LL/SI ≤400 subgroup (or γ = 4.5985 in the LL/SI >400 subgroup), and OReg > ORe × ORg, also indicative of a super-multiplicative model [Table 7].
Table 6.
Interaction of the −420C/G polymorphism and cigarette smoking in NAFLD, n (%)
| Groups | −420C/G genotype and smoking status | |||||
|---|---|---|---|---|---|---|
| CC+CG/nonsmoking | CC+CG/SI ≤400 | CC+CG/SI >400 | GG/nonsmoking | GG/SI ≤400 | GG/SI >400 | |
| Control (n = 900) | 582 (64.67) | 54 (6.00) | 51 (5.67) | 111 (12.33) | 62 (6.89) | 40 (4.44) |
| NAFLD (n = 900) | 361 (40.11) | 40 (4.44) | 53 (5.89) | 92 (10.22) | 104 (11.56) | 250 (27.78) |
| OR* | 1.0000 | 1.1942† (ORe) | 1.6754† (ORe) | 1.3362‡ (ORg) | 2.7032§ (OReg) | 10.0762§ (OReg) |
| 95% CI | 0.8721–1.6285 | 1.2470–2.1649 | 0.9325–2.6457 | 1.9727–3.5193 | 7.0958–13.6125 | |
| β | 0.0771† (βe) | 0.2241† (βe) | 0.1259‡ (βg) | 0.4319§ (βeg) | 1.0033§ (βeg) | |
| P | 0.0849 | 0.0253 | 0.0637 | 0.0076 | 0.0038 | |
*Adjusted for sex and age; †OR (β) value by simple smoking exposure (ORe [βe]); ‡OR (β) value by simple homozygous mutant type exposure (ORg [βg]); §OR (β) value by the interaction of smoking and homozygous mutant (OReg [βeg]). NAFLD: Nonalcoholic fatty liver disease; CI: Confidence interval; OR: Odds ratio; SI: Smoking index.
Table 7.
Interaction of the Pro198Leu polymorphism and cigarette smoking in NAFLD, n (%)
| Groups | Pro198Leu genotype and smoking status | |||||
|---|---|---|---|---|---|---|
| PP+PL/nonsmoking | PP+PL/SI ≤400 | PP+PL/SI >400 | LL/nonsmoking | LL/SI ≤400 | LL/SI >400 | |
| Control (n = 900) | 579 (64.33) | 53 (5.89) | 50 (5.56) | 114 (12.67) | 63 (7.00) | 41 (4.56) |
| NAFLD (n = 900) | 359 (39.89) | 39 (4.33) | 51 (5.67) | 94 (10.44) | 105 (11.67) | 252 (28.00) |
| OR* | 1.0000 | 1.1868† (ORe) | 1.6451† (ORe) | 1.3299‡ (ORg) | 2.6880§ (OReg) | 9.9129§ (OReg) |
| 95% CI | 0.7694–1.8923 | 1.2548–2.2760 | 0.9499–2.0978 | 1.9902–4.1749 | 6.5294–12.2581 | |
| β | 0.0744† (βe) | 0.2162† (βe) | 0.1238‡ (βg) | 0.4294§ (βeg) | 0.9962§ (βeg) | |
| P | 0.0861 | 0.0375 | 0.0708 | 0.0082 | 0.0043 | |
*Adjusted for sex and age; †OR (β) value by simple smoking exposure (ORe [βe]); ‡OR (β) value by simple homozygous mutant type exposure (ORg [βg]); §OR (β) value by the interaction of smoking and homozygous mutant (OReg [βeg]). NAFLD: Nonalcoholic fatty liver disease; CI: Confidence interval; OR: Odds ratio; SI: Smoking index.
DISCUSSION
Resistin is an adipocyte-specific polypeptide hormone that plays an important role in metabolism by desensitizing fat cells, skeletal muscle cells, and liver cells to insulin. It promotes liver fat deposition and is the cause of hepatic insulin resistance.[10] The expression of resistin is controlled by genes and induced by environmental factors. The human resistin gene is located on chromosome 19p13.2 and has highly conserved intron sequence boundaries. Its mRNA is 476 bp in length, with a coding region of 326 bp that encodes 108 amino acids. The resistin gene contains several SNPs, including −420C/G, 299G/A, and −638G/A, of which −420C/G is considered the most important because it affects promoter activity and increases the expression of resistin in blood and tissues. This polymorphism has three genotypes, −420C/G (CC), −420C/G (CG), and −420C/G (GG), which correlate with metabolic syndrome.[11,12] The present study found that the genotype −420C/G (GG) significantly increased the risk of NAFLD (OR = 3.1685, 95% CI: 1.9366–5.2073) and although the mechanism underlying this is unclear, a related study showed that the G allele may activate resistin gene transcription, thus decreasing insulin sensitivity[13] and causing lipid deposition in the liver, which raises the risk of NAFLD.
Oxidative stress has been suggested as participating in the development of NAFLD. GPX1 is one of the antioxidant enzymes responsible for the defense of cells against oxidative damage and thus for protection against oxidative stress-related diseases. Human GPx-1 gene is located on chromosome 3p21.3, containing 2 exons and 1 intron. The GPx-1 candidate variant (rsl050450; Pro198Leu), which resides in the coding region and results in an amino acid substitution of proline with leucine, was reported to be associated with 40% reduction of GPx-1 activity.[14] Studies have shown that the GPx-1 Pro198Leu polymorphism increases the incidence of oxidative stress-related diseases such as prostate cancer and bladder cancer.[15,16] The present study found that GPx-1 Pro198Leu (LL) was significantly more common in NAFLD patients than in healthy controls (OR = 3.1424, 95% CI = 1.7951–5.2367, P = 0.0072).
ALDH2 is a mitochondrial enzyme that plays a key role in the metabolism of acetaldehyde and other toxic aldehydes. Evidence from many studies has revealed a beneficial role for ALDH2 against alcohol, acetaldehyde, and toxic aldehyde-induced reactive oxygen species formation, as well as tissue injury. Human ALDH2 is encoded by ALDH2, which is located on chromosome 12. The ALDH2 G1510A polymorphism causes the replacement of glutamate at position 487 with lysine (Glu487Lys), which eliminates dehydrogenase activity. Moreover, the G1510A polymorphism increases the risk of a variety of alcohol-related cancers.[17,18] The present study found no significant difference between the two groups with respect to ALDH2 (GG + GL) and LL genotype frequency (P = 0.1294), suggesting that ALDH2 polymorphisms may not be associated with NAFLD susceptibility.
We also observed that the interaction between − 420C/G and GPx-1 Pro198Leu polymorphisms increased the risk for NAFLD. −420C/G (GG) and Pro198Leu (LL) genotypes in NAFLD were shown to have a super-multiplicative effect (γ1 = 3.8919, γ2 = 3.8577). Moreover, smoking increased the risk of NAFLD (OR = 1.8990, P = 0.0083 in the SI ≤400 subgroup; OR = 5.0937, P = 0.0051 in the SI >400 subgroup), and had a positive interaction with both −420C/G (GG) (γ = 5.6018 in the SI ≤400 subgroup; γ = 4.4770 in the SI >400 subgroup) and Pro198Leu (LL) genotypes (γ = 5.7715 in the SI ≤400 subgroup; γ = 4.5985 in the SI >400 subgroup). OReg of each of the two genotypes and smoking exposure was greater than ORe × ORg, indicating that the two homozygous mutant genes interact with smoking in a super-multiplicative model in the pathogenesis of NAFLD.
Long-term smoking causes the oxidation of glucose in cells and can significantly weaken the nonoxidative pathway and increase free fatty acid levels in plasma, which can be taken up by the liver and adipose tissue, leading to the development of insulin resistance. The nicotine in tobacco can also cause stimulate the sympathetic nervous system and increase the release of catecholamines and glucagon, which is a potent antagonist of insulin action. This impaired insulin action creates the conditions for the first hit in the pathogenesis of NAFLD.[19,20] Usually the body can remove excess free radicals through the free radical scavenging system, which includes GPx-1. However, in long-term smokers, excessive oxygen free radicals travel through the blood into the liver cells, inducing lipid peroxidation and free radical reactions, eventually leading to fatty liver. This may explain why smoking can promote the risk of NAFLD through its interaction with −420C/G (GG) and GPx-1 Pro198Leu (LL) genotypes.
In summary, NAFLD is a complex process involving interactions between environmental factors and multiple genes. This study suggests that carriers of resistin gene promoter −420C/G (GG) and GPx-1 Pro198Leu (LL) genotypes may have a higher risk of developing NAFLD, and that these genotypes interact with cigarette smoking in the pathogenesis of NAFLD. Therefore, effective prevention measures for NAFLD should consider controlling environmental factors such as smoking cessation.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We acknowledge colleagues from the Department of Gastroenterology at The First Affiliated Hospital of Xinxiang Medical University for their help with statistical analysis.
Footnotes
Edited by: Li-Shao Guo and Peng Lyu
REFERENCES
- 1.do Nascimento JH, Epifanio M, Soder RB, Baldisserotto M. MRI-diagnosed nonalcoholic fatty liver disease is correlated to insulin resistance in adolescents. Acad Radiol. 2013;20:1436–42. doi: 10.1016/j.acra.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Nomura K, Yamanouchi T. The role of fructose-enriched diets in mechanisms of nonalcoholic fatty liver disease. J Nutr Biochem. 2012;23:203–8. doi: 10.1016/j.jnutbio.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Dai M, Bi Y, Xu M, Xu Y, Li M, et al. Active smoking, passive smoking, and risk of nonalcoholic fatty liver disease (NAFLD): A population-based study in China. J Epidemiol. 2013;23:115–21. doi: 10.2188/jea.JE20120067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyraz M, Cekmez F, Karaoglu A, Cinaz P, Durak M, Bideci A. Serum adiponectin, leptin, resistin and RBP4 levels in obese and metabolic syndrome children with nonalcoholic fatty liver disease. Biomark Med. 2013;7:737–45. doi: 10.2217/bmm.13.13. [DOI] [PubMed] [Google Scholar]
- 5.Samy W, Hassanian MA. Paraoxonase-1 activity, malondialdehyde and glutathione peroxidase in non-alcoholic fatty liver disease and the effect of atorvastatin. Arab J Gastroenterol. 2011;12:80–5. doi: 10.1016/j.ajg.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Fatty Liver and Alcoholic Liver Disease Study Group of the Chinese Liver Disease Association. Guidelines for diagnosis and treatment of nonalcoholic fatty liver disease (in Chinese) J Clin Hepatol. 2010;26:120–4. [Google Scholar]
- 7.Hossein-Nezhad A, Varzaneh FN, Mirzaei K, Emamgholipour S, Varzaneh FN, Sahraian MA. A polymorphism in the resistin gene promoter and the risk of multiple sclerosis. Minerva Med. 2013;104:431–8. [PubMed] [Google Scholar]
- 8.Suzen HS, Gucyener E, Sakalli O, Uckun Z, Kose G, Ustel D, et al. CAT C-262T and GPX1 Pro198Leu polymorphisms in a Turkish population. Mol Biol Rep. 2010;37:87–92. doi: 10.1007/s11033-009-9540-4. [DOI] [PubMed] [Google Scholar]
- 9.Khoury MJ, Wagener DK. Epidemiological evaluation of the use of genetics to improve the predictive value of disease risk factors. Am J Hum Genet. 1995;56:835–44. [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang LY, Jin YJ, Jin QS, Lin LY, Zhang DD, Kong LL. Association between resistin+299A/A genotype and nonalcoholic fatty liver disease in Chinese patients with type 2 diabetes mellitus. Gene. 2013;529:340–4. doi: 10.1016/j.gene.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Boumaiza I, Omezzine A, Rejeb J, Rebhi L, Ben Rejeb N, Nabli N, et al. Association between four resistin polymorphisms, obesity, and metabolic syndrome parameters in Tunisian volunteers. Genet Test Mol Biomarkers. 2012;16:1356–62. doi: 10.1089/gtmb.2012.0156. [DOI] [PubMed] [Google Scholar]
- 12.Bik W, Ostrowski J, Baranowska-Bik A, Wolinska-Witort E, Bialkowska M, Martynska L, et al. Adipokines and genetic factors in overweight or obese but metabolically healthy Polish women. Neuro Endocrinol Lett. 2010;31:497–506. [PubMed] [Google Scholar]
- 13.Norata GD, Ongari M, Garlaschelli K, Tibolla G, Grigore L, Raselli S, et al. Effect of the -420C/G variant of the resistin gene promoter on metabolic syndrome, obesity, myocardial infarction and kidney dysfunction. J Intern Med. 2007;262:104–12. doi: 10.1111/j.1365-2796.2007.01787.x. [DOI] [PubMed] [Google Scholar]
- 14.Cao M, Mu X, Jiang C, Yang G, Chen H, Xue W. Single-nucleotide polymorphisms of GPX1 and MnSOD and susceptibility to bladder cancer: A systematic review and meta-analysis. Tumour Biol. 2014;35:759–64. doi: 10.1007/s13277-013-1103-6. [DOI] [PubMed] [Google Scholar]
- 15.Paz-y-Miño C, Muñoz MJ, López-Cortés A, Cabrera A, Palacios A, Castro B, et al. Frequency of polymorphisms pro198leu in GPX-1 gene and ile58thr in MnSOD gene in the altitude Ecuadorian population with bladder cancer. Oncol Res. 2010;18:395–400. doi: 10.3727/096504010x12644422320780. [DOI] [PubMed] [Google Scholar]
- 16.Erdem O, Eken A, Akay C, Arsova-Sarafinovska Z, Matevska N, Suturkova L, et al. Association of GPX1 polymorphism, GPX activity and prostate cancer risk. Hum Exp Toxicol. 2012;31:24–31. doi: 10.1177/0960327111411499. [DOI] [PubMed] [Google Scholar]
- 17.Oze I, Matsuo K, Hosono S, Ito H, Kawase T, Watanabe M, et al. Comparison between self-reported facial flushing after alcohol consumption and ALDH2 Glu504Lys polymorphism for risk of upper aerodigestive tract cancer in a Japanese population. Cancer Sci. 2010;101:1875–80. doi: 10.1111/j.1349-7006.2010.01599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Druesne-Pecollo N, Tehard B, Mallet Y, Gerber M, Norat T, Hercberg S, et al. Alcohol and genetic polymorphisms: Effect on risk of alcohol-related cancer. Lancet Oncol. 2009;10:173–80. doi: 10.1016/S1470-2045(09)70019-1. [DOI] [PubMed] [Google Scholar]
- 19.Jia WP. The impact of cigarette smoking on metabolic syndrome. Biomed Environ Sci. 2013;26:947–52. doi: 10.3967/bes2013.029. [DOI] [PubMed] [Google Scholar]
- 20.Yalcinkaya E, Celik M, Gursoy E. Determining the combined effects of smoking and obesity on insulin resistance and inflammation. Eur Rev Med Pharmacol Sci. 2014;18:760. [PubMed] [Google Scholar]


