Abstract
Contact-dependent inhibition (CDI) toxins, delivered into the cytoplasm of target bacterial cells, confer to host strain a significant competitive advantage. Upon cell contact, the toxic C-terminal region of surface-exposed CdiA protein (CdiA-CT) inhibits the growth of CDI- bacteria. CDI+ cells express a specific immunity protein, CdiI, which protects from autoinhibition by blocking the activity of cognate CdiA-CT. CdiA-CT are separated from the rest of the protein by conserved peptide motifs falling into two distinct classes, the “E. coli”- and “Burkholderia-type”. CDI systems have been described in numerous species except in Pseudomonadaceae. In this study, we identified functional toxin/immunity genes linked to CDI systems in the Pseudomonas genus, which extend beyond the conventional CDI classes by the variability of the peptide motif that delimits the polymorphic CdiA-CT domain. Using P. aeruginosa PAO1 as a model, we identified the translational repressor RsmA as a negative regulator of CDI systems. Our data further suggest that under conditions of expression, P. aeruginosa CDI systems are implicated in adhesion and biofilm formation and provide an advantage in competition assays. All together our data imply that CDI systems could play an important role in niche adaptation of Pseudomonadaceae.
Introduction
Bacteria share habitats and have therefore developed cooperative and antagonistic behaviors that facilitate their struggle for living space and nutrient availability. To compete with each other, bacteria have evolved sophisticated weapons such as the polymorphic toxin systems to impede the proliferation of other microorganisms [1,2]. Polymorphic toxin systems are characterized by the presence of an immunity gene immediately downstream of the toxin gene whose product protects bacteria against suicide and fratricide. One of the major pathways for delivery of polymorphic toxins is the Contact-Dependent growth Inhibition (CDI) system, which is implicated in inter-bacterial competition and allows growth inhibition of neighboring competitors via direct cell-cell contact [3].
CDI systems are composed of the CdiA toxin secreted by CdiB, an outer membrane β-barrel transporter, and a CdiI immunity protein. CdiA and CdiB are part of the Two-Partner Secretion (TPS) system family. CdiA are very large filamentous proteins predicted to extend far from the cell surface [4]. CdiA homologs share large N-terminal sequences of homology that exhibit haemagglutinin repeats while their C-terminal domains, called CdiA-CT, are highly variable [5]. CdiA-CT carry toxin activity and are delivered to the cytoplasm of the target cell where they degrade DNA or specific tRNAs leading to growth inhibition or death. When present in target cells, cognate CdiI immunity proteins protect bacteria by specifically interacting with CdiA-CT and subsequently blocking their nuclease activity [5–7]. CdiI proteins are also highly variable and share no sequence homology, thus they confer immunity to their cognate toxin but not to toxins deployed by other CDI systems. The peptide motifs delimiting the CdiA-CT from the rest of the protein fall into two distinct classes, the “E. coli-type” and the “Burkholderia-type” restricted to Burkholderia species and a few closely related species of Ralstonia and Cupriavidus [5,8,9]. The genetic organization of the “E. coli”-type CDI locus is cdiBAI and the CdiA-CT regions diverge abruptly after a common VENN motif (PF04829) [5]. In Burkholderia, the CDI system operon is organized as bcpAIOB (Burkholderia CDI proteins A, I, O and B), with the bcpO gene encoding a predicted lipoprotein required for BcpA function. The sequence divergence of the BcpA-CT toxic domain is observed after a conserved Nx(Q/E)LYN motif instead of the VENN motif [8,9].
In addition to mediating inter-bacterial competition, CDI systems are thought to play a role in biofilms establishment. Indeed, the CDI systems of Burkholderia thailandensis E264 and E. coli EC93 are implicated in auto-aggregation, adherence to abiotic surfaces and are required for biofilm formation [8,10,11]. The contribution to biofilm development might facilitate cooperative bacterial behaviors and eventually lead to an exclusion of non-self competitors from the community [12].
Mechanisms and functions of CDI systems have been well studied in Enterobacteria and Burkholderia species, but remain poorly characterized in Pseudomonadaceae. The Pseudomonadaceae includes a heterogeneous set of microorganisms that are able to colonize diverse niches, ranging from terrestrial and aquatic environments to tissues of eukaryotic hosts. To date over 100 different strains have been described with Pseudomonas aeruginosa being the best characterized so far. P. aeruginosa is the primary agent of opportunistic infection in humans, especially in cystic fibrosis (CF) patients, causing both acute and chronic infections. Ghequire and de Mot recently proposed putative CDI gene loci in Pseudomonadaceae through in silico analysis [13]. In this work, we show that predicted CDI loci are indeed widely distributed in Pseudomonas strains and that the pre-toxin motifs fall into at least five distinct classes including the “E. coli-type”. We functionally characterize two CDI systems present in P. aeruginosa PAO1 and show their negative regulation by the RNA-binding protein RsmA. We demonstrate that besides contributing to biofilm formation, these systems also mediate inter-bacterial competition between Pseudomonas. Furthermore we were able to demonstrate that the CdiA-CT domains carry toxic activities and are specifically blocked by their cognate CdiI immunity proteins. Finally, we extend this study showing that P. aeruginosa PA7, PA14 and P. syringae pv. tomato DC3000 strains also produce functional CdiA-CT toxin / CdiI immunity pairs.
Results
Identification of two potential CDI systems in P. aeruginosa PAO1 strain
In silico analysis of P. aeruginosa PAO1 genome revealed that the PA2463-PA2462 and PA0040-PA0041 loci contain characteristic features of a CDI locus. First, PA2463 and PA0040 are predicted to encode TpsB homologues whereas PA2462 and PA0041 contain the conserved TPS domain of TpsA proteins and encode putative large proteins of 570 and 360 kDa respectively. Moreover, PA2462 C-terminal (CT) domain shares 60% identity with MafBMGI-NEM8013, the bacterial EndoU ribonuclease recently identified in Neisseria meningitidis NEM8013 (S1 Fig and [14]) while 3D modelization of PA0041 CT domain revealed a predicted structure similar to the Burkholderia pseudomallei 1026b CdiA-CT ribonuclease domain [6]. We further noticed the presence of small non-annotated ORFs immediately downstream of PA2462 and PA0041, which potentially encode immunity proteins (S2 Fig). Additionally, both loci are flanked by transposable elements (data not shown) and the GC content decreases abruptly below 50% in the region encoding the CT domains and potential immunity proteins (S2 Fig) indicating that these regions might have been acquired by horizontal gene transfer, a characteristic of CDI systems. Interestingly, these clusters share high sequence identity (>95%) in the center and at the 5’ border (S2 Fig). By contrast, CT domain and immunity-encoding sequences share no homology in accordance with the high variability of CdiA-CT/CdiI modules. Based on the similarities with identified CDI systems, we renamed PA2463/PA0040, PA2462/PA0041, and predicted immunity proteins as CdiBPA2463/CdiBPA0040, CdiAPA2462/CdiAPA0041 and CdiIPA2462/CdiIPA0041 respectively.
Expression and regulation of the cdiAPA2462 and cdiAPA0041 genes
A genome-wide study showed that the cdiAPA0041 mRNA level of P. aeruginosa PAK strain increases in ΔrsmA (regulator of secondary metabolism) cells [15]. In order to determine whether RsmA influences cdiA mRNA levels in P. aeruginosa PAO1, we performed qRT-PCR in a ΔrsmA strain. The deletion of rsmA resulted in a ~3.5- to 5-fold increase level of both cdiA mRNAs indicating that RsmA acts as a negative regulator (Fig 1). RsmA is a post-transcriptional regulator that blocks the interaction between the ribosome-binding site (RBS) and the 30S ribosomal subunit and subsequently prevents the initiation of translation leading eventually to destabilization and degradation of the transcript [16]. As translation stabilizes mRNA and can affect the accumulation of transcript in cells, the difference in mRNA levels observed between wild-type and ΔrsmA could reflect a difference in translation levels. Notably putative RsmA binding sequence overlaps with cdiAPA0041 and cdiAPA2462 RBS indicating that CdiAPA0041 and CdiAPA2462 are probably directly regulated by RsmA at post-transcriptional level. Interestingly, RsmA downregulates the expression of several genes involved in biofilm development [16]. To investigate whether growth conditions impact the regulation of cdiA genes, we compared the level of mRNAs extracted from cells grown under static (in LB medium without agitation) and planktonic conditions. In wild-type cells, qRT-PCR analyses showed that cdiAPA2462 and cdiAPA0041 mRNA levels increase by a 4.5- to 8-fold in static conditions compared to planktonic conditions (Fig 1). Interestingly, in static conditions, we observed that rsmA deletion has no longer an effect on transcript levels as mRNA quantities were almost equal in wild-type and ΔrsmA cells (Fig 1).
Fig 1. Analysis of cdiAPA2462 and cdiAPA0041 gene expression profile.
Expression of cdiAPA2462 and cdiAPA0041 in WT and ΔrsmA strains grown 7 h under agitation (planktonic) or without agitation (static) in LB medium. For each gene, expression was normalized to 16S expression and is shown relative to the WT planktonic condition level. Error bars represent the standard error of the mean from three independent experiments. Data were analyzed for significance using a two-tailed Student’s t-test. ** 0.025<p-value <0.01; *** p-value<0.01; ns: non-significant.
Wild-type cells inhibit the growth of strains lacking the entire cdiPA2462 or cdiPA0041 loci
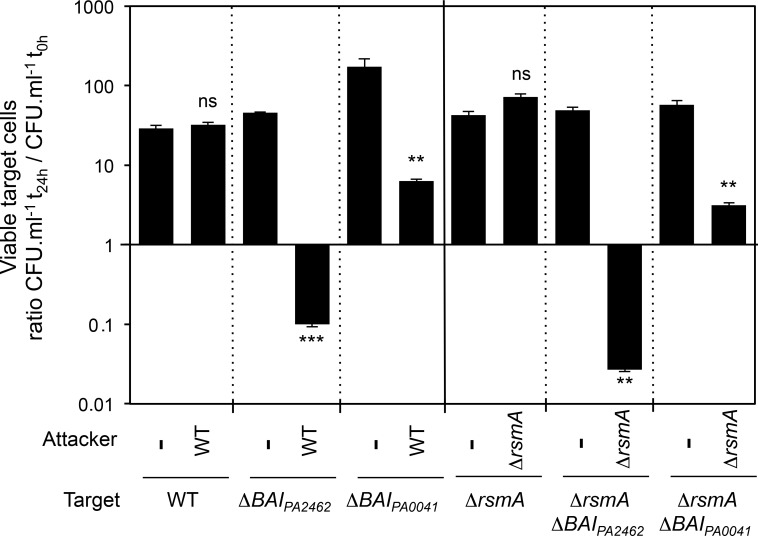
To assess the role of cdiPA2462 and cdiPA0041 loci in bacterial growth inhibition, we performed competition assays. Attacker and gentamycin-resistant target cells were grown separately 4 h in static condition prior to being mixed at an attacker/target ratio of 4:1. Interestingly, no competition occurred with agitated mixed cultures (data not shown). At the beginning of the experiment (t0) and after 24 h, mixed bacteria were serially diluted and plated on gentamycin to determine the CFU/ml of the target. A wild-type target had no growth inhibition when mixed with a wild-type attacker (Fig 2). By contrast, the growth of targets lacking the entire cdiBAIPA2462 or cdiBAIPA0041 locus was inhibited after contact with a wild-type attacker suggesting that at least one gene present in each locus protects from the attack of the wild-type strain (Fig 2). Since our qRT-PCR experiments showed that RsmA is a negative regulator of cdiAPA2462 and cdiAPA0041, we next carried out the competition assays in a ΔrsmA background. In this context, both ΔrsmAΔcdiBAIPA2462 and ΔrsmAΔcdiBAIPA0041 mutant targets still showed a growth defect when mixed with a ΔrsmA attacker (Fig 2). Although the rsmA mutation does not increase the cdiAPA2462 and cdiAPA0041 mRNA levels in static condition (Fig 1), we noticed a larger extent of growth inhibition in the ΔrsmA background (Fig 2). Consequently, as growth inhibition was reproducibly higher in the ΔrsmA background, only results obtained in this condition are shown below.
Fig 2. The growth of cdiBAIPA2462 and cdiBAIPA0041 mutants is inhibited by contact with a wild-type strain.
Gentamycin-resistant wild-type and mutant target bacteria were mixed with or without wild-type or ΔrsmA attackers and the number of viable target cells was calculated as the number of CFU/ml after contact (t24h) divided by the number of CFU/ml before contact (t0h). Error bars represent the standard error of the mean from three independent experiments.
CdiAPA2462 and CdiAPA0041 are responsible for the growth inhibition phenotype
In order to identify the growth inhibition factor, we next generated deletions in the attacker strain genome. Inter-bacterial competition assays showed that attacker strains lacking the entire cdiBAIPA2462 locus, cdiBAPA2462 or only cdiAPA2462 did not inhibit the growth of a ΔBAIPA2462 target strain anymore (Fig 3A). By contrast, a ΔAPA0041 attacker still inhibited ΔBAIPA2462 target cells, although it should be noted that the level of inhibition was lower than what we observed with a wild-type attacker (Fig 3A). We concluded that CdiAPA2462 is the main protein able to mediate growth inhibition of ΔBAIPA2462 mutant cells. Similarly, deletion of the entire cdiBAIPA0041 locus, cdiBAPA0041 or cdiAPA0041 genes in an attacker cell altered its capacity to inhibit a ΔBAIPA0041 strain (Fig 3B). Interestingly, similarly to what we observed with ΔAPA0041, the ΔAPA2462 strain did not impede growth of ΔBAIPA0041 target bacteria (Fig 3B). As contact between bacteria is important to observe CDI activity [3], we hypothesized that a defect in adhesion between the ΔAPA2462 attacker and target bacteria might explain the absence of growth inhibition (Fig 3B). To study cell attachment, we visualized bacteria grown in static chambers using confocal laser-scanning microscopy (CLSM) (Fig 4A). Importantly, we first verified that cdiA genes were expressed under the minimal media growth conditions used in this assay (S3 Fig). After 8 h, wild-type bacteria attached as a sparse monolayer onto the coverslip. By contrast, cdiAPA2462 or cdiAPA0041 mutants adhered significantly less than wild-type cells (Fig 4A). A growth defect could not explain this decrease of adhesion since the wild-type, cdiAPA2462 and cdiAPA0041 mutant strains showed identical growth curves (data not shown). Next, to determine whether CdiA proteins also play a role in biofilm formation, we quantified biofilm biomass in 96-well polystyrene plates over a 24 h time period. Both cdiA mutants generated significantly less biofilm mass than the wild-type strain after 24 h of culture (Fig 4B), showing that CdiAPA2462 and CdiAPA0041 contribute to the formation of biofilm. The fact that ΔAPA2462 is defective in adhesion and biofilm formation (Fig 4A and 4B), could explain the incapacity of ΔAPA2462 to inhibit ΔBAIPA0041 growth (Fig 3B), suggesting that adhesive activities are essential for inhibiting ΔBAIPA0041. By contrast, although ΔAPA0041 is impaired in process of adhesion (Fig 4A and 4B), it still retained an inhibiting activity against ΔBAIPA2462 target strains (Fig 3A), implying that an adhesion defect does not interfere with the capacity to inhibit the growth ΔBAIPA2462 cells. To strengthen our hypotheses, we tested a mutant with defects in cell adhesion/biofilm formation speculating that it would display similar competition defects. We used an attacker mutant devoid of type IVa pili and of flagella (ΔpilAΔfliC) and confirmed that it displayed a defect in biofilm formation (Fig 4C). Competition assays showed that the ΔpilAΔfliC mutant still inhibited the growth of ΔBAIPA2462 target bacteria but to a lesser extent than a wild-type attacker (Fig 4D). By contrast, no growth defect of ΔBAIPA0041 target cells was observed in contact with ΔpilAΔfliC mutant (Fig 4D). Altogether these results confirmed that adhesion process seems essential to the inhibition of ΔBAIPA0041 cells and that ΔBAIPA2462 bacteria are less sensitive to a cell adhesion/biofilm formation defect.
Fig 3. Roles of CdiAPA2462, CdiAPA0041 in growth competition.
All experiments were performed in a ΔrsmA background. A) ΔBAIPA2462 and B) ΔBAIPA0041 target strains were mixed with different attacker cells. Error bars represent the standard error of the mean from three independent experiments.
Fig 4. Roles of adhesion and biofilm formation in competition assays.
(A). CLSM of adhesive structures formed by WT, cdiAPA2462 and cdiAPA0041 mutants (ΔAPA2462 and ΔAPA0041 respectively) at 8 h post-inoculation. All strains carry a gfp gene inserted at the glmS locus on the P. aeruginosa PAO1 chromosome. White scale bar = 10 μm. (B) and (C). After staining with crystal violet, biofilm was quantified over time by measuring the optical density at 550 nm (OD550). Each strain was tested in triplicate experiment. Data were examined for significance using a two-tailed Student’s t-test. *** p-value<0.01; ns: non-significant. D. Competition experiments were performed in a ΔrsmA background. WT or ΔpilAΔfliC attacker strains were mixed with ΔBAIPA2462 or BAIPA0041 target bacteria. Error bars represent the standard error of the mean from three independent experiments.
Identification of potential immunity proteins: roles of CdiIPA2462 and CdiIPA0041
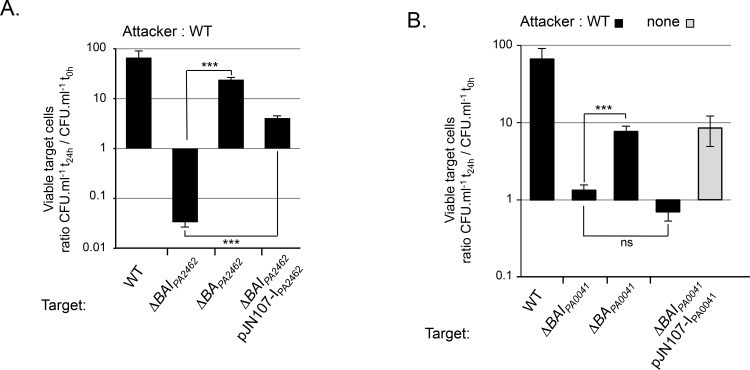
To determine whether the cdiI genes encode immunity proteins, we performed competition experiments with target cells that only produce CdiI proteins. As shown in Fig 5A, wild-type bacteria inhibit the growth of a ΔBAIPA2462 (CdiI-) target strain but not the one of ΔBAPA2462 (CdiI+) strain or a ΔBAIPA2462 strain producing the CdiIPA2462 protein in trans. These results show that wild-type cells can only inhibit bacteria that do not express cdiIPA2462 and hence suggest that CdiIPA2462 alone protects from the attack. Similarly, a ΔBAPA0041 (CdiI+) target is unaffected by the growth inhibitory activity of a wild-type strain. However, a ΔBAIPA0041 strain containing a plasmid carrying cdiIPA0041 remains sensitive to a wild-type attack (Fig 5B). This sensitivity could be due to a growth defect associated with CdiIPA0041 production or a lack of cdiIPA0041 expression. We ruled out the first hypothesis as the growth of ΔBAIPA0041 cells producing CdiIPA0041 is not affected in absence of attacker strains (Fig 5B, grey bar) and propose that the observed sensitivity comes from a defect in cdiIPA0041 expression. Altogether, these data indicate that a wild-type strain inhibits the growth of cdiIPA0041 mutant cells but do not directly prove that the CdiIPA0041 is an immunity protein.
Fig 5. CdiIPA2462 and CdiIPA0041 play a protective role.
All experiments were carried out in a ΔrsmA background. Wild-type (WT) attacker was mixed with different gentamycin-resistant mutant targets: (A) ΔBAIPA2462 carrying or not the pJN107 plasmid encoding the CdiIPA2462 protein or the BAPA2462 mutant. No arabinose was added to produce CdiIPA2462 from pJN107-IPA2462; (B) BAIPA0041 carrying or not the pJN107 plasmid encoding the CdiIPA0041 protein or the BAPA0041 mutant. As control, ΔBAIPA0041 expressing cdiIPA0041 was tested alone in inter-bacterial competition (grey bar). 0.5% arabinose was used to induce CdiIPA0041.
CdiA-CTPA2462/CdiIPA2462 and CdiA-CTPA0041/CdiIPA0041 encode specific toxin/immunity pairs
CT domains of CdiAPA2462 and CdiAPA0041 show homologies with nuclease domains. To assess their putative toxic function, we cloned the DNA fragments encoding both CT-domains under the arabinose inducible PBAD promoter in E. coli using pBAD33. While we readily cloned cdiA-CTPA0041, we only obtained one clone with cdiA-CTPA2462 albeit with a deletion in the Shine Dalgarno sequence likely decreasing the production of the CT domain (S1 Table). This result strongly suggests that the production of CdiA-CTPA2462 in the cytoplasm is toxic. In presence of arabinose, the number of CFU/ml of E. coli strain producing either CdiA-CTPA2462 or CdiA-CTPA0041 decreased drastically by 5 and 6 logs respectively (Fig 6A and 6B). Thus, the production of these domains in E. coli is toxic.
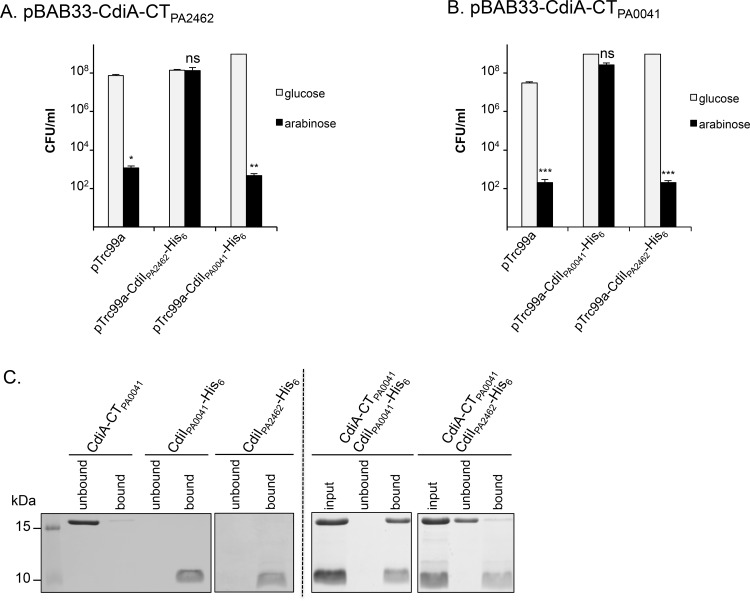
Fig 6. Intracellular toxicity of CdiA-CTPA2462 and CdiA-CTPA0041 in E. coli cells and protection by their cognate CdiI immunity proteins.
Production of (A) CdiA-CTPA2462 and (B) CdiA-CTPA0041 were repressed with 0.5% glucose (grey bars) or induced with 1% arabinose (black bars). Error bars represent the standard error of the mean from three independent experiments. C. Purified CdiA-CTPA0041 and CdiI-His6 proteins were mixed in vitro with Ni2+-NTA resin. Fractions were analyzed by SDS-PAGE and Coomassie blue staining. Control experiments with purified proteins incubated alone are shown on the left panel while CdiA-CT/CdiI-His6 mixtures are shown on the right panel. Marker lane (kDa) is shown on the left.
CdiIPA2462 and potentially CdiIPA0041 protect from P. aeruginosa PAO1 attack, raising the possibility that these proteins would protect E. coli from the toxicity of the CT-domains. In order to test this hypothesis, the cdiIPA2462 and cdiIPA0041 genes were fused to a 6xHis epitope-encoding sequence downstream the IPTG-inducible Ptrc promoter. The production of CdiIPA2462-His6 and CdiIPA0041-His6 protected E. coli from CdiA-CTPA2462 and CdiA-CTPA0041 toxicity respectively as CFU/ml counts with arabinose and glucose were comparable (Fig 6A and 6B). By contrast, the number of CFU/ml of E. coli co-producing CdiA-CTPA2462 with CdiIPA0041-His6 or CdiA-CTPA0041 with CdiIPA2462-His6 decreased by 5 and 6 logs respectively, showing that these systems are not interchangeable. The inability to rescue E. coli growth was not due to the absence of CdiI production as shown by western-blot analyses (S4A and S4B Fig). Altogether, these results demonstrate that CdiA-CTPA2462/CdiIPA2462 and CdiA-CTPA0041/CdiIPA0041 function as toxin/immunity systems.
We speculated that the CdiI proteins prevent the toxicity of CdiA-CT through specific binding inhibition. To test pair-wise interaction, the CdiI and CdiA proteins were purified and their interactions assessed using Ni2+-affinity pull-down experiments. While the CdiIPA2462-His6, CdiIPA0041-His6 and CdiA-CTPA0041 proteins were readily produced and purified, we failed to clone a non-mutated cdiA-CTPA2462-endoding DNA even in the presence of the cognate cdiIPA2462 gene, presumably due to high toxicity of CdiA-CTPA2462. Therefore, only the CdiIPA0041/CdiA-CTPA0041 interaction was assayed. CdiIPA0041-His6 but not CdiIPA2462-His6 specifically retained CdiA-CTPA0041 (Fig 6C) showing that CT-domain of CdiAPA0041 interacts specifically to its cognate CdiIPA0041 immunity protein in vitro.
CDI systems are widely distributed among Pseudomonas species
To identify other potential Pseudomonas CDI-encoding loci, we performed a BLAST search using the CdiAPA2462 and CdiAPA0041 proteins from P. aeruginosa PAO1 and analyzed the sequences flanking each of the identified cdiA homologues. We considered as part of a potential CDI locus (listed in Table 1) cdiA homologues identified immediately downstream of a cdiB homologue and upstream of a small non-annotated or unknown ORF.
Table 1. Predicted Pseudomonas CDI-encoding locus.
| Strain | CdiB | CdiA / CT domaina | motif | CdiI* | Stop-Start /lenghtb | Class |
|---|---|---|---|---|---|---|
| P. syringae pv.tomato DC3000 | PSPTO_3230 | PSPTO_3229 | PT-HINT | CdiI- PSPTO_3229* | TAGCTATG /132 | I |
| P. fluorescens A506 | PflA506_0158 | PflA506_0159 | PT-HINT/DUF637 | PflA506_0160 | I | |
| P. fluorescens SBW25 | PFLU_3192 | PFLU_3191 / Tox-REase-7 (PF15649) | PT-HINT | PFLU_3190 | I | |
| P. protegens Pf-5 | PFL_1551 | PFL_1552 | PT-HINT/DUF637 | CdiI-PFL_1552* | TGATTTATG /133 | I |
| P. fluorescens SBW25 | PFLU_0148 | PFLU_0148 | PT-VENN | CdiI-PFLU_0148* | ATGAAAAAGTGA /72 | II |
| P. fluorescens SBW25 | PFLU_3246 | PFLU_3246 / Tox-HNH-HHH (PF15637) | PT-VENN | CdiI-PFLU_3246* | TAATG /143 | II |
| P. aeruginosa NCGM2.S1 | NCGM2_0040 | NCGM2_0041 / MafB19-deam (PF14437) | PT-VENN | CdiI-NCGM2_0041* | AATATGA /133 | II |
| P. fulva 12-X | Psefu_0120 | Psefu_0121/ Tox-REase-7 (PF15649) | none | Psefu_0122 | III | |
| P. fluorescens F113 | PSF113_0792 | PSF113_0793 / AHH (PF14412) | none | PSF113_0794 | III | |
| P. fluorescens A506 | PflA506_2790 | PflA506_2789 | none | CdiI-PflA506_2789* | TAGAGAAATTTTATATG /68 | III |
| P. entomophila L48 | PSEEN_3947 | PSEEN_3946 | DUF637 | CdiI-PSEEN_3946* | GCAGTGAA /129 | IV |
| P. brassicacearum DF41 | CD58_03745 | CD58_03750 / Ribonuclease/ribotoxin (IPR016191) | DUF637 | CD58_03755 | IV | |
| P. protegens CHA0 | PFLCHA0_c15900 | PFLCHA0_c15910 | DUF637 | CdiI-PFLCHA0_c15910* | TAGAGGAGGACGGTTATG /160 | IV |
| P. aeruginosa PAO1 | PA2463 | PA2462 /RNAse EndoU fold (PF14436) | none | CdiI- PA2462* | TAGGTCTTTTATG /84 | V |
| P. aeruginosa PA14 | PA14_32780 | PA14_32790 | DUF637 | CdiI-PA14_32790* | AGTGAA /113 | V |
| P. aeruginosa PA7 | PSPA7_2776 | PSPA7_2777 / CDI_toxin_Bp1026b_like | DUF637 | CdiI- PSPA7_2777* | GATGAGC /130 | V |
| P. aeruginosa LESB58 | PALES_2831 | PALES_28341 / Toxin deaminase (PF14424) | none | CdiI-PALES_28341* | GTAATGG /117 | V |
| P. aeruginosa PAO1 | PA0040 | PA0041 / CDI_toxin_Bp1026b_like | DUF637 | CdiI-PA0041* | CTGATTATG /84 | V |
| P. aeruginosa PA7 | PSPA7_0042 | PSPA7_0044 | DUF637 | PSPA7_0045 | V | |
| P. aeruginosa LESB58 | PALES_00391 | PALES_00401 | DUF637 | CdiI- PALES_00401* | AAAATGATT /106 | V |
| P. aeruginosa PACS2 | PaerPA_01000042 | PaerPA_01000043 /Toxin_49 (PF15529) | DUF637 | CdiI- PaerPA_01000043* | CATGAA /156 | V |
| P. aeruginosa PA14 | PA14_00490 | PA14_00510 | DUF637 | CdiI-PA14_00510* | CTGATGA/155 | V |
| P. aeruginosa NCGM2.S1 | NCGM2_3465 | NCGM2_3464 | DUF637 | CdiI- NCGM2_3464* | TGAGTTAATATG /106 | V |
| P. aeruginosa DK2 | PADK2_00205 | PADK2_00210 / Pyocin large subunit (COG5529) | DUF637 | CdiI-PADK2_00210* | TAAGGAGAGCCCAATG /80 | V |
| P. syringae pv.tomato DC3000 | PSPTO_3230 | PSPTO_3229 | PT-HINT | CdiI- PSPTO_3229* | TAGCTATG /132 | |
| P. fluorescens A506 | PflA506_0158 | PflA506_0159 | PT-HINT/DUF637 | PflA506_0160 | ||
| P. fluorescens SBW25 | PFLU_3192 | PFLU_3191 / Tox-REase-7 (PF15649) | PT-HINT | PFLU_3190 | ||
| P. protegens Pf-5 | PFL_1551 | PFL_1552 | PT-HINT/DUF637 | CdiI-PFL_1552* | TGATTTATG /133 | |
| P. fluorescens SBW25 | PFLU_0148 | PFLU_0148 | PT-VENN | CdiI-PFLU_0148* | ATGAAAAAGTGA /72 | |
| P. fluorescens SBW25 | PFLU_3246 | PFLU_3246 / Tox-HNH-HHH (PF15637) | PT-VENN | CdiI-PFLU_3246* | TAATG /143 | |
| P. aeruginosa NCGM2.S1 | NCGM2_0040 | NCGM2_0041 / MafB19-deam (PF14437) | PT-VENN | CdiI-NCGM2_0041* | AATATGA /133 | |
| P. fulva 12-X | Psefu_0120 | Psefu_0121/ Tox-REase-7 (PF15649) | none | Psefu_0122 | ||
| P. fluorescens F113 | PSF113_0792 | PSF113_0793 / AHH (PF14412) | none | PSF113_0794 | ||
| P. fluorescens A506 | PflA506_2790 | PflA506_2789 | none | CdiI-PflA506_2789* | TAGAGAAATTTTATATG /68 | |
| P. entomophila L48 | PSEEN_3947 | PSEEN_3946 | DUF637 | CdiI-PSEEN_3946* | GCAGTGAA /129 | |
| P. brassicacearum DF41 | CD58_03745 | CD58_03750 / Ribonuclease/ribotoxin (IPR016191) | DUF637 | CD58_03755 | ||
| P. protegens CHA0 | PFLCHA0_c15900 | PFLCHA0_c15910 | DUF637 | CdiI-PFLCHA0_c15910* | TAGAGGAGGACGGTTATG /160 | |
| P. aeruginosa PAO1 | PA2463 | PA2462 /RNAse EndoU fold (PF14436) | none | CdiI- PA2462* | TAGGTCTTTTATG /84 | |
| P. aeruginosa PA14 | PA14_32780 | PA14_32790 | DUF637 | CdiI-PA14_32790* | AGTGAA /113 | |
| P. aeruginosa PA7 | PSPA7_2776 | PSPA7_2777 / CDI_toxin_Bp1026b_like | DUF637 | CdiI- PSPA7_2777* | GATGAGC /130 | |
| P. aeruginosa LESB58 | PALES_2831 | PALES_28341 / Toxin deaminase (PF14424) | none | CdiI-PALES_28341* | GTAATGG /117 | |
| P. aeruginosa PAO1 | PA0040 | PA0041 / CDI_toxin_Bp1026b_like | DUF637 | CdiI-PA0041* | CTGATTATG /84 | |
| P. aeruginosa PA7 | PSPA7_0042 | PSPA7_0044 | DUF637 | PSPA7_0045 | ||
| P. aeruginosa LESB58 | PALES_00391 | PALES_00401 | DUF637 | CdiI- PALES_00401* | AAAATGATT /106 | |
| P. aeruginosa PACS2 | PaerPA_01000042 | PaerPA_01000043 /Toxin_49 (PF15529) | DUF637 | CdiI- PaerPA_01000043* | CATGAA /156 | |
| P. aeruginosa PA14 | PA14_00490 | PA14_00510 | DUF637 | CdiI-PA14_00510* | CTGATGA/155 | |
| P. aeruginosa NCGM2.S1 | NCGM2_3465 | NCGM2_3464 | DUF637 | CdiI- NCGM2_3464* | TGAGTTAATATG /106 | |
| P. aeruginosa DK2 | PADK2_00205 | PADK2_00210 / Pyocin large subunit (COG5529) | DUF637 | CdiI-PADK2_00210* | TAAGGAGAGCCCAATG /80 |
a CT domain: identified conserved domains using Interproscan5 program and NCBI database
b stop codon of cdiA gene and start codon of cdiI gene are in bold and underlined respectively, number indicate the length of the predicted CdiI protein
*: non-annotated ORF encoding potential CdiI proteins
Interestingly, we identified CDI systems in P. aeruginosa species, generally found in human infection, but also in Pseudomonas species associated with plants (Table 1). While P. protegens, P. fluorescens and P. brassicacearum are plant-protecting bacteria, others, such as P. syringae, are phytopathogens. All predicted Pseudomonas CdiA (pCdiA) sequences contain a Sec signal peptide allowing translocation across the inner membrane, a conserved TPS domain recognized by cognate CdiB transporter and repetitive regions with haemagglutinin-type repeats (Fig 7). In addition, all pCdiA exhibit a C-terminal variable region (Fig 7, CT domain) with no amino-acid similarities except the CT domains of CdiAPFLU_3191 and CdiAPsefu_0121 from P. fluorescens SBW25 and P. fulva 12-X respectively, sharing 70% identity. Similarly to the high variability of CT domains, the sequences of small ORFs, immediately downstream of pcdiA, are also variable, except for PFLU_3191 and Psefu_0122, the predicted immunity proteins of CdiAPFLU_3191 and CdiAPsefu_0121, which also share 70% identity.
Fig 7. Representative CdiA proteins of each class of pre-toxin motif.
The signal peptide (SP) is a sequence potentially recognized by the sec machinery and cleaved during the export through the inner membrane. TPS domain found in N-terminal part of TpsA allows targeting to the TpsB transporter and subsequent translocation through the outer membrane. Haemagglutinin repeats are highly divergent repeats found in FHA-like proteins. DUF637 is a conserved region found in bacterial haemagglutinins or hemolysins. WVHN, VENN, LYVT, DAMV, NEALV are the pre-toxin motifs delimiting the conserved N-terminal and the variable CT domains.
Based on the amino-acid alignment of pCdiA proteins and the observed sequence identity between them, we classified the pre-toxin motifs at the transition between the N-termini and CT-variable domains into five distinct classes (Fig 7 and S5 Fig). Proteins with the class I peptide motif contain a bacterial intein-like domain (BIL domain) [17] sandwiched between a conserved motif (WVHN) and the CT variable domain. Class II pre-toxin motif represents the highly conserved VENN motif (PF04829) found in “E. coli-type” CDI systems [5]. In class III, IV and V pre-toxin motifs, the peptides that separate the N-terminus domain from the variable CT region are LYVT, DAMV and NEVLA respectively. Class I, IV and V exhibit a small-helical DUF637 domain of unknown function (Table 1 and Fig 7). Some CT domains of pCdiA proteins carry potential nuclease activities (Table 1) but ~ 50% of the CT domains have no predicted function. Sequence analyses of predicted immunity proteins revealed that CdiIPSPTO_3229 and CdiIPALES_28341 contains respectively SMI1 and Imm2 domains, typically found in immunity proteins of bacterial polymorphic toxin systems [2,18]. In addition, the CD58_03755 protein encoded by P. brassicacearum DF41 contains a barstar domain predicted to inhibit specifically the Barnase protein, an extracellular ribonuclease of Bacillus amyloliquefaciens [19]. Except for these 3 proteins, no known domains have been found in other predicted immunity proteins.
P. aeruginosa PA7, PA14 and P. syringae pv. tomato DC3000 encode functional toxin/immunity pairs
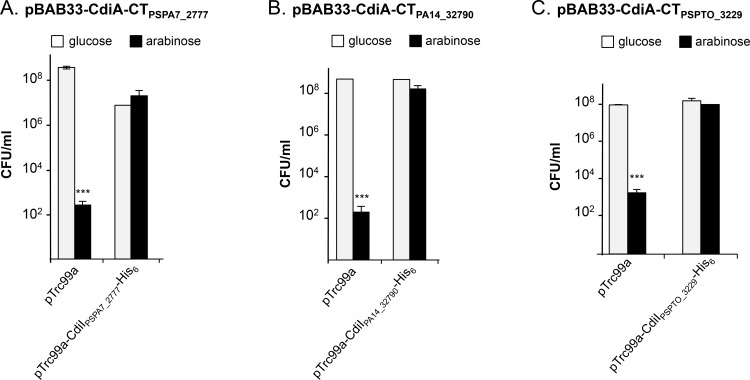
To investigate the functionality of some of the newly discovered CDI toxins (Table 1), we focused on three CdiA-CT domains with predicted or unknown activities (Fig 7). CdiA-CTPSPA7_2777 found in P. aeruginosa PA7 potentially carries nuclease activity based on the homology with the B. pseudomallei 1026b CT domain (Table 1 and [6]). On the other hand, no predicted functions have been attributed to the CdiA-CTPA14_32790 and CdiA-CTPSPTO_3229 respectively produced by P. aeruginosa PA14 and P. syringae pv. tomato DC3000. To assess putative toxic function, CT-domains were separately produced in E. coli with or without their cognate immunity proteins. Production of each CT-domains with arabinose significantly inhibited E. coli cell growth (Fig 8A, 8B and 8C) while co-production of the cognate CdiI protein completely alleviated the cell growth inhibition showing that CdiA-CTPSPA7_2777/CdiIPSPA7_2777 and CdiA-CTPA14_32790/CdiIPA14_32790 constitute specific toxin/immunity systems. In addition, these results confirm the predicted toxic activity of CdiA-CTPSPA7_2777 and strongly suggest that CdiA-CTPA14_32790 and CdiA-CTPSPTO_3229 act in the cytoplasm of the target cells.
Fig 8. CdiA-CT encoded by P. aeruginosa PA7, PA14 and P. syringae pv. tomato DC3000 inhibit E. coli cell growth.
Production of (A) CdiA-CTPSPA7_2777, (B) CdiA-CTPA14_32790 and (C) CdiA-CTPSPTO_3229 was repressed with 0.5% glucose (grey bars) and induced with 1% arabinose (black bars). CdiI-His6 proteins were produced from pTrc99a with 1 mM IPTG. The number of CFU/ml was calculated after 6 h of culture and the graphs show the mean of three independent experiments.
Discussion
The results presented demonstrate that Pseudomonas species encode CDI systems: (i) CT domains of Pseudomonas CdiA proteins are highly variable and toxic when produced intracellularly in E. coli cells; (ii) Pseudomonas cdiI genes encode immunity proteins that neutralize the toxicity in an allele-specific manner; (iii) Pseudomonas CdiA function in growth competition and wild-type P. aeruginosa can outcompete a ΔBAI mutant that does not express the cognate immunity gene; and (iv) such as in Burkholderia and E. coli systems [10,11], CdiA proteins contribute to biofilm formation.
In this study we defined four novel classes of peptide motif separating the conserved N-terminal and variable CT regions, in addition to the common characteristics found in E. coli- and Burkholderia-type CDI systems. Indeed, we identified CdiA proteins with class I peptide motif harboring a bacterial intein-like (BIL) domain [17] related to the Hedgehog/Intein (Hint) domain implicated in the maturation of inteins and Hogs proteins [20]. The BIL domain of P. syringae CdiAPSPTO_3230 is active in autoproteolytic cleavage [17] suggesting that CdiA proteins might have the intrinsic ability to auto-cleave before being delivered across the target-cell envelope. Our analyses suggest that the BIL domain is restricted to CdiA encoded by Pseudomonas phytopathogens (Table 1). Similarly, class V peptide motif has been identified only in CdiAs encoded by P. aeruginosa species. It has been suggested that the VENN sequence might be important for homologous recombination in order to acquire new cdiA-CT/cdiI pairs by horizontal gene transfer and consequently to participate in the variability of secreted toxins. The diversity of motifs that define the variable CT-region in Pseudomonas species could explain the various predicted toxin activities of the CdiA-CT domain (Table 1) but also reflects the high genomic plasticity and capacity of Pseudomonas to capture genes from its environment. We identified at least 10 different putative functions for the CT-domains, all of them possibly targeting nucleic acids. However, half of the identified CT-domains do not belong to a defined protein family domain and characterization of their biological functions remains a challenging question.
While the role and mechanisms of delivery of CDI toxins in enterobacteria and Burkholderia species are now better understood, the regulatory pathways underlying CDI systems expression remain unknown for most of these systems. Except E. coli EC93 which constitutively expresses its CDI system [3], bacteria tightly regulate the expression of CDI-encoding genes. For example, Burkholderia express bcp genes in a stochastic manner, with only a subset of bacteria expressing these genes to high level [12] and bcp expression is strongly induced by cell density in cells grown on solid media and reduced in cells grown in liquid [10]. Similarly, Dickeya dadantii 3937 and probably Erwinia chrysanthemi EC16 activate their CDI systems only when in contact with plant hosts [5]. In this study, we identified RsmA as a negative regulator of the expression of P. aeruginosa PAO1 CDI systems. In P. aeruginosa, the activity of RsmA is controlled by a complex regulatory system. This includes the RetS and LadS sensor kinases and the histidine phosphotransfer protein HptB which all converge on the master two-component regulator system GacS/GacA to influence the levels of the RsmY and RsmZ sRNAs, which ultimately modulates the expression of target genes by titrating RsmA [16]. Putative RsmA binding sequences overlap the RBS of cdiAPA2462 and cdiAPA0041 ([15] and our study) suggesting that these genes are directly regulated by RsmA at a post-transcriptional level. Remarkably, all 5’unstranslated region of P. aeruginosa cdiA genes identified in this study contain identical RsmA boxes suggesting a conserved post-transcriptional regulation by RsmA. However, the contribution of RetS, HptB, LadS and GacS in expression of cdi genes remains to be determined. Additional experiments will be necessary to identify the regulatory circuits controlling other Pseudomonas cdi gene expression.
In contrast to the CDI-mediated competition that occurs in shaking conditions in E. coli [3], we observed inter-bacterial competition in Pseudomonas only when bacteria were mixed in static conditions, either on solid surface or in liquid. The gene expression patterns cannot explain the disparity observed in competition experiments since the maximum mRNA levels were obtained in rsmA mutant grown either in shaking or static conditions (Fig 1). CDI-mediated growth inhibition occurs upon direct cell-cell contact and static conditions would favor stable contacts and close proximity between bacteria. Indeed, we observed that one P. aeruginosa PAO1 CDI system we studied is not able to mediate competition upon a defect in cell adhesion (Fig 3 and Fig 4).
It is now accepted that CDI systems are also involved in biofilm formation and self-recognition by inhibiting the incorporation of non-self competitors in the community. These mechanisms have been observed for B. thailandensis and E. coli EC93 CDI systems [8,10–12]. Our results demonstrate that Pseudomonas CdiA proteins also play a role in biofilm formation. Confirming the implication of the CDI systems in Pseudomonas biofilm architecture is a main goal of future studies. Interestingly, the CdiA proteins characterized in this study are among the five most abundant proteins found in the extracellular matrix of colony biofilms as well as in outer membrane vesicles (OMVs) [21]. OMVs are spherical structures derived from outer membranes of gram-negative bacteria [22], which have specific virulence-associated activities but are also implicated in cell-cell communication, horizontal gene transfer and competition with other organisms to promote bacterial colonization [23], activities surprisingly close to those assigned to CDI systems. CdiA-associated vesicles could be advantageous acting as bridging factors in biofilms and/or targeting distant bacteria in a competition context. While the role of OMVs in inter-bacterial interactions is well documented, no implication of CDI systems in this context has been described so far.
Most Pseudomonas genomes encode multiple CDI systems (Table 1) and in the case of P. aeruginosa species, our study indicates that CDI systems undergo the same regulatory pathway suggesting that each strain coordinately activates their CDI systems. The determination of the actor(s) implicated in cell inhibition is complex. Indeed, we found that cdi loci contain additional orphan cdiA-CT/cdiI modules located downstream to cdiBAI genes (S2 Table). Interestingly, the presence of such orphan modules is limited to the P. aeruginosa species. We showed that some of these modules are functional and have growth inhibition activities in E. coli but we have no clues on their synthesis under physiological conditions. Identified orphan cdiA-CT sequences lack initiation codon and RBS sequence indicating that they might not be produced in contrast to cdiI genes that have translation initiation signal. It is then conceivable that these cdiI genes are expressed as the orphan cdiIo1EC93 gene in E. coli EC93 [24]. The function of orphan toxin-immunity modules is unknown but they may represent a reservoir of diverse toxins or may be implicated in discerning kin versus non-kin within a population [25,26]. The co-existence of different P. aeruginosa lineages observed in chronic CF lung infections is in line with this concept [27]. A further complication in studying the polymicrobial interactions is that other factors contribute to inter-species competition such as type VI secretion systems (T6SS), which target microorganisms from the same or a different genus [28]. Interestingly, one of the P. aeruginosa T6SS, H1-T6SS, is also under the control of RsmA [29–31]. The co-activation of the T6SS and CDI system as anti-bacterial weapons in P. aeruginosa species would ensure a fitness advantage against other pathogens especially in lung infections of CF patients, where the bacteria are thought to reside within biofilm-like structures. In conclusion, the finding that a considerable number of strains encode diverse putative CDI systems suggests that these systems play a significant role in the social life of Pseudomonadaceae.
Experimental procedures
Growth condition and strain constructions
Plasmids (listed in S1 Table) were maintained in E. coli strains and transferred into P. aeruginosa PAO1 strain by electroporation [32] or by tri-parental mating using the helper plasmid pRK2013 [33]. P. aeruginosa was cultured in solid or liquid Luria-Bertani (LB) medium supplemented with appropriate antibiotics: 30 μg/ml gentamycin (Gm), 50 μg.ml-1 carbenicillin (300 μg.ml-1 on plates), 2 mg.ml-1 streptomycin. M63 minimal medium (15 mM (NH4)2SO4, 22 mM KH2PO4, 40 mM K2HPO4, pH6.8) supplemented with 1 mM MgCl2, 0.5% casamino acids and 0.2% glucose was used for growth in static chambers.
The P. aeruginosa PAO1 cdi or rsmA genes deletion strains were engineered by allelic exchange. Briefly, the DNA regions upstream and downstream of the gene to be deleted were PCR amplified (Expand High Fidelity DNA polymerase, Roche), combined by overlapping extension PCR and cloned into the pKNG101 suicide vector [33]. The resulting plasmids were used to create deletion strains.
All Gm-resistant strains and green fluorescent protein (GFP)-tagged bacteria were created using respectively the pUC18T-mini-Tn7T-Gm and pUC18T-mini-Tn7T-Gm-gfpmut3 [32]. The pJN107 plasmid was constructed by cloning the ClaI-AraC-PBAD-EcoRI fragment from pJN105 vector [34] into pBBR-MCS4 plasmid digested by ClaI/EcoRI [35]. cdiIPA0041 and cdiIPA2462 genes were amplified with an artificial Shine-Dalgarno (AAGAAG) and cloned into pJN107 using the SLIC method [36].
Isolation of total RNA and quantitative Real Time-PCR
qRT-PCR have been performed as previously described [37]. Briefly, total cellular RNAs from 1×1010 bacteria were isolated using the PureYield RNA Midiprep System (Promega) and contaminating DNA eliminated by TURBO DNase (Ambion) treatment. cDNA synthesis was performed on 1.6 μg of RNA using the SuperScriptIII first strand synthesis system (Invitrogen). The mRNAs level of cdiAPA2462 and cdiAPA0041 were determined using primers listed in S3 Table. To determine the amplification kinetics of each product during the quantitative reverse transcription, the fluorescence derived from the incorporation of EvaGreen into the double-stranded PCR products was measured at the end of each cycle using the SoFast EvaGreen Supermix (Biorad). The 16S gene was used as control to normalize the results.
Adhesion and biofilm assays
GFP-tagged strains were cultured overnight in LB medium, then washed in M63 medium and inoculated to an OD600 of ~ 0.1 in 400 μl of M63 medium in chambered cover glass dishes (Thermo scientific) and incubated at 30°C for 8 h, washed 2 times with 1X PBS, overlaid with 400 μl of 1X PBS and imaged by CLSM with a olympus FV-1000 microscope. Biofilm assays were performed following the O’Toole protocol [38]. Briefly, overnight cultures were inoculated into LB at a final OD600 of 0.05. 200 μl of culture were added to 96-well polystyrene plates and incubated at 37°C over a 24h period. At the indicated times, cell in suspension were removed and 200 μl of 0.1% crystal violet was added to the wells and incubated for 10 min. Wells were washed three times with 1X PBS, then crystal violet was solubilized with 200 μl 95% ethanol and the absorbance was measured at 550 nm.
Growth competitions assays
Overnight cultures were washed, diluted to an OD600 of ~ 0.1 in LB medium and grown in static condition at 30°C to an OD600 ~ 0.3. Strains were mixed at an attacker to target cell ratio of 4 to 1 and 10 μl of mixed strains were used for determination of target initial CFU by plating on Gm-containing LB agar plates (t0h). 25 μl of the mix was spotted on 1.5% agar plate and incubated at 30°C for 24 h. Spotted bacteria were then resuspended in 500 μl LB and plated on Gm-containing LB agar plates to determine the target’s CFU after incubation with attacker (t24h). The viable target cells were calculated as the CFU.ml-1 (t24h)/CFU.ml-1 (t0h) ratio. Data were examined for significance using a two-tailed Student’s t-test. * 0.05<p-value<0.025; ** 0.025<p-value<0.01; *** p-value<0.01; ns: non-significant.
Vectors construction and toxicity assays in E. coli DH5α
CT domains delimited from their transition motif (S5 Fig) were cloned with the artificial AGGAGG Shine-Dalgarno sequence into the pBAD33 plasmid using SacI and SalI. The cdiI genes were PCR-amplified with a reverse primer encoding a 6xHis C-terminal tag and cloned into the pTrc99a plasmid using NcoI and BamHI. To assess CT-domain toxicity, bacteria were grown overnight with 0.5% glucose to repress CT-domain expression, washed twice in LB medium and diluted to an OD600 ~ 0.05 in LB with 1 mM IPTG to induce immunity protein production and 0.5% glucose or 1% arabinose to repress or induce CT-domain expression respectively. After 6 h of culture at 37°C, cells were washed twice in LB medium and plated onto LB agar plates supplemented with 0.5% glucose to determine the CFU/ml. Data were analyzed for significance using a two-tailed Student’s t-test. * 0.05<p-value<0.025; ** 0.025<p-value<0.01; *** p-value<0.01. Production of immunity proteins was verified in parallel by western-blot analyses by resuspending the bacterial pellet in 1X SDS-PAGE loading buffer. 0.1 OD600 units of boiled samples were separated on 18% SDS-PAGE gel, transferred onto nitrocellulose membranes and probed with primary mouse anti-pentaHis (Qiagen) or anti-EF-Tu (Hycult Biotech) antibodies and anti-mouse IgG conjugated to HRP (Sigma).
Vector construction and protein production in E. coli BL21(DE3)
The cdiIPA0041 gene was PCR amplified with a reverse primer encoding a 6xHis C-terminal tag and cloned into the pRSF-Duet1 vector using NcoI and BamHI. The CT domain of cdiAPA0041 was cloned into pRSF-Duet-CdiIPA0041 with NdeI and XhoI. cdiIPA2462, PCR-amplified with a reverse primer encoding a 6xHis C-terminal tag, was cloned into the pET19b vector with NcoI and BamHI.
Proteins were produced in E. coli BL21(DE3). Overnight cultures were diluted in LB medium to an OD600 ~ 0.05, grown at 37°C to OD600 ~ 0.5 and proteins produced for 2 h with 1 mM IPTG. The bacterial pellets were resuspended in lysis buffer (50 mM Tris-HCl pH8, 150 mM NaCl, 0.1% Triton X-100, 1X protease inhibitor cocktail (Complete EDTA-free; Roche), and 10% glycerol), lysed by sonication and centrifuged for 1 hr at 100,000×g at 4°C. Supernatants were loaded on Ni-NTA resin columns equilibrated in buffer A (50 mM Tris-HCl pH8, 300 mM NaCl, 0.1% Triton X-100 and 10% glycerol). After washing the resin with buffer A supplemented with 20 mM imidazole, CdiIPA0041-His6 and CdiIPA2462-His6 were eluted using a linear gradient of 50–400 mM imidazole in buffer A. Peak fractions were pooled, dialyzed against storage buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM β-mercaptoethanol and 30% glycerol) and stored at -80°C. CdiA-CTPA0041 was eluted with denaturating buffer (buffer A supplemented with 6 M guanidine-HCl) and peak fractions were pooled, dialyzed twice against storage buffer and frozen to -80°C.
Pull down experiments
100 pmol of each protein were incubated at room temperature for 30 min. An aliquot of the mixture was removed to analyze the input fraction. Mixtures were then incubated 1 h at 4°C with Ni2+-NTA resin pre-equilibrated with buffer A. Unbound fraction was removed before washing the resin with buffer A supplemented with 25 mM imidazole. Bound complexes were eluted with buffer A supplemented with 300 mM imidazole. Prior 18% SDS-PAGE running, proteins were precipitated with 12% (w/v) trichloroacetic acid (TCA) and resuspended in 1X SDS-PAGE loading buffer.
Supporting Information
The 117 last C-terminal residues of PA2462 were aligned with the EndoU nuclease domain of MafBMGI-1NEM8013 (C9X2Z7) using ClustalW (1.8) multiple sequence alignment with default parameters and identical and similar residues were shaded with black or grey respectively using the BoxShade server.
(TIF)
White triangles represent the predicted immunity genes non-annotated on the P. aeruginosa PAO1 genome. Sequence alignment was generated using Webact (http://www.webact.org/WebACT/home) and implemented in Easyfig 2.1 with no cutoff value. Blue vertical and orange blocks represent normal and inverted blast matches respectively, with a gradient scale showing the level of nucleotide identity. GC content greater and lower than 50% are represented in red and blue respectively. The orientation of region including PA2463-PA2462 locus has been reversed for clarity.
(TIFF)
The level of mRNA was measured by qRT-PCR in wild-type strain grown 7 h at 30°C in M63 medium under planktonic or static condition. For each gene, expression was normalized to 16S expression and is shown relative to the planktonic condition level. Error bars represent the standard error of the mean from three independent experiments. Data were analyzed for significance using a two-tailed Student’s t-test. *** p-value<0.01.
(TIFF)
After 6 h of culture with glucose (Glc) or arabinose (Ara), E. coli cell extracts containing indicated set of plasmids (A) and (B) were subjected to 18% SDS-PAGE and the production of CdiI-His6 and the cytoplasmic EF-Tu control were confirmed by western blot experiments using the anti-pentaHis (top panels) and anti-EF-Tu monoclonal antibodies (bottom panels). Molecular weight markers (in kDa) are indicated on the right.
(TIFF)
Protein sequences were aligned using ClustalW (1.8) multiple sequence alignment, with default parameters and shaded using the BoxShade server. Identical and similar residues are shaded with black or grey respectively. Numbers represent the amino acid position of each CdiA proteins.
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Acknowledgments
We thank E. Cascales for critical reading of this manuscript and for P. syringae pv. tomato DC3000 genomic DNA, pBAD33, pTrc99a, pET19b and pRSF-Duet-1 vectors. We thank Members of R. Voulhoux lab (CNRS UMR7255 and Aix-Marseille University, France) for providing genomic DNA of P. aeruginosa PA14 and PA7. We also thank S. Bleves (CNRS UMR7255 and Aix-Marseille University, France) for critical analysis, I. Bringer, A. Brun and O. Uderso for technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the CNRS, Aix-Marseille University, by a grant from the French cystic fibrosis foundation (http://www.vaincrelamuco.org/) -SB- grant number: RF20120600719 and a FINOVI grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jamet A, Nassif X. New players in the toxin field: polymorphic toxin systems in bacteria. mBio. 2015;6: e00285–00215. 10.1128/mBio.00285-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7: 18 10.1186/1745-6150-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki SK. Contact-Dependent Inhibition of Growth in Escherichia coli. Science. 2005;309: 1245–1248. 10.1126/science.1115109 [DOI] [PubMed] [Google Scholar]
- 4.Mazar J, Cotter PA. New insight into the molecular mechanisms of two-partner secretion. Trends Microbiol. 2007;15: 508–515. 10.1016/j.tim.2007.10.005 [DOI] [PubMed] [Google Scholar]
- 5.Aoki SK, Diner EJ, de Roodenbeke C t’Kint, Burgess BR, Poole SJ, Braaten BA, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468: 439–442. 10.1038/nature09490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morse RP, Nikolakakis KC, Willett JLE, Gerrick E, Low DA, Hayes CS, et al. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proc Natl Acad Sci. 2012;109: 21480–21485. 10.1073/pnas.1216238110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck CM, Morse RP, Cunningham DA, Iniguez A, Low DA, Goulding CW, et al. CdiA from Enterobacter cloacae Delivers a Toxic Ribosomal RNase into Target Bacteria. Structure. 2014;22: 707–718. 10.1016/j.str.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson MS, Garcia EC, Cotter PA. The Burkholderia bcpAIOB Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems. Guttman DS, editor. PLoS Genet. 2012;8: e1002877 10.1371/journal.pgen.1002877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolakakis K, Amber S, Wilbur JS, Diner EJ, Aoki SK, Poole SJ, et al. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol Microbiol. 2012;84: 516–529. 10.1111/j.1365-2958.2012.08039.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia EC, Anderson MS, Hagar JA, Cotter PA. BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition: CDI system protein-mediated biofilm. Mol Microbiol. 2013;89: 1213–1225. 10.1111/mmi.12339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruhe ZC, Townsley L, Wallace AB, King A, Van der Woude MW, Low DA, et al. CdiA promotes receptor-independent intercellular adhesion. Mol Microbiol. 2015; 10.1111/mmi.13114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson MS, Garcia EC, Cotter PA. Kind Discrimination and Competitive Exclusion Mediated by Contact-Dependent Growth Inhibition Systems Shape Biofilm Community Structure. Mougous JD, editor. PLoS Pathog. 2014;10: e1004076 10.1371/journal.ppat.1004076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghequire MGK, De Mot R. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev. 2014;38: 523–568. 10.1111/1574-6976.12079 [DOI] [PubMed] [Google Scholar]
- 14.Jamet A, Jousset AB, Euphrasie D, Mukorako P, Boucharlat A, Ducousso A, et al. A New Family of Secreted Toxins in Pathogenic Neisseria Species. Skaar EP, editor. PLoS Pathog. 2015;11: e1004592 10.1371/journal.ppat.1004592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72: 612–632. 10.1111/j.1365-2958.2009.06670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vakulskas CA, Potts AH, Babitzke P, Ahmer BMM, Romeo T. Regulation of Bacterial Virulence by Csr (Rsm) Systems. Microbiol Mol Biol Rev MMBR. 2015;79: 193–224. 10.1128/MMBR.00052-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amitai G, Belenkiy O, Dassa B, Shainskaya A, Pietrokovski S. Distribution and function of new bacterial intein-like protein domains. Mol Microbiol. 2003;47: 61–73. [DOI] [PubMed] [Google Scholar]
- 18.Zhang D, Iyer LM, Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011;39: 4532–4552. 10.1093/nar/gkr036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartley RW. Barnase and barstar: two small proteins to fold and fit together. Trends Biochem Sci. 1989;14: 450–454. [DOI] [PubMed] [Google Scholar]
- 20.Perler FB. Protein splicing of inteins and hedgehog autoproteolysis: structure, function, and evolution. Cell. 1998;92: 1–4. [DOI] [PubMed] [Google Scholar]
- 21.Toyofuku M, Roschitzki B, Riedel K, Eberl L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J Proteome Res. 2012;11: 4906–4915. 10.1021/pr300395j [DOI] [PubMed] [Google Scholar]
- 22.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64: 163–184. 10.1146/annurev.micro.091208.073413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev MMBR. 2010;74: 81–94. 10.1128/MMBR.00031-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poole SJ, Diner EJ, Aoki SK, Braaten BA, t’ Kint de Roodenbeke C, Low DA, et al. Identification of Functional Toxin/Immunity Genes Linked to Contact-Dependent Growth Inhibition (CDI) and Rearrangement Hotspot (Rhs) Systems. Achtman M, editor. PLoS Genet. 2011;7: e1002217 10.1371/journal.pgen.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t’ Kint de Roodenbeke C, Kaplan MD, Low DA, et al. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci. 2013;110: 7032–7037. 10.1073/pnas.1300627110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koskiniemi S, Garza-Sánchez F, Sandegren L, Webb JS, Braaten BA, Poole SJ, et al. Selection of Orphan Rhs Toxin Expression in Evolved Salmonella enterica Serovar Typhimurium. Søgaard-Andersen L, editor. PLoS Genet. 2014;10: e1004255 10.1371/journal.pgen.1004255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams D, Evans B, Haldenby S, Walshaw MJ, Brockhurst MA, Winstanley C, et al. Divergent, Coexisting, Pseudomonas aeruginosa Lineages in Chronic Cystic Fibrosis Lung Infections. Am J Respir Crit Care Med. 2015; 10.1164/rccm.201409-1646OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RRS, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7: 25–37. 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312: 1526–1530. 10.1126/science.1128393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei X, Huang X, Tang L, Wu D, Xu Y. Global control of GacA in secondary metabolism, primary metabolism, secretion systems, and motility in the rhizobacterium Pseudomonas aeruginosa M18. J Bacteriol. 2013;195: 3387–3400. 10.1128/JB.00214-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li K, Xu C, Jin Y, Sun Z, Liu C, Shi J, et al. SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. mBio. 2013;4: e00419–00413. 10.1128/mBio.00419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi K-H, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc. 2006;1: 153–161. 10.1038/nprot.2006.24 [DOI] [PubMed] [Google Scholar]
- 33.Kaniga K, Delor I, Cornelis GR. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109: 137–141. [DOI] [PubMed] [Google Scholar]
- 34.Newman JR, Fuqua C. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227: 197–203. [DOI] [PubMed] [Google Scholar]
- 35.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166: 175–176. [DOI] [PubMed] [Google Scholar]
- 36.Jeong J-Y, Yim H-S, Ryu J-Y, Lee HS, Lee J-H, Seen D-S, et al. One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl Environ Microbiol. 2012;78: 5440–5443. 10.1128/AEM.00844-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faure LM, Llamas MA, Bastiaansen KC, de Bentzmann S, Bigot S. Phosphate starvation relayed by PhoB activates the expression of the Pseudomonas aeruginosa σvreI ECF factor and its target genes. Microbiol Read Engl. 2013;159: 1315–1327. 10.1099/mic.0.067645-0 [DOI] [PubMed] [Google Scholar]
- 38.O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp JoVE. 2011; 10.3791/2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 117 last C-terminal residues of PA2462 were aligned with the EndoU nuclease domain of MafBMGI-1NEM8013 (C9X2Z7) using ClustalW (1.8) multiple sequence alignment with default parameters and identical and similar residues were shaded with black or grey respectively using the BoxShade server.
(TIF)
White triangles represent the predicted immunity genes non-annotated on the P. aeruginosa PAO1 genome. Sequence alignment was generated using Webact (http://www.webact.org/WebACT/home) and implemented in Easyfig 2.1 with no cutoff value. Blue vertical and orange blocks represent normal and inverted blast matches respectively, with a gradient scale showing the level of nucleotide identity. GC content greater and lower than 50% are represented in red and blue respectively. The orientation of region including PA2463-PA2462 locus has been reversed for clarity.
(TIFF)
The level of mRNA was measured by qRT-PCR in wild-type strain grown 7 h at 30°C in M63 medium under planktonic or static condition. For each gene, expression was normalized to 16S expression and is shown relative to the planktonic condition level. Error bars represent the standard error of the mean from three independent experiments. Data were analyzed for significance using a two-tailed Student’s t-test. *** p-value<0.01.
(TIFF)
After 6 h of culture with glucose (Glc) or arabinose (Ara), E. coli cell extracts containing indicated set of plasmids (A) and (B) were subjected to 18% SDS-PAGE and the production of CdiI-His6 and the cytoplasmic EF-Tu control were confirmed by western blot experiments using the anti-pentaHis (top panels) and anti-EF-Tu monoclonal antibodies (bottom panels). Molecular weight markers (in kDa) are indicated on the right.
(TIFF)
Protein sequences were aligned using ClustalW (1.8) multiple sequence alignment, with default parameters and shaded using the BoxShade server. Identical and similar residues are shaded with black or grey respectively. Numbers represent the amino acid position of each CdiA proteins.
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.