Abstract
FAS rs2234767 (−1377 G>A), rs1800682 (−670 A>G) and FASLG rs763110 (−844 C>T) promoter polymorphisms can influence transcriptional activities of the genes and thus multiple tumors susceptibility. To investigate their association with risk of colorectal cancer (CRC), the three SNPs were genotyped in 878 cases and 884 controls and the results showed that the FAS rs2234767 and rs1800682 were in a high linkage disequilibrium (LD) with each other (D’ = 0.994) and jointly contributed to an increased risk of CRC (without vs. with rs2234767 GG/rs1800682 AA genotypes, adjusted OR = 1.30, 95% CI = 1.05 − 1.61). In vivo ChIP assays evaluated the effect of rs2234767 and rs1800682 on recruitment of SP1 and STAT1, respectively, to chromatin. The results showed SP1 interacting specifically with STAT1 recruited to their respective motifs for transcriptional activation. The mutant alleles rs2234767 A and rs1800682 G jointly affected coupled SP1 and STAT1 recruitment to chromatin. The interplay between SP1 and STAT1 was critical for the functional outcome of rs2234767 and rs1800682 in view of their high LD. In conclusion, the FAS rs2234767 and rs1800682 polymorphisms were in high LD with each other, and they jointly contributed to an increased risk of CRC by altering recruitment of SP1/STAT1 complex to the FAS promoter for transcriptional activation.
Colorectal cancer (CRC) is the third most commonly diagnosed cancers worldwide, accounting for roughly 1.2 million new cases and 600,000 deaths per year1. CRC is a complex disease resulting from both genetic and epigenetic alterations, including genetic variants2 and abnormal DNA methylation patterns3,4,5, among others. Two decades of research on genetic architecture of CRC has revealed that inherited susceptibility is a major component of CRC predisposition, with genetic factors accounting for 12–35% risk of CRC2.
A large number of studies have implicated the involvement of deregulated apoptosis pathway in CRC carcinogenesis6,7. Defect, dysfunction or altered expression of genes encoding key apoptotic proteins modify risk of CRC8. FAS, also known as CD95, encoded by FAS gene, is a cell surface factor and important inducer of the extrinsic apoptosis signaling pathway. FAS ligand, FASLG, also known as CD95L, encoded by FASLG gene, is a member of the tumor necrosis factor superfamily. FASLG binding to FAS triggers apoptosis through activation of CASP89. Accumulating evidence suggests that altered expression of FAS and/or FASLG contributes to development of CRC8.
Functional SNPs within the promoter region of gene are capable of affecting transcription and subsequently modulating risk of disease10,11. It has been reported that there are two functional SNPs in the promoter of FAS gene (FAS −1377 G>A, rs2234767; −670 A>G, rs1800682), which located within the consensus sequences of the SP1 and STAT1 transcription factors (TF) binding sites, respectively12. Sibley et al.13 reported that the rs2234767 A had a greatly reduced ability to bind SP1 compared with rs2234767 G and people with A allele had a significantly increased risk of acute myeloid leukemia (AML); however, both the rs1800682 A and rs1800682 G alleles could bind STAT1 and have no detectable difference in binding affinity.
Wu et al.14 first identified a T to C substitution at position −844 in the promoter of FASLG gene (FASLG −844 C>T, rs763110), which located in a putative binding motif for CAAT/enhancer-binding protein β (C/EBPβ). Functional study revealed that −844 C allele could increase basal FASLG expression, suggesting the −844 C>T polymorphism may affect the FASLG-mediated apoptotic signaling. A number of studies have been conducted to investigate the association between the three SNPs and a variety of tumors, including esophageal squamous-cell carcinoma (ESCC)15,16, squamous cell carcinoma of the head and neck (HNSCC)17, bladder cancer18, and gastric cancer19.
In this study, we aimed to determine the association of the FAS rs2234767, rs1800682, and FASLG rs763110 polymorphisms with risk of CRC in a Chinese population and the molecular mechanism underlying the association.
Methods and Materials
Ethics statement
The study was approved by the institutional review board of Southeast University. Each subject signed an informed consent. The research protocol was carried out in accordance with the approved guidelines.
Patients and samples
A total of 878 CRC patients and 884 healthy controls were enrolled in this study. The detailed information on the subjects has been described elsewhere20. Briefly, all patients were recruited from the First Affiliated Hospital of Nanjing Medical University between September 2010 and October 2013. The pathological stage of CRC at the time of diagnosis was classified into Dukes A, B, C and D. All controls were genetically unrelated to the cases and recruited from those who were seeking for health care in the same hospital. After signed the informed consent, all subjects donated 5 ml of venous blood sample for genomic DNA extraction.
Genotyping
The genotyping of FAS rs2234767, rs1800682 and FASLG rs763110 was performed by TaqMan allelic discrimination method equipped with ABI 7900 HT Real Time PCR System (Applied Biosystems, CA, USA). The reaction conditions were set as follows: 95 °C for 10 min followed by 40 cycles of 95 °C for 15 sec, and 60 °C for 1 min. At least 10% of the samples were randomly selected for genotyping confirmation, and the results were 100% concordant.
Chromatin immunoprecipitation assay (ChIP)
Human peripheral white blood cells (5 × 107 per sample) were fixed for 10 min at 37 °C with 4% formaldehyde. After incubation, fresh glycine was added to a final concentration of 125 mM to stop cross-linking. After 5 min at room temperature, the samples were pelleted in an centrifuge at 3600 rpm (2000 g) for 2 min at 4 °C, washed once with cold PBS plus protease inhibitors, and then repelleted. The pellet was resuspended in 1 ml of PBS and ground the cells using a micro-tissue grinder on ice. Cells were pelleted again as above at 4 °C. ChIP was performed using the ChIP-ITTM Express Magnetic assay kit (Cat. No. 53009, Active Motif). The antisera for ChIP reaction was normal mouse IgG (Cat. No. 2027, Santa Cruz Biotechnology, Inc.), normal rabbit IgG (Cat. No. NI01, EMD Chemicals, Inc., Gibbstown, NJ), anti-human SP1 (Cat. No. 9389, Cell Signaling Technology), and anti-human STAT1 (Cat. No. 9172, Cell Signaling Technology). Precipitated genomic DNA was analyzed by quantitative PCR in triplicate measurements for each sample using the following human FAS promoter primers: 5′-ACCATCCTCCTTATCCCACT-3′ (forward) and 5′-GTAGGTGTTGATAGGCTTGA-3′ (reverse) for rs2234767; 5′-CTAAGGGGCCCTCCCTTTT-3′ (forward) and 5′-ACTTGCGGGGCATTTGACT-3′ (reverse) for rs1800682. Captured genomic DNA was normalized to input material and the samples with different genotypes compared.
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) of the controls’ genotype frequencies was evaluated by a goodness-of-fit chi-square test (χ2 test). Bonferroni correction for multiple testing was also applied. Crude and adjusted odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to analyze the magnitude of the association between the genotypes and risk of CRC by univariate and multivariate unconditional logistic regression models, respectively. A P-value < 0.05 was considered statistically significant.
Results
Association between the FAS and FASLG polymorphisms and risk of CRC
The genotype frequencies of FAS rs2234767, rs1800682 and FASLG rs763110 among the controls were all in agreement with Hardy-Weinberg equilibrium (P = 0.11 for rs2234767, 0.060 for rs1800682 and 0.53 for rs763110). As shown in Table 1, the frequencies of rs2234767 mutant A allele was higher in the cases than the controls (39% and 34%, P = 0.013 after Bonferroni correction). Moreover, the frequencies distribution of rs2234767 genotypes were significantly different between the cases and controls (P = 0.0051), which remained significant after Bonferroni correction (P = 0.015). When the rs2234767 GG genotype used as the reference, the heterozygous GA and AA genotypes were both associated with significantly increased risk of CRC (adjusted OR = 1.39, 95% CI = 1.13 − 1.71 for the GA genotype; 1.37, 1.02 − 1.84 for the AA genotype), and the risk did not change substantially under the assumption of a dominant genetic model (adjusted OR = 1.38, 95% CI = 1.14 − 1.68).
Table 1. Association between FAS rs2234767, rs1800682 and FASLG rs763110 genotypes and risk of CRC.
| SNPs | Genotype | Cases (n = 878) |
Controls (n = 884) |
Pa | Pb | Adjusted OR (95% CI)c | ||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | |||||
| FASLG | CC | 462 | 53 | 470 | 53 | 0.71 | 1.0 | 1.00 (Ref) |
| rs763110 | CT | 354 | 40 | 344 | 39 | 1.06 (0.87–1.28) | ||
| TT | 62 | 7 | 70 | 8 | 0.90 (0.62–1.30) | |||
| Ptrend | 0.92 | |||||||
| T allele | 478 | 27 | 484 | 27 | 0.92 | 1.0 | ||
| FAS | GG | 305 | 37 | 385 | 44 | 0.0051 | 0.015 | 1.00 (Ref) |
| rs2234767 | GA | 407 | 49 | 372 | 43 | 1.39 (1.13–1.71) | ||
| AA | 124 | 15 | 114 | 13 | 1.37 (1.02–1.84) | |||
| Ptrend | 0.0051 | |||||||
| A allele | 655 | 39 | 600 | 34 | 0.0042 | 0.013 | ||
| GG | 305 | 36 | 385 | 44 | 1.00 (Ref) | |||
| GA/AA | 531 | 64 | 486 | 56 | 1.38 (1.14–1.68) | |||
| FAS | AA | 301 | 34 | 348 | 40 | 0.060 | 0.18 | 1.00 (Ref) |
| rs1800682 | AG | 435 | 50 | 392 | 44 | 1.30 (1.06–1.60) | ||
| GG | 142 | 16 | 144 | 16 | 1.16 (0.88–1.53) | |||
| Ptrend | 0.14 | |||||||
| G allele | 719 | 41 | 680 | 38 | 0.13 | 0.39 | ||
| AA | 301 | 34 | 348 | 39 | 1.00 (Ref) | |||
| AG/GG | 577 | 66 | 536 | 61 | 1.26 (1.04–1.53) | |||
Bold indicated statistically significant.
aχ2 test for either genotype distributions or allele frequencies between the cases and controls.
bAdjusted for multiple comparisons by Bonferroni correction.
cAdjusted for age, sex, smoking and drinking status in logistic regression model.
The genotype and allele frequencies distribution of the FASLG rs763110 were not significantly different between the cases and controls (Pgenotype = 0.71 and Pallele = 0.92; Pgenotype = 1.0 and Pallele = 1.0 after Bonferroni correction). The allele frequency distribution of the FAS rs1800682 was not significantly different between the cases and controls (Pallele = 0.13 and Pallele = 0.39 after Bonferroni correction). The difference of the rs1800682 genotype distribution was Quasi significant between the cases and controls (P = 0.060); however, the difference did not remain significant after Bonferroni correction (P = 0.18). Further analysis showed that the rs1800682 polymorphism was associated with an increased risk of CRC under the dominant genetic model (AG/GG vs. AA, adjusted OR = 1.26, 95% CI = 1.04 − 1.53).
Association between the combined genotypes of the FAS polymorphisms and risk of CRC
Linkage disequilibrium analysis (LD) revealed a strong LD between the two FAS polymorphisms among the controls (D’ = 0.994 and r2 = 0.848, P < 0.001), suggesting a joint effect between the two FAS polymorphisms. To evaluate the genotype- genotype interaction, we dichotomized the FAS genotypes as either rs2234767 GG or rs2234767 GA/AA and rs1800682 AA or rs1800682 AG/GG. When the rs2234767 GG/rs1800682 AA genotypes used as the reference, the (rs2234767 GA/AA)/(rs1800682 AA) genotypes and the (rs2234767 GA/AA)/(rs1800682 AG/GG) genotypes were associated with a significantly higher risk of CRC (adjusted OR = 15.49, 95% CI = 2.01 − 119.25 for the (rs2234767 GA/AA)/(rs1800682 AA) genotypes; and 1.30, 1.06 − 1.59 for the (rs2234767 GA/AA)/(rs1800682 AG/GG) genotypes; Table 2).
Table 2. Combined genotype frequencies of the FAS polymorphisms among the cases and controls and their association with risk of CRC.
| Combined genotypes | Cases (n = 836) | Controls (n = 871) | Pa | Adjusted OR (95% CI)b | |
|---|---|---|---|---|---|
| n (%) | n (%) | ||||
| FAS rs2234767 | FAS rs1800682 | <0.001 | |||
| GG | AA | 287 (34) | 347 (40) | 1.00 (Reference) | |
| GG | AG/GG | 18 (2) | 38 (3.9) | 0.60 (0.33–1.07) | |
| GA/AA | AA | 13 (2) | 1 (0.1) | 15.49 (2.01–119.25) | |
| GA/AA | AG/GG | 518 (62) | 485 (56) | 1.30 (1.06–1.59) | |
| Trend test | 0.0051 |
Bold indicated statistically significant.
aχ2 test for the combined genotype distributions between the cases and controls.
bAdjusted for age, sex, smoking and drinking status in logistic regression model.
Stratification analysis of the association of FAS combined genotypes with CRC susceptibility by demographic variables
To control the impact of confounders on the genetic association, we performed stratification analysis. The combined genotypes were dichotomized into two groups, i.e., with rs2234767 GG/rs1800682 AA and without rs2234767 GG/rs1800682 AA, to facilitate further analysis. As shown in Table 3, compared with the rs2234767 GG/rs1800682 AA genotypes, the individuals carrying the combined genotypes without rs2234767 GG/rs1800682 AA had a higher risk of CRC (adjusted OR = 1.28, 95% CI = 1.05 − 1.56), and the risk was more pronounced among the subgroups of age >60 years, female, never smokers or drinkers, having no family history of cancer (adjusted OR = 1.50, 95% CI = 1.13 − 1.99 for >60 years, 1.77; 1.26 − 2.47 for female; 1.39, 1.09 − 1.77 for never smokes; 1.33, 1.05 − 1.67 for never drinkers; and 1.34, 1.08 − 1.67 for people having no family history of cancer).
Table 3. Stratified analysis of the FAS combined genotypes associated with CRC risk by demographic variables.
| Variables | Case/control (n) | Combined genotypes (case/control) |
Pa | Adjusted OR (95% CI)b | |||
|---|---|---|---|---|---|---|---|
| With rs2234767 GG/rs1800682 AA |
Without rs2234767 GG/rs1800682 AA |
||||||
| n | % | n | % | ||||
| Total | 836/871 | 287/347 | 34/40 | 549/524 | 66/60 | 0.019 | 1.28 (1.05–1.56) |
| Age (years) | |||||||
| ≤60 | 436/401 | 156/156 | 36/39 | 280/245 | 64/61 | 0.35 | 1.09 (0.82–1.45) |
| >60 | 400/470 | 131/191 | 33/41 | 269/279 | 67/59 | 0.016 | 1.50 (1.13–1.99) |
| Sex | |||||||
| Male | 510/507 | 198/207 | 39/41 | 312/300 | 61/59 | 0.51 | 1.08 (0.84–1.39) |
| Female | 326/364 | 89/140 | 27/38 | 237/224 | 73/62 | 0.0019 | 1.77 (1.26–2.47) |
| Smoking status | |||||||
| Never | 558/602 | 185/244 | 33/40 | 373/358 | 67/60 | 0.0093 | 1.39 (1.09–1.77) |
| Ever | 278/269 | 102/103 | 37/38 | 176/166 | 63/62 | 0.70 | 1.08 (0.76–1.53) |
| Drinking status | |||||||
| Never | 610/655 | 207/264 | 34/40 | 403/391 | 66/60 | 0.019 | 1.33 (1.05–1.67) |
| Ever | 226/216 | 80/83 | 35/38 | 146/133 | 65/62 | 0.51 | 1.16 (0.78–1.72) |
| Family history of cancer | |||||||
| No | 642/786 | 222/323 | 35/41 | 420/463 | 65/59 | 0.012 | 1.34 (1.08–1.67) |
| Yes | 194/85 | 65/24 | 34/28 | 129/61 | 66/72 | 0.39 | 0.78 (0.44–1.37) |
Bold indicated statistically significant.
aχ2 test for the combined genotype distributions between the cases and controls.
bAdjusted for age, sex, smoking and drinking status in logistic regression model.
Association between the combined genotypes of the FAS polymorphisms and progression of CRC
We further evaluated the association between the FAS combined genotypes and grade and stage of CRC. When compared with the rs2234767 GG/rs1800682 AA genotypes, the combined genotypes without rs2234767 GG/rs1800682 AA were associated with a significantly increased risk of CRC with intermediate grade (adjusted OR = 1.30, 95% CI = 1.05 − 1.61; Table 4). However, the combined genotypes were not significantly associated with CRC with low or high grade, which was likely due to the reduced number of subjects. In the stratification of stage, a significantly increased risk was only found between the combined genotypes without rs2234767 GG/rs1800682 AA and CRC with Dukes C and D stage (adjusted OR = 1.33, 95% CI = 1.04 − 1.71; Table 4).
Table 4. Association between the FAS combined genotypes and progression of CRC.
| Variables | Combined genotypes |
Pa | Adjusted OR (95% CI)b | |||
|---|---|---|---|---|---|---|
| With rs2234767 GG/rs1800682 AA |
Without rs2234767 GG/rs1800682 AA |
|||||
| n | % | n | % | |||
| Controls (n = 871) | 347 | 40 | 524 | 60 | 1.00 (reference) | |
| Cases (n = 836) | ||||||
| Tumor grade | ||||||
| Low | 24 | 40 | 36 | 60 | 0.98 | 0.99 (0.58–1.70) |
| Intermediate | 219 | 34 | 427 | 66 | 0.018 | 1.30 (1.05–1.61) |
| High | 44 | 34 | 86 | 66 | 0.19 | 1.30 (0.88–1.91) |
| Dukes stage | ||||||
| A + B | 150 | 35 | 275 | 65 | 0.11 | 1.22 (0.96–1.55) |
| C + D | 137 | 33 | 274 | 67 | 0.025 | 1.33 (1.04–1.71) |
Bold indicated statistically significant.
aχ2 test for the combined genotype distributions between the cases and controls.
bAdjusted for age, sex, smoking and drinking status in logistic regression model.
rs2234767 A coordinates with rs1800682 G to attenuate binding affinity of SP1 and STAT1 to FAS promoter in vivo
To determine the molecular mechanisms underlying the association of FAS polymorphisms with an increased risk of CRC, the effect of the rs2234767 and rs1800682 polymorphisms on the ability of SP1 and STAT1 transcription factors to bind the endogenous FAS promoter region was assessed by ChIP assays. Because the rs2234767 G and rs1800682 A alleles were in high LD with each other as mentioned above, there existed only three combined genotypes in our samples for ChIP assays. Finally, a total of 9 people with three different combined genotypes, i.e., rs2234767 GG/rs1800682 AA, rs2234767 GA/rs1800682 AG, and rs2234767 AA/rs1800682 GG, were selected for ChIP assays.
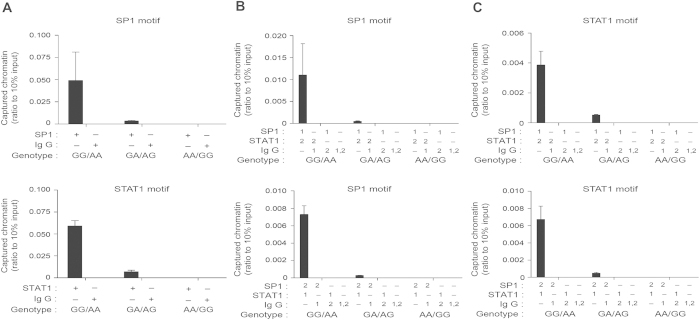
As shown in Fig. 1A, the region with rs2234767 G and rs1800682 A alleles were readily immunoprecipitated by anti-SP1 and anti-STAT1, respectively, but not IgG, demonstrating SP1 and STAT1 interact specifically with the FAS promoter in chromatin (left). Interestingly, the ability of SP1 and STAT1 to bind the FAS promoter was dramatically decreased with the increase of mutant alleles. The amount of chromatin with the rs2234767 GA/rs1800682 AG genotypes captured by anti-SP1 and anti-STAT1 was 6.9% and 12%, respectively, of that with the rs2234767 GG/rs1800682 AA genotypes (middle). The homozygous mutant genotypes rs2234767 AA/rs1800682 GG eliminated both the SP1 and STAT1 recruitment to the FAS promoter (right). Furthermore, as assessed by sequential ChIP (Re-ChIP), the SP1 interacting with STAT1 was recruited to the SP1 (Fig. 1B, left) and STAT1 motifs (Fig. 1C, left) for transcriptional regulation. The mutant alleles could also eliminate the SP1/STAT1 complex recruitment to the SP1 (Fig. 1B, middle and right) and STAT1 motifs (Fig. 1C, middle and right). We also selected 9 other people with three combined genotypes for reproducibility of ChIP assays, and found the results were consistent with those mentioned above (Fig. S1). Our results indicated that SP1 and STAT1 contributed equally to activate the transcription of FAS in CRC and the interplay between these factor was critical for the functional outcome of FAS rs2234767 and rs1800682 in view of their high LD.
Figure 1. FAS rs2234767 A and rs1800682 G alleles affect coupled SP1 and STAT1 recruitment to chromatin.
(A) Chromatin immunoprecipitation (ChIP) of the FAS promoter with three different genotypes (rs2234767 GG/rs1800682 AA, rs2234767 GA/rs1800682 AG and rs2234767 AA/rs1800682 GG) using antibody for SP1 and STAT1(single pool generated from triplicate biological samples/manipulation; triplicate measurements/pool; mean ± SE). (B) Sequential ChIP of the FAS promoter containing SP1 motif immunoprecipitated first using antibody for SP1 followed by antibody for STAT1 (upper) or first using antibody for STAT1 followed by antibody for SP1 (lower). (C) Sequential ChIP of the FAS promoter containing STAT1 motif immunoprecipitated first using antibody for SP1 followed by antibody for STAT1 (upper) or first using antibody for STAT1 followed by antibody for SP1 (lower). For all combined genotypes in the figure, left genotype arises from the rs2234767 and right from the rs1800682 polymorphism.
Discussion
In the present study, we analyzed the association of FAS rs2234767, rs1800682 and FASLG rs763110 polymorphisms with risk of CRC in a Chinese population. The FAS rs2234767 and rs1800682 polymorphisms had effect on increasing risk of CRC and a joint effect on risk and progression, but not for the FASLG rs763110 polymorphism. The joint effects of the two FAS polymorphisms on risk of CRC were more pronounced among the subgroups with age >60 years, female, never smokers, never drinkers, having no family history of cancer, and CRC with intermediate grade and with Dukes C and D stage. Functional studies revealed that the SP1 interacting with STAT1 was recruited to the SP1 and STAT1 motifs within the promoter of FAS for transcriptional regulation, and the interplay between these factor was critical for the functional outcome of FAS rs2234767 and rs1800682 in view of their high LD. Given the role of FAS/FASLG pathway in carcinogenesis, it is biologically plausible that the rs2234767 and rs1800682 polymorphisms may modulate the risk of CRC by attenuating SP1/STAT1 complex-mediated transcriptional activation of FAS, which in turn dampening FAS apoptotic pathway.
Colorectal cancer is a complex disease and develops through a multistage process21,22. During the process, colorectal epithelial cells accumulate a number of molecular changes and eventually become fully malignant cells. These molecular changes involve mutations in the well-defined genes or pathways such as APC, mismatch repair genes like MLH1, and SMAD4, and epigenetic changes such as global DNA hypomethylation in repetitive sequences (satellite and LINE repeats) and promoter hypermethylation of tumor suppressor genes like MLH1, RUNX3 and SEPT93,4,5. It is now well accepted that genes that regulate apoptosis are important variables in cancer development. A lot of studies have shown that alteration of FAS and FASLG expression decreases the apoptotic activity and facilitates tumor cells evading or suppressing the immune system23,24. Deregulated FAS and FASLG expression are common features of most human malignancies and associated with progression of a variety of tumors, including CRC24,25,26. Therefore, the functional variants of the FAS and FASLG genes which were capable of influencing their expression could be expected to have effect on cell death and thus, carcinogenesis.
FAS rs2234767 and rs1800682 polymorphisms have been reported to be able to alter the SP1 and STAT1 binding site, respectively, leading to dysregulated FAS expression13,14. A lot of studies have investigated the relationship between these functional SNPs and risk of diseases including tumors such as esophageal cancer16, HNSCC17, leukemia27, and gastric cancer28. However, the studies on the association between these SNPs and risk of CRC are limited. Our data showed that the functional FAS rs2234767 G>A and rs1800682 A>G polymorphisms were associated with significantly increased risk of CRC. Yang et al.29 also reported that the FAS rs2234767 polymorphism was associated with an risk of CRC, which was consistent with our results. In a meta-analysis of 52 studies, Xu et al.30 found that the carriers of the FAS rs2234767 A are more susceptible to the majority of cancers than non-carriers, which further corroborated our conclusions.
Although the FAS polymorphisms have been widely investigated their association with multiple tumors susceptibility30,31,32, the studies on mechanisms underlying the association were limited. The rs2234767 and rs1800682 polymorphisms can alter the SP1 and STAT1 binding site, leading to downregulated FAS expression13,14. It is reported that occupation of contiguous DNA-binding sites for STAT1 and SP1 are both required for full activation of the ICAM-1 by IFN-γ33. Similarly, we also found that the SP1 interacting with STAT1 was recruited to the SP1 and STAT1 motifs within the promoter of FAS for transcriptional regulation by ChIP assays. Moreover, the mutant alleles rs2234767 A and rs1800682 G could eliminate the SP1/STAT1 complex recruitment to the SP1 and STAT1 motifs. Our ChIP assays indicated that the interplay between the two transcription factors was critical for the functional outcome of FAS rs2234767 and rs1800682 in view of their high LD.
Recently, a relatively new field of epidemiology named molecular pathological epidemiology (MPE) has emerged as an integrative interdisciplinary field of molecular pathology and epidemiology, the concept of which has been consolidated by Ogino and Stampfer34,35. In MPE, a particular exposure including genetic factor is evaluated in relation to a specific somatic molecular change to better understanding its role in the carcinogenic and pathologic process. Moreover, an interactive effect of tumorous molecular features and the exposures of interest on tumor behavior can gain insights into tumor molecular changes, which may be predictive or prognostic tissue biomarkers35. Therefore, MPE can shed light on the pathogenic process and help optimize personalized prevention and therapy. MPE requires multidisciplinary collaboration between epidemiology, pathology, bioinformatics, biostatistics, and computational biology. Hence, the Second International MPE Meeting was held in Boston in December 2014 to discuss measures to address challenges and move this field forward, especially initiating the effort of specifying guidelines for MPE (“STROBE-MPE”) as first proposed in 201236,37. Making consensus guidelines facilitating study reporting contributes to building a field and enhancing its contributions, such as the development of the guidelines for clinical trials38.
Our study represents MPE research that we examined the relationship between the FAS susceptibility alleles and its aberrant transcriptional activities by ChIP assays and finally revealed the function of these alleles and gained insight into whether susceptibility alleles were truly causal. Recently, a new direction of MPE where researchers investigate the interactive effects of tumorous molecular features and the exposures of interest on tumor behavior (prognosis or clinical outcome) has emerged. These studies will help us to attribute the effects of exposure of interest to a specific molecular subtype of cancer.
In conclusion, the FAS rs2234767 and rs1800682 polymorphisms were in high LD with each other, and they jointly contributed to an increased risk and progression of CRC. The interaction between them altered the the ability of SP1 and STAT1 transcription factors to bind the endogenous FAS promoter region as assessed by ChIP assays. These findings suggest that the functional promoter polymorphisms of FAS may jointly contribute to the etiology of CRC.
Additional Information
How to cite this article: Wang, S. et al. FAS rs2234767 and rs1800682 polymorphisms jointly contributed to risk of colorectal cancer by affecting SP1/STAT1 complex recruitment to chromatin. Sci. Rep. 6, 19229; doi: 10.1038/srep19229 (2016).
Supplementary Material
Acknowledgments
This study was partly supported by National Natural Science Foundation of China (81302502 and 81472938), Natural Science Foundation of Jiangsu Province (BK20130641 and BK20151418), Specialized Research Fund for the Doctoral Program of Higher education of China (20130092120063), the Open Research Fund of State Key Laboratory of Bioelectronics, the Fundamental Research Funds for the Central Universities, the Fund of the Distinguished talents of Jiangsu Province (BK20150021), the Fund of the Distinguished Professor of Jiangsu Province, and the National Program for Support of Topnotch Young Professionals from the Organization Department of the CPC Central Committee.
Footnotes
Author Contributions R.C. and M.W. conceived and designed the experiments. S.W., S.W. and Q.M. performed the experiments. S.W., and X.L. analyzed the data. R.C. and M.W. contributed reagents/materials/analysis tools. S.W. and R.C. wrote the paper.
References
- Brenner H., Kloor M. & Pox C. P. Colorectal cancer. Lancet 383, 1490–1502 (2014). [DOI] [PubMed] [Google Scholar]
- Peters U., Bien S. & Zubair N. Genetic architecture of colorectal cancer. Gut 64, 1623–1636 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardhan K. & Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers (Basel) 5, 676–713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J. M. & Yu J. Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int J Mol Sci 16, 2472–2496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi D., Brandi G., Bazzoli F. & Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci 14, 16365–16385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. Y., Sales K. M., Fuller B., Seifalian A. M. & Winslet M. C. Apoptosis and colorectal cancer: implications for therapy. Trends Mol Med 15, 225–233 (2009). [DOI] [PubMed] [Google Scholar]
- Mehlen P. & Tauszig-Delamasure S. Dependence receptors and colorectal cancer. Gut 63, 1821–1829 (2014). [DOI] [PubMed] [Google Scholar]
- Hoogwater F. J., Steller E. J., Westendorp B. F., Borel Rinkes I. H. & Kranenburg O. CD95 signaling in colorectal cancer. Biochim Biophys Acta 1826, 189–198 (2012). [DOI] [PubMed] [Google Scholar]
- Ashkenazi A. & Salvesen G. Regulated cell death: signaling and mechanisms. Annu Rev Cell Dev Biol 30, 337–356 (2014). [DOI] [PubMed] [Google Scholar]
- Lee W., Yue P. & Zhang Z. Analytical methods for inferring functional effects of single base pair substitutions in human cancers. Hum Genet 126, 481–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. et al. Genetic variant in APE1 gene promoter contributes to cervical cancer risk. Am J Obstet Gynecol 209, 360 e361-367 (2013). [DOI] [PubMed] [Google Scholar]
- Huang Q. R., Morris D. & Manolios N. Identification and characterization of polymorphisms in the promoter region of the human Apo-1/Fas (CD95) gene. Mol Immunol 34, 577–582 (1997). [DOI] [PubMed] [Google Scholar]
- Sibley K. et al. Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res 63, 4327–4330 (2003). [PubMed] [Google Scholar]
- Wu J. et al. A novel polymorphic CAAT/enhancer-binding protein beta element in the FasL gene promoter alters Fas ligand expression: a candidate background gene in African American systemic lupus erythematosus patients. J Immunol 170, 132–138 (2003). [DOI] [PubMed] [Google Scholar]
- Sun T. et al. Polymorphisms of death pathway genes FAS and FASL in esophageal squamous-cell carcinoma. J Natl Cancer Inst 96, 1030–1036 (2004). [DOI] [PubMed] [Google Scholar]
- Zhao H., Zheng L., Li X. & Wang L. FasL gene −844T/C mutation of esophageal cancer in South China and its clinical significance. Sci Rep 4, 3866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. et al. Polymorphisms of FAS and FAS ligand genes involved in the death pathway and risk and progression of squamous cell carcinoma of the head and neck. Clin Cancer Res 12, 5596–5602 (2006). [DOI] [PubMed] [Google Scholar]
- Li C. et al. Functional polymorphisms in the promoter regions of the FAS and FAS ligand genes and risk of bladder cancer in south China: a case-control analysis. Pharmacogenet Genomics 16, 245–251 (2006). [DOI] [PubMed] [Google Scholar]
- Wang M. et al. FAS and FAS ligand polymorphisms in the promoter regions and risk of gastric cancer in Southern China. Biochem Genet 47, 559–568 (2009). [DOI] [PubMed] [Google Scholar]
- Du H. et al. Association study between XPG Asp1104 His polymorphism and colorectal cancer risk in a Chinese population. Sci Rep 4, 6700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B. et al. Genetic alterations during colorectal-tumor development. N Engl J Med 319, 525–532 (1988). [DOI] [PubMed] [Google Scholar]
- Kinzler K. W. & Vogelstein B. Lessons from hereditary colorectal cancer. Cell 87, 159–170 (1996). [DOI] [PubMed] [Google Scholar]
- Fouque A., Debure L. & Legembre P. The CD95/CD95L signaling pathway: a role in carcinogenesis. Biochim Biophys Acta 1846, 130–141 (2014). [DOI] [PubMed] [Google Scholar]
- Peyvandi S. et al. Fas Ligand Deficiency Impairs Tumor Immunity by Promoting an Accumulation of Monocytic Myeloid-Derived Suppressor Cells. Cancer Res 75, 4292–4301 (2015). [DOI] [PubMed] [Google Scholar]
- Liu F. et al. NF-kappaB directly regulates Fas transcription to modulate Fas-mediated apoptosis and tumor suppression. J Biol Chem 287, 25530–25540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajate C. & Mollinedo F. Lipid raft-mediated Fas/CD95 apoptotic signaling in leukemic cells and normal leukocytes and therapeutic implications. J Leukoc Biol 98, 739–759 (2015). [DOI] [PubMed] [Google Scholar]
- Chen Y. et al. Association between Fas/FasL polymorphism and susceptibility to leukemia: a meta-analysis. Int J Clin Exp Med 8, 3817–3824 (2015). [PMC free article] [PubMed] [Google Scholar]
- Gu D. et al. Functional polymorphisms in apoptosis pathway genes and survival in patients with gastric cancer. Environ Mol Mutagen 55, 421–427 (2014). [DOI] [PubMed] [Google Scholar]
- Yang S. et al. [Genetic polymorphisms of apoptosis-associated genes FAS and FASL and risk of colorectal cancer]. Zhonghua Yi Xue Za Zhi 85, 2132–2135 (2005). [PubMed] [Google Scholar]
- Xu Y. et al. Association of the polymorphisms in the Fas/FasL promoter regions with cancer susceptibility: a systematic review and meta-analysis of 52 studies. PLoS One 9, e90090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Hu L. & Pan Y. Lack of association between the FAS/FASL polymorphisms and cervical cancer risk: A meta-analysis. Biomed Rep 1, 269–274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H. P., Liu Q. D., Li G. Q. & Cong Y. Q. Fas −670A/G (rs1800682) polymorphism and digestive cancer risk in Asians: a meta-analysis. Genet Test Mol Biomarkers 18, 482–488 (2014). [DOI] [PubMed] [Google Scholar]
- Look D. C., Pelletier M. R., Tidwell R. M., Roswit W. T. & Holtzman M. J. Stat1 depends on transcriptional synergy with Sp1. J Biol Chem 270, 30264–30267 (1995). [DOI] [PubMed] [Google Scholar]
- Ogino S. & Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: the evolving field of molecular pathological epidemiology. J Natl Cancer Inst 102, 365–367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S., Chan A. T., Fuchs C. S. & Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut 60, 397–411 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S. & Giovannucci E. Commentary: Lifestyle factors and colorectal cancer microsatellite instability--molecular pathological epidemiology science, based on unique tumour principle. Int J Epidemiol 41, 1072–1074 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S. et al. Interdisciplinary education to integrate pathology and epidemiology: towards molecular and population-level health science. Am J Epidemiol 176, 659–667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz K. F., Altman D. G., Moher D. & Group C. CONSORT 2010 statement: updated guidelines for reporting parallel group randomised trials. PLoS Med 7, e1000251 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.