Abstract
Protein ADP-ribosylation is an important posttranslational modification that plays versatile roles in multiple biological processes. ADP-ribosylation is catalyzed by a group of enzymes known as ADP-ribosyltransferases (ARTs). Using nicotinamide adenine dinucleotide (NAD+) as the donor, ARTs covalently link single or multiple ADP-ribose moieties from NAD+ to the substrates, forming mono ADP-ribosylation or poly ADP-ribosylation (PARylation). Novel functions of ARTs and ADP-ribosylation have been revealed over the past few years. Here we summarize the current knowledge on ARTs and PARylation.
Keywords: ADP-ribosylation, ADP-ribosyltransferase, ART, ARTD, PAR, PARylation
1. INTRODUCTION
ADP-ribosylation has been identified for over 50 years. In early1960s, Collier, Honjo and their colleagues found that the toxicity of diphtheria toxin required NAD+ to inhibit mammalian protein synthesis [1-3]. Subsequently, single ADP-ribose moiety from NAD+ was revealed to covalently bind to elongation factor 2 or aminoacyl transferase 2, which leads to the identification of mono ADP-ribosylation. In 1966, Mandel’s group discovered a DNA-dependent NAD+ consuming reaction in which poly ADP-ribose (PAR) chains were synthesized [4, 5]. The identification of ADP-ribosylation opened a new avenue in the areas of DNA damage repair, cell proliferation, apoptosis, gene transcription, signal transduction and etc. [6]. The reactions are catalyzed by a group of enzymes, namely ADP-ribosyltransferases (ARTs), which transfer single or multiple ADP-ribose moieties from NAD+ to their targets, including proteins, nucleotides, antibiotics and other small molecules [6-13].
Among these reactions, ADP-ribosylation on proteins is well studied. Protein ADP-ribosylation is initiated with transferring one ADP-ribose moiety from NAD+ to the side chain of asparagine, aspartic acid, glutamic acid, arginine, lysine and cysteine resides [14-25]. Poly ADP-ribosyltransferases continue to add ADP-ribose moieties from NAD+ to the initial ADP-ribose through glycosidic bonds to form linear chain of PAR. Branched ADP-ribose chain is also generated by α(1′′′−2′′)-ADP-ribose linkage. Up to 200 ADP-ribose residues can be immediately linked to form a single PAR chain in vitro [26]. With phosphate moieties in each ADP-ribose, the polymer of ADP-ribose brings huge amount of negative charges to local environment and regulates different biological processes. Moreover, ADP-ribosylation is a reversible posttranslational modification. Mono ADP-ribose and PAR are recognized and degraded by ADP-ribosylhydrolases including Poly (ADP-ribose) glycohydrolase (PARG), Terminal ADP-ribose protein glycohydrolase 1 (TARG1), ADP-ribosylhydrolase 1 (ARH1), ARH3, Macro D1 and D2, and Nudix-Type Motif 9 and 16 (NUDT9 and NUDT16) [27-31]. Thus, this dynamically regulated posttranslational modification allows cells to response to external stimuli as well as to participate in a variety of cellular activities.
In this review, we will focus on the protein ADP-ribosylation catalyzed by ARTs. ADP-ribosylation on other substrates, such as nucleotides, antibiotics and other small molecules will not be discussed here.
2. ADP-RIBOSYLTRANSFERASE
To date, there are 22 human gene products possessing ADP-ribosyltransferase activity [32]. These enzymes were previously named as poly ADP-ribose polymerases (PARPs) and ADP-ribosyltransferases (ARTs). However, since most PARPs only catalyze mono ADP-ribosylation, Hottiger et al. have suggested that ADP-ribosyltransferase should be subjected to a unified nomenclature and categorized into two sub-families [32]. First, the ADP-ribosyltransferases sharing homology with bacterial diphtheria toxin, are named as ARTDs. Second, the rest enzymes with homology to clostridial C2 and C3 toxins are named as ARTCs. Besides these ADP-ribosyltransferases, sirtuin family enzymes, known as the NAD+-dependent deacetylases, are also able to transfer mono ADP-ribose moiety [33-36], although the detailed catalytic mechanism is unclear.
ARTC family enzymes, aka ecto-ARTs, are extracellular, membrane-bound or secretory proteins that only mediate mono ADP-ribosylation. Although five ARTC genes have been cloned, only four of them are expressed in human due to a pseudo ARTC2 gene. These ARTCs catalyze protein mono ADP-ribosylation extracellularly or at cell surface, which regulates cell-cell communication and activates signaling transduction [37].
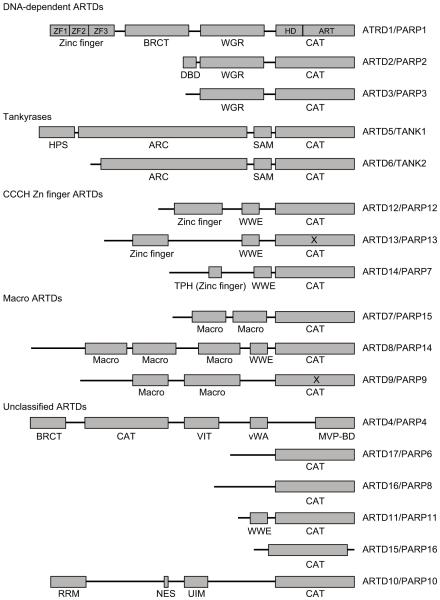
ARTD family members are intracellular enzymes with either mono or poly ADP-ribosyltransferase activities. Based on the domain architectures, ARTDs are categorized into five subgroups: DNA-dependent ARTDs, Tankyrases, CCCH Zn Finger ARTDs, ARTDs with the Macro domain, and other unclassified ARTDs (Table 1 and Fig. 1). Among DNA-dependent ARTDs, both ARTD1 (aka PARP1) and ARTD2 (aka PARP2) contain DNA-binding domain at the N-terminus. Upon binding DNA ends, the activation sites in their enzymatic domains are fully exposed, which activates PARylation [38]. Different from ARTD1 and 2, ARTD3 does not have an obvious DNA-binding domain. But ATRD3 is still able to be activated by DNA in vitro [39, 40], although the detailed activation mechanism is unclear. The tankyrases contain several ankyrin repeats clusters, serving as protein-protein interaction platform, and facilitating substrate recognition [41]. The CCCH Zn Finger ARTDs contain both the Cys-Cys-Cys-His zinc fingers and the WWE domains. The zinc fingers are able to interact with RNA, while the WWE domains recognize PAR. The Macro domains of ARTDs also bind to ADP-ribose and mediate substrate ADP-ribosylation.
All the ARTs contain the catalytic domain (CAT) with similar secondary structure. These catalytic domains share a common NAD+ binding motif similar to those in bacterial exotoxins, such as diphtheria toxin and exotoxin A [32]. In these NAD+ binding motifs, a His residue and a Tyr residue are crucial for positioning the A-ribose moiety and the N-ribose moiety of NAD+ in a correct orientation [42, 43]. A conserved Glu residue in these motifs is responsible for transferring of ADP-ribose. However, not all the ARTs retain these critical residues. Substitution of the key NAD+-binding His residue in ARTD9 and ARTD13 abolish the activities ADP-ribose transferring. Substitution of the Glu residue to other residues may alter the activities of ARTs from poly to mono ADP-ribosyltransferase. Recent studies suggest that only ARTD1, 2, 5 and 6 possess PARylation activity, while the rest of ARTs are likely to be mono ADP-ribosyltransferases.
2.1. DNA-Dependent ARTDs
2.1.1. ARTD1
ARTD1 (PARP1) is a highly abundant chromatin-associated protein important for the maintenance of genomic integrity, chromatin remodeling and gene transcription. All these diverse functions of ARTD1 are carried out by its multiple functional domains, including three zinc fingers motifs (ZF1-3), a BRCT domain, a WGR domain, and a catalytic domain (CAT containing two sub-domains: helical domain-HD and ADP-ribose transferase domain-ART). The majority of ARTD1 exists in nucleus and associates with nucleosome with a very low activity [44]. However, upon DNA damage, ARTD1 is significantly activated in cells [45, 46]. Interaction between ARTD1 and DNA ends is mediated by the N-terminal three zinc finger motifs. Since the zinc finger motifs primarily contact the ribose-phosphate back-bone of DNA, ARTD1 can be activated by DNA breaks regardless the sequence of DNA fragments [38, 47]. It has been suggested that ZF1 and ZF2 function together to recognize DNA single-strand breaks (SSBs), whereas ZF1 and ZF3 mediate the interaction with DNA double-strand breaks (DSBs). In addition, ZF3 plays an important role for the homodimer formation of ARTD1 [48]. Besides DNA fragment, RNA could also be recognized by ZF3 and WGR domains of ARTD1, which induces the conformational change in CAT and activates CAT in vitro [49]. Thus, ARTD1 could recognize diverse oligonucleotide structures. Upon oligonucleotide binding, ARTD1 forms a network of interdomain contacts, which results a global conformational change in CAT. It distorts HD to significantly increase the flexibility of CAT and expose the activation sites to substrates [38]. This model has been further confirmed by mutations within the hydrophobic core of HD domain, which also increase the flexibility of catalytic domain and thus activate enzymatic activity in the absence of oligonucleotide-binding [38].
Besides oligonucleotide, ARTD1 could also be activated by nucleosomes in vitro [50, 51]. Moreover, posttranslational modifications on ARTD1, such as phosphorylation by extracellular signal-regulated kinases 1/2 and acetylation by PCAF, may facilitate the activation of ARTD1 in vitro [52, 53].
2.1.2. ARTD2
Like ARTD1, ARTD2 (PARP2) has the C-terminal WGR domain and CAT domain. Different from ARTD1, ARTD2 has a very short DNA-binding domain (DBD) at the N-terminus. Due to the presence of basic residues in this region, the N-terminal DBD domain of ARTD2 has been proposed to bind nucleic acid. Indeed, like ARTD1, the activity of ARTD2 is largely increased in the presence of DNA and RNA in vitro [39, 54], especially with the phosphate group at 5’ end [55]. However, compared to the essential role of the N-terminal zinc finger domains in ARTD1 for its activation, the N-terminal DNA-binding domain of ARTD2 only contributes partially for enzymatic activation. The WGR domain is also important for DNA-dependent activation of ARTD2 [55]. Due to lacking of structural analysis of ARTD2, the detailed mechanism of ARTD2 activation is still unclear.
2.1.3. ARTD3
Compared to ARTD1 and 2, ARTD3 (PARP3) also has a WRG domain and a CAT domain. However, the N-terminus of ARTD3 lacks a canonical DNA-binding domain, although it can also be activated by DNA [39, 40]. Different from ARTD1 and ARTD2, ARTD3 only has mono ADP-ribosyltransferase activity even with conserved catalytic motif (H-Y-E triad). It is probably because of the difference of donor site loop (D-loop, NAD+ binding site) in ARTD3 [56]. Vyas et al. swapped the D-loops of ARTD1 and ARTD3, and found that ARTD1 with ARTD3’s D-loop could only catalyze mono ADP-ribosylation, while ARTD3 with the D-loop of ARTD1 was enzymatically inactivated, suggesting that D-loop in catalytic domain plays a key role for PARylation. However, simple replacement of D-loop is insufficient for PAR synthesis [39].
The current studies on ARTD3 are mainly focused on its role in DNA damage response. ARTD3 has been shown to associate with Ku70/80, DNA-PKcs and DNA ligase IV [57], which all participate in non-homologous end joining pathway (NHEJ) for DSB repair. Moreover, ARTD3 associates with both ARTD1 and DNA ligase III, and was found to mediate the recruitment of APLF to the site of DNA damage [40, 58]. ARTD1, DNA ligase III and APLF are crucial SSB repair factors. Thus, accumulated evidence suggests that ARTD3 participates in both DSB and SSB repair.
2.2. Tankyrases
In 1998, Smith and de Lange identified an ankyrin repeats-containing ADP-ribose transferase, named as Tankyrase 1 (ARTD5/TANK1), interacting with Telomeric Repeat-binding Factor1 (TRF1) [59]. ARTD5 contains N-terminal homopolymeric stretches of His, Pro and Ser residues (HPS domain), Ankyrin Repeat Clusters (ARCs), a sterile alpha module (SAM), and a C-terminal ADP-ribose transferase (CAT) domain. With similar searching for the binding partner of TRF1, ARTD6 (TANK2) was identified [60]. It shares 85 % primary sequence identity with ARTD5 in the ARCs, SAM and CAT but has a unique N-terminal domain. Deletion of either Artd5 or Artd6 gene does not affect development in mice. However, lacking both Tankyrases cause early embryonic lethality, suggesting that they have redundant function during prenatal development [61].
At each end of a chromosome, a telomere shelterin complex is responsible for chromosome end protection. This complex consists of TRF1, TRF2, protection of telomeres protein 1 (POT1), TERF-interacting nuclear factor 2 (TINF2), tripeptidyl peptidase 1 (TPP1) and TRF2-interacting protein (TERF2IP, aka RAP1). Both ARTD5 and 6 recognize TRF1 and TRF2 via their ARCs and poly ADP-ribosylate TRF1 and TRF2, which facilitates proteasome-dependent degradation of TRF1 and TRF2 [62, 63].
In addition, Tankyrase-dependent PARylation of DNA-PKcs may also play a role in telomere maintenance as DNA-PKcs-dependent NHEJ is involved in telomere capping. In contrast to TRF1, PARylation of DNA-PKcs stabilizes itself from proteasome-mediated degradation [64].
Another pathway regulated by Tankyrases is the WNT signaling pathway. In this pathway, β-catenine, the key transcription factor, is regulated by the cytoplasmic β-catenine destruction complex. This complex consists of adenomatous polyposis coli (APC), GSK3β and AXIN1/2. Interestingly, AXIN1/2 are poly ADP-ribosylated by Tankyrases. Poly ADP-ribosylated AXIN1/2 is recognized by the WWE domain of E3 ligase RNF146, which facilitates RNF146-dependent ubiquitination and degradation of AXINs. Therefore, the suppression of Tankyrases rescues AXIN from proteasome-dependent degradation, and promotes the destruction of β-catenine [65].
2.3. CCCH Zn Finger ARTDs
ARTD12 (PARP12) is a putative anti-viral factor, whose expression is induced during viral infections [66, 67]. Five CCCH-type Zn-fingers at N-terminus were known to bind viral RNA for degradation [68, 69]. Recently, ARTD12 is reported to accumulate in cytoplasmic stress-granules in the presence of oxidative stress, and inhibits the mRNA translation [70]. Also, in response to LPS, ARTD12 relocates to autophagosomes by interacting with p62/SQSTM1, and facilitates NF-κB-dependent signaling, suggesting a role of ARTD12 in inflammation [70].
ARTD13 (PARP13/ZAP/ZC3HAV1) is another anti-viral factor, actively against specific RNA viruses such as murine leukemia virus, Sindbis virus, human immunodeficiency virus, Epstein–Barr virus as well as the RNA intermediate of hepatitis B DNA virus [71-76]. During viral infection, ARTD13 binds viral RNA via its four CCCH-type zinc finger motifs and targets viral RNA for degradation by recruiting RNA decay factors [68, 77]. ARTD12 and ARTD13 co-localize in the stress granules [78]. Recently, ARTD13 has been shown to regulate mRNA degradation without viral infection [79].
ARTD14 (PARP7/tiPARP) contains a TPH domain, a WWE domain and a CAT. Inside of the TPH domain, it contains a CCCH-type zinc finger that is a putative RNAbinding module. Moreover, ARTD14 shares 27.5% and 26.0% primary sequence identity with ARTD12 and ARTD13, respectively [80]. The WWE domain is known as an ADP-ribose-binding motif. Thus, it is likely that ARTD14 recognizes ADP-ribosylated targets and catalyzes additional ADP-ribosylation. ARTD14 was identified as a gene product that was strongly up-regulated upon exposure to halogenated aromatic hydrocarbons, a type of industrial contaminate [81]. ARTD14 exhibits auto mono ADP-ribosyltransferase activity and mono ADP-ribosylates core histones as well [81, 82]. In addition, ARTD14 may function together with ARTD1 for maintaining the naive state of pluripotency of ES cells [83].
2.4. Macro Domain ARTs
Macro domain ARTDs contain multiple macro domains at the N-terminus and an ADP-ribosyltransferase domain at the C-terminus. The Macro domain is known as an ADP-ribose binding module [84]. Thus, it is likely that this type of ARTD recognizes ADP-ribosylated substrates and catalyzes additional ADP-ribosylation. Whereas ARTD7 (PARP15) remains largely uncharacterized, both ARTD8 (PARP14) and ARTD9 (PARP9) have been reported to regulate gene transcription [85, 86].
ARTD8 has three tandem Macro domains at the N-terminus and exists in both cytoplasm and nucleus [87]. It has been shown that ARTD8 regulates gene transcription following the stimulation of IL-4. ARTD8 mono ADP-ribosylates HDAC2 and HDAC3 at gene promoters for reprogramming histone codes and facilitating transcription [85]. Recent study also shows that ARTD8 is involved in mRNA stability. ARTD8 is found to form a complex with tristetraprolin (TTP) and 3’ untranslated region of mRNA, which suppresses the degradation of mRNA [88].
ARTD9 is originally identified as a transcription modulator that is overexpressed in chemoresistant, diffuse large B-cell lymphomas [86]. Although it has two tandem Macro domains for binding ADP-ribose, the CAT domain of ARTD9 lacks key residues for NAD+-binding and transferring ADP-ribose [89]. The tandem Macro domains of ARTD9 have been shown to mediate the recruitment of ARTD9 to the sites of DNA damage [90, 91]. Since ARTD9 is an enzymatically inactive enzyme, it is likely to behave as a negative regulator for DNA damage-induced PARylation.
2.5. Other ARTs
ARTD4 (PARP4/vPARP) has been identified as a protein subunit in the vault ribonucleoprotein particle that might be involved in mRNA transportation [92]. ARTD4 contains multiple functional domains including a BRCT domain, a CAT domain, a VIT (vault inter-α-trypsin) domain, a vWA (von Willebrand factor type A) domain and a MVP-BD (major vault protein binding domain). However, the function of each domain is unclear. Since ARTD4 has been shown to localize at mitotic spindle, ARTD4 may regulate mitosis [92]. ARTD4-deficient mice have been generated. However, no obvious developmental defect has been observed. It has been shown that ARTD4-deficient mice were prone to carcinogen-induced tumors, suggesting ARTD4 may protect genomic stability from chemically induced neoplasia [93].
ARTD16 (PARP8) and ARTD17 (PARP6) have not been well characterized. Both of them contain a mono ADPribosyltransferase domain and a uncharacterized N-terminal region. Over expression of ARTD16 suppresses cell proliferation. Moreover, in human tumor samples, ARTD17 expression is negatively correlated with the Ki-67 proliferation index, suggesting ARTD17 is a possible negative regulator for cell growth [94].
ARTD15 (PARP16) is the only ARTD family member with a C-terminal transmembrane domain. It localizes at nuclear envelope and the membrane of endoplasmic reticulum (ER) [97]. It has mono ADP-ribosylation activity. Recently, ARTD15 was reported to bind and ADP-ribosylate Karyopherin-β1/importin-β1 for nuclear protein transportation [97]. It has also been found that the enzymatic activity of ARTD15 is upregulated during ER stress when it is auto ADP-ribosylated or ADP-ribosylates PERK and IRE1α [98]. The C-terminal luminal tail of ARTD15 is required for its function during ER stress, suggesting that ARTD15 transduces stress signals to the cytoplasmic catalytic domain [98].
ARTD10 (PARP10) contains an N-terminal RRM (RNA recognition motif), a NES (nuclear export sequence), two tandem UIMs (ubiquitin interaction motifs) and a C-terminal CAT domain. It is conceivable that some of these domains may mediate the ADP-ribosylation of specific substrates in different compartments, which renders ARTD10 suitable for wide set of processes in cells. Interestingly, the UIMs of ARTD10 bind to K63-linked poly-ubiquitin, a modification that is essential for NF-κB signaling. ARTD10 has been shown to inhibit the activation of NF-κB and downstream target genes in response to interleukin-1β and tumor necrosis factor-α [99]. The molecular mechanism of the activation of ARTD10 remains elusive as oligonucleotide is not required for its activation. As ARTD10 shuttles between cytoplasm and nucleus, multiple substrates have already been revealed, such as histones, c-MYC, GSK3β, NEMO and PCNA [99-102].
3. FUNCTION OF PARYLATION IN DNA DAMAGE
Although ADP-ribosylation has been identified for more than half a century, the biological function of ADP-ribosylation, especially mono ADP-ribosylation, remains elusive. Compared to mono ADP-ribosylation, PARylation is a bulky and highly negatively charged posttranslational modification. PAR has been shown to regulate protein-protein or protein-nucleic acid interaction, function as a scaffold to mediate the recruitment of DNA damage response factors, and regulate protein degradation via PAR-binding E3 ligases.
PARylation has been shown to participate in DNA damage response, gene transcription, DNA replication, cell cycle regulation, ageing, intercellular transport and apoptosis/ necrosis [6]. Especially upon DNA binding, PAR synthesis could be magnified to 1700 fold above basal level in vitro [48]. Thus, the role of PARylation has been most extensively studied. Here, we summarize the identified function of PARylation in DNA damage response, including chromatin remodeling surround the sites of DNA damage, recruiting DNA damage repair machineries, and crosstalk with other DNA damage-induced posttranslational modifications.
3.1. Decondensation of Chromatin
As a founder member of ARTDs, ARTD1 is an abundant protein and associated with chromatin. Enzymatically silent ARTD1 suppresses gene transcription by contributing to the condensation of chromatin, which creates a barrier against gene transcription. This is supported by the evidence from an in vitro assay in which purified chromatin become more compacted in the presence of recombinant ARTD1 [103]. Moreover, the presence of inactive ARTD1 in heterochromatin regions, such as telomeres, facilitates the maintenance of the repressive DNA state [104]. How-ever, immediately following DNA damage, ARTD1 and ARTD2 are activated by DNA ends, and could use up to 90% cellular NAD+ to generate a huge amount of PAR at the sites of DNA damage [105, 106]. The highly negatively charged PAR chains repulse nucleotide and relax chromatin surround DNA lesions, mimicking the effects of linker histone H1 depletion [107]. Electron microscopic studies further demonstrated that PAR results in opening and relaxation of the tightly coiled nucleosome structure in the isolated chromatin [107]. Similar to DNA damage response, activation of ARTD1 also relaxes chromatin during gene transcription. In the study of stress-induced gene activation in Drosophila, it has been reported that ARTD1 was activated by steroids and environmental stress [108]. Synthesized PAR facilitates the removal of transcription barriers and induces chromatin remodeling for active gene transcription during development [108].
3.2. Recruitment of DNA Damage Response Factors
Besides directly inducing chromatin remodeling through negative charges, PARylation mediates the recruitment of DNA damage response factors to DNA lesions via PAR-binding domains. To date, 7 PAR recognition modules have been found to mediate PAR-dependent protein interaction: the Macro domain, the PAR-binding zinc finger (PBZ), the WWE domain, the BRCT domain, the FHA domain, the OB-fold domain and the RRM domain. Although a putative PAR-binding motif (PBM) has been suggested to interact with PAR, the PBM with a short putative peptide sequence is much degenerated and is unlikely to form a folded structure to interact with PAR. Moreover, the putative PBM in XRCC1 is actually a part of the BRCT domain. The short peptide alone does not fold well into any secondary structure. Additional study suggests that the BRCT domain of XRCC as a whole is a PAR-binding module [109]. Thus, in this review, we do not include PBM as a PAR-binding module.
3.2.1. The Macro Domain
The Macro domain is the first identified PAR-binding module. It contains 140–190 residues and is able to recognize one ADP-ribose residue [84]. The Marco domain was initially identified in histone variant macroH2A [110]. Other proteins containing the Macro domain include ARTD7, 8, 9, MacroD1, MacroD2, MacroD3, ALC1 and TARG1 [111]. ALC1 (aka CHD1L) is a SNF2-like ATPase, and is rapidly recruited to the sites of DNA damage following PARylation. As an ATP-dependent chromatin remodeler, ALC1 forms nucleosomal intermediate with ARTD1 and histones, and promotes PAR-dependent chromatin relaxation at DNA lesions [112]. Depletion of ALC1 sensitizes cells to DNA damage reagents [113]. MarcoD1/2 and TARG1 have terminal ADP-ribose glycohydrolase activity and are recruited to the sites of DNA damage rapidly [29, 30]. Although the molecular mechanism of these enzymes in DNA damage repair have not been extensively studied, it is likely that these enzymes participate in PAR removal from DNA lesions.
3.2.2. The PAR-Binding Zinc Finger (PBZ)
The PBZ domain is another small PAR-binding module, which recognizes tandem ADP-ribose residues [114]. It has been found in various proteins, such as DNA damage repair machineries APLF and CHFR [114, 115]. APLF (Aprataxin-PNK-like factor) is an apurinic-apyrimidinic (AP) endonuclease with a high binding affinity for PAR [116]. Depletion of APLF impaired DNA damage repair following ionizing radiation [116]. Checkpoint with Forkhead-associated and Ring finger domain protein (CHFR), is an E3 ubiquitin ligase and found to be recruited to DSBs by PAR [114, 117]. Via PAR recognition, CHFR ubiquitinates ARTD1 for its displacement from the sites of DNA damage. Thus, CHFR is an important E3 ligase which links PARylation and ubiquitination pathways during DNA damage response [117].
3.2.3. The WWE Domain
Recently, a set of WWE domain-containing proteins have been identified to bind the linker region between two ADP-ribose residues [118-120]. One of them is an E3 ubiquitin ligase RNF146/Iduna. RNF146 recognizes Tankyrase-induced PARylation on AXIN1/2 and ubiquitinates AXIN1/2 for proteasome-dependent degradation [65]. Moreover, RNF146 is recruited to the site of DNA damage and participate in DNA damage repair [121, 122].
3.2.4. The BRCT Domain and the FHA Domain
The BRCT domain and FHA domain are originally known as phospho-protein binding domain. It has been suggested that the FHA domains recognize phosphor-Thr motifs [123-125] and BRCT domains recognize phosphor-Ser motifs [126, 127].
Unexpectedly, we found that the BRCT domains of BARD1 recognize ADP-ribose [128]. Upon DNA damage, the interaction between the BARD1 BRCT domain and PAR mediates the fast recruitment of the BRCA1/BARD1 complex to the sites of DNA damage thus promoting the homologous recombination repair. Similarly, we found that the BRCT domains in Ligase IV can also bind to ADP-ribose and mediate the fast recruitment of Ligase IV to DNA lesions, which is likely to promote NHEJ [109]. Another BRCT domain containing protein, NBS1 is a subunit in the MRN complex that activates ATM in response to DSBs. We find that the BRCT domain of NBS1 binds to DNA damage-induced PAR, which mediates the fast recruitment of the MRN complex to DSBs and facilitates the early activation of ATM-dependent signal transduction as well as cell-cycle checkpoints [109].
Besides BRCT domains, we also find that the FHA domains in PNKP and APTX bind to PAR, which is important for their early recruitment to DNA lesions. Interestingly, these two FHA domains recognize the iso-ADP-ribose, the linker of PAR, instead of ADP-ribose [109]. A chemical feature of iso-ADP-ribose is that it has phosphate group on each site. Since the BRCT domain and FHA domain are originally known as phospho-amino acid binding domains, it is likely that the BRCT domain and the FHA domain recognize the phosphate group in ADP-ribose and iso-ADP-ribose respectively.
3.2.5. The OB-Fold Domain and the RRM Domain
Since chemical structure of PAR is similar to oligonucleotide, it is possible that oligonucleotide-binding domains may recognize PAR. The oliganucleotide/oligosaccharide-binding (OB) fold is a single-strand DNA/RNA binding motif in prokaryotes and eukaryotes [129]. We find that the OB-fold domain of hSSB1 recognizes iso-ADP-ribose and meditates the fast recruitment of hSSB1 to the DNA lesions. We also screened other OB-folds and found that several of them could also recognize PAR, suggesting a set of OB-fold domains sever as a PAR-binding domain [130].
Recently, a RNA-binding protein NONO was identified as a novel PAR-binding protein [131]. It has two tandem RNA recognition motifs (RRM) at the N-terminus, while only the first RRM has strong affinity for PAR. Its fast recruitment to the DNA damage sites through PAR-binding is essential for its function in NHEJ, although the detailed mechanism is still unclear. Another example of PAR-binding RRM is the RRM in Alternative Splicing Factor/Splicing Factor 2 (ASF/SF2) [132]. It also contains two tandem RRMs at the N-terminus, but only the first RRM is able to bind PAR. ASF is a RNA splicing factor, which is regulated by the kinase activity of DNA topoisomerase I. When binding PAR, ASF becomes a poor substrate of DNA topoisomerase I, which suppresses its function in RNA splicing.
3.3. Crosstalk Between ADP-Ribosylation and Other Posttranslational Modifications
ARTs are able to transfer ADP-ribose to lysine, arginine, glutamate, aspartate, cysteine, and serine residues [14-25]. However, besides ADP-ribosylation, other post-translational modifications, including phosphorylation, ubiquitination, sumoylation, methylation, and acetylation may also modify the same sites or adjacent sites on the same targets. Thus, crosstalk between these post-translational modifications may occur during different biological processes.
Different modifications carry our different functions during gene transcription, DNA damage repair and chromatin remodeling. With improved mass spectrometry, more and more ADP-ribosylation sites have been revealed. Recent studies showed that H2AK13, H2BK30, H3K27, K37, and H4K16 could be ADP-ribosylated [21]. Interestingly, H4K16 could also be acetylated, which induces chromatin relaxation. Since acetylation competes with ADP-ribosylation at H4K16 [21], deacetylation by SIRT1 [133] may facilitate ARTD1-dependent PARylation of H4K16.
As mentioned above, PARylation also regulates protein ubiquitination and proteasome-dependent degradation. Both CHFR and RNF146 are PAR-binding E3 ubiquitin ligases. Both E3 ligases recognize poly ADP-ribosylated substrates and catalyze ubiquitination following PARylation. Thus, PARylation is an activator for protein ubiquitination and degradation.
4. SUMMARY
Studies over the past few years have gradually uncovered the molecular mechanisms and biological functions of ARTs in multiple biologic processes. ADP-ribosylation changes the chemical and physical characters of the substrate, and regulates protein-protein or protein-DNA interaction. The function of PARylation is relatively better understood, but the function of mono ADP-ribosylation is still largely unknown. Studies on writers, readers and erasers of mono ADP-ribosylation are required for elucidate its role in a variety of biological processes.
Fig. (1).
The classification and domain architecture of human ARTDs. X: CAT domain without ADP-ribosyltransferase activity; ZF: zinc finger; BRCT: BRCA1 C-terminus domain; WGR: Trp-Gly-Arg domain; CAT: catalytic ADP-ribosyltransferase domain; DBD: DNA-binding domain; HPS: His-Pro-Ser domain; ARC: ankyrin repeats cluster; SAM: sterile alpha motif; TPH: TIPARP homologous domain; VIT: vault inter-α-trypsin domain; vWA: von Willebrand factor type A domain; MVP-BD: major vault protein binding domain; RRM: RNA recognition motif; NES: nuclear exporting sequence; UIM: ubiquitin interaction motif.
Table 1.
Summary of human ARTs.
| Subfamily | Name | Aliases | Size (aa) | Subcellular Localization | Triad Motif |
Key Domains | Activity |
|---|---|---|---|---|---|---|---|
| DNA dependent | ARTD1 ARTD2 ARTD3 |
PARP1 PARP2 PARP3 |
1014 583 533 |
Nucleus Nucleus and cytoplasm Nucleus and cytoplasm |
H-Y-E H-Y-E H-Y-E |
zinc fingers, BRCT, WGR WGR, DBD WGR |
P P M |
| Tankyrase | ARTD5 ARTD6 |
PARP5a, TANK1 PARP5b, TANK2 |
1327 1166 |
cytoplasm cytoplasm | H-Y-E H-Y-E |
Ankyrin repeat Ankyrin repeat |
P P |
| CCCH Zn Finger | ARTD14 ARTD12 ARTD13 |
PARP7, tiPARP PARP12 PARP13, ZC3HAV1 |
657 701 902 |
Nucleus and cytoplasm cytoplasm cytoplasm |
H-Y-I H-Y-I Y-Y-V |
zinc fingers, WWE zinc fingers, WWE zinc fingers, WWE |
M M I |
| Macro | ARTD9 ARTD8 ARTD7 |
PARP9, BAL1 PARP14, BAL2 PARP15, BAL3 |
854 1801 678 |
Nucleus and cytoplasm Nucleus and cytoplasm ND |
Q-Y-T H-Y-L H-Y-L |
macro domain macro domain, WWE macro domain |
I M M |
| unclassified | ARTD4 ARTD17 ARTD16 ARTD10 ARTD11 ARTD15 |
PARP4 PARP6 PARP8 PARP10 PARP11 PARP16 |
1724 630 854 1025 331 322 |
Nucleus and cytoplasm cytoplasm cytoplasm Nucleus and cytoplasm Nucleus and cytoplasm cytoplasm |
H-Y-E H-Y-Y H-Y-I H-Y-I H-Y-I H-Y-I |
BRCT RRM, UIM WWE |
M M M M M M |
| tRNA phosphotransferase |
ARTD18 | TRPT1, TpT1 | 253 | cytoplasm | H-H-V | ||
| ecto-ARTs | ARTC1 ARTC2P ARTC3 ARTC4 ARTC5 |
ART1 ART2P ART3 ART4 ART5 |
327 389 314 291 |
ER, plasma membrane plasma membrane, ER plasma membrane extracellular |
R-S-E K-L-V G-S-E R-S-E |
M M M M |
Note: P, PARylation; M, mono ADP-ribosylation; I, inactive enzyme; ND, not determined.
ACKNOWLEDGEMENTS
This work was supported by grants from National Institutes of Health (CA132755, CA130899 and CA187209 to X.Y.). X.Y. is a recipient of Era of Hope Scholar Award from the Department of Defense and a research scholar of Leukemia and Lymphoma Society.
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- [1].Collier RJ, Pappenheimer AM., Jr. Studies on the mode of action of diphtheria toxin. II. Effect of toxin on amino acid incorporation in cell-free systems. J. Exp. Med. 1964;120:1019–1039. doi: 10.1084/jem.120.6.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Collier RJ. Effect of diphtheria toxin on protein synthesis: inactivation of one of the transfer factors. J. Mol. Biol. 1967;25(1):83–98. doi: 10.1016/0022-2836(67)90280-x. [DOI] [PubMed] [Google Scholar]
- [3].Honjo T, Nishizuka Y, Hayaishi O. Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J. Biol. Chem. 1968;243(12):3553–3555. [PubMed] [Google Scholar]
- [4].Chambon P, Weill JD, Doly J, Strosser MT, Mandel P. On the formation of a novel adenylic compound by enzymatic extracts of liver nuclei. Biochem. Biophys. Res. Commun. 1966;25(6):638–643. [Google Scholar]
- [5].Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- [6].Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADPribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70(3):789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26(8):882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- [8].Koch-Nolte F, Kernstock S, Mueller-Dieckmann C, Weiss MS, Haag F. Mammalian ADP-ribosyltransferases and ADPribosylhydrolases. Front. Biosci. 2008;13:6716–6729. doi: 10.2741/3184. [DOI] [PubMed] [Google Scholar]
- [9].Moss J, Zolkiewska A, Okazaki I. ADP-ribosylarginine hydrolases and ADP-ribosyltransferases. Partners in ADPribosylation cycles. Adv. Exp. Med. Biol. 1997;419:25–33. doi: 10.1007/978-1-4419-8632-0_3. [DOI] [PubMed] [Google Scholar]
- [10].Zolkiewska A. Ecto-ADP-ribose transferases: cell-surface response to local tissue injury. Physiology (Bethesda) 2005;20:374–381. doi: 10.1152/physiol.00028.2005. [DOI] [PubMed] [Google Scholar]
- [11].Di Girolamo M, Dani N, Stilla A, Corda D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005;272(18):4565–4575. doi: 10.1111/j.1742-4658.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- [12].Holbourn KP, Shone CC, Acharya KR. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 2006;273(20):4579–4593. doi: 10.1111/j.1742-4658.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- [13].Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22(9):1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sekine A, Fujiwara M, Narumiya S. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. J. Biol. Chem. 1989;264(15):8602–8605. [PubMed] [Google Scholar]
- [15].Manning DR, Fraser BA, Kahn RA, Gilman AG. ADP-ribosylation of transducin by islet-activation protein. Identification of asparagine as the site of ADP-ribosylation. J. Biol. Chem. 1984;259(2):749–756. [PubMed] [Google Scholar]
- [16].Moss J, Stanley SJ. Amino acid-specific ADP-ribosylation. Identification of an arginine-dependent ADP-ribosyltransferase in rat liver. J. Biol. Chem. 1981;256(15):7830–7833. [PubMed] [Google Scholar]
- [17].Geipel U, Just I, Schering B, Haas D, Aktories K. ADP-ribosylation of actin causes increase in the rate of ATP exchange and inhibition of ATP hydrolysis. Eur. J. Biochem. 1989;179(1):229–232. doi: 10.1111/j.1432-1033.1989.tb14545.x. [DOI] [PubMed] [Google Scholar]
- [18].Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J. Biol. Chem. 1980;255(16):7616–7620. [PubMed] [Google Scholar]
- [19].Hoshino S, Kikkawa S, Takahashi K, Itoh H, Kaziro Y, Kawasaki H, Suzuki K, Katada T, Ui M. Identification of sites for alkylation by N-ethylmaleimide and pertussis toxin-catalyzed ADP-ribosylation on GTP-binding proteins. FEBS Lett. 1990;276(1-2):227–231. doi: 10.1016/0014-5793(90)80548-w. [DOI] [PubMed] [Google Scholar]
- [20].Just I, Wollenberg P, Moss J, Aktories K. Cysteine-specific ADP-ribosylation of actin. Eur. J. Biochem. 1994;221(3):1047–1054. doi: 10.1111/j.1432-1033.1994.tb18823.x. [DOI] [PubMed] [Google Scholar]
- [21].Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38(19):6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37(11):3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods. 2013;10(10):981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- [24].Daniels CM, Ong SE, Leung AK. Phosphoproteomic approach to characterize protein mono- and poly(ADP-ribosyl) ation sites from cells. J. Proteome Res. 2014;13(8):3510–3522. doi: 10.1021/pr401032q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML. Proteome-wide identification of poly(ADP-ribosyl) ation targets in different genotoxic stress responses. Mol. Cell. 2013;52(2):272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- [26].Alvarez-Gonzalez R, Jacobson MK. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26(11):3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- [27].Ueda K, Oka J, Naruniya S, Miyakawa N, Hayaishi O. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem. Biophys. Res. Commun. 1972;46(2):516–523. doi: 10.1016/s0006-291x(72)80169-4. [DOI] [PubMed] [Google Scholar]
- [28].Niere M, Mashimo M, Agledal L, Dolle C, Kasamatsu A, Kato J, Moss J, Ziegler M. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose) J. Biol. Chem. 2012;287(20):16088–16102. doi: 10.1074/jbc.M112.349183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32(9):1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, Ladurner AG. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013;20(4):508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Palazzo L, Thomas B, Jemth AS, Colby T, Leidecker O, Feijs K, Zaja R, Loseva O, Puigvert JC, Matic I, Helleday T, Ahel I. Processing of Protein ADP-ribosylation by Nudix Hydrolases. Biochem. J. 2015 doi: 10.1042/BJ20141554. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35(4):208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- [33].Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999;260(1):273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- [34].Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 2005;280(22):21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- [35].Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- [36].Garcia-Salcedo JA, Gijon P, Nolan DP, Tebabi P, Pays E. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. EMBO J. 2003;22(21):5851–5862. doi: 10.1093/emboj/cdg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Seman M, Adriouch S, Haag F, Koch-Nolte F. Ecto-ADP-ribosyltransferases (ARTs): emerging actors in cell communication and signaling. Curr. Med. Chem. 2004;11(7):857–872. doi: 10.2174/0929867043455611. [DOI] [PubMed] [Google Scholar]
- [38].Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336(6082):728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, Chang P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell. 2011;41(1):33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- [41].Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008;90(1):83–92. doi: 10.1016/j.biochi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- [42].Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bell CE, Eisenberg D. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry. 1996;35(4):1137–1149. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- [44].Ji Y, Tulin AV. The roles of PARP1 in gene control and cell differentiation. Curr. Opin. Genet. Dev. 2010;20(5):512–518. doi: 10.1016/j.gde.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Benjamin RC, Gill DM. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J. Biol. Chem. 1980;255(21):10493–10501. [PubMed] [Google Scholar]
- [46].Jacobson EL, Antol KM, Juarez-Salinas H, Jacobson MK. Poly(ADP-ribose) metabolism in ultraviolet irradiated human fibroblasts. J. Biol. Chem. 1983;258(1):103–107. [PubMed] [Google Scholar]
- [47].Ali AA, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, Ladurner AG, Pearl LH, Oliver AW. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 2012;19(7):685–692. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 2010;285(24):18877–18887. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Huambachano O, Herrera F, Rancourt A, Satoh MS. Doublestranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J. Biol. Chem. 2011;286(9):7149–7160. doi: 10.1074/jbc.M110.175190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Clark NJ, Kramer M, Muthurajan UM, Luger K. Alternative modes of binding of poly(ADP-ribose) polymerase 1 to free DNA and nucleosomes. J. Biol. Chem. 2012;287(39):32430–32439. doi: 10.1074/jbc.M112.397067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+− dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119(6):803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- [52].Kauppinen TM, Chan WY, Suh SW, Wiggins AK, Huang EJ, Swanson RA. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc. Natl. Acad. Sci. U. S. A. 2006;103(18):7136–7141. doi: 10.1073/pnas.0508606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, Gupta MP. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol. Cell Biol. 2009;29(15):4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Leger K, Bar D, Savic N, Santoro R, Hottiger MO. ARTD2 activity is stimulated by RNA. Nucleic Acids Res. 2014;42(8):5072–5082. doi: 10.1093/nar/gku131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Langelier MF, Riccio AA, Pascal JM. PARP-2 and PARP-3 are selectively activated by 5' phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014;42(12):7762–7775. doi: 10.1093/nar/gku474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Oncu D, Kull B, Robertson GM, Pellicciari R, Schüler H, Weigelt J. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat. Biotechnol. 2012;30(3):283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- [57].Rouleau M, McDonald D, Gagne P, Ouellet ME, Droit A, Hunter JM, Dutertre S, Prigent C, Hendzel MJ, Poirier GG. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J. Cell Biochem. 2007;100(2):385–401. doi: 10.1002/jcb.21051. [DOI] [PubMed] [Google Scholar]
- [58].Fenton AL, Shirodkar P, Macrae CJ, Meng L, Koch CA. The PARP3- and ATM-dependent phosphorylation of APLF facilitates DNA double-strand break repair. Nucleic Acids Res. 2013;41(7):4080–4092. doi: 10.1093/nar/gkt134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282(5393):1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- [60].Kaminker PG, Kim SH, Taylor RD, Zebarjadian Y, Funk WD, Morin GB, Yaswen P, Campisi J. TANK2, a new TRF1-associated poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J. Biol. Chem. 2001;276(38):35891–35899. doi: 10.1074/jbc.M105968200. [DOI] [PubMed] [Google Scholar]
- [61].Chiang YJ, Hsiao SJ, Yver D, Cushman SW, Tessarollo L, Smith S, Hodes RJ. Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PLoS One. 2008;3(7):e2639. doi: 10.1371/journal.pone.0002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ. The Fbox protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J. Biol. Chem. 2006;281(2):759–768. doi: 10.1074/jbc.M509855200. [DOI] [PubMed] [Google Scholar]
- [63].Her YR, Chung IK. Ubiquitin Ligase RLIM Modulates Telomere Length Homeostasis through a Proteolysis of TRF1. J. Biol. Chem. 2009;284(13):8557–8566. doi: 10.1074/jbc.M806702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Dregalla RC, Zhou J, Idate RR, Battaglia CL, Liber HL, Bailey SM. Regulatory roles of tankyrase 1 at telomeres and in DNA repair: suppression of T-SCE and stabilization of DNA-PKcs. Aging (Albany NY) 2010;2(10):691–708. doi: 10.18632/aging.100210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- [66].de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001;69(6):912–920. [PubMed] [Google Scholar]
- [67].Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. U. S. A. 2012;109(11):4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 2004;78(23):12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. U. S. A. 2007;104(1):151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Welsby I, Hutin D, Gueydan C, Kruys V, Rongvaux A, Leo O. PARP12, an interferon-stimulated gene involved in the control of protein translation and inflammation. J. Biol. Chem. 2014;289(38):26642–26657. doi: 10.1074/jbc.M114.589515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297(5587):1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- [72].Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 2003;77(21):11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhu Y, Chen G, Lv F, Wang X, Ji X, Xu Y, Sun J, Wu L, Zheng YT, Gao G. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc. Natl. Acad. Sci. U. S. A. 2011;108(38):15834–15839. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Muller S, Moller P, Bick MJ, Wurr S, Becker S, Gunther S, Kummerer BM. Inhibition of filovirus replication by the zinc finger antiviral protein. J. Virol. 2007;81(5):2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xuan Y, Liu L, Shen S, Deng H, Gao G. Zinc finger antiviral protein inhibits murine gammaherpesvirus 68 M2 expression and regulates viral latency in cultured cells. J. Virol. 2012;86(22):12431–12434. doi: 10.1128/JVI.01514-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, Cuconati A, Block TM, Guo JT, Guo H. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog. 2013;9(7):e1003494. doi: 10.1371/journal.ppat.1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chen S, Xu Y, Zhang K, Wang X, Sun J, Gao G, Liu Y. Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nat. Struct. Mol. Biol. 2012;19(4):430–435. doi: 10.1038/nsmb.2243. [DOI] [PubMed] [Google Scholar]
- [78].Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell. 2011;42(4):489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Todorova T, Bock FJ, Chang P. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat. Commun. 2014;5:5362. doi: 10.1038/ncomms6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Katoh M. Identification and characterization of human TIPARP gene within the CCNL amplicon at human chromosome 3q25.31. Int. J. Oncol. 2003;23(2):541–547. [PubMed] [Google Scholar]
- [81].Ma Q, Baldwin KT, Renzelli AJ, McDaniel A, Dong L. TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 2001;289(2):499–506. doi: 10.1006/bbrc.2001.5987. [DOI] [PubMed] [Google Scholar]
- [82].MacPherson L, Tamblyn L, Rajendra S, Bralha F, McPherson JP, Matthews J. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 2013;41(3):1604–1621. doi: 10.1093/nar/gks1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Roper SJ, Chrysanthou S, Senner CE, Sienerth A, Gnan S, Murray A, Masutani M, Latos P, Hemberger M. ADP-ribosyltransferases Parp1 and Parp7 safeguard pluripotency of ES cells. Nucleic Acids Res. 2014;42(14):8914–8927. doi: 10.1093/nar/gku591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24(11):1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Mehrotra P, Riley JP, Patel R, Li F, Voss L, Goenka S. PARP-14 functions as a transcriptional switch for Stat6-dependent gene activation. J. Biol. Chem. 2011;286(3):1767–1776. doi: 10.1074/jbc.M110.157768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Aguiar RC, Yakushijin Y, Kharbanda S, Salgia R, Fletcher JA, Shipp MA. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood. 2000;96(13):4328–4334. [PubMed] [Google Scholar]
- [87].Vyas S, Chesarone-Cataldo M, Todorova T, Huang YH, Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013;4:2240. doi: 10.1038/ncomms3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Iqbal MB, Johns M, Cao J, Liu Y, Yu SC, Hyde GD, Laffan MA, Marchese FP, Cho SH, Clark AR, Gavins FN, Woollard KJ, Blackshear PJ, Mackman N, Dean JL, Boothby M, Haskard DO. PARP-14 combines with tristetraprolin in the selective posttranscriptional control of macrophage tissue factor expression. Blood. 2014;124(24):3646–3655. doi: 10.1182/blood-2014-07-588046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Aguiar RC, Takeyama K, He C, Kreinbrink K, Shipp MA. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 2005;280(40):33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- [90].Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, Shipp MA. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol. Cell. 2009;36(1):110–120. doi: 10.1016/j.molcel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Yan Q, Xu R, Zhu L, Cheng X, Wang Z, Manis J, Shipp MA. BAL1 and its partner E3 ligase, BBAP, link Poly(ADP-ribose) activation, ubiquitylation, and double-strand DNA repair independent of ATM, MDC1, and RNF8. Mol. Cell Biol. 2013;33(4):845–857. doi: 10.1128/MCB.00990-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 1999;146(5):917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Raval-Fernandes S, Kickhoefer VA, Kitchen C, Rome LH. Increased susceptibility of vault poly(ADP-ribose) polymerasedeficient mice to carcinogen-induced tumorigenesis. Cancer Res. 2005;65(19):8846–8852. doi: 10.1158/0008-5472.CAN-05-0770. [DOI] [PubMed] [Google Scholar]
- [94].Tuncel H, Tanaka S, Oka S, Nakai S, Fukutomi R, Okamoto M, Ota T, Kaneko H, Tatsuka M, Shimamoto F. PARP6, a mono(ADP-ribosyl) transferase and a negative regulator of cell proliferation, is involved in colorectal cancer development. Int. J. Oncol. 2012;41(6):2079–2086. doi: 10.3892/ijo.2012.1652. [DOI] [PubMed] [Google Scholar]
- [95].He F, Tsuda K, Takahashi M, Kuwasako K, Terada T, Shirouzu M, Watanabe S, Kigawa T, Kobayashi N, Guntert P, Yokoyama S, Muto Y. Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 2012;586(21):3858–3864. doi: 10.1016/j.febslet.2012.09.009. [DOI] [PubMed] [Google Scholar]
- [96].Meyer-Ficca ML, Ihara M, Bader JJ, Leu NA, Beneke S, Meyer RG. Spermatid Head Elongation with Normal Nuclear Shaping Requires ADP-Ribosyltransferase PARP11 (ARTD11) in Mice. Biol. Reprod. 2015;92(3):80. doi: 10.1095/biolreprod.114.123661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Di Paola S, Micaroni M, Di Tullio G, Buccione R, Di Girolamo M. PARP16/ARTD15 is a novel endoplasmic-reticulumassociated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ss1. PLoS One. 2012;7(6):e37352. doi: 10.1371/journal.pone.0037352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Jwa M, Chang P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1alpha-mediated unfolded protein response. Nat. Cell Biol. 2012;14(11):1223–1230. doi: 10.1038/ncb2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Verheugd P, Forst AH, Milke L, Herzog N, Feijs KL, Kremmer E, Kleine H, Luscher B. Regulation of NF-kappaB signalling by the mono-ADP-ribosyltransferase ARTD10. Nat. Commun. 2013;4:1683. doi: 10.1038/ncomms2672. [DOI] [PubMed] [Google Scholar]
- [100].Feijs KL, Kleine H, Braczynski A, Forst AH, Herzog N, Verheugd P, Linzen U, Kremmer E, Luscher B. ARTD10 substrate identification on protein microarrays: regulation of GSK3beta by mono-ADP-ribosylation. Cell Commun. Signal. 2013;11(1):5. doi: 10.1186/1478-811X-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, Vervoorts J, Walsemann G, Grotzinger J, Kremmer E, Mehraein Y, Mertsching J, Kraft R, Austen M, Lüscher-Firzlaff J, Lüscher B. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005;24(12):1982–1993. doi: 10.1038/sj.onc.1208410. [DOI] [PubMed] [Google Scholar]
- [102].Kaufmann M, Feijs KL, Luscher B. Function and regulation of the mono-ADP-ribosyltransferase ARTD10. Curr. Top. Microbiol. Immunol. 2015;384:167–188. doi: 10.1007/82_2014_379. [DOI] [PubMed] [Google Scholar]
- [103].Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol. Cell Biol. 2007;27(21):7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Gomez M, Wu J, Schreiber V, Dunlap J, Dantzer F, Wang Y, Liu Y. PARP1 Is a TRF2-associated poly(ADP-ribose) polymerase and protects eroded telomeres. Mol. Biol Cell. 2006;17(4):1686–1696. doi: 10.1091/mbc.E05-07-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Berger NA, Sims JL, Catino DM, Berger SJ. Poly(ADP-ribose) polymerase mediates the suicide response to massive DNA damage: studies in normal and DNA-repair defective cells. Princess Takamatsu Symp. 1983;13:219–226. [PubMed] [Google Scholar]
- [106].Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- [107].Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. U. S. A. 1982;79(11):3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299(5606):560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- [109].Li M, Lu LY, Yang CY, Wang S, Yu X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013;27(16):1752–1768. doi: 10.1101/gad.226357.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Pehrson JR, Fried VA. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257(5075):1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- [111].Han W, Li X, Fu X. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat. Res. 2011;727(3):86–103. doi: 10.1016/j.mrrev.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Gottschalk AJ, Trivedi RD, Conaway JW, Conaway RC. Activation of the SNF2 family ATPase ALC1 by poly(ADP-ribose) in a stable ALC1.PARP1.nucleosome intermediate. J. Biol. Chem. 2012;287(52):43527–43532. doi: 10.1074/jbc.M112.401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325(5945):1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451(7174):81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- [115].Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, Ahel I, Neuhaus D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nat. Struct. Mol. Biol. 2010;17(2):241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Iles N, Rulten S, El-Khamisy SF, Caldecott KW. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell Biol. 2007;27(10):3793–3803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Liu C, Wu J, Paudyal SC, You Z, Yu X. CHFR is important for the first wave of ubiquitination at DNA damage sites. Nucleic Acids Res. 2013;41(3):1698–1710. doi: 10.1093/nar/gks1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Aravind L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 2001;26(5):273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- [119].Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, Cong F, Xu W. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26(3):235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, Myer VE, Finan PM, Porter JA, Huang SM, Cong F. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 2011;13(5):623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- [121].Kang HC, Lee YI, Shin JH, Andrabi SA, Chi Z, Gagne JP, Lee Y, Ko HS, Lee BD, Poirier GG, Dawson VL, Dawson TM. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. U. S. A. 2011;108(34):14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Zhou ZD, Chan CH, Xiao ZC, Tan EK. Ring finger protein 146/Iduna is a poly(ADP-ribose) polymer binding and PARsylation dependent E3 ubiquitin ligase. Cell Adh. Migr. 2011;5(6):463–471. doi: 10.4161/cam.5.6.18356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell. 1999;4(3):387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- [124].Mahajan A, Yuan C, Lee H, Chen ES, Wu PY, Tsai MD. Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal. 2008;1(51):re12. doi: 10.1126/scisignal.151re12. [DOI] [PubMed] [Google Scholar]
- [125].Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol. Cell. 2002;9(5):1045–1054. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- [126].Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302(5645):636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- [127].Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302(5645):639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- [128].Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23(5):693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12(3):861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Zhang F, Chen Y, Li M, Yu X. The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc. Natl. Acad. Sci. U. S. A. 2014;111(20):7278–7283. doi: 10.1073/pnas.1318367111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Krietsch J, Caron MC, Gagne JP, Ethier C, Vignard J, Vincent M, Rouleau M, Hendzel MJ, Poirier GG, Masson JY. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012;40(20):10287–10301. doi: 10.1093/nar/gks798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Malanga M, Czubaty A, Girstun A, Staron K, Althaus FR. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J. Biol. Chem. 2008;283(29):19991–19998. doi: 10.1074/jbc.M709495200. [DOI] [PubMed] [Google Scholar]
- [133].Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NADdependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]