Abstract
Staphylococcal biofilms are problematic and play a critical role in the persistence of chronic infections because of their abilities to tolerate antimicrobial agents. Thus, the inhibitions of biofilm formation and/or toxin production are viewed as alternative means of controlling Staphylococcus aureus infections. Here, the antibiofilm activities of 560 purified phytochemicals were examined. Alizarin at 10 μg/ml was found to efficiently inhibit biofilm formation by three S. aureus strains and a Staphylococcus epidermidis strain. In addition, two other anthraquinones purpurin and quinalizarin were found to have antibiofilm activity. Binding of Ca2+ by alizarin decreased S. aureus biofilm formation and a calcium-specific chelating agent suppressed the effect of calcium. These three anthraquinones also markedly inhibited the hemolytic activity of S. aureus, and in-line with their antibiofilm activities, increased cell aggregation. A chemical structure-activity relationship study revealed that two hydroxyl units at the C-1 and C-2 positions of anthraquinone play important roles in antibiofilm and anti-hemolytic activities. Transcriptional analyses showed that alizarin repressed the α-hemolysin hla gene, biofilm-related genes (psmα, rbf, and spa), and modulated the expressions of cid/lrg genes (the holin/antiholin system). These findings suggest anthraquinones, especially alizarin, are potentially useful for controlling biofilm formation and the virulence of S. aureus.
Most bacteria are likely to form biofilms that attach to living or abiotic surfaces using self-produced extracellular polymeric substances, and thus, biofilms are ubiquitous in natural, medical, and engineering environments1. Biofilms exhibit reduced sensitivity to conventional antimicrobial agents, host defenses, and external stresses, and thus, contribute to the bacterial persistence in chronic infections2,3. Since biofilm formation is a mechanism of antibiotic resistance, it is important to identify novel compounds capable of inhibiting biofilms without allowing bacteria to develop drug resistance.
Biofilm formation by Staphylococcus aureus and Staphylococcus epidermidis is of particular concern in the medical field, and S. aureus has caused numerous outbreaks of nosocomial infections4. Furthermore, the emergence of multidrug-resistant strains, such as, methicillin-resistant S. aureus (MRSA) and vancomycin-methicillin-resistant S. aureus has become a serious threat. These bacteria can secrete exotoxins, such as, hemolysin, enterotoxins, coagulase, TSST-1, and protein A, which are associated with specific diseases5, and can form biofilms on a variety of surfaces, including those of catheters, implants, prosthetics, and medical equipment2. Diverse mechanisms and environmental cues, for example, quorum sensing, c-di-GMP, protease, DNase, cis-2-decenoic acid, d-amino acids, phenol-soluble polypeptides, and pH, contribute to biofilm formation by S. aureus6,7. In addition, S. aureus produces α-toxin, which causes hemolysis and contributes to biofilm formation8. Hence, we sought to understand how biofilm inhibitors control biofilm formation by S. aureus.
Plant secondary metabolites are major sources of antimicrobial agents and other pharmaceuticals9, and several plant-derived biofilm inhibitors have been identified and shown to possess antibiofilm activity against S. aureus, examples include; magnolol10, ellagic acid11, tannic acid12, quercetin13, ginkgolic acids14, eugenol15, and flavonoids16. It has also been reported staphylococcal biofilm formation is inhibited by several plant essential oils17,18,19,20,21,22. However, the identification of active compounds in plant extracts and essential oils often requires extensive investigation to identify active components, and thus, only a limited number of organic biofilm inhibitors have been identified.
The goal of this work was to identify novel antibiofilm compounds against Staphylococcus species (including MRSA) from among 560 purified phytochemicals. Structure-activity analysis, confocal microscopy, slime analysis, hemolysis analysis, a cell aggregation assay, and transcriptional analysis were used to elucidate the mechanisms responsible for the inhibition of biofilm formation and toxin production.
Results
Alizarin inhibited biofilm formation by S. aureus and S. epidermidis without affecting planktonic cell growth
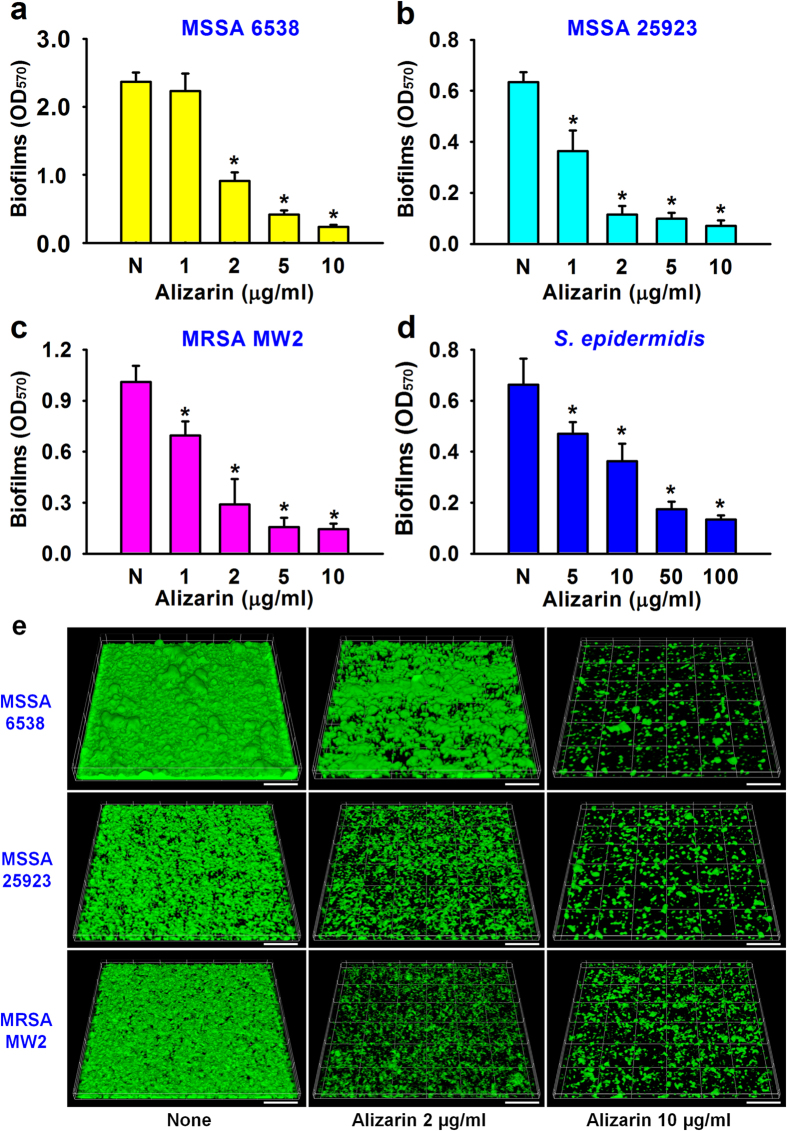
Screening of the 560 phytochemicals for antibiofilm activity against S. aureus MSSA 6538 on 96-well polystyrene plates showed that alizarin at 10 μg/ml most inhibited S. aureus biofilm formation. Twenty of the 560 chemicals inhibited S. aureus biofilm formation by >60% and nine enhanced biofilm formation by >60% (Supplementary Fig. S1). Further experiments showed that the addition of alizarin (0, 1, 2, 5, 10, 50, or 100 μg/ml) at the beginning of bacterial culture dose-dependently inhibited biofilm formation by all three S. aureus strains (MSSA 6538, MSSA 25923, and MRSA MW2) and a S. epidermidis strain (ATCC 14990) (Fig. 1a–d). Specifically, alizarin (at 10 μg/ml) decreased biofilm formation by all three S. aureus strains by ≥90%, whereas in the case of S. epidermidis, 50 μg/ml was required to inhibit biofilm formation by ≥70%. Unlike Gram-positive bacteria, biofilm formation by two Gram-negative bacteria (Escherichia coli O157:H7 and Pseudomonas aeruginosa PAO1) was unaffected by alizarin at concentrations up to 100 μg/ml (Supplementary Fig. S2).
Figure 1. Antibiofilm activities of alizarin against S. aureus and S. epidermidis.
The antibiofilm activities (OD570) of alizarin were determined against two methicillin-sensitive S. aureus strains (MSSA, ATCC 25923 and ATCC 6538), a methicillin-resistant S. aureus strain (MRSA, MW2) (a–c), and S. epidermidis (ATCC 14990) (d). Two independent experiments were conducted (12 wells per sample); error bars indicate standard deviations. *P < 0.05 versus non-treated controls (N or None). Biofilm formation on glass was observed by confocal laser microscopy (e). Scale bars represent 50 μm.
Confocal laser microscopy was used to analyze changes in biofilm formation on glass, and in-line with biofilm data obtained using 96-well polystyrene plates (Fig. 1a–c), fluorescent images indicated alizarin (0, 2, or 10 μg/ml) dose-dependently inhibited S. aureus biofilm formation (Fig. 1e). Biofilm inhibition was further confirmed by COMSTAT biofilm analysis, which showed alizarin (at 10 μg/ml) reduced all three measured biofilm parameters (biomass, mean thickness, and substratum coverage) of the three S. aureus strains by ≥80% versus untreated controls (Supplementary Table S1). For example, S. aureus MSSA 6538 biofilm biomass was reduced from 12 μm3 μm−2 to 0.9 μm3 μm−2 in the presence of alizarin at 10 μg/ml.
Counts of viable biofilm cells were performed to confirm biofilm inhibition by alizarin. In agreement with the results of other biofilm assays, alizarin dose-dependently reduced viable cell numbers in the biofilms of the four Staphylococcus strains. For example, alizarin at 10 μg/ml reduced the number of viable cells in MSSA 6538 and MRSA MW2 biofilms by more than 7-fold versus untreated controls (Supplementary Table S2).
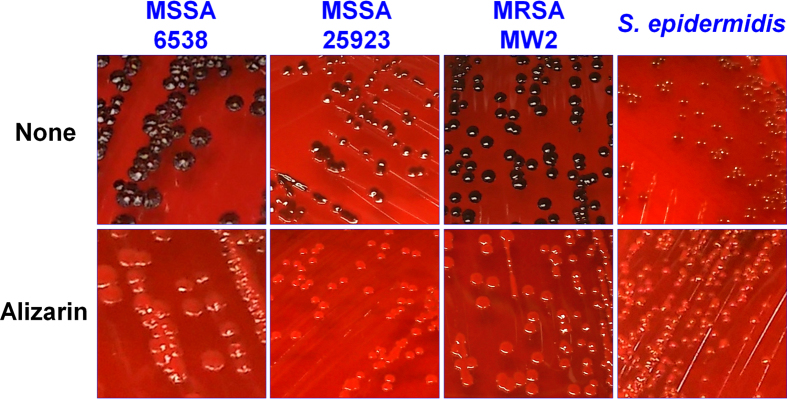
Slime detection using Congo red plates is conventionally used to detect biofilm-forming staphylococci23, and consistent with the 96-well plate and microscopic results, slime production by all four staphylococci strains was markedly reduced by alizarin at 20 μg/ml (Fig. 2). Noticeably, S. epidermidis produced least slime, whereas the two S. aureus strains (MSSA 6538 and MRSA MW2) produced large amounts.
Figure 2. Inhibition of slime production by alizarin.
Slime production was analyzed using Congo red agar plates. Three S. aureus strains (MSSA 25923, MSSA 6538, and MRSA) and a S. epidermidis strain were cultured with and without alizarin (20 μg/ml) on Congo red agar plates for 24 h at 37 °C. Four independent experiments were conducted and one set of representative results is shown. None represents non-treated controls.
The antimicrobial activity of alizarin was investigated by measuring minimum inhibitory concentration (MICs), and the MICs of alizarin against S. aureus MSSA 6538 and S. epidermidis were found to be >1000 μg/ml, which were consistent with previously reported values24. Notably, its MIC against S. aureus was 100-times higher than the concentration (10 μg/ml) required for antibiofilm activity. Furthermore, alizarin at concentrations up to 20 μg/ml did not retard the growth of S. aureus planktonic cells, although at 200 μg/ml it had a slight inhibitory effect (Supplementary Fig. S3). These findings show the reduced biofilm formation caused by alizarin was due to its antibiofilm activity and not to its antimicrobial activity.
Antibiofilm activities of anthraquinone derivatives against S. aureus
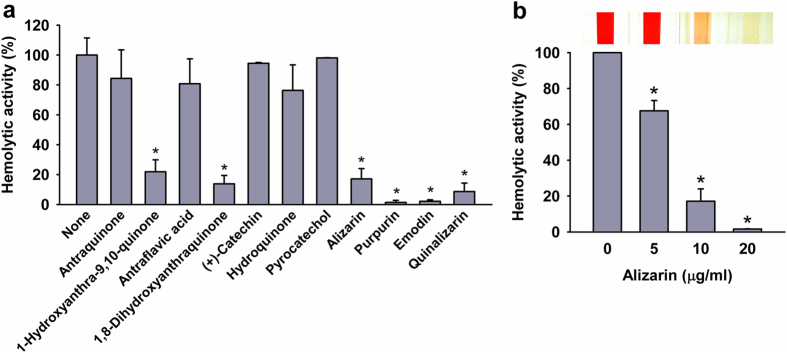
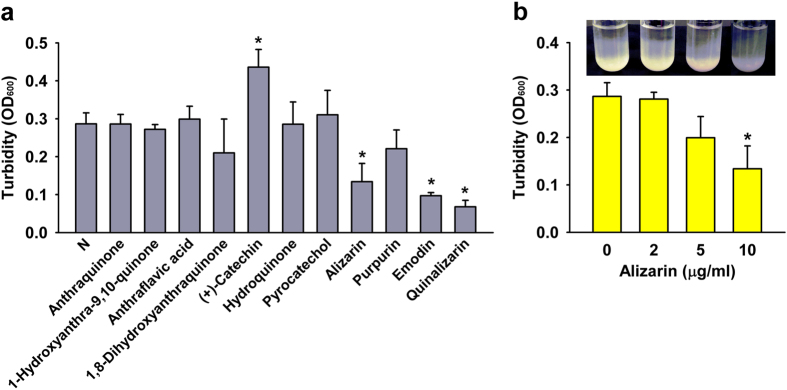
Since alizarin (1,2-dihydroxyanthraquinone) is an anthraquinone, we also investigated the antibiofilm activities of eleven other anthraquinone-related compounds (Fig. 3a). It was found alizarin, emodin, purpurin, and quinalizarin at 10 μg/ml markedly inhibited S. aureus MSSA 6538 biofilm formation by ≥70% versus untreated controls, whereas the other seven compounds had no significant effect (Fig. 3b).
Figure 3. Inhibition of biofilm formation by alizarin-related chemicals.
Chemical structures are shown (a). Hydroxyl groups are shown in red and the two hydroxyls at C-1 and C-2 of anthraquinone are shown in blue. Biofilm formation by MSSA 6538 was quantified in the presence of selected chemicals after incubation for 24 h in 96-well polystyrene plates without shaking (b). At least two independent experiments were conducted (6 wells per sample). Error bars indicate standard deviations. *P < 0.05 versus non-treated controls (None).
Interestingly, antibiofilm activity against S. aureus was found to be closely related to the number and position of hydroxyl units (Fig. 3). Hydroxyls at the C-1 and C-2 positions of the anthraquinone skeleton appeared to be important for antibiofilm activity, because alizarin, purpurin, and quinalizarin possess a hydroxyl group at both positions (Fig. 3a). However, pyrocatechol (1,2-dihydroxybenzene), which has two hydroxyl units in a benzene structure lacked inhibitory activity, indicating that the anthraquinone backbone and the C-1 and C-2 hydroxyl units are required for antibiofilm activity. Emodin, which has a methyl group at the C-6 position, inhibited planktonic growth, as previously reported25, and the additional hydroxyl units at positions other than C-1 and C-2 of purpurin and quinalizarin had minor effects on antibiofilm activity. The same pattern of antibiofilm activities was observed for the other two S. aureus strains, MSSA 25923 and MRSA MW2 (Supplementary Fig. S4). Because alizarin reduced biofilm formation most, we focused on alizarin for further study.
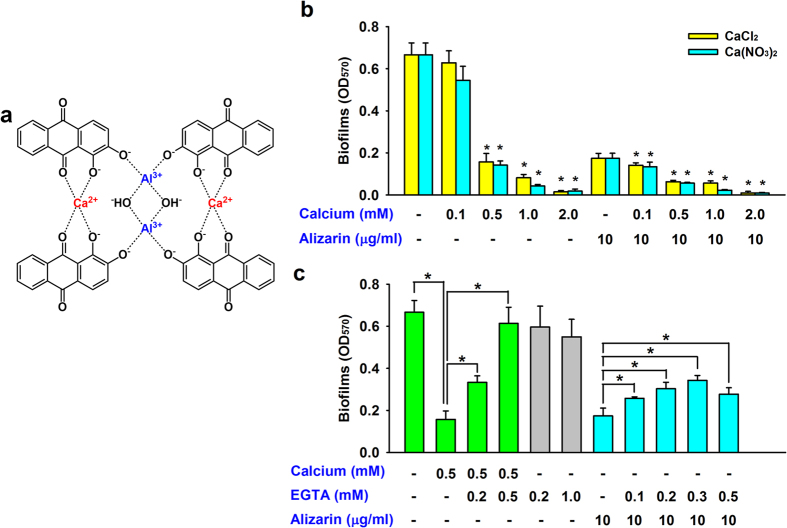
The inhibitory activity of alizarin and the effect of Ca2+
Alizarin forms a calcium/aluminum complex (Fig. 4a)26, and it has been previously shown that Ca2+ participates in27 and inhibits S. aureus biofilm formation28. When we investigated the effect of calcium and alizarin on S. aureus MSSA 6538 biofilm formation, two sources of Ca2+ (CaCl2 and Ca(NO3)2) were found to dose-dependently and similarly inhibit S. aureus biofilm formation (Fig. 4b). In addition, the antibiofilm activity of alizarin was augmented by Ca2+ (Fig. 4b). Furthermore, the inhibitory effect of Ca2+ disappeared in the presence of EGTA (ethylene glycol tetraacetic acid; a calcium-specific chelating agent), whereas EGTA alone at concentrations up to 1 mM did not influence biofilm formation (Fig. 4c). Furthermore, the addition of EGTA in the presence of alizarin partially decreased the antibiofilm effect of alizarin (Fig. 4c). These results suggest alizarin inhibits S. aureus biofilm formation with the involvement of Ca2+.
Figure 4. The effect of Ca2+ and alizarin on biofilm formation by S. aureus.
Structures of the alizarin/calcium/aluminum complex (a)26. Biofilm formation by MSSA 6538 was quantified in M9 minimal medium supplemented with Ca2+ (b). EGTA (a calcium-specific chelator) complements the effect of calcium and decreases the effect of alizarin (c). At least two independent experiments were conducted (6 wells for each sample). Error bars indicate standard deviations. *P < 0.05 for the indicated pairs of groups.
Inhibition of the hemolytic effects of S. aureus by alizarin and other anthraquinones
S. aureus produces α-toxin that causes hemolysis29 and contributes to biofilm formation8, and thus, we investigated the effects of alizarin and of 10 other anthraquinone-related compounds on blood hemolysis by S. aureus (Fig. 5). In accord with observed antibiofilm activities, alizarin, emodin, purpurin, and quinalizarin at 10 μg/ml inhibited the hemolytic activity of S. aureus MSSA 6538 by ≥70% versus untreated controls (Fig. 5a). 1-Hydroxyanthra-9,10-quinone and 1,8-dihydroxyanhraquinone also showed anti-hemolytic activity, indicating that the hydroxyl unit at C-1 plays an important role in the inhibition of hemolysis by S. aureus. This result also suggests that biofilm inhibitions by alizarin, purpurin, and quinalizarin are associated with the inhibition of hemolytic activity. Furthermore, alizarin (0, 5, 10, or 20 μg/ml) dose-dependently reduced hemolysis by S. aureus, and at 20 μg/ml (84 μM) alizarin completely abolished the hemolytic activity of S. aureus (Fig. 5b).
Figure 5. Anti-hemolytic activities of alizarin and other anthraquinones.
The hemolysis of human blood by S. aureus MSSA 6538 was quantified in the presence of anthraquinone-related compounds at 10 μg/ml (a) or alizarin (0, 5, 10, or 20 μg/ml) (b) after incubation for 20 h. Pictures of spectrophotometer cuvettes are shown. At least two independent experiments were conducted. *P < 0.05 versus non-treated controls (None or 0).
Alizarin, purpurin, and quinalizarin increased cell aggregation
Since polyphenols bind to proteins and cause the formation of insoluble aggregates30, we investigated the abilities of the 11 anthraquinone-related compounds to induce aggregation. Interestingly, alizarin and quinalizarin caused obvious aggregation of S. aureus MSSA 6538, while the other compounds did not (Fig. 6a). In the case of emodin, low optical density was due to low cell growth, but not due to cell aggregation. On the other hand, alizarin dose-dependently increased cell aggregation (Fig. 6b). Furthermore, aggregation results were generally in-line with the observed antibiofilm and anti-hemolytic activities of alizarin and quinalizarin.
Figure 6. Effects of alizarin-related compounds on cell aggregation.
S. aureus MSSA 6538 cells were grown for 20 h in the presence of alizarin-related compounds (10 μg/ml) (a) or in the presence of alizarin (0, 2, 5, or 10 μg/ml) (b). Absorbances of the top 1 ml portions of test tubes were measured at OD600. Tested tubes are shown. At least two independent experiments were conducted. *P < 0.05 versus non-treated controls (N or 0).
Alizarin modulated the expressions of biofilm- and toxin-related genes
To investigate the molecular mechanism underlying the antibiofilm and anti-hemolytic activities of alizarin in S. aureus MSSA 6538, we examined the differential expressions of 22 biofilm- and toxin-related genes using planktonic S. aureus cells by real-time qRT-PCR. As shown in Fig. 7, alizarin altered the expressions of many genes. Of particular note, and in accord with its observed inhibition of hemolytic activity (Fig. 5), alizarin repressed expression of the α-hemolysin gene (hla) by 9-fold (Fig. 7a), and significantly repressed the expressions of the biofilm-related genes, psmα (phenol soluble modulins α), rbf (clumping factor B), and spa (surface protein A) (Fig. 7a). In addition, alizarin altered the expressions of the cid and lrg genes, which constitute the holin-antiholin system. Notably, alizarin induced cidB, which encodes for a holin-like protein, by more than 13-fold, but repressed lrgAB, which encode for antiholin proteins. However, the expressions of other biofilm-related genes, such as, intercellular adhesion locus genes (icaA, icaD, and icaR), proteases genes (aur and clp9), and other biofilm regulators (clfB, coa isaA, and sarA) were relatively unaffected by alizarin. In addition, alizarin repressed the expressions of agrA and of the nuclease genes (nuc1 and nuc2) (Fig. 7). Although it has been established that agrA quorum-sensing causes dispersal31, alizarin did not induce biofilm dispersal (data not shown), which suggests the mode of action of alizarin is less associated with biofilm dispersal systems, such as, agr quorum sensing or the actions of proteases and nucleases.
Figure 7. Transcriptional profiles of S. aureus cells treated with or without alizarin.
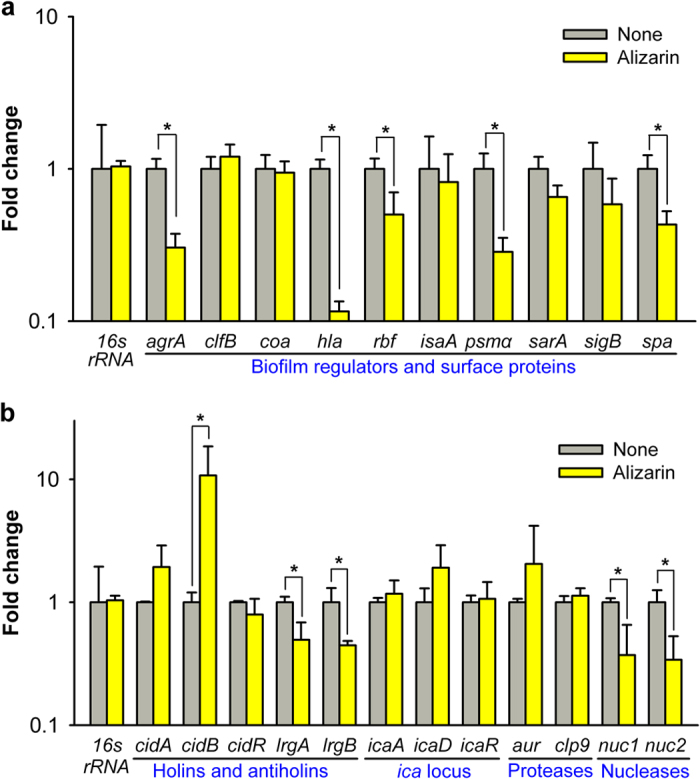
S. aureus MSSA 6538 was cultivated to an A600 of 1 and then incubated with or without alizarin (20 μg/ml) for 2 h with shaking at 250 rpm. Transcriptional profiles were measured by qRT-PCR. The expression level of 16s rRNA was used to normalize the expressions of genes of interest. Fold changes represent changes in transcriptions of treated versus untreated S. aureus. The experiment was performed in triplicate (6 qRT-PCR reactions were performed per gene). *P < 0.05 versus non-treated controls (None).
Discussion
The present study demonstrates for the first time that alizarin, purpurin, and quinalizarin exhibit antibiofilm and anti-hemolytic activity against S. aureus. In addition, it identifies chemical structure-activity relationships and partially reveals the action mechanisms underlying their antibiofilm effects.
Hydroxyanthraquinones are found in various plants, and mixtures of anthraquinones have long been employed in medical preparations as laxatives32. Furthermore, the toxicities of alizarin, purpurin, and quinalizarin are relatively low as compared with many phenolic agents32. Alizarin (also known as Turkey Red) was originally derived from the roots of the madder genus and has been used as a red dye. Alizarin stains ionic calcium in bones and for this reason has been widely used in studies on bone growth26. In the present study, alizarin at 10 μg/ml (100-times lower than its MIC) significantly inhibited biofilm formation by and the hemolytic activity of S. aureus, indicating that it is a non-toxic biofilm inhibitor.
One of objectives of the present study was to identify the structural motif present in anthraquinones responsible for antibiofilm and anti-hemolytic activities against S. aureus. Our results suggest that the anthraquinone backbone and two hydroxyl units at the C-1 and C-2 positions of the anthraquinone skeleton are required for the observed antibiofilm (Fig. 3) and anti-hemolytic effects of anthraquinones on S. aureus (Fig. 5). Many alizarin derivatives can be synthesized, and thus, further investigations of alizarin derivatives are probably worthwhile. Previously, it was reported that emodin reduced biofilm formation by another Gram-positive oral bacterium, Streptococcus mutans33. In the present study, the antimicrobial activity of emodin was found to be responsible for inhibiting biofilm formation by and the hemolytic activity of S. aureus (Figs 3 and 5).
Staphylococcal biofilms are encased in an extracellular matrix composed of proteins, polysaccharides, and extracellular DNA. The mechanism of biofilm formation by S. aureus is complicated and involves environmental factors, quorum sensing, proteases, DNase, several surface proteins, and other global regulators6,7. In the present study, we investigated the transcriptional levels of various biofilm- and toxin-related genes and found positive biofilm regulators (psmα, rbf, and spa) were repressed by alizarin (Fig. 7), which supports its biofilm reducing effect on S. aureus. Phenol-soluble modulins (PSMs) are a novel family of toxins and play multiple roles in the pathogeneses of staphylococcal infections, which typically involve blood cell lysis and biofilm development34,35. On the other hand, Rbf is an activator of biofilm formation by S. aureus36, and was found to promote virulence in a murine model of infection37. Furthermore, surface protein A (SpA) production has been reported to be essential for biofilm formation by S. aureus38. Thus, our findings show that alizarin down-regulates several important biofilm regulators in this bacterium.
S. aureus produces four hemolysins (alpha, beta, gamma, and delta), which have hemolytic, cytotoxic, and dermonecrotic properties39. In particular, α-toxin (Hla) causes hemolysis29 and contributes to biofilm formation8. In the present study, alizarin, purpurin, and quinalizarin showed antibiofilm and anti-hemolytic activities (Figs 3 and 5), and previous studies have shown that several flavonoids16, nerolidol22, stilbenoids40, and thermoresponsive oligo (N-vinylcaprolactam)41 have antibiofilm activity and anti-hemolytic activity against S. aureus. Thus, it appears there is a positive relation between antibiofilm and anti-hemolytic activities.
In the present study, alizarin markedly up-regulated the gene expression of holin-like protein (CidB) and down-regulated those of antiholin proteins (LrgAB) (Fig. 7). Holin (CidA) and antiholin (LrgA) may serve as molecular control elements of bacterial cell lysis and play significant roles during biofilm development42. Like cidA and lrgA, the cidB and lrgB genes encode homologous hydrophobic proteins, but the functions of these have not been well established43. Although the mechanism responsible for modulation of the Cid/Lrg system by alizarin is unclear, alizarin could affect bacterial cell wall integrity. Interestingly, our qRT-PCR data revealed a gene modulation pattern similar to that of a synthetic antibiofilm agent CCG-203592, which temporally down-regulates the expressions of hla, lrgA, psmα, and spa, but temporally up-regulates cidA in S. aureus44. Further genetic studies should provide more detail of the molecular mechanisms responsible for the effects of alizarin and its derivatives.
It is generally believed that cell aggregation is a prerequisite of biofilm development7. However in the present study, alizarin and quinalizarin increased cell aggregation (Fig. 6) but decreased biofilm formation (Fig. 3). Nevertheless, these results are in-line with those of a recent study on the effects of proanthoyanides on biofilm formation by S. epidermidis, in which it was proposed that inhibition of bacterial attachment is based on electrostatic repulsion and changes in hydrophobicity45.
Ca2+ plays a role in S. aureus biofilm formation27 and at millimolar concentrations has an inhibitory effect28. Furthermore, calcium addition has been reported to decrease α-hemolysin-induced hemolytic activity by S. aureus46. Our results suggest that alizarin and complexed Ca2+ at micromolar concentrations effectively inhibit the biofilm and hemolytic activities of S. aureus (Figs 3 and 5). However, it remains to be determined how alizarin and Ca2+ function at the molecular level in S. aureus cells.
Because the long-term use of antibiotics has generated multidrug resistant bacteria like MRSA, novel strategies are urgently required to control antibiotic resistant Staphylococcus strains, and strategies based on inhibiting biofilm formation and toxin production offer an alternative means of reducing bacterial virulence. The present study shows for the first time that the alizarin exhibits antibiofilm and anti-hemolytic activities and down-regulates the expressions of various biofilm- and toxin-related genes, and thus, identifies alizarin and its derivatives as potential antivirulence compounds against recalcitrant S. aureus.
Methods
Ethics statement
Hemolysis experiment was approved by the Ethical Committee of Yeungnam University, Gyeongsan, Korea and the methods were carried out as per the guidelines of the Ethical Committee of Yeungnam University. All participants provided written informed consent for blood collection and research.
Bacterial strains, growth measurements, and materials
The following bacterial strains were used in the present study; methicillin-sensitive S. aureus strains (MSSA; ATCC 25923 and ATCC 6538), a methicillin-resistant S. aureus strain (ATCC BAA-1707, MW2), S. epidermidis (ATCC 14990), Pseudomonas aeruginosa PAO1 (ATCC 15692), and Escherichia coli O157:H7 (ATCC 43895, EDL933). Experiments were conducted on the two MSSA strains and S. epidermidis, P. aeruginosa PAO1, and E. coli O157:H7 at 37 °C in LB medium, and on the MRSA strain in LB medium containing 0.2% glucose. For cell growth measurements, colony forming units (CFUs) were measured by spreading cell cultures on LB agar plates. For the MIC experiment, cells were inoculated with overnight culture at a dilution of 1:100 in LB medium and cultured for 24 h at 37 °C. After serial dilutions, cultures were spread on LB agar plates, incubated for 24 h at 37 °C, and cell colonies were counted. Each experiment was performed using at least two independent cultures.
We have established a library of 560 phytochemicals, and deposited it in the Natural Product Library in the Korea Chemical Bank (http://www.chembank.org, Daejeon, Republic of Korea). These 560 compounds were purified from various plant sources and included terpenoids, flavonoids, polyphenols, and saponins, as we previously described47. All were dissolved in dimethyl sulfoxide (DMSO). Alizarin and ten other anthraquinone-related compounds, namely, anthraflavic acid, anthraquinone, (+)-catechin, 1,8-dihydroxyanthraquinone, emodin, 1-hydroxyanthra-9,10-quinone, hydroquinone, purpurin, pyrocatechol, and quinalizarin were purchased from Sigma-Aldrich (St. Louis, USA).
Crystal-violet biofilm assay
A static biofilm formation assay was performed on six bacterial strains (MSSA 6538, MSSA 25923, MRSA MW2, S. epidermidis, Pseudomonas aeruginosa PAO1, and Escherichia coli O157:H7) in 96-well polystyrene plates (SPL Life Sciences, Korea), as previously reported48. Briefly, cells were inoculated into LB medium (total volume 300 μl) at an initial turbidity of 0.05 at 600 nm. Antibiofilm agents were added at different concentrations at inoculation and cultured for 24 h without shaking at 37 °C. To quantify biofilm formation, biofilms were stained with crystal violet for 20 min, dissolved in 300 μl of 95% ethanol, and absorbances were measured at 570 nm (OD570). Cell growths in 96-well plates were also measured at 620 nm (OD620). Biofilm formation and static cell growth results are presented as the averages of two independent cultures of twelve replicate wells.
Biofilm cell counting assay
To confirm biofilm inhibition, we performed viable counts on biofilm cells. Biofilm cells were formed in 96-well polystyrene plates for 24 h with or without alizarin (as mentioned above), and the biofilms obtained were washed three times with phosphate-buffered saline (PBS). Biofilms were then resuspended in 300 μl PBS, pipetted vigorously for 60 sec, vortexed for 30 sec (to disrupt the biofilms), serially diluted, and plated on LB agar plates. CFUs were counted after overnight incubation at 37 °C. To check complete biofilm disruption had been achieved, we used the crystal-violet biofilm assay after vigorous pipetting. Three independent experiments were conducted.
Slime assay using Congo red agar (CRA)
Colony morphologies and phenotypic changes were investigated using CRA, as previously described23. The CRA was composed of 37 g/L of brain–heart infusion broth (BD Biosciences, Franklin Lakes, NJ, USA), 36 g/L of sucrose (Sigma, St. Louis, MO, USA), 15 g/L of agar (BD Biosciences, Franklin Lakes, NJ, USA), and 0.8 g/L of Congo red (Sigma, St. Louis, MO, USA). Staphylococcus cells (MSSA 6538, MSSA 25923, MRSA MW2, and S. epidermidis) on CRA were incubated with and without alizarin for 24 h at 37 °C before taking images. Four independent experiments were conducted.
Confocal laser microscopy and COMSTAT analysis
Biofilm formations by S. aureus (MSSA 6538, MSSA 25923, and MRSA MW2) on glass were evaluated by confocal laser microscopy (Nikon Eclipse Ti, Tokyo) and compared with S. aureus biofilms grown in medium alone. S. aureus cells were stained with carboxyfluorescein diacetate succinimidyl ester (Catalog #:C34554 Invitrogen, Molecular Probes, Inc, Eugene, USA)49, which is a minimally fluorescent lipophile, but on entering cells esterases remove its acetyl groups to become markedly the fluorescent50. Hence, this fluorescent dye targets viable cells in biofilms. Briefly biofilms were allowed to form by incubating 96-well plates for 24 hr at 37 °C without shaking, washed with PBS twice, stained with carboxyfluorescein diacetate succinimidyl ester (2.8 μg/ml in PBS) for 20 min at 37 °C, and rewashed twice with PBS. Samples were visualized using a 40 x objective and an Ar laser (excitation 488 nm; emission 500 to 550 nm). Confocal images of same strains were captured using the same conditions. Color confocal images were constructed using NIS-Elements C version 3.2 (Nikon eclipse). At least 4 random positions in three independent cultures were subjected to analysis.
To quantify biofilm formation, color confocal images (20 image stacks) were converted to gray scale using ImageJ. COMSTAT biofilm software51 was used to determine biomasses (μm3 per μm2), mean thicknesses (μm), and substratum coverages (%). Thresholding was fixed for all image stacks, and at least 4 positions and 20 planar images were analyzed per position.
Hemolysis assay
Human red blood cell lysis efficacies were measured using whole cultures of S. aureus grown in the presence of biofilm inhibitors, as described previously13. Briefly, S. aureus cells (MSSA 6538) were diluted 1:100 in LB medium and cultured with or without test compounds for 20 h at 250 rpm. Cell cultures (cells and supernatants) were then added to the diluted human red blood cells (previously separated by centrifugation at 890 x g for 2 min and washed 3 times with PBS (330 μl red blood cells/10 ml of PBS buffer). To determine hemolytic activities, mixtures of blood and S. aureus (200 μl of cell culture) were incubated at 250 rpm for 1 h at 37 °C. Supernatants were collected by centrifugation at 16,600 x g for 10 min and optical densities were measured at 543 nm.
Cell aggregation assay
Cell aggregation was analyzed as previously reported52. Briefly, S. aureus cells (MSSA 6538) were inoculated into 2 ml of LB medium in 14-ml test tubes with or without alizarin or alizarin-related compounds and incubated for 20 h with shaking at 250 rpm. Cell cultures (1 ml) were then collected by centrifugation at 16,600 x g for 2 min and cells were washed with PBS 3 times. Washed cells were resuspended in 3 ml of PBS in clean glass tubes and allowed to stand for 20 h at room temperature. Cell turbidities of the top portions of tubes were measured at OD600 using a spectrophotometer (UV/Vis, spectrophotometer, Optizen, Korea).
RNA isolation
For qRT-PCR (quantitative real-time reverse transcription polymerase chain reaction) experiments, the RNAs of S. aureus cells were isolated using the following procedure. S. aureus cells (MSSA 6538) were inoculated into 25 ml of LB medium at 37 °C in 250 ml shake flasks with overnight cultures (1 : 100 dilution) and cultured for 3 h with shaking at 250 rpm. Alizarin was then added to a concentration of 20 μg/ml, at which it showed significant antibiofilm and anti-hemolytic activity, and incubated for 2 h. Before sample collection, RNase inhibitor (Ambion, TX, USA) was added and planktonic cells were immediately chilled for 30 sec with dry ice and 95% ethanol to prevent RNA degradation. Cells were then centrifuged at 16,600 x g for 1 min and the cell pellets obtained were immediately frozen with dry ice and stored at −80 °C. RNA was isolated using a Qiagen RNeasy mini Kit (Valencia, CA, USA). To remove all DNA, purified RNA was treated with 30 units of DNase I for 15 min. RNA quality was assessed using a NanoVue Plus (Biochrom Ltd., Cambridge, UK).
qRT-PCR
qRT-PCR was used to assess the transcription levels of biofilm-related genes (agrA, aur, cidA, cidB, cidR, clfB, clp9, coa, hla, icaA, icaD, icaR, isaA, lrgA, lrgB, nuc1, nuc2, psmα, rbf, sarA, sigB, and spa) in S. aureus (MSSA 6538) cells. Gene specific primers were used for these genes and appropriate primers for 16s rRNA as a housekeeping control (Supplementary Table S3), which was used to normalize the expressions of genes of interest. The qRT-PCR method employed was adapted from a previous study53, and performed using a SYBR Green master mix (Applied Biosystems, Foster City, USA) and an ABI StepOne Real-Time PCR system (Applied Biosystems). Expression levels were determined using three independent cultures and six qRT-PCR reactions for each gene.
Statistical analysis
Sample sizes of all experiments are indicated in ‘Methods’. Average values are expressed as means ± standard deviations, and the Student’s t-test was used to determine the significances differences between samples and non-treated controls. Statistical significance was accepted for p values < 0.05, and significant changes are indicated using asterisks in figures.
Additional Information
How to cite this article: Lee, J.-H. et al. Calcium-chelating alizarin and other anthraquinones inhibit biofilm formation and the hemolytic activity of Staphylococcus aureus. Sci. Rep. 6, 19267; doi: 10.1038/srep19267 (2016).
Supplementary Material
Acknowledgments
All plant secondary metabolites used in this study were kindly provided by the Korean Chemical Bank at the Korean Research Institute of Chemical Technology. This research was supported by National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (#2015R1A2A2A01004542 to J. Lee) and Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2014R1A6A1031189).
Footnotes
Author Contributions J.-H.L. and Y.-G.K. designed the study, performed experiments, and analyzed data. S.Y.R. established a library of phytochemicals. J.-H.L. and J.L. designed experiments and wrote the manuscript. All authors have read and approved the final manuscript.
References
- Potera C. Forging a link between biofilms and disease. Science 283, 1837–1839 (1999). [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S. & Greenberg E. P. Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 (1999). [DOI] [PubMed] [Google Scholar]
- Hoffman L. R. et al. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175 (2005). [DOI] [PubMed] [Google Scholar]
- Lowy F. D. Staphylococcus aureus infections. N Engl J Med 339, 520–532 (1998). [DOI] [PubMed] [Google Scholar]
- Ohlsen K., Koller K. P. & Hacker J. Analysis of expression of the alpha-toxin gene (hla) of Staphylococcus aureus by using a chromosomally encoded hla::lacZ gene fusion. Infect Immun 65, 3606–3614 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles B. R. & Horswill A. R. Staphylococcal biofilm disassembly. Trends Microbiol 19, 449–455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciola C. R., Campoccia D., Speziale P., Montanaro L. & Costerton J. W. Biofilm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for biofilm-resistant materials. Biomaterials 33, 5967–5982 (2012). [DOI] [PubMed] [Google Scholar]
- Caiazza N. C. & O’toole G. A. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J Bacteriol 185, 3214–3217 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. W. H. & Vederas J. C. Drug discovery and natural products: End of an era or an endless frontier? Science 325, 161–165 (2009). [DOI] [PubMed] [Google Scholar]
- Wang D. et al. Transcriptional and functional analysis of the effects of magnolol: inhibition of autolysis and biofilms in Staphylococcus aureus. PloS One 6, e26833 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quave C. L. et al. Ellagic acid derivatives from Rubus ulmifolius inhibit Staphylococcus aureus biofilm formation and improve response to antibiotics. PloS One 7, e28737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D. E. et al. Tannic acid inhibits Staphylococcus aureus surface colonization in an IsaA dependent manner. Infect Immun (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H. et al. Antibiofilm activities of quercetin and tannic acid against Staphylococcus aureus. Biofouling 29, 491–499 (2013). [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Kim Y.-G., Ryu S. Y., Cho M. H. & Lee J. Ginkgolic acids and Ginkgo biloba extract inhibit Escherichia coli O157:H7 and Staphylococcus aureus biofilm formation. Int J Food Microbiol 174, 47–55 (2014). [DOI] [PubMed] [Google Scholar]
- Yadav M. K., Chae S. W., Im G. J., Chung J. W. & Song J. J. Eugenol: a phyto-compound effective against methicillin-resistant and methicillin-sensitive Staphylococcus aureus clinical strain biofilms. PloS One 10, e0119564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H. S., Lee J.-H., Cho M. H. & Lee J. Red wines and flavonoids diminish Staphylococcus aureus virulence with anti-biofilm and anti-hemolytic activities. Biofouling 31, 1–11 (2015). [DOI] [PubMed] [Google Scholar]
- Nostro A. et al. Effects of oregano, carvacrol and thymol on Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Med Microbiol 56, 519–523 (2007). [DOI] [PubMed] [Google Scholar]
- Schillaci D., Arizza V., Dayton T., Camarda L. & Di Stefano V. In vitro anti-biofilm activity of Boswellia spp. oleogum resin essential oils. Lett Appl Microbiol 47, 433–438 (2008). [DOI] [PubMed] [Google Scholar]
- Kwieciński J., Eick S. & Wójcik K. Effects of tea tree (Melaleuca alternifolia) oil on Staphylococcus aureus in biofilms and stationary growth phase. Int J Antimicrob Agents 33, 343–347 (2009). [DOI] [PubMed] [Google Scholar]
- Nuryastuti T. et al. Effect of cinnamon oil on icaA expression and biofilm formation by Staphylococcus epidermidis. Appl Environ Microbiol 75, 6850–6855 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adukwu E. C., Allen S. C. & Phillips C. A. The anti-biofilm activity of lemongrass (Cymbopogon flexuosus) and grapefruit (Citrus paradisi) essential oils against five strains of Staphylococcus aureus. J Appl Microbiol 113, 1217–1227 (2012). [DOI] [PubMed] [Google Scholar]
- Lee K., Lee J.-H., Kim S. I., Cho M. H. & Lee J. Anti-biofilm, anti-hemolysis, and anti-virulence activities of black pepper, cananga, myrrh oils, and nerolidol against Staphylococcus aureus. Appl Microbiol Biotechnol 98, 9447–9457 (2014). [DOI] [PubMed] [Google Scholar]
- Freeman D. J., Falkiner F. R. & Keane C. T. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol 42, 872–874 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Uchida K., Takizawa T., Yamaguchi H. & Abe S. Evaluation of the effect of terpenoid quinones on Trichophyton mentagrophytes by solution and vapor contact. J Infect Chemother 12, 100–104 (2006). [DOI] [PubMed] [Google Scholar]
- Ayo R. G., Amupitan J. O. & Zhao Y. Cytotoxicity and antimicrobial studies of 1,6,8- trihydroxy-3-methyl-anthraquinone (emodin) isolated from the leaves of Cassia nigricans Vahl. Afr J Biotechnol 6, 1276–1279 (2007). [Google Scholar]
- Puchtler H., Meloan S. N. & Terry M. S. On the history and mechanism of alizarin and alizarin red S stains for calcium. J Histochem Cytochem 17, 110–124 (1969). [DOI] [PubMed] [Google Scholar]
- Cucarella C. et al. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun 72, 2177–2185 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. K. & Rao T. S. Effect of calcium on Staphylococcus aureus biofilm architecture: a confocal laser scanning microscopic study. Colloid Surface B 103, 448–454 (2013). [DOI] [PubMed] [Google Scholar]
- Song L. et al. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866 (1996). [DOI] [PubMed] [Google Scholar]
- Charlton A. J. et al. Polyphenol/peptide binding and precipitation. J Agr Food Chem 50, 1593–1601 (2002). [DOI] [PubMed] [Google Scholar]
- Boles B. R. & Horswill A. R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 4, e1000052 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendelbach L. E. A review of the toxicity and carcinogenicity of anthraquinone derivatives. Toxicology 57, 227–240 (1989). [DOI] [PubMed] [Google Scholar]
- Coenye T., Honraet K., Rigole P., Nadal Jimenez P. & Nelis H. J. In vitro inhibition of Streptococcus mutans biofilm formation on hydroxyapatite by subinhibitory concentrations of anthraquinones. Antimicrob Agents Chemother 51, 1541–1544 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy S. et al. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci USA 109, 1281–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med 64, 175–188 (2013). [DOI] [PubMed] [Google Scholar]
- Lim Y., Jana M., Luong T. T. & Lee C. Y. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J Bacteriol 186, 722–729 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T. T., Lei M. G. & Lee C. Y. Staphylococcus aureus Rbf activates biofilm formation in vitro and promotes virulence in a murine foreign body infection model. Infect Immun 77, 335–340 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino N. et al. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol 191, 832–843 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges M. M., Orwin P. M. & Schlievert P. M. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 13, 16–34 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Lee J.-H., Ryu S. Y., Cho M. H. & Lee J. Stilbenes reduce Staphylococcus aureus hemolysis, biofilm formation, and virulence. Foodborne Pathog Dis 11, 710–717 (2014). [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Kim Y.-G., Lee K., Kim S. C. & Lee J. Temperature-dependent control of Staphylococcus aureus biofilms and virulence by thermoresponsive oligo(N-vinylcaprolactam). Biotechnol Bioeng 112, 716–724 (2015). [DOI] [PubMed] [Google Scholar]
- Rice K. C. et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci USA 104, 8113–8118 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles K. W. The biological role of death and lysis in biofilm development. Nat Rev Microbiol 5, 721–726 (2007). [DOI] [PubMed] [Google Scholar]
- Ma Y. et al. Novel inhibitors of Staphylococcus aureus virulence gene expression and biofilm formation. PloS one 7, e47255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentin D. S. et al. Natural Green coating inhibits adhesion of clinically important bacteria. Sci Rep 5, 8287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman S. & Sugg N. Effect of calcium ions on staphylococcal alpha-toxin-induced hemolysis of rabbit erythrocytes. Infect Immun 47, 37–40 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H. et al. Coumarins reduce biofilm formation and the virulence of Escherichia coli O157:H7. Phytomedicine 21, 1037–1042 (2014). [DOI] [PubMed] [Google Scholar]
- Park J. H., Lee J. H., Cho M. H., Herzberg M. & Lee J. Acceleration of protease effect on Staphylococcus aureus biofilm dispersal. FEMS Microbiol Lett 335, 31–38 (2012). [DOI] [PubMed] [Google Scholar]
- Weston S. A. & Parish C. R. New fluorescent dyes for lymphocyte migration studies. Analysis by flow cytometry and fluorescence microscopy. J Immunol Methods 133, 87–97 (1990). [DOI] [PubMed] [Google Scholar]
- Lyons A. B. Analysing cell division in vivo and in vitro using flow cytometric measurements of CFSE dye dilution. J Immunol Methods 243, 147–154 (2000). [DOI] [PubMed] [Google Scholar]
- Heydorn A. et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146, 2395–2407 (2000). [DOI] [PubMed] [Google Scholar]
- Haaber J., Cohn M. T., Frees D., Andersen T. J. & Ingmer H. Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PloS one 7, e41075 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Cho M. H. & Lee J. 3-Indolylacetonitrile decreases Escherichia coli O157:H7 biofilm formation and Pseudomonas aeruginosa virulence. Environ Microbiol 13, 62–73 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.