Abstract
Endogenous formation of the mutagenic DNA adduct 1,N6-ethenoadenosine (εA) originates from lipid peroxidation. Elevated levels of εA in cancer-prone tissues suggest a role for this adduct in the development of some cancers. The base excision repair (BER) pathway has been considered the principal repair system for εA lesions until recently, when it was shown that the Escherichia coli (E. coli) AlkB dioxygenase could directly reverse the damage. We report here kinetic analysis of the recombinant human AlkB homolog 2 (hABH2), which is able to repair εA lesions in DNA. Furthermore, cation exchange chromatography of nuclear extracts from wild-type and mABH2−/− mice indicate that mABH2 is the principal dioxygenase for εA repair in vivo. This is further substantiated by experiments showing that hABH2, but not hABH3, is able to complement the E. coli alkB mutant with respect to its defective repair of etheno adducts. We conclude that ABH2 is active in direct reversal of εA lesions and that ABH2 together with the ANPG glycosylase, which is the most effective enzyme for repair of εA, comprise the cellular defense against εA lesions.
Keywords: DNA repair, ABH2, AlkB, ethenoadenine
INTRODUCTION
Etheno adducts are ubiquitous and have been found in genomic DNA from a variety of rodent and human tissues (1;2). These lesions are formed by oxidative stress through lipid peroxidation, and by the reaction of DNA with vinyl chloride. 1,N6-Ethenoadenine (εA) residues are highly mutagenic in mammalian cells (3-5). Upon transfection of human cells with double-stranded (ds) and single-stranded (ss) plasmid DNA containing a single εA at a defined position, A→T, A→G and A→C mutations were observed (3). The efficient induction of A→T transversions supports the hypothesis that human and rodent tumors induced by vinyl compounds reflects the misincorporation of A opposite εA (6-8). In such studies, A→T transversions were observed both in the p53 tumor suppressor gene and in the ras oncogene. In addition, a decrease in the activities for repairing εA and 3,N4-ethenocytosine (εC) residues has been observed in lung adenocarcinoma patients (9).
In mammalian cells, the alkyl-N-adenine-DNA glycosylase (ANPG; alias MPG, APG and AAG) efficiently excises εA adducts from DNA, leaving behind an abasic site (10-14). Complete repair requires additional enzymes in the base excision repair (BER) pathway (15). Experiments in gene-targeted mice lacking ANPG (16;17) show that ANPG is the major DNA glycosylase for removal of εA as well as for the cytotoxic 3-methyladenine (3meA) and the mutagenic hypoxanthine lesions. However, the incidence of carcinoma following treatment with vinyl carbamate (Vcar) was similar in wild-type and ANPG-deficient mice (18). This may be explained by recent results using new immunoaffinity-LC-MS/MS methodology, which has revealed that the lesion is removed slowly from the genome also in the absence of ANPG (18;19). The recent observation that E. coli AlkB repairs, in addition to deleterious methyl lesions, εA adducts in DNA by direct reversal might explain the slow repair in the absence of ANPG; if a mammalian dioxygenase possesses the same activity (20). Eight mammalian AlkB homologues, ABH1-ABH8 (denoted hABH1-8 in humans; mABH1-8 in mice), have been identified by bioinformatics (21), two of which - ABH2 and ABH3 - have been shown to share the ability of E. coli AlkB to directly reverse damaged nucleic acids in vitro (22;23). In mice, ABH2 is the major, or possible only, dioxygenase for repair of methylated DNA in vivo (24). In previous studies conditions for in vitro repair of εA was optimized for ANPG. Therefore, reactions did not contain iron and oxoglutarate, two essential cofactors for AlkB mediated damage reversal and, consequently, AlkB mediated repair was not discovered (22;23).
We report here that recombinant human ABH2 exhibits robust activity for direct reversal of εA lesions. Furthermore, cation exchange chromatography of nuclear extracts from wild-type and mABH2−/− mice demonstrated that ABH2 is the principal dioxygenase for εA repair in vivo. This is further substantiated by experiments analyzing the ability of hABH2 and hABH3 to complement the E. coli alkB mutant for its defective repair of etheno adducts.
METHODS
Protein purification (hABH2)
The construct pET28a-hABH2 (provided by Dr. Geir Slupphaug, the Norwegian University of Science and Technology) was transferred into the expression host E. coli strain BL21 (DE3) RIL and grown at 37°C in LB (Luria Bertani) medium containing kanamycin (100 µg/ml) until OD600 = 0.8. Expression of recombinant protein was induced by adding isopropyl β-D-thiogalactoside (IPTG) to a final concentration of 1 mM and the cells further incubated at 18°C for 6 h. Pellets were harvested at 4°C by centrifugation at 6000 rpm for 10 min and resuspended in a buffer containing 50 mM NaP (pH 7.5), 300 mM NaCl, 0.01% Tween-20, 1 × Complete-Protease inhibitor EDTA free tablet (Roche) and 2 mM β-merkaptoethanol. Lysozyme was added to 1 μg/μl and the suspension incubated on ice for 30 min. The protein extract was prepared from the cell suspension by sonication (3 × 30 seconds, 60 amplitude) and denaturated proteins and cell debris were removed by centrifugation (12000 rpm, 10 min, 4°C). Precipitation of nucleic acid with protamine sulfate (0.4% w/v final concentration, Sigma Cat. P4020) was done twice with centrifugation after each precipitation to reduce viscosity. The cell lysate was filtered (0.45 μm; Sarstedt) and directly loaded onto a TALON Superflow resin (Clontech), which was washed and eluted as recommended (Clontech). Fractions were analyzed using sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (NuPAGE gel; Invitrogen 10%, SeeBlue Plus 2 Prestained 1 × Standard, Invitrogen Cat. LC5925). Finally, purified protein was dialyzed against a buffer containing 50 mM Tris (pH 7.0), 100 mM NaCl, 10 mM β-merkaptoethanol, and stored at −80°C until use.
Preparation of nuclear extracts
Five livers were collected from wild-type mice. Extracts and fractionations were prepared as described previously (24). Fractions 26-35 were tested for enzymatic DNA repair activity.
Preparation of oligonucleotide substrate
A 49 nt DNA sequence containing the site-specifically inserted εA residue (underlined) (5’[32P]-TAGACATTGCCATTCTCGATAGGεATCCGGTCAAACCTAGACGAATTCCG-3’) or N-3-methyl-dC(3meC) residue (5’[32P]-TAGACATTGCCATTCTCGATAGG-3meC-TCCGGTCAAACCTAGACGAATTCCG-3’) was provided by Midland Certified Reagent Company Inc. and ChemGenes, respectively. The oligonucleotides were 5’ end-labeled, where the 3meC substrate were further processed as previously described (24). The εA-containing oligonucleotide substrates were separated on a denaturing polyacrylamide gel (PAG) and purified by the “Crush-and-Soak” method (25) and ethanol precipitation. Duplex DNA substrates were prepared by annealing labeled oligonucleotides to a twofold molar quantity (56 pmol) of unlabeled complementary oligonucleotides containing a thymine residue opposite to εA and a guanine residue opposite to 3meC. Annealing was achieved by incubation for 2 min at 90°C followed by slow cooling to room temperature. The end-labeled duplex oligonucleotide substrates were purified following separation on a nondenaturing 20% polyacrylamide gel. The concentration of unlabeled oligonucleotides was measured using Nanodrop. The specific activity of substrate (in c.p.m.) was determined in triplicate by scintillation counting (1 ml substrate or [γ-32P]ATP placed in a 6 ml scintillation tube). Substrate (32P-labeled) concentration (fmol/ml) was calculated according to a formula recommended by the manufacturer (Amersham).
DNA repair reactions
Repair reactions were performed by incubating hABH2 or AlkB with εA and 3meC containing DNA at 37°C for 30 min in a total volume of 50 μl, containing 50 mM Tris (pH 8.0), 2 mM ascorbic acid, 100 mM 2-oxoglutarate and 40 mM FeSO4. In typical experiments, 2 nM of ds εA-DNA substrate was incubated with 12 pmol of AlkB or 10.3 pmol of hABH2 (Figure 1). Incubation times, substrate and enzyme concentration varied as indicated in the figures. MgCl2 (10 mM) was added to optimize hABH2 reaction conditions. As a positive control, εA-DNA was incubated with 12 pmol of a 26-kDa truncated human ANPG protein (26) at 37°C for 30 min followed by NaOH-mediated abasic site cleavage (27). AlkB and its homologs oxidize the methyl group, resulting in direct reversal of the damage without cleaving the DNA strand. The restriction enzyme DpnII is methyl- and ethenoA sensitive and was used to distinguish between methylated and repaired oligonucleotides. In reactions containing ss DNA substrate, the complementary DNA strand was added prior to DpnII cleavage. The site of the damage is within the DpnII recognition sequence (5’-GATC-3’). Repaired product DNA were cleaved by incubating the reaction with 20 U DpnII for 30 min at 37°C and resolved by 20% denaturing PAGE. Visualization and quantification were performed by phosphorimaging analysis using ImageQuant Software (Molecular Dynamics Inc.).
Figure 1. DNA substrates for εA and 3meC repair and εA repair activities of AlkB and hABH2.
(A) To create εA- and 3meC-containing substrates, 49 mer oligonucleotides were modified with an εA in position 24, or 3meC in position 26, radioactively labeled at the 5’-end and hybridized to a complementary strand. The modified bases are located in a DpnII restriction site such that this enzyme cleaves the substrate only if the etheno/methyl group is removed. Unrepaired substrate appears as a band of 49 nt, whereas repaired and cleaved substrate will appear as a 22 nt band following denaturing PAGE. (B) Activities of purified AlkB, hABH2 and ANPG on εA in ss DNA and ds DNA. DNA substrates were incubated with purified enzymes as indicated, digested with DpnII, or treated with 0.1M NaOH/30 min 90°C if reacted with ANPG, and separated by 20% denaturing PAGE. Labeled DNA was visualized by phosphorimaging. Untreated DNA substrates were incubated with DpnII as negative control. (C) Activities of purified hABH3 on εA in ss DNA and ds DNA. Same reaction conditions as in (B). (D) Reactions as in B. Prior to incubation, cold DNA substrates were added
Host cell reactivation of MMS- and CAA-treated M13 ss DNA phage
The experiments were performed as previously described (23). The toluic acid-inducible expression plasmid pJB658 (28) carrying genes encoding hABH2, hABH3 or AlkB (29) was transfected into an F-pilus-expressing, AlkB-deficient E. coli strain, HK82/F′. Overnight cultures of bacteria were diluted 100-fold in selective LB medium and grown at 37°C until A600 = 0.1. Expression of recombinant protein was then induced by the addition of m-toluate (Fluka) to a final concentration of 2 mM, and the cultures were grown further until A600 = 1.0. (about 2.5 h). The ss DNA phage M13 was treated at 30°C for 0.5 h with different concentrations of methyl methanesulfonate (MMS) (Sigma-Aldrich) or chloroacetaldehyde (CAA) (Sigma-Aldrich) to introduce methyl lesions or etheno adducts, respectively. A volume of 100 μl of various dilutions of the treated phage was mixed with 300 μl of induced bacteria and 3 ml LB top agar, and plated onto LB plates. Plates were incubated overnight at 37°C, and progeny phages were scored by counting the resulting plaques. M9 minimum salt medium containing 1 mM MgSO4 was added for all treatments and dilutions of the M13 phage.
MTT assay
The MTT (tetrazolium cytotoxicity assay) was performed in mouse embruo fibroblast (MEF) cell lines cultured as described (24). The cells were seeded in 96-well plates with approximately 3000 cells/well for the wild-type, mABH2−/− and ANPG−/− cell lines. Cells were exposed to medium containing chloroacetaldehyde (CAA) for 2 h (0–20 μM, 100 μl/well), 22 h after seeding. The cells were then washed twice with PBS (phosphate buffered saline) and fresh media added. Relative cell numbers were measured after 48 h using the MTT assay (Roche).
Preparation of genomic DNA from mouse livers and MS analysis
Frozen mouse liver tissue was homogenized in cell lysis buffer (Roche) using a Brinkmann Polytron homogenizer and genomic DNA was isolated using a Roche genomic DNA isolation kit, following manufacturer’s directions. The following anti-oxidants and deaminase inhibitors were added to the PBS, cell lysis buffer, and protein precipitation solution prior to use: coformycin (Sigma, 5 μg/ml), desferrioxamine (Sigma, 0.6 mg/ml) and butylated hydroxytoluene (Sigma, 100 μM). εA in DNA was quantified as described previously (30). The GraphPad Prism Software was used to estimate the p-value.
RESULTS
Direct reversal of εA lesions in ds and ss DNA by AlkB and hABH2
Recent in vitro studies using recombinant protein have shown that recombinant E. coli AlkB corrects 1meA and 3meC lesions in DNA by oxidative demethylation, and that two human homologs, hABH2 and hABH3, share this ability (29;31). In vitro, hABH3 displays a strong preference for ssDNA and ssRNA, while hABH2 is exclusively active on DNA, and displays a moderate preference for dsDNA over ssDNA (24;29;31;32). However, studies conducted in ABH3 targeted mice have not revealed substrates for ABH3 in vivo [(24), unpublished data]. Recently, εA was discovered to be a substrate for E. coli AlkB (20).
The current work was initiated by an in vitro assay in which we show that recombinant hABH2 (Figure 1B), but not hABH3 (Figure 1C), produces a DpnII cleavable substrate when incubated with a ss or a ds synthetic oligonucleotide containing an εA lesion in the G-εA-T-C sequence (Figure 1A). As illustrated in Figure 1B, the 49 nt 5’-[32P] end-labeled oligonucleotides were cleaved into 22 nt fragments only when the DpnII digestion was preceded by incubation with AlkB or hABH2, indicating high quality of substrate DNA. As a positive control for εA repair, we used human ANPG protein, which is able to excise εA residues only from ds DNA substrates (12). Minor repair of ss DNA by ANPG might be due to some repair activity on ss DNA or, alternatively, flexible structures of the ss oligonucleotide (Figure 1B). It is difficult to measure the accurate concentration of the 32P-labeled εA-DNA substrate. Therefore we added, in excess, an increasing amount of unlabeled εA-DNA to the specific amount of labeled substrate employed in the reaction mixture, to decide the concentration of substrate DNA being used in subsequent single-turnover kinetic experiments (Figure 1D).
Single-turnover kinetics for repair of εA in DNA by AlkB and hABH2
In contrast to the 1meA and 3meC lesions, which are preferentially introduced into ss regions of DNA, the majority of εA lesions is introduced into ds DNA, and so we investigated the kinetic parameters of AlkB and hABH2 (0-100 pmol) for εA repair in ds DNA substrates (Figure 2). Enzyme activity was measured under single-turnover conditions as described in detail elsewhere (27). The concentration of repairable substrate was determined to be 0.32±0.02 nM for hABH2 and 0.51±0.01 nM for AlkB (Figure 2A), indicating that more εAs are present in an appropriate conformation to be repaired by AlkB than hABH2. The reaction rate was determined by the slopes of the linear regression curves presented in Figure 2B (hABH2) and Figure 2C (AlkB), which divided by the active substrate concentration leads to the first-order rate constant k (Figure 2D) which describes the overall accumulation of product during the reaction. The calculations show that AlkB and hABH2 remove the etheno structure from adenine with essentially identical efficiency (hABH2, k2 = 0.094±0.009 min−1, KD = 210±60 nM; AlkB, k2 = 0.100±0.009 min−1, KD = 260±70) (Figure 2D).
Figure 2. Single-turnover kinetics for repair of εA.
Substrate DNA (2 nM) was incubated with three different (twice to 100 times higher) concentrations of hABH2 and AlkB to measure product formation as a function of time. Each value represents the average of three independent measurements. (A) Plots of product formation as a function of time using 1000 nM enzyme, together with a curve fit (r2 values: hABH2, 0.99651; AlkB, 0.97915) to the experimental data [see (27)], to determine the active substrate concentration. (B and C) Linear regression curves (r2 values hABH2: 20 nM, 0.94361; 200 nM, 0.99617; 1000 nM, 0.96471; r2 values AlkB: 20 nM, 0.94061; 200 nM, 0.97327; 1000 nM, 0.92739) showing the initial product formation as a function of time (where the reaction rate is determined by their slopes) at the different enzyme concentrations. (D) Calculated k values (determined from the value of the slope of the curves presented in B divided by the active enzyme concentration) as a function of total enzyme concentration together with a curve fit (r2 values: hABH2, 0.99533; AlkB, 0.99631) to the experimental data (27).
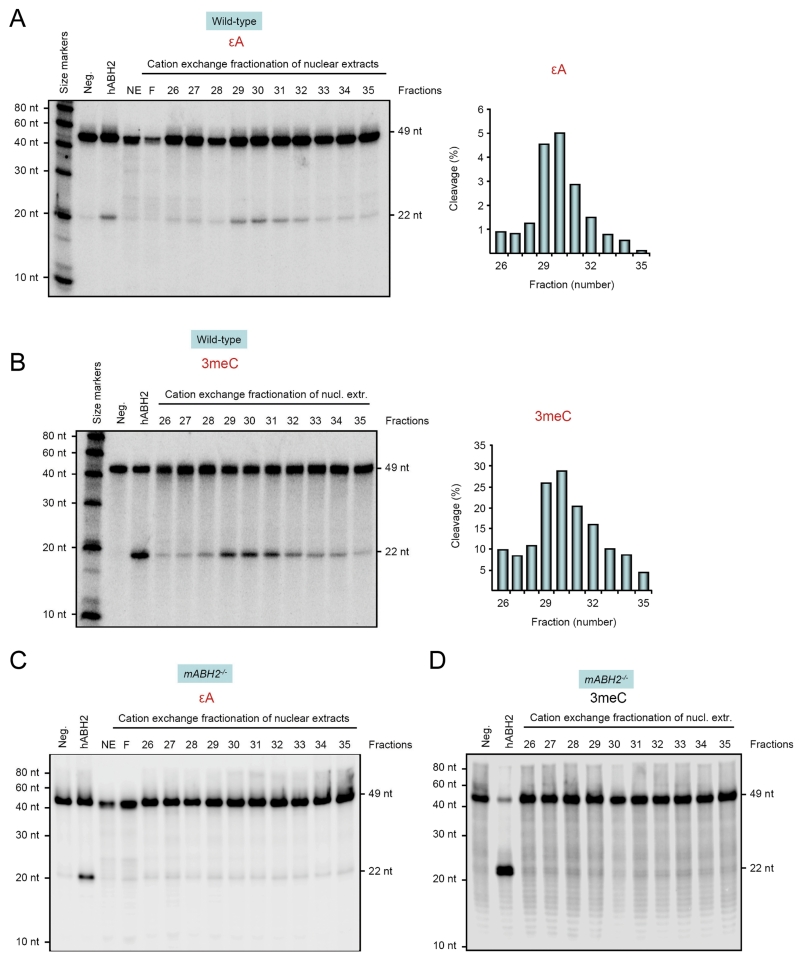
Repair of εA and 3meC by freshly prepared protein fractions from wild-type and mABH2−/− mice
The activity of hABH2 has previously been characterized in vitro and in vivo, measured by the repair of 1meA, 3meC, 1meG and 3meT lesions (24;29;31;32). To determine the properties and relevance of ABH2 in the repair of εA, the ds εA-oligonucleotide was incubated with fractionated nuclear extracts from mouse liver, since the activity of ABH2 is difficult to measure in whole nuclear extracts (24). The repair activity eluted at ~370 mM NaCl, in good agreement with our previous results (24) (Figure 3, panels A and B). The fractionation of the nuclear extracts increased the repair activities for both εA and 3meC by several fold. Repair of εA and 3meC peaked in fractions 29-31 and was dependent on mABH2 activity, since no activity was observed in fractions prepared from mABH2−/− mice (Figure 3, panels C and D). No activity was detected from the eluted fractions nor in the flow-through (Figure 3). The relative repair capacities for εA and 3meC of individual fractions prepared from wild-type mice were quantified (Figure 3, right panels A and B) matched each other very well.
Figure 3. Repair of εA and 3meC in wild-type cell extracts.
Repair of εA (panel A) and 3meC (panel B, positive control) in fractionated wild-type extract is demonstrated with DpnII cleavage, as described in Figure 1. Repair of εA and 3meC was measured in freshly prepared protein fractions corresponding to 0.75 μg total protein. Repair activities peaked in fractions 29–31 and were completely dependent on mABH2 as no activity was observed in fractions from mABH2−/− mice (C and D). Right panels A and B indicate the proportion of the substrate (from the gels shown) that has been repaired and cleaved by DpnII. Details and substrate as in Figure 1. NE: nuclear extract; F: flow through.
hABH2-mediated repair of etheno adducts in M13 ssDNA in E. coli
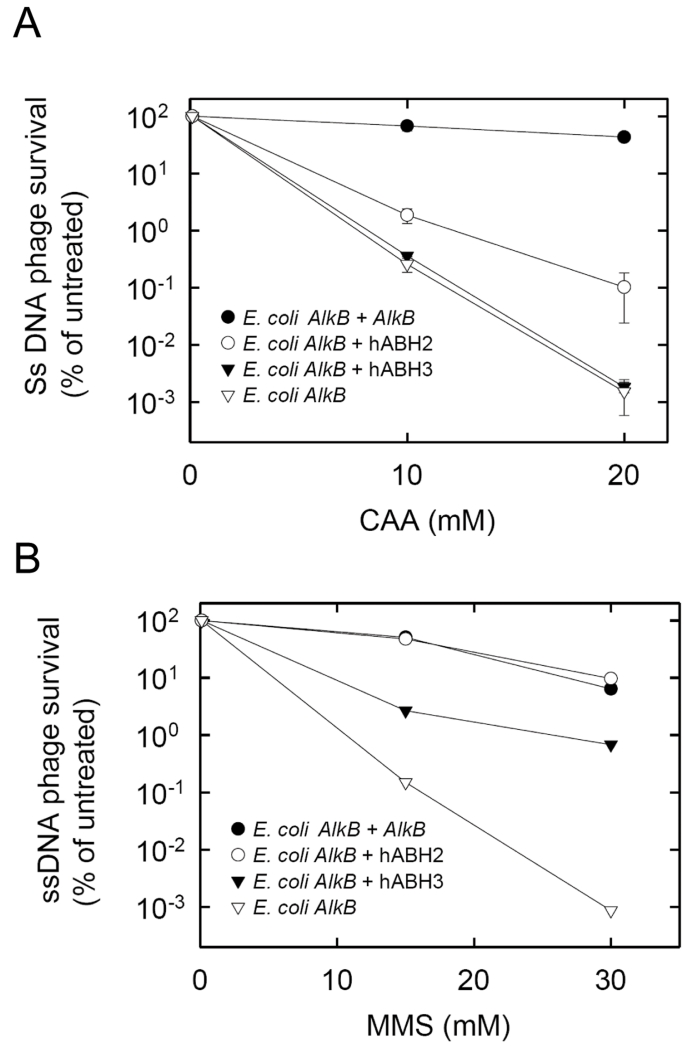
The E. coli alkB mutant is defective in reactivating ss phage DNA containing methyl or etheno lesions (20;33;34). In the case of methyl lesions, this defect has been shown to be suppressed by the expression of functional AlkB homologs (29;31), and we expected the same to be true for etheno adducts. The alkB mutant carrying an expression plasmid for hABH2 was infected with M13 ss DNA phage treated with different concentrations of CAA, a vinyl chloride metabolite known to introduce etheno adducts in DNA. The number of surviving phages decreased with increasing concentrations of CAA (Figure 4A). Higher phage survival was observed in the hABH2-expressing bacteria than in the control, indicating that hABH2-mediated removal of etheno adducts did occur. Compared with hABH2, the E. coli AlkB protein mediated a substantially higher degree of M13 phage survival. Despite almost identical kinetic properties of the two enzymes measured in vitro (Figure 2), there were two important differences between the experimental conditions. Firstly, while ss DNA was substrate in the phage reactivation experiments, ds DNA was used in the in vitro repair reactions, and AlkB actually prefers ss DNA over ds DNA, while the opposite is true for hABH2 (29;35). Secondly, the in vitro reactions used an εA-containing oligonucleotide as substrate, but εC, and not εA, is the etheno lesion introduced most frequenly when DNA is exposed to CAA (36;37).
Figure 4. Survival of CAA- or MMS-treated M13 ssDNA phage in alkB mutant bacteria producing different AlkB proteins.
HK82/F′ alkB bacteria carrying expression plasmids for the proteins indicated were infected with ssDNA phage M13 treated with the indicated concentrations of CAA (A) or MMS (B). Formation of progeny phage was assessed by counting plaques, and values are expressed relative to untreated M13. Error bars represent the standard deviation of triplicate samples (The apparent non-symmetry of the error bars is due to the logarithmic scale).
In these experiments, hABH3 did not display any ability to remove CAA-induced etheno adducts, while in control experiments there was appreciable repair of MMS-induced methyl lesions by hABH3, hABH2 and AlkB (Figure 4B), in accordance with previous reports (29). In summary, our phage reactivation experiments indicate, in accordance with the in vitro data, that CAA-induced etheno lesions are substrates for AlkB and hABH2, but not hABH3.
Cellular sensitivity to CAA
CAA introduces mutagenic and toxic etheno adducts into genomic DNA. M ammalian cells can repair etheno adducts via the ANPG DNA glycosylase of the BER pathway (17;19), and our kinetic analyses indicate that ANPG is six times more efficient than ABH2 for repair of εA (see Discussion). E. coli cells deficient in either AlkB or AlkA are sensitive to CAA treatment, although loss of AlkA causes a more severe phenotype than loss of AlkB (20). AlkA is the E. coli counterpart of ANPG. The relative survival of mABH2−/− cells after 48 h was reduced by approximately 25%, 30% and 40% for CAA concentrations of 2, 5 and 10 μM, whereas the corresponding numbers for wild-type were 5%, 15% and 30% (Figure 5A). Similar experiments with ANPG−/− cells also indicated that the mutant cell lines could be slightly more sensitive than the corresponding wild-type cell line. In general, the differences in relative cell survival between wild-type and mABH2−/− cells, and between wild-type and ANPG−/− cells, were reproducible modest but not significant. Similar data is previously presented for MMS survival (24). We observed no difference in viability and growth for the two cell lines, at CAA concentrations higher than 10 μM (concentrations between 20 and 100 μM are not shown).
Figure 5. Effect of CAA on growth and viability of wild-type, mABH2−/− and ANPG−/− MEF cells.
Wild-type cells, mABH2−/− and ANPG−/− cells were used at passages 16 to 34. Cells were seeded in 96-well plates with approximately 3000 cells/well. After 22 hours the cells were treated with 0-20 μM CAA for 2 hours, washed twice with PBS, before fresh media was added. Cell survival was measured after 48 h. Results are expressed as cell numbers in CAA-treated cultures compared to untreated controls. Cell numbers were measured using the MTT assay (Roche). Untreated wild-type, mABH2−/− (A) and ANPG−/− (B) cell lines had a density of about 60% after 48 hours. Values presented are average of five to ten independent experiments, and the standard deviations are always less than 10%.
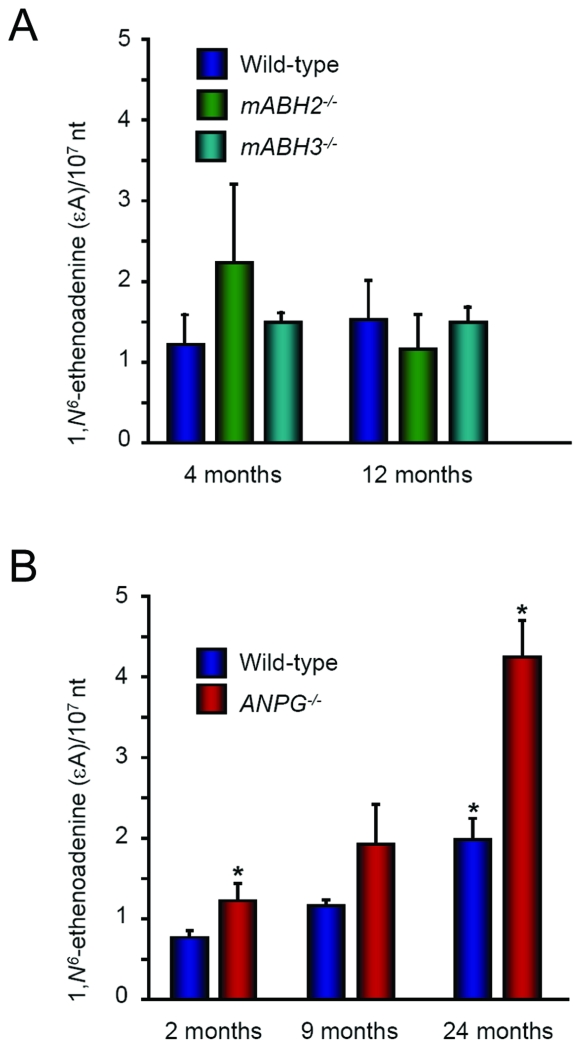
Age-related accumulation of εA lesions in mABH2−/− and ANPG−/− null mice
To study the role played by ABH2 and ANPG in the repair of spontaneously formed εA lesions, genomic DNA was extracted from the livers of untreated wild-type, mABH2−/− and ANPG−/− mice at different ages (Figure 6, panels A and B). Steady-state levels of εA were determined at 4 and 12 months in mABH2−/− and mABH3−/− mice, and at 2, 9 and 24 months for ANPG−/− mice by LC-MS/MS as described in Materials and Methods. Two to five mice of each genotype were sacrificed at each time point. No εA accumulation could be detected in wild-type, mABH2−/− and mABH3−/− null mice up to 12 months of age. By contrast, significant εA accumulation was observed in 9- and 24-months old ANPG−/− mice. These results suggest that a substantial degree of εA production occurs under normal physiological conditions. At 24 months of age, a small increase of εA bases was also observed in wild-type mice. Thus, these data are in keeping with the obtained kinetic parameters of the ABH2 and ANPG enzymes, and the accumulation seen in ANPG−/− null mice may suggest that the activity provided by ABH2 is not sufficient to clear all endogenously generated εA adducts.
Figure 6. Age-dependent accumulation of εA in wild-type, ABH2−/−, ABH3−/− and ANPG−/− mice.
Histogram graph showing the quantification of εA DNA lesions in liver tissue from wild type, mABH2−/− and mABH3−/− mice ranging in age from 4 to 12-months (panel A) and wild-type and ANPG−/− mice ranging in age from 2 to 24 months (panel B). Lesions were quantified using a highly sensitive HPLC-tandem mass spectrometric (MS-MS) method (30). A minimum of 2 and a maximum of 5 mice were used per time point/per genotype. * p<0.05 24 months ANPG−/− mice compared with 24 months old wild-type or 2 months old ANPG−/− mice.
DISCUSSION
Many types of human cancers are linked to oxidative stress conditions resulting from chronic inflammations. Reactive oxygen species (ROS) can damage DNA, RNA, proteins and lipids. Lipid peroxidation stimulates the formation of mutagenic DNA adducts, such as εA and εC. Being the predominant lesions formed in response to inflammation, etheno bases outscore oxidized and deaminated bases (30). Mutation spectra obtained from tumors induced by Vcar in mice are compatible with εA being the initiating lesion (38). The removal of εA and εC from genomic DNA has long been attributed to ANPG and the mismatch-specific thymine-DNA glycosylase (TDG), respectively (12;39;40). TDG removes εC residues from ds DNA without a strict preference for a certain opposite base, although the εC/G mismatch is the best substrate (39). Since an increased formation and persistence of εA in DNA in ANPG knockout mice treated with Vcar was not reported to be associated with higher susceptibility to cancer, the ABH2 dioxygenase could account for the slow but significant repair of εA observed in the ANPG−/− liver (18). The half-life of εA in non-dividing cells increased from 8 h in wild-type to 56 h in ANPG−/− livers. The kinetic parameters of ANPG and ABH2, together with the repair kinetics of εA in wild-type and ANPG−/− mice [this work and (18;41)] indicates that ANPG and ABH2 together might account for the complete repair of εA lesions in vivo.
The development of a mABH2−/−/ANPG−/− double mutant is currently in progress. This might well yield results equivalent to those seen with OGG1−/− and MYH−/− (MutY homolog)-deficient mice. OGG1 removes the highly mutagenic 7,8-dihydro-8-oxoguanine (8-oxoG) from DNA. 8-oxoG lesions (42), which appear in greatly increased numbers in OGG1−/− mice (43;44), can mispair with adenine (A) during replication; this 8-oxoG:A mismatch is handled by the MYH glycosylase which removes the mispaired A. Whereas both OGG1−/− and MYH−/− single mutant mice appear normal with no overt phenotype (43-45), simultaneous deletions of both OGG1 and MYH predisposed two-thirds of the mice to tumors, establishing an obvious link between oxidative DNA damage and tumorigenesis (45) . The MYH−/−/OGG1−/−-related tumors are characterized by unique G → T transversions in codon 12, GGT, of the K-ras oncogene (45;46).
ABH2 is probably the only, or the major, enzyme responsible for the repair of 1meA in the genome. In unexposed mice lacking ABH2, 1meA lesions accumulate roughly at a rate of one per 107 adenines per year (24). A similar value would be obtained by continuous exposure to 20 nM MMS alone, where a possible intracellular source of methylation is SAM (S-adenosyl-methionine) (47). Endogenous formation of εA adducts, probably through lipid peroxidation, has been demonstrated by the presence of εA in genomic DNA prepared from humans and rodents tissues (1;48). The greatly variable levels of εA indicates variable sources for εA generation and/or variable capacity for repair (1). The recent development of ultrasensitive methods for the detection of εA has made it possible to measure this lesion in vivo and to study its formation and role in experimental carcinogenesis. Nevertheless, the endogenous sources and concentration of εA are not well characterized. In our experiments, εA lesions accumulate four times faster in the liver of ANPG−/− mice as compared to wild-type mice. In line with previous experiments where significant amounts of this lesion were detected in the genome of unexposed tissues (1;48;49), these data confirm that the lesion is produced at significant levels under normal cellular conditions. Based on this study we conclude that the activity of ABH2 is not sufficient for removal of spontaneously produced εA lesions in ANPG−/− mouse liver. On the contrary, the ANPG activity is sufficient for removal of spontaneously produced εA lesions in mABH2−/− mouse liver, supporting the notion that ANPG is the most effective enzyme for repair of this lesion.. The significance of the endogenous production of εA, as well as its induction of carcinogenesis following treatment with Vcar, for example, will be further studied in cells and mice with combined mABH2−/−/ANPG−/− deficiency. Such studies, which are in good progress, should also identify potential added effect of combined gene deletion.
Acknowledgements
We thank the Norwegian Transgenic Center (NTS) and the Center for Comparative Studies at Rikshospitalet HF for the excellent services provided. We also acknowledge the financial support provided by the Norwegian Cancer Society, the National Program in Functional Genomics (FUGE) sponsored by the Norwegian Research Council and the University of Oslo (EMBiO) and by the NIH grants ES02109, CA75576, and AI37750 to LDS.
Reference List
- 1.Nair J, Barbin A, Guichard Y, Bartsch H. 1,N6-ethenodeoxyadenosine and 3,N4-ethenodeoxycytine in liver DNA from humans and untreated rodents detected by immunoaffinity/32P-postlabeling. Carcinogenesis. 1995;16:613–7. doi: 10.1093/carcin/16.3.613. [DOI] [PubMed] [Google Scholar]
- 2.Laib RJ. The role of cyclic base adducts in vinyl-chloride-induced carcinogenesis: studies on nucleic acid alkylation in vivo. IARC Sci Publ. 1986;70:101–8. [PubMed] [Google Scholar]
- 3.Levine RL, Yang IY, Hossain M, Pandya GA, Grollman AP, Moriya M. Mutagenesis induced by a single 1,N6-ethenodeoxyadenosine adduct in human cells. Cancer Res. 2000;60:4098–104. [PubMed] [Google Scholar]
- 4.Pandya GA, Moriya M. 1,N6-ethenodeoxyadenosine, a DNA adduct highly mutagenic in mammalian cells. Biochemistry. 1996;35:11487–92. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 5.Bartsch H, Nair J. Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: role of lipid peroxidation, DNA damage, and repair. Langenbecks Arch Surg. 2006;391:499–510. doi: 10.1007/s00423-006-0073-1. [DOI] [PubMed] [Google Scholar]
- 6.Froment O, Boivin S, Barbin A, Bancel B, Trepo C, Marion MJ. Mutagenesis of ras proto-oncogenes in rat liver tumors induced by vinyl chloride. Cancer Res. 1994;54:5340–5. [PubMed] [Google Scholar]
- 7.Hollstein M, Marion MJ, Lehman T, et al. p53 mutations at A:T base pairs in angiosarcomas of vinyl chloride-exposed factory workers. Carcinogenesis. 1994;15:1–3. doi: 10.1093/carcin/15.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Wiseman RW, Stowers SJ, Miller EC, et al. Activating mutations of the c-Ha-ras protooncogene in chemically induced hepatomas of the male B6C3 F1 mouse. Proc Natl Acad Sci U S A. 1986;83:5825–9. doi: 10.1073/pnas.83.16.5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speina E, Zielinska M, Barbin A, et al. Decreased repair activities of 1,N(6)-ethenoadenine and 3,N(4)-ethenocytosine in lung adenocarcinoma patients. Cancer Res. 2003;63:4351–7. [PubMed] [Google Scholar]
- 10.Rydberg B, Qiu ZH, Dosanjh MK, Singer B. Partial purification of a human DNA glycosylase acting on the cyclic carcinogen adduct 1,N6-ethenodeoxyadenosine. Cancer Res. 1992;52:1377–9. [PubMed] [Google Scholar]
- 11.Dosanjh MK, Chenna A, Kim E, Fraenkel-Conrat H, Samson L, Singer B. All four known cyclic adducts formed in DNA by the vinyl chloride metabolite chloroacetaldehyde are released by a human DNA glycosylase. Proc Natl Acad Sci U S A. 1994;91:1024–8. doi: 10.1073/pnas.91.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saparbaev M, Kleibl K, Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–5. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singer B, Antoccia A, Basu AK, et al. Both purified human 1,N6-ethenoadenine-binding protein and purified human 3-methyladenine-DNA glycosylase act on 1,N6-ethenoadenine and 3-methyladenine. Proc Natl Acad Sci U S A. 1992;89:9386–90. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rydberg B, Dosanjh MK, Singer B. Human cells contain protein specifically binding to a single 1,N6-ethenoadenine in a DNA fragment. Proc Natl Acad Sci U S A. 1991;88:6839–42. doi: 10.1073/pnas.88.15.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–70. [PMC free article] [PubMed] [Google Scholar]
- 16.Hang B, Singer B, Margison GP, Elder RH. Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc Natl Acad Sci U S A. 1997;94:12869–74. doi: 10.1073/pnas.94.24.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelward BP, Weeda G, Wyatt MD, et al. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:13087–92. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbin A, Wang R, O’Connor PJ, Elder RH. Increased formation and persistence of 1,N(6)-ethenoadenine in DNA is not associated with higher susceptibility to carcinogenesis in alkylpurine-DNA-N-glycosylase knockout mice treated with vinyl carbamate. Cancer Res. 2003;63:7699–703. [PubMed] [Google Scholar]
- 19.Ham AJ, Engelward BP, Koc H, et al. New immunoaffinity-LC-MS/MS methodology reveals that Aag null mice are deficient in their ability to clear 1,N6-etheno-deoxyadenosine DNA lesions from lung and liver in vivo. DNA Repair (Amst) 2004;3:257–65. doi: 10.1016/j.dnarep.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Delaney JC, Smeester L, Wong C, et al. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol. 2005;12:855–60. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 21.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–8. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 23.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–82. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 24.Ringvoll J, Nordstrand LM, Vagbo CB, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–98. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Ruffner DE. Modified crush-and-soak method for recovering oligodeoxynucleotides from polyacrylamide gel. Biotechniques. 1996;21:820–2. doi: 10.2144/96215bm14. [DOI] [PubMed] [Google Scholar]
- 26.O’Connor TR. Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Res. 1993;21:5561–9. doi: 10.1093/nar/21.24.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leiros I, Nabong MP, Grosvik K, et al. Structural basis for enzymatic excision of N(1)-methyladenine and N(3)-methylcytosine from DNA. EMBO J. 2007;26:2206–17. doi: 10.1038/sj.emboj.7601662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blatny JM, Brautaset T, Winther-Larsen HC, Haugan K, Valla S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl Environ Microbiol. 1997;63:370–9. doi: 10.1128/aem.63.2.370-379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aas PA, Otterlei M, Falnes PO, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–63. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 30.Pang B, Zhou X, Yu H, et al. Lipid peroxidation dominates the chemistry of DNA adduct formation in a mouse model of inflammation. Carcinogenesis. 2007;28:1807–13. doi: 10.1093/carcin/bgm037. [DOI] [PubMed] [Google Scholar]
- 31.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci U S A. 2002;99:16660–5. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–7. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinglay S, Trewick SC, Lindahl T, Sedgwick B. Defective processing of methylated single-stranded DNA by E. coli AlkB mutants. Genes Dev. 2000;14:2097–105. [PMC free article] [PubMed] [Google Scholar]
- 34.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:14051–6. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falnes PO, Bjoras M, Aas PA, Sundheim O, Seeberg E. Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:3456–61. doi: 10.1093/nar/gkh655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guichard Y, el GF, Nair J, Bartsch H, Barbin A. Formation and accumulation of DNA ethenobases in adult Sprague-Dawley rats exposed to vinyl chloride. Carcinogenesis. 1996;17:1553–9. doi: 10.1093/carcin/17.8.1553. [DOI] [PubMed] [Google Scholar]
- 37.Singer B, Grunberger D. Molecular biology of mutagens and carcinogens. Plenum Press; New York: 1983. [Google Scholar]
- 38.Barbin A. Role of etheno DNA adducts in carcinogenesis induced by vinyl chloride in rats. IARC Sci Publ. 1999;150:303–13. [PubMed] [Google Scholar]
- 39.Saparbaev M, Laval J. 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc Natl Acad Sci U S A. 1998;95:8508–13. doi: 10.1073/pnas.95.15.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hang B, Medina M, Fraenkel-Conrat H, Singer B. A 55-kDa protein isolated from human cells shows DNA glycosylase activity toward 3,N4-ethenocytosine and the G/T mismatch. Proc Natl Acad Sci U S A. 1998;95:13561–6. doi: 10.1073/pnas.95.23.13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy R, Biswas T, Hazra TK, et al. Specific interaction of wild-type and truncated mouse N-methylpurine-DNA glycosylase with ethenoadenine-containing DNA. Biochemistry. 1998;37:580–9. doi: 10.1021/bi972313l. [DOI] [PubMed] [Google Scholar]
- 42.Klungland A, Bjelland S. Oxidative damage to purines in DNA: role of mammalian Ogg1. DNA Repair (Amst) 2007;6:481–8. doi: 10.1016/j.dnarep.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 43.Minowa O, Arai T, Hirano M, et al. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc Natl Acad Sci U S A. 2000;97:4156–61. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klungland A, Rosewell I, Hollenbach S, et al. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–5. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie Y, Yang H, Cunanan C, et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 46.Russo MT, De LG, Degan P, et al. Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Ogg1 DNA glycosylases. Cancer Res. 2004;64:4411–4. doi: 10.1158/0008-5472.CAN-04-0355. [DOI] [PubMed] [Google Scholar]
- 47.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–6. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nair J, Barbin A, Velic I, Bartsch H. Etheno DNA-base adducts from endogenous reactive species. Mutat Res. 1999;424:59–69. doi: 10.1016/s0027-5107(99)00008-1. [DOI] [PubMed] [Google Scholar]
- 49.Bartsch H, Nair J. Ultrasensitive and specific detection methods for exocylic DNA adducts: markers for lipid peroxidation and oxidative stress. Toxicology. 2000;153:105–14. doi: 10.1016/s0300-483x(00)00307-3. [DOI] [PubMed] [Google Scholar]