Abstract
Estrogen receptor-α (ESR1) is an important transcriptional regulator in the mammalian oviduct, however ESR1-dependent regulation of the transcriptome of this organ is not well defined, especially at the genomic level. The objective of this study was therefore to investigate estradiol- and ESR1-dependent regulation of the transcriptome of the oviduct using transgenic mice, both with (ESR1KO) and without (wild-type, WT) a global deletion of ESR1. Oviducts were collected from ESR1KO and WT littermates at 23 days of age, or ESR1KO and WT mice were treated with 5 IU PMSG to stimulate follicular development and the production of ovarian estradiol, and the oviducts collected 48 h later. RNA extracted from whole oviducts was hybridized to Affymetrix Genechip Mouse Genome 430–2.0 arrays (n = 3 arrays per genotype and treatment) or reverse transcribed to cDNA for analysis of the expression of selected mRNAs by real-time PCR. Following microarray analysis, a statistical two-way ANOVA and pairwise comparison (LSD test) revealed 2428 differentially expressed transcripts (DEG’s, P < 0.01). Genotype affected the expression of 2215 genes, treatment (PMSG) affected the expression of 465 genes, and genotype x treatment affected the expression of 438 genes. With the goal of determining estradiol/ESR1-regulated function, gene ontology (GO) and bioinformatic pathway analyses were performed on DEG’s in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice. Significantly enriched GO molecular function categories included binding and catalytic activity. Significantly enriched GO cellular component categories indicated the extracellular region. Significantly enriched GO biological process categories involved a single organism, modulation of a measurable attribute and developmental processes. Bioinformatic analysis revealed ESR1-regulation of the immune response within the oviduct as the primary canonical pathway. In summary, a transcriptomal profile of estradiol- and ESR1-regulated gene expression and related bioinformatic analysis is presented to increase our understanding of how estradiol/ESR1 affects function of the oviduct, and to identify genes that may be proven as important regulators of fertility in the future.
Introduction
Gamete storage and maturation, fertilization, and early embryonic development occur in the oviduct. Ovarian-derived estradiol is a known regulator of a oviductal function, modulating contraction of its smooth musculature [1,2] and secretion by its epithelial cells [3,4], with the transcription factors estrogen receptor-α (ESR1) and estrogen receptor-β (ESR2) both expressed within this organ. ESR1 has been localized to ciliated and secretory epithelial cells, stromal cells, as well as smooth muscle cells of the oviduct [5,6,7], whereas ESR2 appears to be largely confined to ciliated epithelial cells [6]. Immunoreactivity of both receptor subtypes to the nucleus, cytoplasm and plasma membranes [8] indicative of both genomic and non-genomic responses. Estradiol also acts as a regulator of ESR1 protein expression [5,6,7], estradiol and ESR1 therefore acting as important transcriptional regulators where hormone bound receptors target the estrogen responsive element (ERE) on the promoter region of their target genes to either enhance or repress transcription [9,10].
Genome-wide reports on estradiol/ESR1-dependent regulation of oviductal function are lacking. Our objective was therefore to identify estradiol/ESR1-dependent transcriptomal changes in the oviduct using a mouse model that is deficient in ESR1 expression (ESR1KO), testing the hypothesis that estradiol, acting through ESR1, affects the expression of mRNAs within this organ. Immature mice were utilized to circumvent the reproductive phenotype observed in ESR1KO mice observed following puberty, where females develop cystic ovaries and dysfunction of the hypothalamic-pituitary axis due to elevated concentrations of circulating estradiol [11,12]. Immature mice (ESR1KO and WT) were left untreated, or were treated with PMSG to stimulate follicular development and the production of estradiol. Microarray-based transcriptional profiling and bioinformatic analyses was therefore performed using oviducts collected from mice bearing a global deletion of ESR1 and their wild-type (WT) littermates, both before and after PMSG-induced production of ovarian estradiol.
To provide the reader with full access to the transcriptomal dataset, the raw data (*.cel files) plus the GCRMA-normalized and log2 transformed transcript data (Park Genomics Suite [13]), have been deposited into the Gene Expression Omnibus (National Center for Biotechnology Information [14]) as accession number GSE72614 (http://www.ncbi.nlm.nih.gov/geo).
Materials and Methods
Animals and Tissue Collection
All animal procedures were approved by the University of Kentucky Institutional Animal Care and Use Committee. Mice with a global deletion of ESR1 (ESR1KO) on a C57BL/6 background were generated as previously described [15,16]. Briefly, two transgenic mouse lines were used; male ESR1flox/flox were bred with female Zp3cre to produce a line expressing Cre recombinase in the oocyte. The F1 heterozygotes (ESR1flox/+Zp3cre) were bred with ESR1flox/flox resulting in ESR1flox/floxZp3cre mice, where females produce oocytes that are ESR1-. ESR1flox/floxZp3cre females were then bred with ESR1flox/- males to produce ESR1KO progeny (ESR1-/- and ESR1-/- ZP3Cre) or sibling controls (ESR1flox/- and ESR1flox/- ZP3Cre). Genomic DNA was extracted from ear punches using the Easy DNA kit (Invitrogen, Carlsbad CA) to confirm genotypes, as previously described [16]. Whole oviducts were collected for extraction of RNA from immature female mice (ESR1KO and WT) killed at 23 days of age, or ESR1KO and WT mice treated i.p. with 5 IU PMSG at 23 days of age and killed 48 h later.
RNA Extraction
Oviducts were pooled from 3–4 mice per treatment group and genotype and total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA) and purified through RNeasy columns (Qiagen, Valencia, CA), as described before [15,17]. RNA was analyzed for quality and quantified by spectrophotometry using an Eppendorf BioPhotometer Plus (Eppendorf, Germany) as well as by visual distinction of 18S and 28S rRNA bands after ethidium bromide staining in an agarose gel. Spectrophotometry revealed a mean 260/280 ratio of 1.75 ± 0.10 for all samples. Aliquots of the same total RNA were used for both microarray and real-time reverse-transcription PCR (real-time RT-PCR).
Microarray Hybridization
A total of 12 microarray hybridizations were performed using the Affymetrix Genechip Mouse Genome 430–2.0 arrays (GeneChip; Affymetrix, Inc., Santa Clara, CA) according to the manufacturer’s instructions at the University of Kentucky Microarray Core Facility, as described before [15,17,18]. Three replicates using different mice were generated for each treatment group.
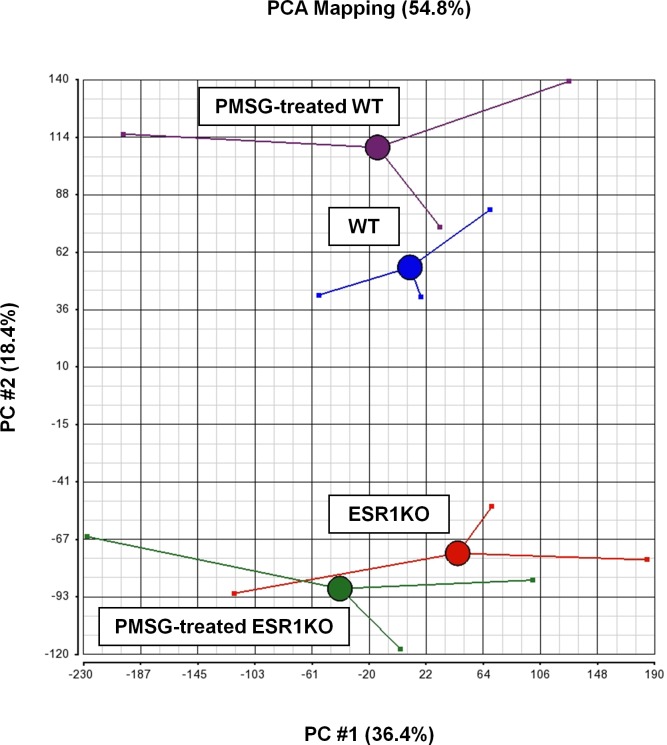
Microarray data were analyzed by importing raw expression intensity values (*.cel files) into Partek Genomics Suite 6.6 (Partek Inc., St. Louis, MO), where the GC-Robust Multiarray Analysis algorithm (GC-RMA), quantile normalization, and Median Polish was applied for GeneChip background correction, log base 2 transformation, conversion of expression values and probeset summarization. Annotation was performed using NetAffx annotation database (Release 34) on December 3rd, 2014. Quality of data was assessed using light intensity expression values on a per chip and per gene basis and visualized as box plots (Fig 1). Principal component analysis (PCA) was conducted to determine the quality of the microarray hybridization and visualize the general data variation among the chips (Fig 2, [13]).
Fig 1. Box plot of the log2 expression signal for each microarray chip.
Fig 2. Principal component analysis (PCA) of the microarray-derived transcriptomal results for oviducts collected from ESR1KO mice and WT littermates at 23 days of age, or treated with 5 IU PMSG at 23 days of age and collected 48 h later.
Red: ESR1KO, Blue: WT, Green: PMSG-treated ESR1KO, Purple: PMSG-treated WT.
Statistical, Gene Ontology and Pathway Analysis
To detect differentially expressed genes (DEG’s) and the interaction between genotype and PMSG treatment, the normalized and background adjusted microarray data were imported into Partek Genomics suite 6.6 (Partek, Inc.) and a two-way ANOVA performed with factor 1 being genotype and factor 2 being PMSG treatment. Statistical significance of difference for each gene was set to P-value <0.01 with Benjamini-Hochberg multiple testing correction for false discovery rate (FDR) < 0.13. Genes considered significant in the overall model (P < 0.01, FDR < 0.13) were then subjected to pairwise comparisons using Fischer’s Least Significant Difference (LSD) test to estimate the significance of difference for each gene in each comparison. Genes showing a mean difference in signal intensity of at least 2-fold change and a P-value < 0.01 were considered differentially expressed.
With our primary objective of identifying estradiol/ESR1-dependent affects on the transcriptome, the 1185 DEG’s identified in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice was subjected to gene ontology and pathway analysis. Differentially expressed genes were interrogated for their gene ontology (GO) classes using Partek Genomics Suite 6.6 (Partek, Inc.). Partek derives gene ontology classifications from geneontology.org and/or the affymetrix database. GO hierarchies leads to the division of the gene list into significant classifications when the observed number of differentially expressed genes in a GO category is greater than expected. Statistical analysis for significant classifications was performed using Fischer’s exact test, right-tailed. A P-value < 0.01 is suggestive of an over representation of genes from within a particular GO category, indicative of a functional effect.
These same DEG’s (PMSG-treated ESR1KO versus PMSG-treated WT) were then subjected to Ingenuity Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity) which uses multiple databases to extrapolate significant pathways based on the number of significant genes within our list and known to be involved in a particular pathway. To determine significant pathways in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT treated mice, a Fischer’s exact test was performed with significance set to P-value < 0.05.
Real-time RT-PCR
Pathway analysis revealed that the most significant pathways were reflective of immune responses. Therefore, to validate the microarray analysis, real-time RT-PCR was performed to quantify the level of expression of a selection of immune-regulating mRNAs: chemokine (C-C motif) ligand 5, Ccl5; cytochrome P450, family 26, subfamily A, polypeptide 1, Cyp26a; hematopoetic prostaglandin D synthase, Hpgds; interleukin 18 receptor accessory protein, Il18rap; prostaglandin-endoperoxide synthase 2, Ptgs2; lecithin retinol acyltransferase (phosphatidylcholine-retinol O-acyltransferase), Lrat; S100 calcium binding protein A8, S100a8; and uroplakin 1A, Upk1a. Real-time RT-PCR was performed using an Eppendorf Mastercycler ep realplex2 system (Eppendorf) using iQ SYBR Green Supermix (Bio-RAD, Hercules, CA), as described before [15,17].
Briefly, cDNA was synthesized using the SuperScript III 1st Strand Synthesis System (Invitrogen), with 0.5 μg of RNA used for each reverse transcription reaction. Real-time RT-PCR was performed with a total volume of 25 μl per reaction, with each reaction containing 5 μL of cDNA, 1 μL of a 10 μm stock of each primer (forward and reverse), 12.5 μL of 2× SYBR Green PCR Master Mix, and 5.5 μL of nuclease-free water. Gene expression was analyzed by the 2−ΔΔCT method [19]. The typical dissociation curves of these cDNA, plus Gapdh as the housekeeping gene was confirmed. Oligonucleotide primer pairs (Integrated DNA technologies, Coralville, IA) are described in Table 1.
Table 1. Primer sequences (forward and reverse) and PCR product sizes used for real-time RT-PCR analyses.
| Name | Accession # | Primer sequence (5'–3') | Product size |
|---|---|---|---|
| Ccl5 | NM_013653.3 | F: CCT CAC CAT ATG GCT CGG AC | 121 |
| R: ACG ACT GCA AGA TTG GAG CA | |||
| Cyp26a1 | NM_007811.2 | F: AGC TCC TGA TTG AGC ACT CG | 292 |
| R: GGA GGA TTC AAT CGC AGG GT | |||
| Hpgds | NM_019455.4 | F: CAC TAG TTT CCT GGC TAG GGT | 383 |
| R: TGT CAC AGC TCC TTT CCT TGT | |||
| Il18rap | NM_010553.3 | F: TGC AAT GAA GCG GCA TCT GT | 133 |
| R: CCG GTG ATT CTG TTC AGG CT | |||
| Lrat | NM_023624.4 | F: GTC GCC CAT CTA ATG CCT GA | 324 |
| R: CTG TGG ACT GAT CCG AGA GC | |||
| Ptgs2 | NM_011198.4 | F: CAT CCC CTT CCT GCG AAG TT | 178 |
| R: CAT GGG AGT TGG GCA GTC AT | |||
| S100a8 | NM_013650.2 | F: CTT TCG TGA CAA TGC CGT CTG | 99 |
| R: AGA GGG CAT GGT GAT TTC CT | |||
| Upk1a | NM_026815.2 | F: TGA GCA AGA GTG TTG TGG CA | 240 |
| R: CAC GAT ATG CCC CAC GTG TA | |||
| Gapdh | GU214026.1 | F: CCC CCA ATG TGT CCG TCG TGG | 201 |
| R: TGA GAG CAA TGC CAG CCC CG |
For statistical analysis of real-time RT-PCR results, datasets were first tested for normality and equal variance. When appropriate, data were transformed before statistical analysis. A one-way ANOVA using SigmaStat 3.5 (Systat Software, Inc., Point Richmond, CA, USA) was used to determine differences in levels of mRNA. When differences were detected a Fischer’s Least Significant Difference (LSD) test was used to determine which genes differed.
Results
Detection of DEG’s by Microarray Analysis
After chip normalization, a statistical two-way ANOVA and pairwise comparison (LSD test) was performed to generate a list of 2428 differentially expressed genes (P < 0.01, FDR < 0.13). Genotype affected the expression of 2215 genes, PMSG affected the expression of 465 genes, and Genotype x PMSG affected the expression of 438 genes (Table 2). Following removal of unannotated and duplicate probesets, DEG’s were further subdivided between up- and down-regulated genes. The identity of the 20 most highly up- and down-regulated genes in the oviducts of ESR1KO versus WT mice, and PMSG-treated ESR1KO versus PMSG-treated WT mice are provided in Tables 3–6. The identity of all genes determined to be differentially expressed by two-way ANOVA is provided in S1 Table.
Table 2. Number of differentially expressed genes (DEG's) identified by microarray analysis and pair-wise comparisons between genotypes and treatments.
| Parameter | No. of DEG's | ||
|---|---|---|---|
| Model | 2428 | ||
| Genotype | 2215 | ||
| PMSG treatment | 465 | ||
| Genotype by PMSG interaction | 438 | ||
| Pairwise comparisons | No. of DEG's | Up-regulated | Down-regulated |
| PMSG-treated ESR1KO vs. ESR1KO | 37 | 31 (84%) | 6 (16%) |
| PMSG-treated WT vs. WT | 318 | 164 (52%) | 154 (48%) |
| PMSG-treated ESR1KO vs. PMSG-treated WT | 1185 | 689 (58%) | 496 (42%) |
| ESR1KO vs. WT | 664 | 328 (49%) | 336 (51%) |
Significance set to P-value < 0.01 with FDR determined from the Benjamini-Hochberg multiple testing correction < 0.13. For pairwise comparisons, unannotated and duplicate probe sets were removed from gene lists, and only genes with at least a 2-fold change in level of expression were considered differentially expressed.
Table 3. Top 20 most highly up-regulated mRNAs in the oviducts of ESR1KO versus WT mice.
Overall Model: P < 0.01 and at least a 2-fold change in gene expression.
| Gene Symbol | Gene Description | P-value | Fold-Change |
|---|---|---|---|
| Sult1e1 | sulfotransferase family 1E, member 1 | < 0.001 | 33.1834 |
| Chodl | chondrolectin | < 0.001 | 25.6824 |
| Avpr1a | arginine vasopressin receptor 1A | < 0.001 | 24.7639 |
| Synpr | synaptoporin | < 0.001 | 22.5126 |
| Glb1l3 | galactosidase, beta 1 like 3 | < 0.001 | 21.5821 |
| BC048679 | cDNA sequence BC048679 | < 0.001 | 21.0171 |
| Ager | advanced glycosylation end product-specific receptor | < 0.001 | 17.8374 |
| 2310043J07Rik | RIKEN cDNA 2310043J07 gene | < 0.001 | 15.0383 |
| Pcdh8 | protocadherin 8 | < 0.001 | 14.7291 |
| Lemd1 | LEM domain containing 1 | < 0.001 | 14.6033 |
| Slc47a1 | solute carrier family 47, member 1 | < 0.001 | 14.215 |
| Adamts16 | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 mo | < 0.001 | 13.9153 |
| Tnfrsf21 | tumor necrosis factor receptor superfamily, member 21 | < 0.001 | 12.0567 |
| 9330159F19Rik | RIKEN cDNA 9330159F19 gene | < 0.001 | 11.991 |
| S100a8 | S100 calcium binding protein A8 (calgranulin A) | 0.002 | 11.9115 |
| Mmp7 | matrix metallopeptidase 7 | < 0.001 | 11.8858 |
| Kcnd2 | potassium voltage-gated channel, Shal-related family, member 2 | < 0.001 | 11.8108 |
| AA986860 | expressed sequence AA986860 | < 0.001 | 11.5917 |
| Cdh16 | cadherin 16 | < 0.001 | 11.5566 |
| Trank1 | tetratricopeptide repeat and ankyrin repeat containing 1 | < 0.001 | 11.4235 |
Fold-Change in gene expression and P-Values are indicated. Positive changes in fold-change represent increased expression in the oviducts of ESR1KO mice.
Table 6. Top 20 most highly down-regulated mRNAs in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice.
Overall Model: P < 0.01 and at least a 2 fold-change in gene expression.
| Gene Symbol | Gene Description | P-value | Fold-Change |
|---|---|---|---|
| Dcpp3 | demilune cell and parotid protein 3 | <0.001 | -770.92 |
| 2300002M23Rik | RIKEN cDNA 2300002M23 gene | <0.001 | -524.94 |
| Cyp26a1 | cytochrome P450, family 26, subfamily a, polypeptide 1 | <0.001 | -131.13 |
| Tshr | thyroid stimulating hormone receptor | <0.001 | -121.71 |
| Dcpp1/2/3 | demilune cell and parotid protein 1/demilune cell and parotid protein 2/demilune cell and parotid protein 3 | <0.001 | -106.94 |
| Syn2 | synapsin II | <0.001 | -91.45 |
| Slc6a2 | solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 | <0.001 | -77.2 |
| Upk1a | uroplakin 1A | <0.001 | -77.14 |
| Hpgds | hematopoietic prostaglandin D synthase | <0.001 | -73.37 |
| Klk1b24 | kallikrein 1-related peptidase b24 | <0.001 | -73.19 |
| Greb1 | gene regulated by estrogen in breast cancer protein | <0.001 | -64.6 |
| Klk1b1 | kallikrein 1-related peptidase b1 | <0.001 | -62.94 |
| Klk1b21 | kallikrein 1-related peptidase b21 | <0.001 | -53.58 |
| Lrat | lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) | <0.001 | -53.53 |
| Gria1 | glutamate receptor, ionotropic, AMPA1 (alpha 1) | <0.001 | -46.45 |
| Akr1c14 | aldo-keto reductase family 1, member C14 | <0.001 | -45.65 |
| Stat5a | signal transducer and activator of transcription 5A | <0.001 | -44.22 |
| Col6a4 | collagen, type VI, alpha 4 | <0.001 | -43.1 |
| Rasd1 | RAS, dexamethasone-induced 1 | <0.001 | -42.74 |
| Adh7 | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | <0.001 | -42.46 |
Fold-Change in gene expression and P-Values are indicated. Negative changes in fold-change represent decreased expression in the oviducts of PMSG-treated ESR1KO mice.
Table 4. Top 20 most highly down-regulated mRNAs in the oviducts of ESR1KO versus WT mice.
Overall Model: P < 0.01 and at least a 2-fold change in gene expression.
| Gene Symbol | Gene Description | P-value | Fold-Change |
|---|---|---|---|
| Pcdh17 | protocadherin 17 | < 0.001 | -10.6901 |
| Csf3 | colony stimulating factor 3 (granulocyte) | 0.002 | -11.5917 |
| Tshr | thyroid stimulating hormone receptor | < 0.001 | -11.8455 |
| Col6a4 | collagen, type VI, alpha 4 | < 0.001 | -12.206 |
| Akr1c14 | aldo-keto reductase family 1, member C14 | < 0.001 | -12.2614 |
| Upk1a | uroplakin 1A | 0.001 | -12.9078 |
| Slc6a2 | solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 | < 0.001 | -13.1394 |
| Lrat | lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) | < 0.001 | -13.3562 |
| Ano4 | anoctamin 4 | 0.002 | -13.6892 |
| Gp1bb | glycoprotein Ib, beta polypeptide | < 0.001 | -15.0412 |
| Stat5a | signal transducer and activator of transcription 5A | < 0.001 | -16.1053 |
| Rtn1 | reticulon 1 | < 0.001 | -18.0311 |
| Syn2 | synapsin II | < 0.001 | -23.8067 |
| Ramp3 | receptor (calcitonin) activity modifying protein 3 | < 0.001 | -24.6301 |
| Mlc1 | megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) | < 0.001 | -29.3425 |
| Hpgds | hematopoietic prostaglandin D synthase | 0.001 | -32.2313 |
| Cyp26a1 | cytochrome P450, family 26, subfamily a, polypeptide 1 | 0.001 | -39.5376 |
| Dcpp1 /// Dcpp2 /// Dcpp3 | demilune cell and parotid protein 1 /// demilune cell and parotid protein 2 /// demilune cell and parotid protein 3 | < 0.001 | -47.7492 |
| 2300002M23Rik | RIKEN cDNA 2300002M23 gene | 0.003 | -71.6374 |
| Dcpp3 | demilune cell and parotid protein 3 | < 0.001 | -77.6391 |
Fold-Change in gene expression and P-Values are indicated. Negative changes in fold-change represent decreased expression in the oviducts of ESR1KO mice.
Verification of selected DEG’s
The expression of mRNA for Ccl5, Cyp26a1, Hpgds, Il18rap, Lrat, Ptgs2, S100a8, and Upk1a in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice was determined by real-time RT-PCR. A comparison of the results obtained by real-time RT-PCR and microarray analysis is presented in Table 7 as a validation of the microarray platform. Overall, real-time RT-PCR revealed the same directional trends in gene expression that were observed by the microarray analysis.
Table 7. Comparison of gene expression for selected mRNAs by microarray and real-time RT-PCR in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice.
| Microarray | Real-time RT-PCR | |||
|---|---|---|---|---|
| Gene Symbol | Fold-Change | P-Value | Fold-Change | P-Value |
| Ccl5 | 2 | 0.166 | 4.2 | < 0.001 |
| Cyp26a1 | -131.1 | < 0.001 | -27.4 | < 0.001 |
| Hpgds | -73.4 | < 0.001 | -13.4 | < 0.001 |
| Il18rap | 1 | 0.22 | 6.2 | < 0.001 |
| Lrat | -53.5 | < 0.001 | -21.8 | < 0.001 |
| Ptgs2 | 1.57 | 0.24 | 2.19 | < 0.001 |
| S100a8 | 20.7 | 0.0006 | 18.02 | < 0.001 |
| Upk1a | -77.1 | < 0.001 | -38.9 | < 0.001 |
Fold-Change in gene expression and P-Values are indicated after analysis by microarray and by independent real-time RT-PCR. Positive changes in fold change represent increased expression in the oviducts of PMSG-treated ESR1KO mice.
Gene Ontology Analysis of DEG’s in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice
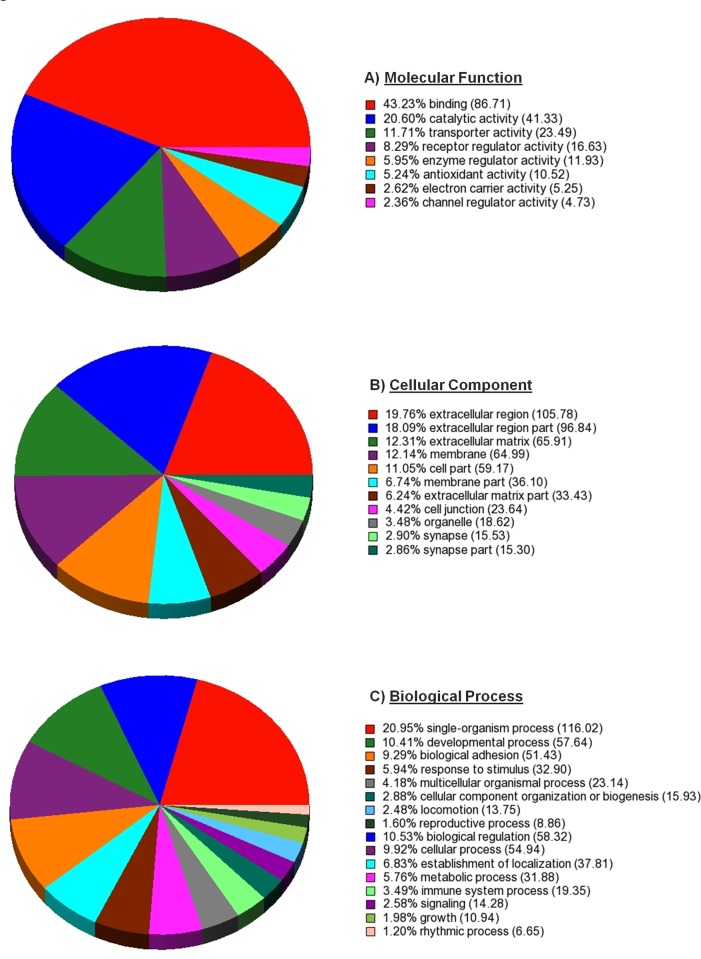
Consistent with our overall goal of identifying estradiol/ESR1-dependent affects on the oviduct, the molecular functions, cellular components, and biological processes of DEG’s expressed in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice were determined by Gene Ontology (GO) Analyses with significance set to enrichment P-value < 0.01. The significantly enriched molecular function categories using GO are shown in Fig 3A. The categories with the highest enrichment score within molecular functions were binding, catalytic and transporter activities. Significantly enriched cellular component categories are shown in Fig 3B. The most highly enriched cellular component categories were the extracellular region/matrix and the cell membrane. Significantly enriched biological processes are indicated in Fig 3C, with the most highly enriched categories including those involving a single organism, modulating a measurable attribute (biological regulation) and specific outcome (developmental process).
Fig 3. Gene Ontology (GO) analysis with Molecular Function, Cellular Component and Biological Processes categories.
Pie chart shows the distribution of the DEG’s in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice that were matched to A) a Molecular Function, B) a Cellular Component, and C) a Biological Process, using GO.
Ingenuity Pathway Analysis of DEG’s in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice
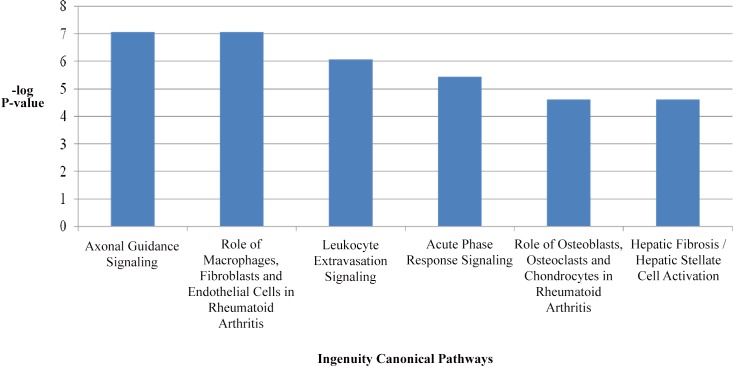
Canonical pathway analysis of DEG’s from PMSG-treated ESR1KO versus PMSG-treated WT mice was performed using QIAGEN’S Ingenuity Pathway Analysis (IPA, QIAGEN, Redwood City, www.qiagen.com/ingenuity). The six most significant pathways identified by Ingenuity Pathway Analysis software are provided in Fig 4, and are reflective of ESR1-dependent regulation of the immune response. The top upstream regulators were tumor necrosis factor (TNF), interferon gamma (IFNG), interleukin 1β (IL1B), amyloid β (A4) precursor protein (APP) and interleukin 13 (IL13). The top regulator effect networks included a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 (ADAMTS12), homeodomain interacting protein kinase 2 (HIPK2), interleukin 22 (IL22), interleukin 27 (IL27), toll-like receptor 3 (TLR3), toll-like receptor 4 (TLR4) and conserved helix-loop-helix ubiquitous kinase (CHUK) as their primary regulators. With pathway analysis indicating the immune response as a primary canonical pathway, a listing of differentially expressed mRNAs specifically encoding chemokines, interleukins and their receptors in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice is provided as Table 8. All the significant pathways (P-value < 0.05) and the corresponding molecules differentially expressed within the pathways from PMSG-treated ESR1KO versus PMSG-treated WT mice are provided in S2 Table.
Fig 4. Most highly significant canonical pathways identified in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT identified using QIAGEN’S Ingenuity Pathway Analysis.
Table 8. Differentially expressed mRNAs encoding chemokines, interleukins and their receptors in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice.
Overall Model: P < 0.01 and at least a 2-fold change in gene expression.
| Gene Symbol | Gene Description | P-value | Fold-Change |
|---|---|---|---|
| Cx3cl1 | chemokine (C-X3-C motif) ligand 1 | 0.004 | 2.25 |
| Cxcl12 | chemokine (C-X-C motif) ligand 12 | < 0.001 | 4.21 |
| Cxcl14 | chemokine (C-X-C motif) ligand 14 | 0.001 | -3.31 |
| Cxcl16 | chemokine (C-X-C motif) ligand 16 | 0.011 | 1.73 |
| Cxcl17 | chemokine (C-X-C motif) ligand 17 | 0.003 | 3.68 |
| Cxcr4 | chemokine (C-X-C motif) receptor 4 | 0.063 | 1.61 |
| Cxcr7 | chemokine (C-X-C motif) receptor 7 | < 0.001 | -3.32 |
| Il13ra2 | interleukin 13 receptor, alpha 2 | 0.003 | 4.48 |
| Il15 | interleukin 15 | < 0.001 | 2.70 |
| Il15ra | interleukin 15 receptor, alpha chain | < 0.001 | 5.16 |
| Il16 | interleukin 16 | 0.002 | 1.62 |
| Il17ra | interleukin 17 receptor A | < 0.001 | -2.25 |
| Il17rb | interleukin 17 receptor B | < 0.001 | 14.38 |
| Il17re | interleukin 17 receptor E | 0.002 | 3.00 |
| Il18 | interleukin 18 | 0.002 | 1.82 |
| Il18bp | interleukin 18 binding protein | < 0.001 | -20.01 |
| Il18r1 | interleukin 18 receptor 1 | < 0.001 | 19.45 |
| Il1r1 | interleukin 1 receptor, type I | 0.001 | 2.14 |
| Il1rap | interleukin 1 receptor accessory protein | < 0.001 | 1.53 |
| Il33 | interleukin 33 | < 0.001 | 11.69 |
| Il7 | interleukin 7 | 0.001 | 3.94 |
| Ilf2 | interleukin enhancer binding factor 2 | 0.012 | 1.31 |
Fold-Change in gene expression and P-Values are indicated. Positive changes in fold-change represent increased expression in the oviducts of PMSG-treated ESR1KO mice.
Discussion
The objective of this study was to determine estradiol/ESR1-dependent changes to the transcriptome of the mouse oviduct, with the overall goals of increasing our understanding of steroidal regulation of this often overlooked reproductive organ, and to provide the identity of ESR1-regulated genes that may prove to be important modulators of oviductal function and fertility in the future. While our focus was on the identification and bioinformatic analysis of DEG’s in the oviducts of PMSG-treated WT versus PMSG-treated ESR1KO mice, the identity of all DEG’s identified by this analysis have been provided (S1 Table), and the raw data (*.cel files) plus the GC-RMA-normalized and log2 transformed transcript data have been deposited into the Gene Expression Omnibus. Important to note, whole oviducts were collected for transcriptomal analysis from WT and ESR1KO mice. Future research of targeted mRNAs identified by this analysis will therefore need to include determination of potential differences in spatial location of a gene or protein between the ampulla and isthmus, as well as cellular localization within a specific section of the oviduct. Furthermore, this study was performed to determine estradiol/ESR1-dependent regulation; genomic signaling via ESR2 and non-genomic effects of estradiol on the oviduct via activation of G-protein-coupled receptor 30 (GPR30, [20]) should not be overlooked, nor potential interactions. Indeed, regardless of treatment with PMSG, we observed that ablation of ESR1 resulted in a 1.6-fold increase in the expression of mRNA for Gpr30 in the oviduct (S1 Table).
ESR1-dependent regulation of immune function was a leading canonical pathway identified. Of note, inflammation and the immune response is a required physiological occurrence within the oviduct as this organ is exposed to freshly ovulated cumulus-oocyte complexes, associated follicular debris, spermatozoa, seminal fluids and possibly an array of foreign pathogens at ovulation and/or mating [21,22,23,24]. However, salpingitis or aberrant inflammation is also one of the most common forms of pelvic inflammatory disease (PID) and is one of the most important components of the PID spectrum due to its impact on female fertility (reviewed in [25]). This uncontrolled inflammation results in oviductal epithelial cell death, tubal scarring and eventually occlusion [26,27,28,29,30], making identification of the specific transcripts involved in ESR1-dependent regulation of immune function a salient finding of this transcriptomal analysis.
Overall, with significance set to P < 0.01, greater than two thousand transcripts were determined to be differentially regulated. A pairwise comparison of DEG’s in 23 day old ESR1KO and WT mice (i.e. without PMSG-stimulated production of ovarian estradiol) revealed 664 DEG’s, and a pairwise comparison of DEG’s in PMSG-treated WT versus PMSG-treated ESR1KO oviducts revealed 1185 differentially regulated genes, which were subsequently analyzed for gene ontology as well as with Ingenuity Pathway Analysis (IPA®, QIAGEN), which uses multiple databases to extrapolate significant canonical pathways based on the number of genes expected to be expressed within each pathway. With the exception of axonal guidance signaling (reviewed in [31]), the other top canonical pathways (Fig 4) were all directly related to immune function, as were the 5 top upstream regulators (TNF, IFNG, IL1B, APP and IL13).
We have previously reported that the expression of the hematopoetic form of prostaglandin D synthase (HPGDS), a putative regulator of inflammation in the oviduct, is dependent upon ESR1 [15]. In that study, genetic deletion of ESR1 reduced the expression of mRNA encoding Hpgds and inhibition of HPGDS in wild-type mice by treatment with HQL-79 (Cayman Chemical, Ann Arbor, MI) resulted in a 2.3-fold increase in the expression of mRNA for one of the upstream regulators identified herein (IL13), a 2.9-fold increase in the expression of mRNA for chemokine (C-X-C motif) ligand 12 (Cxcl12), as well as a 1.8-fold increase in the expression of mRNA for TNF receptor superfamily, member 1b (Tnfrsf1b) which is also known as TNF receptor 2 (TNFR2), one of the two receptors that bind TNFα. Herein, the expression of mRNA for Cxcl12 and Tnfrsf1b was 4.2- and 2.5-fold higher in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice (Table 8 and S1 Table). Taken together, our results are consistent with regulation of inflammation within the oviduct acting, in part, through ESR1-dependent HPGDS signaling. Of physiological relevance, the expression of TNFα is reported to increase after infection of human oviducts with Neisseria gonorrhoeae in vitro [32] and genetic deletion of IL13 in mice improves the rate of clearance after genital infection with Chlamydia muridarum [33], two bacterial pathogens known to induce an inflammatory response within the oviduct [34,35]. Interestingly, the expression of mRNA encoding IL13 receptor, alpha 2 (Il13ra2), but not Il13, was increased in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice (Table 8).
Of the mRNAs selected for independent analysis by real-time RT-PCR, directional trends were consistent among microarray and RT-PCR analyses (Table 7). Analysis by real-time RT-PCR also revealed that the expression of Ptgs2 and Il18rap was increased in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice. Importantly, for these two transcripts, microarray analysis revealed the same directional trend and a similar magnitude or fold-change in expression. Estradiol is a known regulator of PTGS2 in the oviduct [36], and IL18RAP together with the receptor IL18R1 (interleukin 18 receptor 1) mediates IL18-dependent activation [37,38]. IL18 is a Caspase-1-dependent inflammatory cytokine induced by infection with C. trachomatis [39]. We observed a 19-fold increase in the expression of Il18r1 in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice (Tables 5–8), which is also consistent with the regulation of the IL18 receptor by estradiol, as reported in the uterine endometrium [40]. Among the other DEG’s encoding interleukins, mRNA encoding IL17 receptors A, B and E (Il17ra, Il17rb and Il17re) as well as interleukin 1 receptor, type 1 (Il1r1) and interleukin 1 receptor accessory protein (Il1rap) differed in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice (Table 8). Mice deficient in IL17 display an attenuated response to genital infection with C. muridarium [41], whereas IL1 is an established regulator of C. trachomatis-induced inflammation in the oviduct [42]. Our results therefore revealing estradiol/ESR1-dependent changes within the oviduct of transcripts reported to affect inflammation in response to targeted bacterial challenges by others.
Table 5. Top 20 most highly up-regulated mRNAs in the oviducts of PMSG-treated ESR1KO versus PMSG-treated WT mice.
Overall Model: P < 0.01 and at least a 2 fold-change in gene expression.
| Gene Symbol | Gene Description | P-value | Fold-Change |
|---|---|---|---|
| BC048679 | cDNA sequence BC048679 | <0.001 | 222.15 |
| Apod | apolipoprotein D | <0.001 | 98.02 |
| Cdh16 | cadherin 16 | <0.001 | 50.13 |
| Chodl | Chondrolectin | <0.001 | 48.14 |
| Sult1e1 | sulfotransferase family 1E, member 1 | <0.001 | 46.32 |
| G6pc2 | glucose-6-phosphatase, catalytic, 2 | <0.001 | 42.88 |
| Pla2g10 | phospholipase A2, group X | <0.001 | 40.8 |
| Fgf18 | fibroblast growth factor 18 | <0.001 | 38.17 |
| Avpr1a | arginine vasopressin receptor 1A | <0.001 | 35.78 |
| Aldh1a3 | aldehyde dehydrogenase family 1, subfamily A3 | <0.001 | 34.76 |
| Lrrtm1 | leucine rich repeat transmembrane neuronal 1 | <0.001 | 29.44 |
| Serpina1b | serine (or cysteine) preptidase inhibitor, clade A, member 1B | <0.001 | 27.57 |
| Ager | advanced glycosylation end product-specific receptor | <0.001 | 22.77 |
| Synpr | Synaptoporin | <0.001 | 22.32 |
| S100a8 | S100 calcium binding protein A8 (calgranulin A) | <0.001 | 20.77 |
| Adamts16 | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif 16 | <0.001 | 19.82 |
| Il18r1 | interleukin 18 receptor 1 | <0.001 | 19.45 |
| S100a9 | S100 calcium binding protein A9 (calgranulin B) | 0.0015 | 17.86 |
| Atp6v1b1 | ATPase, H+ transporting, lysosomal V1 subunit B1 | <0.001 | 17.8 |
| Wnt7a | wingless-related MMTV integration site 7A | <0.001 | 17.49 |
Fold-Change in gene expression and P-Values are indicated. Positive changes in fold-change represent increased expression in the oviducts of PMSG-treated ESR1KO mice.
Differences in the magnitude of change were observed for some transcripts by microarray analysis versus real-time RT-PCR, as expected [43], whereas technique did not affect the magnitude of change for others. For example, microarray analysis revealed that the S100 calcium binding proteins A8 and A9 were increased by 21-and 18-fold, respectively, in PMSG-treated ESR1KO versus PMSG-treated WT oviducts (Table 5). Real-time RT-PCR confirmed the increased expression of S100A8, with the relative expression for this mRNA increased by 18-fold in the PMSG-treated ESR1KO oviduct (Table 7). The S100 calcium binding proteins are pro-inflammatory, inducing chemotaxis and adhesion of neutrophils [44] and increasing IL1β secretion by IFNG-primed monocytes [45]. Again, consistency with targeted bacterial studies is observed; IL1 signaling and IL1β secretion are involved in the response of the oviduct to infection with C. muridarium [46].
To conclude, this transcriptomal analysis can provide us with great insight into estradiol/ESR1-dependent regulation of oviductal gene expression and presumably function. Pathway analysis illustrated the complex role of estradiol and ESR1 in regulating oviductal function and identified putative ESR1-dependent molecules involved. This dataset can now be examined in greater detail by others with the hope of expanding our understanding of ESR1-dependent regulation of physiological function in this key reproductive organ.
Supporting Information
Gene symbol, gene title and all pair-wise comparisons, P-values and fold changes in expression are indicated.
(XLS)
All the significant pathways (P-value ≤ 0.05) and the corresponding molecules differentially expressed within the pathways from PMSG-treated ESR1KO versus PMSG-treated WT mice are indicated.
(XLS)
Acknowledgments
This work was supported by National Institutes of Health Grants P20 RR15592 (P.B., C.K.), K12 DA014040 (P.B.), and P01 HD071875 (P.B., C.K), the University of Kentucky, and Kentucky Agricultural Experiment Station. The information reported in this paper (publication no. 16-07-008) is part of a project of the Kentucky Agricultural Experiment Station and is published with approval of the Director.
Data Availability
The data (*.cel files) plus the GCRMA-normalized and log2 transformed transcript data (Park Genomics Suite), have been deposited into the Gene Expression Omnibus (National Center for Biotechnology Information) as accession number GSE72614 (http://www.ncbi.nlm.nih.gov/geo).
Funding Statement
This work was supported by National Institutes of Health Grants P20 RR15592 (P.B., C.K.), K12 DA014040 (P.B.), and P01 HD071875 (P.B., C.K.), the University of Kentucky, and Kentucky Agricultural Experiment Station.
References
- 1.Huang N, Liu B, Dong Z, Mao W, Zhang N, Li C, et al. (2015) Prostanoid receptors EP, EP, and FP are regulated by estradiol in bovine oviductal smooth muscle. Prostaglandins Other Lipid Mediat 121(Pt B): 170–175. 10.1016/j.prostaglandins.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 2.Wijayagunawardane MP, Choi YH, Miyamoto A, Kamishita H, Fujimoto S, Takagi M, et al. (1999) Effect of ovarian steroids and oxytocin on the production of prostaglandin E2, prostaglandin F2alpha and endothelin-1 from cow oviductal epithelial cell monolayers in vitro. Anim Reprod Sci 56: 11–17. [DOI] [PubMed] [Google Scholar]
- 3.Dong Z, Zhang N, Mao W, Liu B, Huang N, Li P, et al. (2015) Kinetic effect of oestrogen on secretion of prostaglandins E2 and F2? in bovine oviduct epithelial cells. Reprod Fertil Dev 10.1071/RD15246. [DOI] [PubMed] [Google Scholar]
- 4.Verhage HG, Fazleabas AT, Mavrogianis PA, O'Day-Bowman MB, Donnelly KM, Arias EB, et al. (1997) The baboon oviduct: characteristics of an oestradiol-dependent oviduct-specific glycoprotein. Hum Reprod Update 3: 541–552. [DOI] [PubMed] [Google Scholar]
- 5.Ulbrich SE, Kettler A, Einspanier R (2003) Expression and localization of estrogen receptor alpha, estrogen receptor beta and progesterone receptor in the bovine oviduct in vivo and in vitro. J Steroid Biochem Mol Biol 84: 279–289. [DOI] [PubMed] [Google Scholar]
- 6.Shao R, Egecioglu E, Weijdegard B, Kopchick JJ, Fernandez-Rodriguez J, Andersson N, et al. (2007) Dynamic regulation of estrogen receptor-alpha isoform expression in the mouse fallopian tube: mechanistic insight into estrogen-dependent production and secretion of insulin-like growth factors. Am J Physiol Endocrinol Metab 293: E1430–1442. [DOI] [PubMed] [Google Scholar]
- 7.Okada A, Ohta Y, Inoue S, Hiroi H, Muramatsu M, Iguchi T (2003) Expression of estrogen, progesterone and androgen receptors in the oviduct of developing, cycling and pre-implantation rats. J Mol Endocrinol 30: 301–315. [DOI] [PubMed] [Google Scholar]
- 8.Orihuela PA, Zuniga LM, Rios M, Parada-Bustamante A, Sierralta WD, Velasquez LA, et al. (2009) Mating changes the subcellular distribution and the functionality of estrogen receptors in the rat oviduct. Reprod Biol Endocrinol 7: 139 10.1186/1477-7827-7-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tremblay GB, Tremblay A, Copeland NG, Gilbert DJ, Jenkins NA, Labrie F, et al. (1997) Cloning, chromosomal localization, and functional analysis of the murine estrogen receptor beta. Mol Endocrinol 11: 353–365. [DOI] [PubMed] [Google Scholar]
- 10.Walter P, Green S, Greene G, Krust A, Bornert JM, Jeltsch JM, et al. (1985) Cloning of the human estrogen receptor cDNA. Proc Natl Acad Sci U S A 82: 7889–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon D, Couse JF, Korach KS (1997) Disruption of the estrogen receptor gene in mice. Toxicol Pathol 25: 518–520. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton KJ, Arao Y, Korach KS (2014) Estrogen hormone physiology: reproductive findings from estrogen receptor mutant mice. Reprod Biol 14: 3–8. 10.1016/j.repbio.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Partek D (2009) Partek documentation: turning data into discovery Partek Incorporated, St Louis. [Google Scholar]
- 14.Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridges PJ, Jeoung M, Shim S, Park JY, Lee JE, Sapsford LA, et al. (2012) Hematopoetic prostaglandin D synthase: an ESR1-dependent oviductal epithelial cell synthase. Endocrinology 153: 1925–1935. 10.1210/en.2011-1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gieske MC, Kim HJ, Legan SJ, Koo Y, Krust A, Chambon P, et al. (2008) Pituitary gonadotroph estrogen receptor-alpha is necessary for fertility in females. Endocrinology 149: 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeoung M, Lee S, Hawng HK, Cheon YP, Jeong YK, Gye MC, et al. (2010) Identification of a novel role for endothelins within the oviduct. Endocrinology 151: 2858–2867. 10.1210/en.2009-1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bridges PJ, Jeoung M, Kim H, Kim JH, Lee DR, Ko C, et al. (2011) Methodology matters: IVF versus ICSI and embryonic gene expression. Reprod Biomed Online 23: 234–244. 10.1016/j.rbmo.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 20.Popli P, Sirohi VK, Manohar M, Shukla V, Kaushal JB, Gupta K, et al. (2015) Regulation of cyclooxygenase-2 expression in rat oviductal epithelial cells: Evidence for involvement of GPR30/Src kinase-mediated EGFR signaling. J Steroid Biochem Mol Biol 154: 130–141. 10.1016/j.jsbmb.2015.07.019 [DOI] [PubMed] [Google Scholar]
- 21.Kowsar R, Hambruch N, Marey MA, Liu J, Shimizu T, Pfarrer C, et al. (2014) Evidence for a novel, local acute-phase response in the bovine oviduct: progesterone and lipopolysaccharide up-regulate alpha 1-acid-glycoprotein expression in epithelial cells in vitro. Mol Reprod Dev 81: 861–870. 10.1002/mrd.22355 [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Marey MA, Kowsar R, Hambruch N, Shimizu T, Haneda S, et al. (2014) An acute-phase protein as a regulator of sperm survival in the bovine oviduct: alpha 1-acid-glycoprotein impairs neutrophil phagocytosis of sperm in vitro. J Reprod Dev 60: 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marey MA, Liu J, Kowsar R, Haneda S, Matsui M, Sasaki M, et al. (2014) Bovine oviduct epithelial cells downregulate phagocytosis of sperm by neutrophils: prostaglandin E2 as a major physiological regulator. Reproduction 147: 211–219. 10.1530/REP-13-0375 [DOI] [PubMed] [Google Scholar]
- 24.Kowsar R, Hambruch N, Liu J, Shimizu T, Pfarrer C, Miyamoto A (2013) Regulation of innate immune function in bovine oviduct epithelial cells in culture: the homeostatic role of epithelial cells in balancing Th1/Th2 response. J Reprod Dev 59: 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sweet RL (2011) Treatment of acute pelvic inflammatory disease. Infect Dis Obstet Gynecol 2011: 561909 10.1155/2011/561909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perfettini JL, Darville T, Gachelin G, Souque P, Huerre M, Dautry-Varsat A, et al. (2000) Effect of Chlamydia trachomatis infection and subsequent tumor necrosis factor alpha secretion on apoptosis in the murine genital tract. Infect Immun 68: 2237–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Ulloa AC, Arrieta O (2005) Tubal occlusion causing infertility due to an excessive inflammatory response in patients with predisposition for keloid formation. Med Hypotheses 65: 908–914. [DOI] [PubMed] [Google Scholar]
- 28.Steffl M, Schweiger M, Sugiyama T, Amselgruber WM (2008) Review of apoptotic and non-apoptotic events in non-ciliated cells of the mammalian oviduct. Ann Anat 190: 46–52. 10.1016/j.aanat.2007.04.003 [DOI] [PubMed] [Google Scholar]
- 29.Donnez J, Casanas-Roux F (1988) Histology: a prognostic factor in proximal tubal occlusion. Eur J Obstet Gynecol Reprod Biol 29: 33–38. [DOI] [PubMed] [Google Scholar]
- 30.Punnonen R, Soderstrom KO, Alanen A (1984) Isthmic tubal occlusion: etiology and histology. Acta Eur Fertil 15: 39–42. [PubMed] [Google Scholar]
- 31.Bashaw GJ, Klein R (2010) Signaling from axon guidance receptors. Cold Spring Harb Perspect Biol 2: a001941 10.1101/cshperspect.a001941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maisey K, Nardocci G, Imarai M, Cardenas H, Rios M, Croxatto HB, et al. (2003) Expression of proinflammatory cytokines and receptors by human fallopian tubes in organ culture following challenge with Neisseria gonorrhoeae. Infect Immun 71: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asquith KL, Horvat JC, Kaiko GE, Carey AJ, Beagley KW, Hansbro PM, et al. (2011) Interleukin-13 promotes susceptibility to chlamydial infection of the respiratory and genital tracts. PLoS Pathog 7: e1001339 10.1371/journal.ppat.1001339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briceag I, Costache A, Purcarea VL, Cergan R, Dumitru M, Sajin M, et al. (2015) Fallopian tubes—literature review of anatomy and etiology in female infertility. J Med Life 8: 129–131. [PMC free article] [PubMed] [Google Scholar]
- 35.Maxion HK, Liu W, Chang MH, Kelly KA (2004) The infecting dose of Chlamydia muridarum modulates the innate immune response and ascending infection. Infect Immun 72: 6330–6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez Martinez S, Hermoso M, Farina M, Ribeiro ML, Rapanelli M, Espinosa M, et al. (2006) 17-beta-Estradiol upregulates COX-2 in the rat oviduct. Prostaglandins Other Lipid Mediat 80: 155–164. [DOI] [PubMed] [Google Scholar]
- 37.Hedl M, Zheng S, Abraham C (2014) The IL18RAP region disease polymorphism decreases IL-18RAP/IL-18R1/IL-1R1 expression and signaling through innate receptor-initiated pathways. J Immunol 192: 5924–5932. 10.4049/jimmunol.1302727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boraschi D, Dinarello CA (2006) IL-18 in autoimmunity: review. Eur Cytokine Netw 17: 224–252. [PubMed] [Google Scholar]
- 39.Cheng W, Shivshankar P, Li Z, Chen L, Yeh IT, Zhong G (2008) Caspase-1 contributes to Chlamydia trachomatis-induced upper urogenital tract inflammatory pathologies without affecting the course of infection. Infect Immun 76: 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otsuki M, Kusumoto K, Murakami Y, Kanayama M, Takeuchi S, Takahashi S (2007) Expression of interleukin-18 receptor mRNA in the mouse endometrium. J Reprod Dev 53: 59–68. [DOI] [PubMed] [Google Scholar]
- 41.Andrew DW, Cochrane M, Schripsema JH, Ramsey KH, Dando SJ, O'Meara CP, et al. (2013) The duration of Chlamydia muridarum genital tract infection and associated chronic pathological changes are reduced in IL-17 knockout mice but protection is not increased further by immunization. PLoS One 8: e76664 10.1371/journal.pone.0076664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hvid M, Baczynska A, Deleuran B, Fedder J, Knudsen HJ, Christiansen G, et al. (2007) Interleukin-1 is the initiator of Fallopian tube destruction during Chlamydia trachomatis infection. Cell Microbiol 9: 2795–2803. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Barbacioru C, Hyland F, Xiao W, Hunkapiller KL, Blake J, et al. (2006) Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics 7: 59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA (2003) Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol 170: 3233–3242. [DOI] [PubMed] [Google Scholar]
- 45.Koy M, Hambruch N, Hussen J, Pfarrer C, Seyfert HM, Schuberth HJ (2013) Recombinant bovine S100A8 and A9 enhance IL-1beta secretion of interferon-gamma primed monocytes. Vet Immunol Immunopathol 155: 162–170. 10.1016/j.vetimm.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 46.Nagarajan UM, Sikes JD, Yeruva L, Prantner D (2012) Significant role of IL-1 signaling, but limited role of inflammasome activation, in oviduct pathology during Chlamydia muridarum genital infection. J Immunol 188: 2866–2875. 10.4049/jimmunol.1103461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene symbol, gene title and all pair-wise comparisons, P-values and fold changes in expression are indicated.
(XLS)
All the significant pathways (P-value ≤ 0.05) and the corresponding molecules differentially expressed within the pathways from PMSG-treated ESR1KO versus PMSG-treated WT mice are indicated.
(XLS)
Data Availability Statement
The data (*.cel files) plus the GCRMA-normalized and log2 transformed transcript data (Park Genomics Suite), have been deposited into the Gene Expression Omnibus (National Center for Biotechnology Information) as accession number GSE72614 (http://www.ncbi.nlm.nih.gov/geo).