Abstract
Biomarkers of transplant tolerance would enhance the safety and feasibility of clinical tolerance trials and potentially facilitate management of patients receiving immunosuppression. To this end, we examined blood from spontaneously tolerant renal transplant recipients and patients enrolled in two interventional tolerance trials using flow cytometry and gene expression profiling. Using a previously reported tolerant cohort as well as newly identified tolerant patients we confirmed our previous finding that tolerance was associated with increased expression of B cell-associated genes relative to immunosuppressed patients. This was not accounted for merely by an increase in total B cell numbers, but was associated with the increased frequencies of transitional and naïve B cells. Moreover, serial measurements of gene expression demonstrated that this pattern persisted over several years although patients receiving immunosuppression also displayed an increase in the two most dominant tolerance-related B cell genes, IGKV1D-13 and IGLL-1, over time. Importantly, patients rendered tolerant via induction of transient mixed chimerism, and those weaned to minimal immunosuppression, showed similar increases in IGKV1D-13 as did spontaneously tolerant individuals. Collectively, these findings support the notion that alterations in B cells may be a common theme for tolerant kidney transplant recipients, and a useful monitoring tool in prospective trials.
Introduction
Transplantation is the preferred treatment for appropriately selected patients with end-stage renal disease as it confers a superior quality of life as well as a survival benefit relative to dialysis for the vast majority of affected individuals (1). However transplantation is not a panacea as it is associated with significant risks and toxicities, primarily those accompanying the need for long-term immunosuppression. Registry data underscore the importance of these side effects, as cardiovascular disease, infection, and malignancy account for 60% of deaths in patients with functioning allografts after renal transplantation (USRDS: 2010 Annual Report, Vol 2, Chapter 7, http://www.usrds.org/atlas10.aspx). In addition to these concerns, calcineurin inhibitors, which form the backbone of most commonly used immunosuppressive regimens, are nephrotoxic, a side effect that likely contributes to both the premature failure of renal allografts and the development of end-stage renal disease in individuals who have received non-renal transplants (2, 3). Finally and perhaps most importantly, despite life-long administration of current immunosuppressive regimens, interstitial fibrosis and tubular atrophy continues to develop in a significant proportion of allograft recipients.
Transplantation tolerance, which we define here operationally as stable maintenance of good graft function for at least one year in the absence of immunosuppression in an immunocompetent individual, could, as has been recently reported, improve long-term outcomes following transplantation by minimizing or avoiding the side effects of maintenance immunosuppression (4) (5). Tolerance to renal allografts has been achieved in small numbers of patients enrolled in early phase clinical protocols, however the applicability of these protocols to a broader population is limited at present (6) (7) (8) (9). Development of reliable biomarkers of tolerance would not only greatly enhance the safety and feasibility of such protocols, but also potentially have a large impact on the care of transplant recipients treated with standard immunosuppressive drugs, some of whom may be candidates for minimization, and perhaps eventual withdrawal of, immunosuppression. To this end several groups including our own have recently described biomarkers present in spontaneously tolerant kidney and liver transplant recipients following discontinuation of all immunosuppression (10) (11) (12) (13) (14) (15). In the case of renal transplants, functionally tolerant recipients are characterized by increased numbers of B cells and overexpression of B cell-associated genes in their peripheral blood and urine (11) (13) (14). Interestingly the increase in B cell numbers reflects a specific expansion of transitional B cells (14) and B cells that express inhibitory receptors (12) suggesting that these B cells may actively regulate the immune response to the transplanted kidney. This hypothesis is intriguing given recent reports demonstrating the effects of regulatory B cells in experimental models of transplantation and autoimmunity (16) (17) (18).
In this manuscript, we extend our previous observations from the Immune Tolerance Network (ITN) registry of tolerant renal transplant recipients in several important ways. First, we have analyzed additional tolerant recipients newly recruited to the registry and also have provided a substantially more extensive analysis of B cell subsets. Second, we demonstrate that the B cell-focused gene signature, indicating over-expression of selected B cell genes, is not simply a result of increased circulating total B cell numbers. A third important observation is that both cellular and gene expression changes noted in tolerant kidney transplant recipients, are, in large part, maintained over time. However, we also observed that the B cell related genes that were highly associated with tolerance also increased with time in transplant recipients maintained on conventional immunosuppression, such that the differences between the groups diminished over time. Finally, we report that kidney transplant recipients developing tolerance as a result of a prospectively applied mixed chimerism protocol display the same B cell related gene expression changes as observed in spontaneously tolerant kidney transplant recipients. Collectively, these findings support the notion that alterations in B cells may be a common theme for tolerant kidney transplant recipients, and could potentially provide a useful immune monitoring tool in prospective trials.
Materials and Methods
Patients
Patients enrolled under 4 separate ITN protocols are included in this analysis (please see Table 1 for detailed demographic and clinical information):
Table 1.
Demographic and clinical characteristics
| ITN507 | ITN013 | ITN010 & ITN036 | |||||
|---|---|---|---|---|---|---|---|
| TOL (n=25) | New Tol (n=7) | SI (n=34) | Sirolimus (n=7) | SI Multi-agent (n=2) | TOL (n=7) | Return to SI (n=1) | |
| Race, n | |||||||
| Asian | 1 | 1 | |||||
| Black or African American | 4 | ||||||
| White | 24 | 7 | 29 | 7 | 1 | 7 | 1 |
| American Indian or Alaska Native | 1 | ||||||
| Ethnicity, n | |||||||
| Hispanic or Latino | 2 | 1 | 1 | ||||
| Not Hispanic or Latino | 23 | 6 | 33 | 7 | 2 | 7 | 1 |
| Gender, n | |||||||
| Female | 10 | 4 | 15 | 2 | 1 | 3 | |
| Male | 15 | 3 | 19 | 5 | 1 | 4 | 1 |
| Donor type, n | |||||||
| Living-related | 18 | 3 | 15 | 6 | 2 | 7 | 1 |
| Living-unrelated | 1 | 3 | 9 | 1 | |||
| Deceased donor | 5 | 8 | |||||
| Data missing | 1 | 1 | 2 | ||||
| Age at Enrollment, yrs, mean (SD) | 52 (11.5) | 56 (7.2) | 47 (12.8) | 42 (10.2) | 54 (1.1) | 31 (10.6) | 27 |
| Age at Transplantation, yrs, mean (SD) | 31 (12.8) | 36 (9.9) | 37 (15.1) | 42 (10.2) | 54 (1.1) | 31 (10.6) | 27 |
| Interval between transplant and enrollment, yrs, mean (SD) | 21 (9.6) | 20 (10.9) | 10 (10.3) | 0 | 0 | 0 | 0 |
| Donor Age, yrs, mean (SD) | 52 (8.9) | 52 (10.1) | 53 (9.6) | 45 (7.5) | 43 (18.4) | 50 (9.0) | 49 |
| Primary cause for renal failure1, n | |||||||
| Genetic | 2 | 1 | 3 | 2 | 1 | 4 | |
| Diabetes mellitus | 2 | 1 | 1 | 1 | |||
| Etiology uncertain | 3 | 2 | |||||
| HIV nephropathy | 1 | ||||||
| Hypertension | 4 | ||||||
| Immune mediated | 13 | 1 | 18 | 2 | 2 | ||
| Pyelonephritis/interstitial nephritis | 1 | 1 | |||||
| Structural | 3 | 2 | 1 | 1 | |||
| Other | 6 | 4 | 5 | ||||
| HLA mismatch (A, B, and DR loci) 2 | |||||||
| mean (SD) | 1.1 (1.71) | 2 | 3.3 (1.96) 3 | 2.3 (0.76) | 3 | 2.3 (0.76) 4 | 2 5 |
| Data missing, n | 10 | 6 | 6 | ||||
| Years off IS | |||||||
| yrs, mean (SD) | 16 (10.4) | 14 (17.1) | NA | NA | 5 (1.2) | NA | |
| Data missing, n | 2 | 2 | |||||
| Reason for Discontinuing IS, n | |||||||
| Medical condition | 2 | 1 | |||||
| Non-compliance | 20 | 3 | |||||
| Data missing | 3 | 3 | |||||
| Documented episodes of acute rejection, n | |||||||
| No acute rejection | 23 | 6 | 24 | 7 | 2 | 7 | 1 |
| Mild acute cellular rejection (Grade IA) | 2 | 4 | |||||
| Mild acute cellular rejection (Grade IB) | 3 | ||||||
| Moderate acute cellular rejection (Grade IIA) | 1 | 1 | |||||
| Moderate acute cellular rejection (Grade IIB) | 2 | ||||||
| Renal Function, mean (SD) | |||||||
| Creatinine level, mg/dl | 1.5 (1.77) | 1.7 (0.56) | 1.4 (0.46) | 1.4 (0.29) | 2.9 (0.07) | 1.4 (0.50) | 3.2 |
| Proteinuria (>30 mg/dl), n | |||||||
| < 30 mg/24hrs | 2 | 1 | |||||
| >=30 mg/24hrs | 20 | 6 | 28 | 2 | |||
| Data missing | 5 | 1 | 4 | 7 | 2 | 4 | 1 |
Two ITN507 TOL and 3 SI patients had multiple primary causes for renal failure
Allele or antigen level mismatch analysis was performed based on the resolution of available HLA data. Synonymous mutations were not considered mismatches.
Unable to resolve one HLA-B mismatch; counted as a match
Unable to resolve one HLA-DRB mismatch; counted as a match
HLA-DR typing missing for one participant; this participant is analyzed on four present antigens
ITN507 (FACTOR)
This study is the ITN-sponsored renal transplant registry to identify pre-existing tolerant renal transplant recipients. Some of the patients enrolled under this protocol were initially studied in an earlier publication (14), with limited flow cytometric analysis, and without longitudinal sampling. Since publication of that manuscript, we have identified and added 7 new spontaneously tolerant patients who met the enrollment criteria for the study. Centers participating in enrollment are Emory University (Atlanta, GA), National Institutes of Health (Bethesda, MD), Swedish Medical Center (Seattle, WA), and University of Wisconsin (Madison, WI). The protocol was approved by the IRB of each participating center, and by a DSMB convened by the National Institutes of Allergy and Infectious Diseases. Written informed consent was obtained from each patient. Specimens and clinical information were collected annually, with up to 3 years follow-up now available for the majority of participants.
Renal allograft recipients from ITN507 were enrolled into 2 groups: tolerant (TOL; n = 39), defined as individuals who, for at least 1 year prior to enrollment, had not taken immunosuppressive medications and had stable renal function and serum creatinine within 25% of baseline (as evaluated by 3 experienced transplant physicians); and standard immunosuppression (SI; n = 38), defined as patients with clinically stable renal function (using the same criteria as TOL) while on a maintenance triple-drug immunosuppressive regimen (including a calcineurin or mechanistic target of rapamycin inhibitor, an antiproliferative agent, and corticosteroids). As published previously and shown in Table 1, most of the TOL patients received living related allografts and were well HLA matched. An additional group of normal healthy control (HC) participants (n = 42) with no known history of renal disease/dysfunction or evidence of acute medical illness was enrolled. Of these ITN507 patients, we analyzed samples as follows: TOL = 32 (which includes 25 patients originally reported in this study plus 7 newly identified tolerant patients), SI = 34, and HC = 15 (only for flow cytometry analysis). Note 20 more HC samples from an IRB approved assay registry study were also included for flow cytometry analysis.
ITN010 (formerly NKD03) and ITN036
These were closely related studies of conditioning regimens designed to induce tolerance in recipients of primary live-donor one haplotype HLA-matched renal allografts using mixed chimerism induced by bone-marrow transplantation. The clinical outcomes of these patients have been reported previously (7) (19).
ITN013
This was a study of CAMPATH-1H induction therapy with maintenance tacrolimus/sirolimus immunosuppression, followed by serial immunosuppression withdrawal in primary live donor one-haplotype or zero-mismatched cadaveric adult renal transplant recipients. The clinical outcomes of these patients have been reported (20).
Gene expression
Whole blood was collected in Tempus™ Blood RNA tubes and isolated using the Applied Biosystems® PRISM™ 6100 Nucleic Acid PrepStation. Samples and RNA were stored at −80°C and cDNA was generated at Expression Analysis. The MassARRAY QGE (Sequenom) multiplexed primer and competitive template designs and analysis were reported previously (14). All samples were run in a single batch. Over 94% of the samples had RIN scores > 6.0, and the small number of samples that had lower RIN scores did not display aberrant results.
Flow cytometric analysis
Previously frozen PBMCs were incubated in 1 mL of pre-warmed complete media (RPMI 1640 supplemented with 10% FBS and 2mM L-glutamine) containing 20 nM of MitoTracker Green (Invitrogen) at 37°C for 30 mins. Cells were collected by centrifugation then resuspended in 10 μL of warm complete medium and incubated at 37°C for 30 mins. Next, the cells were stained on ice for 30 mins in 100 μLs FACS buffer (PBS + 0.5% BSA) containing 5% normal mouse serum, 5% normal rat serum and the following fluorochrome-conjugated mouse anti-human monoclonal antibodies: PE-IgD (IA6-2, BD Biosciences), PE-A610-CD24 (SN3, Invitrogen), PE-Cy5-CD21 (B-ly4, BD), PerCP-Cy5.5-CD38 (HIT2, BD), PE-Cy7-CD23 (EBVCS2, eBioscience), Pacific Blue-CD3 (SP34-2, BD), Qdot605-CD27 (CLB-27/1, Invitrogen), allophycocyanin-CD95 (DX2, BD), allophycocyanin-Cy7-CD19 (SJ25C1, BD Biosciences), as well as the biotinylated 9G4 rat anti-human Ig idiotype antibody, which was detected by a further staining with 100 μL of streptavidin-Alexa680 (Invitrogen) at a 1:500 dilution in FACS buffer on ice for 30 mins. Cells were washed with 3 mLs PBS and stained with 1 mL LIVE/DEAD aqua-fluorescent reactive dye (Invitrogen) at a 1:1000 dilution in PBS on ice for 30 mins. After resuspension in FACS buffer, cells were acquired on a LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star).
Measurements of BAFF levels
B-cell activating factor (BAFF) assays were performed by Aushon Biosystems (www.aushonbiosystems.com).
Statistical analysis
For flow cytometry data, sample group differences for each B cell subset (as a fraction of CD19+ cells) were identified using a one-way ANOVA, followed by pair-wise comparisons between groups with multiple testing correction (Tukey-Kramer method). Unsupervised, hierarchical clustering of B cell profiles was accomplished using complete linkage and Euclidean distance applied to centered log-ratio transformed frequency data (“Aitchison distance”) (21)and was performed using Matlab (The Mathworks Inc., Natick MA). MassArray QGE (Sequenom) data was normalized to housekeeping genes (GAPDH, UBC and YWHAZ) and log2 normalized. All gene expression visualizations and t-tests for group comparisons were performed in the R statistical programming language. Hierarchical clustering for MassArray QGE data were generated based on Euclidian distance. All figures, data and programming code associated with this work can be found on the ITN data sharing portal: www.itntrialshare.org
Results
Increased naïve/transitional and decreased memory B cells in tolerant renal allograft recipients
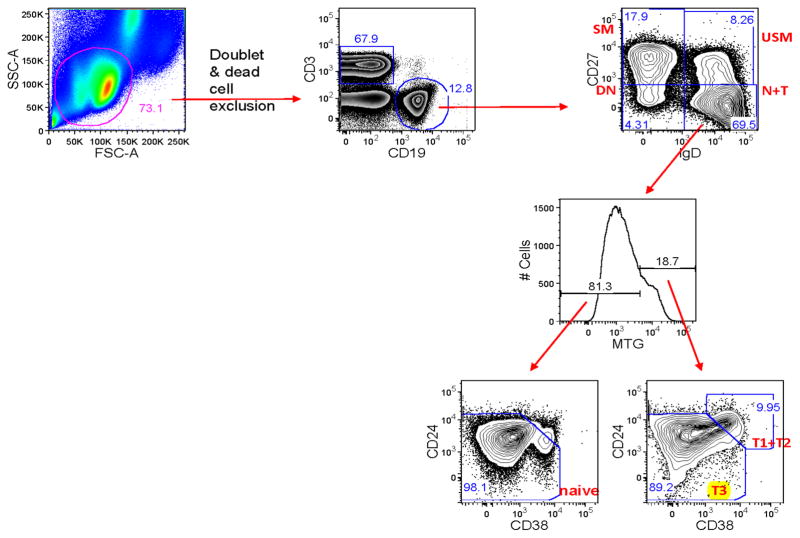
We and others reported previously that spontaneously tolerant renal transplant recipients had increased numbers of peripheral blood B cells compared with a group of patients displaying stable function while receiving standard immunosuppression (12) (13) (14). Moreover, there was a consistently observed increase in the percentage of immature/transitional B cells, defined as CD19+CD38+CD24+IgD+. To extend this analysis further, we performed more detailed multi-parameter flow cytometry in both the previously studied group of patients, as well as 6 newly identified spontaneously tolerant renal allograft recipients. As shown by the example in Figure 1, we defined four canonical B cell subsets using IgD and CD27 as markers: switched memory, IgD−CD27+; unswitched memory, IgD+CD27+; double negative memory, IgD−CD27−; and IgD+CD27− subset. The IgD+CD27− subset contains both naïve and transitional B cells and can be further divided into T1 + T2, T3 and naïve subsets based on MitoTracker Green extrusion and CD24/CD38 expression as follows: naïve, IgD+CD27−MTG−; T3, IgD+CD27−MTG+CD24+/−CD38+/− and T1+ T2, IgD+CD27−MTG+CD24++CD38++ (22) (23).
Figure 1.
B cell subset definitions by flow cytometry. Switched memory (SM) IgD−CD27+; Unswitched memory (UM) IgD+CD27+; Naïve + Transitional (N+T) IgD+CD27− and Double Negative memory (DN) IgD−CD27−. The Naïve + Transitional cells were further subset into T1 + T2, T3 and naïve subsets based on MitoTracker Green (MTG) extrusion and CD24/CD38 expression as follows: Naïve IgD+CD27−MTG− ; T3: IgD+CD27−MTG+CD24+/−CD38+/− ; T1 + T2: IgD+CD27−MTG+CD24++CD38++. The bimodal distribution of MTG staining from total B cells (inset) allowed positioning of the MTG cutoff for analysis of the IgD+CD27− B cells. https://www.itntrialshare.org/FACTOR_fig1.url
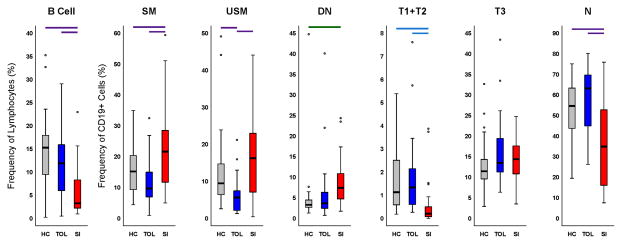
Tolerant recipients had significantly increased frequencies of total B cells, and T1 + T2 early transitional B cells and naïve B cells compared with stable patients on standard immunosuppression. Conversely, the tolerant patients had significantly decreased numbers of both switched and unswitched memory B cells (Fig. 2). It should be noted that in our earlier report on tolerant patients (14), staining was done on fresh whole blood, and the results were reported as absolute cell counts per uL of whole blood. This analysis showed that memory B cells were present in higher absolute numbers in tolerant participants than in patients on standard immunosuppression, because tolerant patients had much higher total B cell counts. In that study, as in the current analysis (done on frozen cells, which does not allow for determination of absolute cell numbers) the percentage of memory B cells within the total B cell population is lower in tolerant patients. The fact that the naïve B cell frequency decreases while the memory B cell frequency increases in participants on standard immunosuppression, suggests that we are not observing a generalized non-specific decrease in B cells caused by immunosuppression, but that tolerance and/or SI therapy may have disparate effects on different B cell subsets. Consistent with our previous results, tolerant recipients were very similar to the group of healthy controls, with the only significant difference between the healthy controls and tolerant patients being that the tolerant group had a lower frequency of unswitched memory cells than did healthy controls.
Figure 2.
Distributions of B cell populations for each study group. Each plot summarizes distribution of frequencies, where: B Cell = CD19+ lymphocytes; SM = switched memory; USM = unswitched memory; DN = double negative memory; T1 + T2 = transitional type 1 and type 2; T3 = transitional type 3; and N = naïve. n = 35 for HC, n = 26 for SI, and n = 27 for TOL. Boxes reflect 25th through 75th percentile of data, black line indicates the median, circles are outliers and whiskers indicate range of non-outlier data. Outliers are data that are 1.5 times the interquartile range beyond the 25th or 75th percentiles. A one-way ANOVA was performed for each subset. Pairwise differences are indicated by bars above the plots as a result of a multiple comparisons test performed after the ANOVA; color reflects the p-value for the ANOVA: purple, p<0.001; blue, p<0.005; green, p<0.05. https://www.itntrialshare.org/FACTOR_fig2.url
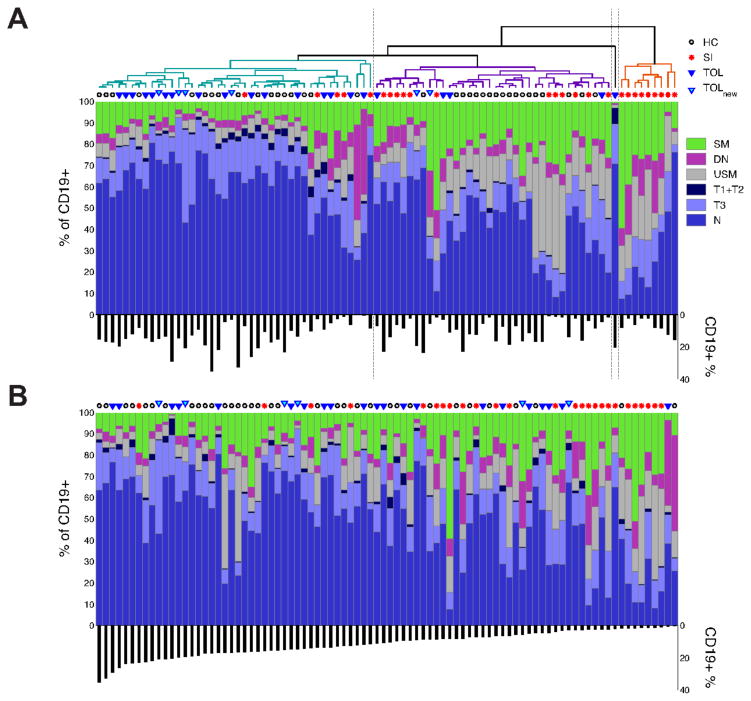
An unsupervised hierarchical cluster analysis based on B cell profiles yielded three primary clusters and one singleton (Fig. 3A). One cluster was comprised predominantly of tolerant patients and healthy controls (with a small number of patients on standard immunosuppression). A second cluster contained primarily healthy controls and patients on standard therapy, and the third (and smallest) cluster had only patients on immunosuppression. We also constructed a stacked bar plot of memory and transitional B cell subset frequencies ordered on the single parameter of peripheral B cell percentage (%CD19+ cells in the total lymphocyte gate – Fig. 3B). There appears to be a relatively greater “mixing” of the tolerant patients among the other cohorts in this type of analysis, suggesting that in addition to the number of B cells, their maturational status also may correlate with the tolerant state. While clearly only limited conclusions can be drawn from this type of analysis, combined with the data in Figure 2, it suggests that tolerant patients may differ from healthy controls, and confirms the presence of a B cell, and B cell subset, bias, in tolerant patients. Of note, we found no significant correlation between time post-transplant and B cell number in either tolerant or drug treated patients (p=0.33 and p=0.51 respectively).
Figure 3.
Panel A. Clustered, stacked bar plot of memory and transitional B cell subset frequencies comprising a 6-part composition. Each column of data is derived from a single sample from one of four sample groups (indicated by a symbol above each column). Each color represents the proportion of a particular cell subset in the sample (of CD19+ cells). The percentages of CD19+ (of Lymphocytes) are shown beneath the stacked bar plot. 88 B cell profiles are hierarchically clustered (see Methods) resulting in three clusters and a singleton. Panel B. Same as A except samples are ordered by the percentages of CD19+ cells in the lymphocyte gate. https://www.itntrialshare.org/FACTOR_fig3.url
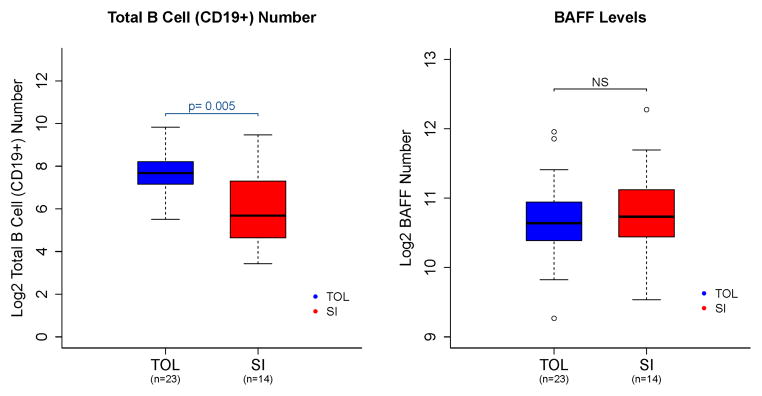
BAFF, a member of the TNF superfamily expressed in multiple cell types, plays an important role in the proliferation and maturation of transitional B cells and the subsequent survival of mature B cells and plasma cells (24). As increased levels of circulating BAFF could explain the increase in B cell numbers observed in tolerant patients, we measured BAFF levels in the serum of our patients from blood draws obtained at the same time points as for B cell analyses. As shown in Figure 4, increased numbers of B cells were not the result of alterations in BAFF levels. Indeed, when normalized for B cell numbers, patients on standard immunosuppression had higher levels of BAFF, perhaps due to lower B cell counts in that cohort.
Figure 4.
B cell numbers and BAFF levels. Absolute number of CD19+ B cells (left panel) and BAFF levels (right panel), on a log2 scale, for tolerant (TOL) patients and subjects on standard immunosuppression (SI). https://www.itntrialshare.org/FACTOR_fig4.url
B cell biased gene expression profiles in tolerant renal transplant recipients
In an earlier manuscript (14), we reported that overexpression of 31 genes, 26 of which were specific to B cells, distinguished tolerant renal transplant recipients from patients who were receiving immunosuppression. Notably, a linear discriminant analysis algorithm utilizing three of the most highly differentially expressed genes identified in a training set, (IGKV1D-13, IGKV4-1, and IGLL-1) was able to accurately predict the status of patients (tolerant vs. stable on immunosuppression) with a high degree of accuracy in a very small test cohort of patients (n=12). In that study, differentially expressed genes were initially identified by microarray and were subsequently assessed by PCR using the Sequenom methodology. Retesting of the original samples (Figure S1) revealed that while the actual numbers of detected mRNA molecules were 30–100 times lower in the repeat assay compared with the original study, differences in the expression of the two genes (IGKV1D-13 and IGLL-1) that most “robustly” differentiated tolerant patients from those receiving immunosuppression in the original study remained significantly different (p≤0.001 and p<0.05 respectively). In contrast the expression levels of the third gene, IGKV4-1, were no longer different between the groups.
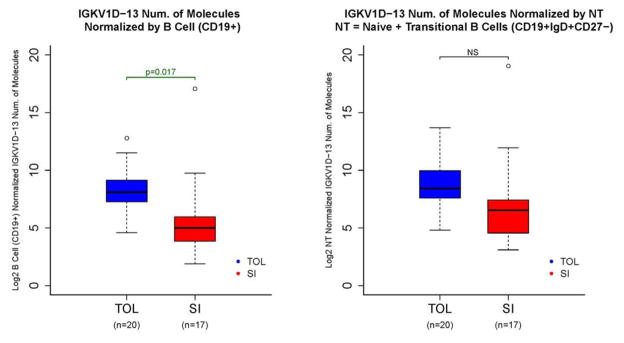
Importantly, transcript levels of the single most predictive gene, IGKV1D-13, did not merely reflect alterations in B cell numbers as even after correcting for total B cells, a significant difference in transcript numbers remained (Fig. 5). However we did find that after normalization for naïve and transitional B cell numbers, while a difference remained between tolerant and drug-treated patients, it was not statistically significant (Fig. 5). This suggests that although the naïve and/or transitional B cell compartments contribute to some portion of the altered gene expression seen in tolerant patients, the gene expression levels also differ on a per cell basis or that other cell subsets contribute as well.
Figure 5.
IGKV1D-13 expression normalized by CD19+ cells (left panel) and naïve plus transitional B cells (right panel). IGKV1D-13 levels as measured by Sequenom analysis in 2011 (see Figure S1) in tolerant patients (TOL) and subjects on standard immunosuppression (SI). Left panel shows expression levels normalized by total number of CD19+ cells, and right panel shows levels normalized by numbers of naïve plus transitional B cells (CD19+IgD+CD27−). https://www.itntrialshare.org/FACTOR_fig5.url
In addition to our focused analysis of the 3 genes originally reported to be robust predictors of clinical tolerance, we also took the opportunity of having recruited new patients to the tolerance registry to examine the expression of the 232 genes shown in Table S2 using Sequenom methodology. These genes were selected based on our original microarray data (14) as well as genes that had been reported by others to be associated with tolerance. As shown in Figure S2, 15 of the top 20 genes differentially expressed between tolerant and standard immunotherapy participants were B cell specific, confirming our findings that B cells and B cell expressed genes are preferentially represented in drug-free tolerant kidney transplant recipients. While these additional genes are potentially of interest, IGKV1-D13 and IGLL-1 remain the most discriminating markers.
Longitudinal IGKV1D-13 and IGLL-1 expression in the ITN registry
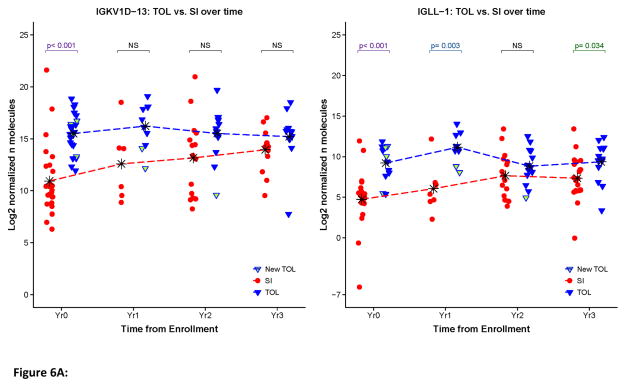
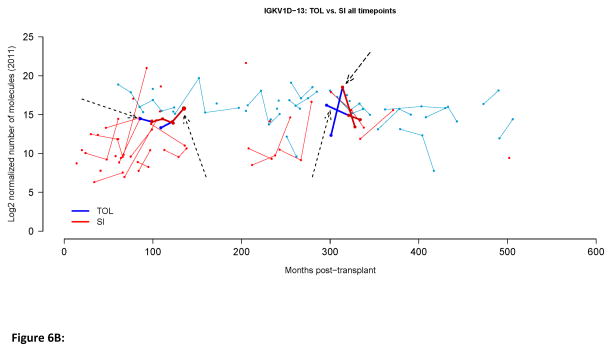
Biomarkers of tolerance should be stable over time. In the case of transplantation tolerance changes in any putative biomarker should occur only with the loss of the tolerant phenotype. We examined the stability of IGKV1D-13 and IGLL-1 expression over time in both tolerant individuals and those continuing to receive immunosuppression. As displayed in Figure 6, at the population level (panel A) and the individual level (panel B) the expression of IGKV1D-13 and IGLL-1 (shown only in panel A) remained relatively stable over time in tolerant patients. Interestingly, the expression of IGKV1D-13 and IGLL-1 tended to increase over time in the cohort of patients receiving immunosuppression, as a result of which, the difference between IGKV1D-13 and IGLL-1 expression in tolerant vs. standard immunosuppression lost statistical significance (Fig. 6A). Whether this relates to changes in immunosuppressive agents, the development of pro-tolerance immune mechanisms in immunosuppressed patients, or is a simple function of time following transplantation remains to be determined.
Figure 6.
Longitudinal IGKV1D-13 and IGLL-1 expression. A. Scatter plots for normalized IGKV1D-13 and IGLL-1 expression in all available samples. The time points indicate the point at which the sample was obtained relative to the start of the study, i.e., year 0 is the first blood sample we obtained, year 1 is one year later, etc. Mean values for each group at each time point are represented by * and connected by dashed lines. P-values represent comparisons between the groups at each time point. B. Longitudinal IGKV1D-13 expression as a function of months post-transplant. Tolerant patients (TOL) are shown in blue, patients on standard immunosuppressive therapy (SI) are shown in red. Points joined by a line depict sequential samples from the same participant. Arrows indicate four patients that were operationally tolerant when first studied, but later went back on immunosuppressive therapy.
IGKV1D-13 expression in patients enrolled in prospective interventional tolerance trials
Previously we have reported on two patient cohorts enrolled in ITN sponsored trials designed to induce tolerance in renal transplant recipients. In ITN010 and ITN036 transient mixed hematopoietic chimerism was induced by donor bone marrow transplantation in conjunction with a non-myeloablative conditioning regimen. Seven of 10 patients enrolled were tolerant for at least five years with excellent graft function. In ITN013 (20), 10 patients were treated with CAMPATH-1H, followed by tacrolimus plus sirolimus, with the intention of withdrawing immunosuppression by 2 years in individuals meeting eligibility criteria. Ultimately, eight of the 10 were weaned down to sirolimus monotherapy.
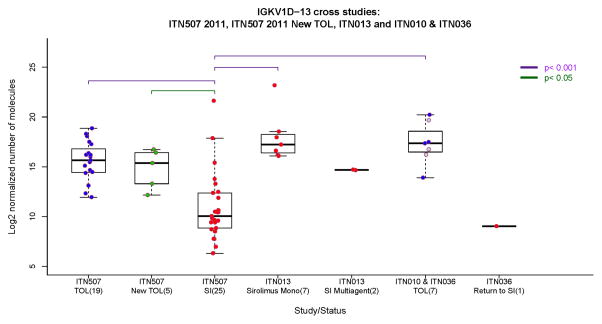
We compared IGKV1D-13 expression levels in “spontaneously” tolerant patients (ITN507 – using the first available samples) with patients enrolled in the interventional trials above (ITN010, ITN036, and ITN013) (Fig. 7). In the latter studies, samples were obtained from the mixed chimerism patients at ~12 months post-transplant, and for the CAMPATH-1H treated patients at 48 months post-transplant, times at which lymphoid reconstitution was complete and B cell numbers had returned to baseline levels.
Figure 7.
Cross-sectional IGKV1D-13 expression in renal transplant recipients enrolled in multiple ITN studies. ITN507 is the tolerant renal transplant registry (TOL n=19; New-TOL n=5; SI n=25); ITN010 and 036 are the mixed chimerism trials with combined bone marrow and renal transplants. Samples from 7 operationally tolerant patients (TOL) were collected 0.33 – 7 years after discontinuation of immunosuppression (no differences in IGKV1D-13 expression were seen in relation to time, not shown). The three patients that eventually had to restart immunosuppression are indicated by light blue circles. One patient who was returned to SI (Return to SI) had their sample obtained one year after transplantation (which was 2 months after the patient rejected and was returned to immunosuppression). ITN013 is the Campath-1H followed by tacrolimus plus sirolimus trial where samples were obtained 4 years after transplantation. In ITN013, 7 patients were weaned and stable on sirolimus monotherapy (Sirolimus Mono), while 2 patients required re-institution of tacrolimus plus mycophenolate (SI Multiagent). https://www.itntrialshare.org/FACTOR_fig7.url
A number of findings emerge from this analysis. First, IGKV1D-13 expression levels in the 5 new patients recruited into the registry for whom expression data were available, are indistinguishable from those originally reported (14), further confirming the validity of this finding. Second, in the combined bone marrow and kidney transplantation trials (ITN010 and ITN036) the expression of IGKV1D-13 in the seven patients who were operationally tolerant was indistinguishable from spontaneously tolerant patients in the ITN registry (ITN507). Also of note is that while three of the 7 operationally tolerant patients eventually required reinstitution of immunosuppression (two with clinical and histologic features of chronic alloimmune-mediated graft injury and one for recurrence of membranoproliferative glomerulonephritis), their IGKV1D-13 expression at early time points when they displayed operational tolerance is similar to that of the other 4 who remain tolerant (light blue circle symbols in Fig. 7). Unfortunately as there are no samples available at later time points we cannot determine if IGKV1D-13 expression was altered at the time tolerance was lost. Third, although only a single sample was available from a patient who experienced early acute rejection and consequently returned to maintenance immunosuppression, the expression of IGKV1D-13 (at the time that patient was on drug therapy) was similar to ITN registry patients who displayed stable renal function while receiving immunosuppression.
Interpretation of the data from ITN013 is complex. As noted above, 8 of the 10 patients were weaned to sirolimus monotherapy (samples were available from 7 of these 8 patients). Three of these 7 met the criteria for sirolimus withdrawal which included reduction of sirolimus to a dose of 1 mg/day, the absence of DSA, and a protocol biopsy that showed no deposition of C4d, however the patients elected to remain on this low dose of sirolimus for non-immunologic reasons. The other four patients all had DSA with one patient also showing C4d deposition on a biopsy. These four individuals were not considered eligible for drug withdrawal. The remaining two patients in the trial were converted from sirolimus to tacrolimus and mycophenolate at ~12–24 months due to development of donor specific antibodies and biopsies showing both histologic evidence of alloimmune injury (Banff grade IIA and borderline) and positive C4d staining (focally positive and diffusely positive).
We observed that the two patients on tacrolimus plus mycophenolate had the lowest levels of IGKV1D-13 at 48 months post-transplant (albeit still higher than most ITN507 patients on standard immunosuppression), while the 7 on sirolimus monotherapy had levels of IGKV1D-13 expression similar to tolerant patients. This point is particularly important and indicates that low levels of IGKV1D-13 expression seen in our previous cohort of stable drug-treated allograft recipients are unlikely to be an artifact of immunosuppression per se, although the influence of the level and/or type of immunosuppression used remains to be determined. Of note however, among the 7 sirolimus monotherapy patients, the 3 which were DSA−/C4d− had indistinguishable IGKV1D-13 levels from the 4 who were DSA+/C4d+ (data not shown).
Discussion
The goal of this study was to further explore the association between B cells and tolerance following renal transplantation. Key aspects of our work include: (1) a detailed phenotypic analysis of B cell subsets; (2) longitudinal data on the expression of key B cell genes in patients who are operationally tolerant as well as those who are stable on immunosuppression; and (3) analysis of relevant B cell genes in patients from two different tolerance-induction studies.
Our data confirm previous findings from own our group and others showing that renal transplant recipients displaying “spontaneous” operational tolerance, i.e., patients who were not enrolled in tolerance protocols, are distinguished from recipients maintained on immunosuppression by increased numbers of B cells in the blood as well as the increased expression of selected B cell-associated genes in peripheral blood mononuclear cells (25). Beyond simple increases in B cell numbers the B cell population in the tolerant patients was characterized by an increased prevalence of transitional and naïve B cells, and reciprocal decreases in both switched and unswitched memory B cells. Recent studies support a role for transitional B cells in renal allograft tolerance. Chesneau et al, demonstrated that tolerant recipients had an elevated frequency of transitional and naïve B cells and a decreased frequency of plasma cells, and moreover that B cells from tolerant patients produced more IL-10 and were less likely to differentiate into plasma cells in vitro than did B cells from patients receiving immunosuppression (26). These investigators have also found that B cells from tolerant recipients can suppress effector T cell responses in vitro in a granzyme B-dependent fashion (27). Finally, it is intriguing that the intentional depletion of B cells using rituximab at the time of renal transplantation may result in high rates of acute rejection in patients receiving a conventional immunosuppressive regimen (28). When taken in aggregate, data from experimental transplant models, clinical transplantation and our studies of tolerance following renal transplantation all suggest that B cells may play a functional role in suppressing alloimmunity (16, 29) (30) (31) (32)
As part of the current study we conducted additional analyses of gene expression in both newly identified as well as previously identified tolerant kidney transplant recipients. As might be predicted, expression of selected B cell associated genes by newly identified tolerant recipients was indistinguishable from that of the previously studied individuals. Importantly however, we now have a longitudinal analysis of gene expression in our patient cohorts, and find that the increased level of expression of the most predictive differentially expressed B cell-related genes, IGKV1D-13 and IGLL-1, remained stable over a period of 3 years in the cohort of tolerant recipients for whom serial samples were available.
We are aware of only a single prior longitudinal study of biomarkers of clinical tolerance following kidney transplantation (33). In that paper, 2 samples separated by 0.8 to 4 years in time from each of 4 tolerant kidney transplant recipients showed stable increases in the number of CD19+ cells in the peripheral blood as well as stable increases in the B cell-associated genes CD19, CD20 and Bank1. Notably, our data show that over-expression of selected B cell genes is not simply a result of increased circulating B cell numbers; much of the perturbation in B cells associated with clinical tolerance appears to be related to the expansion of T1 and T2 transitional B cells and the contraction of memory B cells. We speculate that the increased B cell transcripts we detect in peripheral blood are a result of their overexpression by immature/transitional B cells. Although we were unable to do so in the current study, this could be directly tested by determining the gene expression profiles of freshly sorted transitional B cells from patients and healthy controls.
It is important to note that transplants patients stable on immunosuppression had increases in both IGKV1D-13 and IGLL-1 over time, to the extent that by 3 years after the first entry into the registry, the differences in these parameters between this cohort and the tolerant cohort were no longer statistically significant. Increases in the expression of B cell-related genes over time in the cohort of patients receiving immunosuppression may not be entirely unexpected. In liver transplant recipients one of the most powerful predictors of the tolerant phenotype is time since transplantation (34). While the mechanisms of tolerance induction appear to be different following liver and kidney transplantation, it is conceivable that over time the immune system undergoes adaptations that favor immune tolerance to the transplanted kidney. The increase in B cell-related genes seen over time in at least some patients with stable function who are receiving immunosuppression may be consistent with a model of acquired tolerance that is more prevalent in kidney transplantation than previously appreciated. As well, we cannot exclude the possibility that changes over time (most likely reductions) in doses of immunosuppression may contribute to alterations in B cell profiles and/or B cell expressed genes.
One factor confounding our analysis is that the time since transplantation was much greater for the tolerant cohort than for the group of patients still receiving immunosuppression. This raises the possibility that the increase in B cells observed in the tolerant cohort simply reflects the immune system’s adaptation to the transplanted allograft or that B cell synthesis eventually recovers as immunosuppression is tapered over time in many patients. This notion is also consistent our finding that the peripheral expression of B cell-related genes increases with time following transplantation even for those patients continuing to receive immunosuppression.
Among the most important results reported in this paper are the data on patients enrolled in tolerance-induction protocols. We found that patients who were tolerant following a protocol of thymic irradiation, non-myeloablative conditioning, and combined bone marrow and kidney transplantation had levels of B cell-related gene expression that were similar to our spontaneously tolerant cohort. Of further note, the sole patient treated with this protocol who lost tolerance had levels of B cell-related gene expression (at a time when he/she was returned to immunosuppression) that were similar to patients in the current study who had been maintained on immunosuppression. These findings suggest that patients developing tolerance either spontaneously or as a result of purposeful perturbation of the immune system may share similar markers, or perhaps even mechanisms, of tolerance.
In this vein, the patients in ITN013 also are informative as they displayed increased expression of selected B cell-associated genes at a time when they remained stable on low-dose sirolimus-based immunosuppression. This implies that the association of increased B cells numbers and gene expression seen in tolerance may not solely be the result of the absence of immunosuppression, albeit the numbers of patients in this study are small, and perhaps more notably, all were on single agent sirolimus. However also arguing against a simple drug effect is the observation that tolerant liver transplant recipients do not show the same increase in B cells relative to those receiving immunosuppression as do tolerant kidney transplant recipients (33). The generalizability of these findings to larger numbers of renal allograft recipients, including those on multi-drug immunosuppressive regimens, is under active investigation.
A cautionary finding from our analysis of the patients enrolled in ITN013 that requires further study is the observation that 4 of the 7 patients on sirolimus monotherapy, all of whom had high expression of IGKV1D-13, also had detectable donor specific antibodies and in one case, a biopsy that was C4d+. While the demonstration of donor-specific humoral sensitization is generally incompatible with the tolerant phenotype, in rare instances, DSA has been reported in individuals meeting the definition of operational tolerance, highlighting both the limitations of how clinical tolerance is defined and our understanding of the pathogenic impact of different types of DSA (35). Nonetheless, current clinical practice would preclude drug minimization in the face of a positive DSA and thus any proposed tolerance signature that was observed in DSA+ patients would have to be interpreted with great care.
While several groups including our own have demonstrated an association between B cells and renal allograft tolerance, other cell populations have been implicated as well, including myeloid-derived dendritic cells and regulatory T cells (36, 37). Although it is not yet possible to integrate these various findings into a comprehensive model describing the development of tolerance, these data suggest that factors beyond B cells alone are likely to play an important role.
A final consideration is how these findings may be applied to clinical transplantation. It has recently been suggested that the association of B cells with tolerance following kidney transplantation is robust enough to warrant clinical trials using markers related to B cells to guide the minimization of immunosuppression (38). Several factors must be addressed in contemplating trials of this type including safety, defining an appropriate control group, and their feasibility. Despite the theoretical importance for an individual of having a tool to detect or predict tolerance, it is self-apparent that if the frequency of tolerance is vanishingly low, such a tool will have a very limited impact on the field as a whole. A previous report indicated that only 3.5% of 144 renal transplant recipients with a stable clinical status at 5 or more years following transplantation displayed a B cell-based gene expression profile that was consistent with the tolerant phenotype (11). In addition it is worth noting that patients rendered tolerant as a result of a protocol specifically designed to promote tolerance (ITN010 and ITN036) displayed B cell profiles consistent with spontaneously tolerant kidney transplant recipients. This implies that a predictive tool, if identified, might be used to monitor patients in protocols designed to induce tolerance. It would not be unexpected that the proportion of tolerant patients in these protocols would be much larger than in the group of kidney transplant recipients who spontaneously develop tolerance. Indeed, despite the success of tolerance-induction regimens in a significant proportion of the patients, immunologic failures have occurred with each protocol. This highlights our need to identify biomarkers that can be used to identify or predict tolerance as a means to individualize therapy.
Supplementary Material
Acknowledgments
This research was performed as a project of the Immune Tolerance Network (NIH N01 AI15416 and UM1 AI-109565), an international clinical research consortium headquartered at the Benaroya Research Institute and supported by the National Institute of Allergy and Infectious Diseases. We thank the many clinicians, staff, and particularly the patients, who participated in the clinical trials that made this study possible.
List of Abbreviations
- BAFF
B-cell activating factor
- BD
Becton, Dickinson and Company
- BSA
bovine serum albumin
- cDNA
complementary DNA
- FACS
fluorescence-activated cell sorting
- FBS
fetal bovine serum
- HC
healthy control
- HLA
human leukocyte antigen
- ITN
Immune Tolerance Network
- PBMC
peripheral blood mononuclear cell
- PBS
phosphate-buffered saline
- QGE
Quantitative Gene Expression
- RIN
RNA integrity number
- SI
standard immunosuppression
- TOL
tolerant
- USRDS
United States Renal Data System
Footnotes
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Wolfe RA, Ashby VB, Milford EL, Ojo AO, Ettenger RE, Agodoa LY, et al. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. The New England journal of medicine. 1999;341(23):1725–30. doi: 10.1056/NEJM199912023412303. [DOI] [PubMed] [Google Scholar]
- 2.Casey MJ, Meier-Kriesche HU. Calcineurin inhibitors in kidney transplantation: friend or foe? Current opinion in nephrology and hypertension. 2011;20(6):610–5. doi: 10.1097/MNH.0b013e32834b4343. [DOI] [PubMed] [Google Scholar]
- 3.Ojo AO, Held PJ, Port FK, Wolfe RA, Leichtman AB, Young EW, et al. Chronic renal failure after transplantation of a nonrenal organ. The New England journal of medicine. 2003;349(10):931–40. doi: 10.1056/NEJMoa021744. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Sachs DH, Sprangers B, Spitzer TR, Saidman SL, Zorn E, et al. Long-term results in recipients of combined HLA-mismatched kidney and bone marrow transplantation without maintenance immunosuppression. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2014;14(7):1599–611. doi: 10.1111/ajt.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scandling JD, Busque S, Shizuru JA, Lowsky R, Hoppe R, Dejbakhsh-Jones S, et al. Chimerism, graft survival, and withdrawal of immunosuppressive drugs in HLA matched and mismatched patients after living donor kidney and hematopoietic cell transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2015;15(3):695–704. doi: 10.1111/ajt.13091. [DOI] [PubMed] [Google Scholar]
- 6.Leventhal J, Abecassis M, Miller J, Gallon L, Ravindra K, Tollerud DJ, et al. Chimerism and tolerance without GVHD or engraftment syndrome in HLA-mismatched combined kidney and hematopoietic stem cell transplantation. Science translational medicine. 2012;4(124):124ra28. doi: 10.1126/scitranslmed.3003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai T, Cosimi AB, Spitzer TR, Tolkoff-Rubin N, Suthanthiran M, Saidman SL, et al. HLA-mismatched renal transplantation without maintenance immunosuppression. The New England journal of medicine. 2008;358(4):353–61. doi: 10.1056/NEJMoa071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scandling JD, Busque S, Dejbakhsh-Jones S, Benike C, Sarwal M, Millan MT, et al. Tolerance and withdrawal of immunosuppressive drugs in patients given kidney and hematopoietic cell transplants. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2012;12(5):1133–45. doi: 10.1111/j.1600-6143.2012.03992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leventhal JR, Mathew JM, Salomon DR, Kurian SM, Suthanthiran M, Tambur A, et al. Genomic biomarkers correlate with HLA-identical renal transplant tolerance. Journal of the American Society of Nephrology : JASN. 2013;24(9):1376–85. doi: 10.1681/ASN.2013010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohne F, Martinez-Llordella M, Lozano JJ, Miquel R, Benitez C, Londono MC, et al. Intra-graft expression of genes involved in iron homeostasis predicts the development of operational tolerance in human liver transplantation. The Journal of clinical investigation. 2012;122(1):368–82. doi: 10.1172/JCI59411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brouard S, Le Bars A, Dufay A, Gosselin M, Foucher Y, Guillet M, et al. Identification of a gene expression profile associated with operational tolerance among a selected group of stable kidney transplant patients. Transplant international : official journal of the European Society for Organ Transplantation. 2011;24(6):536–47. doi: 10.1111/j.1432-2277.2011.01251.x. [DOI] [PubMed] [Google Scholar]
- 12.Pallier A, Hillion S, Danger R, Giral M, Racape M, Degauque N, et al. Patients with drug-free long-term graft function display increased numbers of peripheral B cells with a memory and inhibitory phenotype. Kidney international. 2010;78(5):503–13. doi: 10.1038/ki.2010.162. [DOI] [PubMed] [Google Scholar]
- 13.Sagoo P, Perucha E, Sawitzki B, Tomiuk S, Stephens DA, Miqueu P, et al. Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. The Journal of clinical investigation. 2010;120(6):1848–61. doi: 10.1172/JCI39922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, et al. Identification of a B cell signature associated with renal transplant tolerance in humans. The Journal of clinical investigation. 2010;120(6):1836–47. doi: 10.1172/JCI39933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Khatri P, Sigdel TK, Tran T, Ying L, Vitalone MJ, et al. A peripheral blood diagnostic test for acute rejection in renal transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2012;12(10):2710–8. doi: 10.1111/j.1600-6143.2012.04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Q, Yeung M, Camirand G, Zeng Q, Akiba H, Yagita H, et al. Regulatory B cells are identified by expression of TIM-1 and can be induced through TIM-1 ligation to promote tolerance in mice. The Journal of clinical investigation. 2011;121(9):3645–56. doi: 10.1172/JCI46274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshizaki A, Miyagaki T, DiLillo DJ, Matsushita T, Horikawa M, Kountikov EI, et al. Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. 2012 Mar 19;491(7423):264–8. doi: 10.1038/nature11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mauri C, Blair PA. Regulatory B cells in autoimmunity: developments and controversies. Nature reviews Rheumatology. 2010;6(11):636–43. doi: 10.1038/nrrheum.2010.140. [DOI] [PubMed] [Google Scholar]
- 19.Kawai T, Sachs DH, Sykes M, Cosimi AB, Immune Tolerance N. HLA-mismatched renal transplantation without maintenance immunosuppression. The New England journal of medicine. 2013;368(19):1850–2. doi: 10.1056/NEJMc1213779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knechtle SJ, Pascual J, Bloom DD, Torrealba JR, Jankowska-Gan E, Burlingham WJ, et al. Early and limited use of tacrolimus to avoid rejection in an alemtuzumab and sirolimus regimen for kidney transplantation: clinical results and immune monitoring. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2009;9(5):1087–98. doi: 10.1111/j.1600-6143.2009.02581.x. [DOI] [PubMed] [Google Scholar]
- 21.Aitchison J. The Statistical Analysis of Compositional Data. Blackburn Press; 1986. [Google Scholar]
- 22.Wirths S, Lanzavecchia A. ABCB1 transporter discriminates human resting naive B cells from cycling transitional and memory B cells. European journal of immunology. 2005;35(12):3433–41. doi: 10.1002/eji.200535364. [DOI] [PubMed] [Google Scholar]
- 23.Palanichamy A, Barnard J, Zheng B, Owen T, Quach T, Wei C, et al. Novel human transitional B cell populations revealed by B cell depletion therapy. J Immunol. 2009;182(10):5982–93. doi: 10.4049/jimmunol.0801859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackay F, Schneider P, Rennert P, Browning J. BAFF AND APRIL: a tutorial on B cell survival. Annual review of immunology. 2003;21:231–64. doi: 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- 25.Baron D, Ramstein G, Chesneau M, Echasseriau Y, Pallier A, Paul C, et al. A common gene signature across multiple studies relate biomarkers and functional regulation in tolerance to renal allograft. Kidney international. 2015;87(5):984–95. doi: 10.1038/ki.2014.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chesneau M, Pallier A, Braza F, Lacombe G, Le Gallou S, Baron D, et al. Unique B cell differentiation profile in tolerant kidney transplant patients. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2014;14(1):144–55. doi: 10.1111/ajt.12508. [DOI] [PubMed] [Google Scholar]
- 27.Chesneau M, Michel L, Dugast E, Chenouard A, Baron D, Pallier A, et al. Tolerant Kidney Transplant Patients Produce B Cells with Regulatory Properties. Journal of the American Society of Nephrology : JASN. 2015 doi: 10.1681/ASN.2014040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clatworthy MR, Watson CJ, Plotnek G, Bardsley V, Chaudhry AN, Bradley JA, et al. B-cell-depleting induction therapy and acute cellular rejection. The New England journal of medicine. 2009;360(25):2683–5. doi: 10.1056/NEJMc0808481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Texier L, Thebault P, Lavault A, Usal C, Merieau E, Quillard T, et al. Long-term allograft tolerance is characterized by the accumulation of B cells exhibiting an inhibited profile. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11(3):429–38. doi: 10.1111/j.1600-6143.2010.03336.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu C, Noorchashm H, Sutter JA, Naji M, Prak EL, Boyer J, et al. B lymphocyte-directed immunotherapy promotes long-term islet allograft survival in nonhuman primates. Nature medicine. 2007;13(11):1295–8. doi: 10.1038/nm1673. [DOI] [PubMed] [Google Scholar]
- 31.Fehr T, Wang S, Haspot F, Kurtz J, Blaha P, Hogan T, et al. Rapid deletional peripheral CD8 T cell tolerance induced by allogeneic bone marrow: role of donor class II MHC and B cells. J Immunol. 2008;181(6):4371–80. doi: 10.4049/jimmunol.181.6.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fehr T, Haspot F, Mollov J, Chittenden M, Hogan T, Sykes M. Alloreactive CD8 T cell tolerance requires recipient B cells, dendritic cells, and MHC class II. J Immunol. 2008;181(1):165–73. doi: 10.4049/jimmunol.181.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozano JJ, Pallier A, Martinez-Llordella M, Danger R, Lopez M, Giral M, et al. Comparison of transcriptional and blood cell-phenotypic markers between operationally tolerant liver and kidney recipients. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11(9):1916–26. doi: 10.1111/j.1600-6143.2011.03638.x. [DOI] [PubMed] [Google Scholar]
- 34.Benitez C, Londono MC, Miquel R, Manzia TM, Abraldes JG, Lozano JJ, et al. Prospective multicenter clinical trial of immunosuppressive drug withdrawal in stable adult liver transplant recipients. Hepatology. 2013;58(5):1824–35. doi: 10.1002/hep.26426. [DOI] [PubMed] [Google Scholar]
- 35.Brouard S, Pallier A, Renaudin K, Foucher Y, Danger R, Devys A, et al. The natural history of clinical operational tolerance after kidney transplantation through twenty-seven cases. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2012;12(12):3296–307. doi: 10.1111/j.1600-6143.2012.04249.x. [DOI] [PubMed] [Google Scholar]
- 36.Braza F, Dugast E, Panov I, Paul C, Vogt K, Pallier A, et al. Central Role of CD45RA-Foxp3hi Memory Regulatory T Cells in Clinical Kidney Transplantation Tolerance. Journal of the American Society of Nephrology : JASN. 2015 doi: 10.1681/ASN.2014050480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roedder S, Li L, Alonso MN, Hsieh SC, Vu MT, Dai H, et al. A Three-Gene Assay for Monitoring Immune Quiescence in Kidney Transplantation. Journal of the American Society of Nephrology : JASN. 2014 doi: 10.1681/ASN.2013111239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baron D, Giral M, Brouard S. Reconsidering the detection of tolerance to individualize immunosuppression minimization and to improve long-term kidney graft outcomes. Transplant international : official journal of the European Society for Organ Transplantation. 2015 doi: 10.1111/tri.12578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.