Abstract
PRPF31-associated retinitis pigmentosa presents a fascinating enigma: some mutation carriers are blind, while others are asymptomatic. We identify the major molecular cause of this incomplete penetrance through three cardinal features: (1) there is population variation in the number (3 or 4) of a minisatellite repeat element (MSR1) adjacent to the PRPF31 core promoter; (2) in vitro, 3-copies of the MSR1 element can repress gene transcription by 50 to 115-fold; (3) the higher-expressing 4-copy allele is not observed among symptomatic PRPF31 mutation carriers and correlates with the rate of asymptomatic carriers in different populations. Thus, a linked transcriptional modifier decreases PRPF31 gene expression that leads to haploinsufficiency. This result, taken with other identified risk alleles, allows precise genetic counseling for the first time. We also demonstrate that across the human genome, the presence of MSR1 repeats in the promoters or first introns of genes is associated with greater population variability in gene expression indicating that copy number variation of MSR1s is a generic controller of gene expression and promises to provide new insights into our understanding of gene expression regulation.
Incomplete penetrance is an almost universal feature of monogenic disorders, yet the molecular basis underlying this phenomenon remains obscure in all but a few cases. Beyond the scientific importance of understanding how gene mutations can be exacerbated or rescued, its clinical importance should not be underestimated. Even for monogenic disorder, the lack of prediction of disease penetrance and severity frustrates genetic counseling. A sentinel example of incomplete penetrance is PRPF31-associated retinitis pigmentosa, where presence of symptomatic and asymptomatic mutation carriers is a universal feature in affected families. It has been proposed that phenotypic discordance does not only arise as a result of coding mutations with differential loss-of-function, but also from variation in the regulatory sequences of the same gene1. In this work, we provide proof of this theory.
Retinitis pigmentosa is a genetically heterogenous group of retinal degenerations, characterized by progressive cell death of the rod, and then cone, photoreceptors. This leads to progressive nyctalopia, constriction of visual fields and debilitating visual impairment. Autosomal dominant retinitis pigmentosa (adRP) accounts for over one third of cases and mutations in at least 25 genes have been implicated. Intriguingly, mutations in six ubiquitously expressed splicing factors are responsible for adRP (PRPF3, PRPF6, PRPF8, PRPF31, snRNP200 and RP9). Why mutations in such “housekeeping” genes produce a retina-specific phenotype is currently unknown. Mutations in PRPF31 underlie a major adRP locus, termed RP11 (chromosome 19q13.4, OMIM#60138)2,3. PRPF31 protein interacts with the U4/U6.U5 tri-snRNP, the central ribonucleoprotein complex of the spliceosomal machinery. PRPF31 contains a domain that binds the U4 snRNP within the U4/U6 di-snRNP and bridges the U4/U6 di-snRNP with the U5 snRNP to form the U4/U6.U5 tri-snRNP4,5,6,7.
Mutations in PRPF31 are the second most common cause of adRP, accounting for up to 10% of cases8,9,10,11. A diversity of mutations has been described, including nonsense, missense, frameshift and large deletions. The most intriguing feature of PRPF31-associated adRP is incomplete penetrance, whereby even within families, some mutation carriers are blind whereas others are fully sighted and asymptomatic. Previous work has shown that the asymptomatic trait is strongly linked to inheritance of the wildtype PRPF31 allele from the unaffected parent12,13. Haplotype analysis indicates that symptomatic and asymptomatic siblings consistently inherited different wildtype 19q13.4/PRPF31 parental alleles, suggesting the existence of an inherited parental factor that can “rescue” the disease phenotype12,13. The expression of the wildtype PRPF31 allele is more than two-fold higher in asymptomatic individuals as compared to their symptomatic relatives across families14,15. We hypothesized, therefore, that the major rescue factor (allele) must be common and act in cis to increase PRPF31 transcription. Identifying this element is a priority since it provides insight into “natural gene therapy.” Like all genes, a continuous distribution of PRPF31 expression is observed in the population and so PRPF31 transcription must be controlled by multiple genetic elements16. Nevertheless, for analyses, we focused on the core promoter region of PRPF31, termed BiP (−397 to +539 bp of the annotated transcription start site, TSS, which controls basal gene expression) and a cluster of MSR1 elements located ~630 bp upstream to the TSS (~200 bp upstream to the start of the core promoter region) that exhibits copy number variation (CNV)17.
We now show that: (1) the human population harbors 3 or 4 copies of the MSR1 element with the 4 repeat allele conferring increased PRPF31 expression; (2) the 3 repeat allele quenches gene expression in vitro; (3) symptomatic individuals do not carry the higher-expressing 4 repeat allele; and (4) the frequency of asymptomatic individuals in different populations are correlated with the frequency of the 4 repeat allele. This study demonstrates how variation in a minisatellite repeat, widely distributed across the genome, is a major determinant of disease penetrance and suggests that it is a candidate for similar phenomenon at other loci.
Results
MSR1 element CNV controls level of gene expression
A father-daughter pair were selected for further molecular analysis, as these individuals harboured a 112 kb deletion at the chromosome 19q13 site and so are hemizygous for PRPF31 and the upstream region of interest18. The symptomatic daughter was hemizygous for 3-copies of MSR1 while the asymptomatic father was hemizygous for 4-copies of MSR1. This suggested that the MSR1 CNV might alter gene expression and be the basis for the observed phenotypic difference. We designed the fragment BiP-MSR, encompassing the MSR1 cluster and the full experimentally defined PRPF31 promoter (Fig. 1A), which was amplified using DNA from the symptomatic daughter harbouring the reference sequence (3x MSR1; BiP-MSR-3x) and her asymptomatic father (4x MSR1; BiP-MSR-4x).
Figure 1. MSR1 element copy number variation was demonstrated to be the major factor controlling incomplete penetrance in RP11.
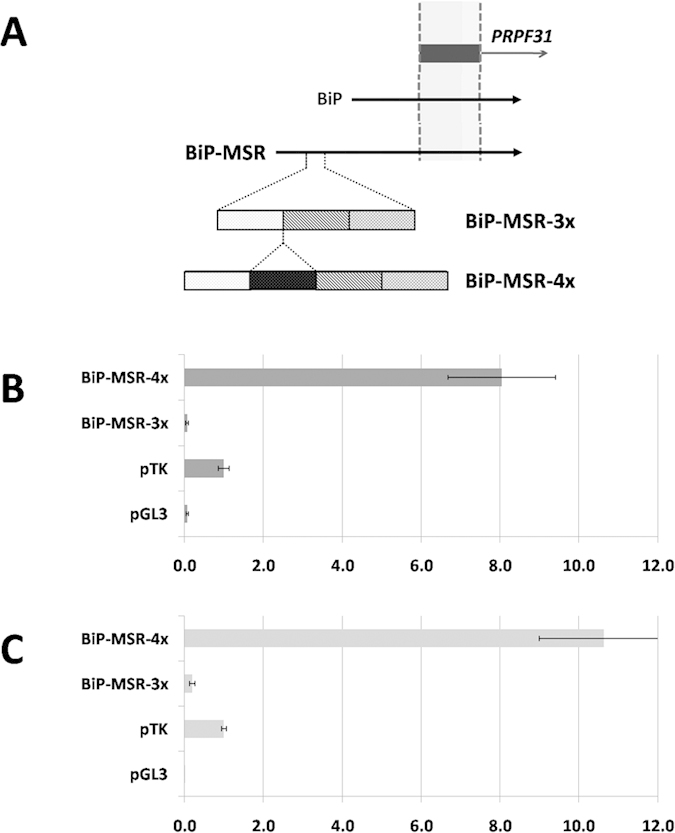
(A) Schematic representation of BiP-MSR. The location of PRPF31 exon 1 is shown, alongside BiP (the previously defined core promoter fragment15) and the relative location of the MSR1 elements. (B) Dual-luciferase reporter assay of BiP-MSR in RPE-1 cells, presented as the mean ratio of the vector construct to pTK, a basic promoter vector, together with one standard deviation. The positive (pTK) and negative (pGL3) controls are shown; (x-axis – fold-induction over pTK; y-axis – vector). (C) Dual-luciferase reporter assay of BiP-MSR in HeLa cells, presented as the mean ratio of the vector construct to pTK, a basic promoter vector, together with one standard deviation. The positive (pTK) and negative (pGL3) controls are shown; (x-axis – fold-induction over pTK; y-axis – vector).
The PRPF31 core promoter, previously defined as BiP, had ~8-fold induction over the pTK control17. The MSR1-containing fragments of interest were cloned into the pGL3 vector and dual-luciferase reporter assays were performed in HeLa and RPE-1 cell lines. In both cell lines, BiP-MSR-4x had strong reporter activity [10.63 ± 1.63 (HeLa); 8.05 ± 1.36 (RPE-1)], and was similar to the BiP promoter (Fig. 1B,C, Table 1)17. Strikingly, BiP-MSR-3x had no luciferase reporter activity in vitro [0.20 ± 0.07 (HeLa); 0.07 ± 0.03 (RPE-1)] (Fig. 1B,C, Table 1). In the HeLa cell line, BiP-MSR-3x had 53 times lower reporter activity than BiP-MSR-4x, a highly significant difference (Mann-Whitney U = 168, n1 = 14, n2 = 12, p < 2 × 10−7). An even greater difference in activity was observed in the RPE-1 cell line, where BiP-MSR-3x had 115 times lower activity than BiP-MSR-4x (Mann-Whitney U = 306, n1 = 18, n2 = 17, p < 4 × 10−10). Thus, in vitro and in the natural relation to the PRPF31 core promoter, decreased copy number of MSR1 elements is associated with almost complete abrogation of PRPF31 transcription.
Table 1. Luciferase assay results of tested fragments in two cell lines, presented as the mean ratio of the constructs to pTK (a basic promoter vector), together with one standard deviation.
| Fold induction over pTK |
||
|---|---|---|
| HeLa | RPE-1 | |
| BiP-MSR-3x | 0.20 ± 0.07 | 0.07 ± 0.03 |
| BiP-MSR-4x | 10.63 ± 1.63 | 8.05 ± 1.36 |
| pTK-MSR-2x (+) | 2.85 ± 0.49 | 4.53 ± 0.64 |
| pTK-MSR-3x (+) | 1.27 ± 0.26 | 3.11 ± 0.35 |
| pTK-MSR-4x (+) | 0.56 ± 0.16 | 1.96 ± 0.24 |
| pTK-MSR-2x (−) | 2.05 ± 0.49 | 3.45 ± 0.36 |
| pTK-MSR-3x (−) | 1.14 ± 0.20 | 1.78 ± 0.24 |
| pTK-MSR-4x (−) | 0.79 ± 0.10 | 0.97 ± 0.19 |
| Δ1-MSR-2x | 0.042 ± 0.012 | 0.065 ± 0.013 |
| Δ1-MSR-3x | 0.041 ± 0.010 | 0.069 ± 0.012 |
| Δ1-MSR-4x | 0.040 ± 0.009 | 0.086 ± 0.024 |
| pTK (positive control) | 1.00 ± 0.06 | 1.00 ± 0.14 |
| pGL3 (negative control) | 0.008 ± 0.004 | 0.074 ± 0.023 |
MSR1 copy number is a major genetic determinant of the PRPF31-associated adRP phenotype
In the general population, in individuals of European ancestry, the minor allele frequency (MAF) of the 4 MSR1 allele is 0.154 with substantial numbers of heterozygous (22.2% are 3/4) and homozygous (4.2% are 4/4) individuals. Consequently, we could search for associations between this common allele and disease penetrance with high statistical power. We studied 42 symptomatic and 29 asymptomatic individuals of European ancestry carrying a PRPF31 mutation and discovered that all symptomatic individuals had the 3/3 genotype, a highly significant increase as compared to ethnically-matched controls (z = 3.84; p = 0.0001). In contrast, among asymptomatic PRPF31 mutation bearers, the 4-copy allele was present in the heterozygous or hemizygous state in 8/29 individuals (27.6%; 7 individuals were heterozygous, 1 individual was hemizygous), the remaining individuals being 3/3: the proportion of heterozygotes was significantly higher as compared to ethnically matched controls (z = 2.31; p = 0.02). The pedigrees of asymptomatic individuals carrying the 4-copy allele confirmed that the 4-copy allele was carried on wildtype chromosomes in all cases.
We evaluated a diagnostic test model where a positive result (symptomatic) was having only 3-copy alleles (homozygous or hemizygous) and a negative result (asymptomatic) was having at least one 4-copy allele (homozygous, heterozygous or hemizygous). Our data estimated the positive predictive value at 67% (95% CI = 53.7–78.0%) and, most usefully, the negative predictive value at 100% (95% CI = 63.0–100%). This diagnostic test would be improved by analysis of a larger number of PRPF31 mutation carrying families. Furthermore, a diagnostic test that uses MSR1 genotype, together with other identified risk alleles, will greatly improve genetic counseling to families.
Disease penetrance correlates with population frequencies of MSR1 alleles
To further investigate the role of MSR1 in the incomplete penetrance of PRPF31-associated adRP, we examined the pedigrees of all families of European and Asian (Japanese/Chinese) origin available to us for asymptomatic mutation carriers. A total of 28 families of European origin with 270 mutation carriers were analysed, of which 92 (34.1%) were asymptomatic (Supplementary Data 1; Figure S1). Twelve families of Asian origin have been published, containing 140 mutation-carriers, of whom 14 (10.0%) were asymptomatic (Supplementary Data 1; Figure S2). Thus, the rate of asymptomatic mutation carriers is significantly lower in Asian as compared to European cases (z = 5.28; p < 0.0001). We postulated that this difference could be accounted for by the allele frequency differences at the MSR1 CNV.
Control samples from unrelated, unaffected individuals were tested for the MSR1 CNV at PRPF31 (71 Asians and 283 Europeans). We demonstrated that only 5 Asians (7.0%) carried the 4-copy allele in the heterozygous state while no homozygotes were observed, corresponding to a minor allele frequency (MAF) of 0.035 (Fig. 2). In concordance with previously published data17, 22.2% of European controls were heterozygous (3/4) and 4.2% were homozygous (4/4), corresponding to a minor allele frequency (MAF) of 0.154 (Fig. 2). Thus, the frequency of the 4-copy allele is significantly lower in Asian than European individuals (z = 3.76; p < 0.0001). These results are consistent with the hypothesis that the 4-copy allele confers higher PRPF31 gene expression and acts as the major protective factor. Therefore, in Asia, where asymptomatic mutation carriers are rare, the protective 4-copy allele is correspondingly rare. Although only a statistical correlation in the two populations studied, the principles of causation are maintained by this correlation. Taken as supporting evidence for the functional in vitro work, and the study in a large patient cohort, this finding strongly supports the conclusion that CNV of the MSR1 is the major cis-acting genetic element controlling incomplete penetrance in PRPF31-associated adRP.
Figure 2. The frequency of PRPF31 MSR1 cluster genotypes varies significantly between ethnic populations.
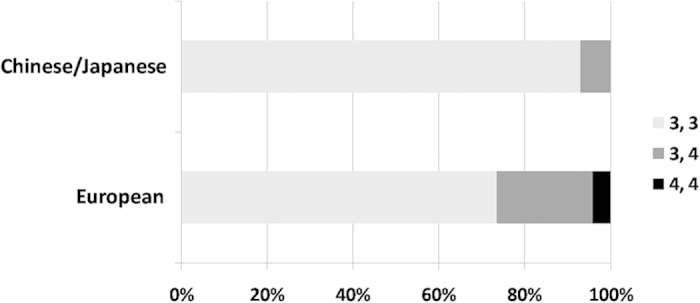
In concordance with the differing rates of incomplete penetrance, the 4-copy allele was much rarer in Oriental populations compared to Caucasian populations.
The MSR1 CNV is a generic modulator of basal promoter activity
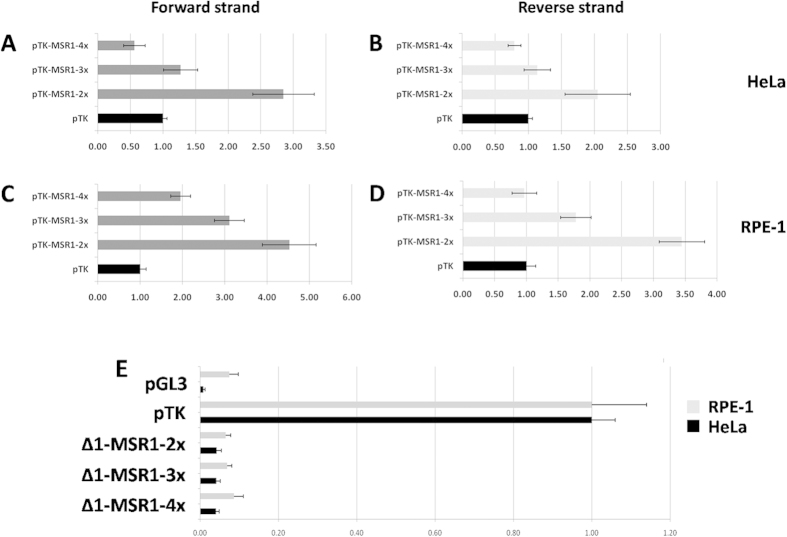
To further assess the copy number effect of MSR1 on gene expression, we cloned DNA segments with 2, 3 or 4 copies of MSR1 (pTK-MSR1-2x, pTK-MSR1-3x and pTK-MSR1-4x respectively) upstream of the thymidine kinase (TK) minimal promoter in a pGL3 vector. In both cell lines assayed, increasing MSR1 copy number resulted in a significant decrease in luciferase reporter activity (Fig. 3A–D, Table 1). Importantly, this effect was independent of MSR1 element orientation. In the forward strand direction, pTK-MSR1-2x had 1.5 to 1.8-fold higher activity than pTK-MSR1-3x; the difference was significant in both cell lines tested (HeLa t = 8.94, p < 0.00001; RPE-1 t = 7.80 p < 0.00001). Also, pTK-MSR1-3x had 1.4 to 1.6 fold higher activity than pTK-MSR1-4x; significant in both cell lines tested (HeLa t = 7.39, p < 0.00001; RPE-1 t = 9.37, p < 0.00001). In the reverse strand direction, pTK-MSR1-2x had 1.9 to 2.2-fold higher activity than pTK-MSR1-3x, the difference being significant in both cell lines tested (HeLa t = 5.69, p < 0.00001; RPE-1 t = 13.36 p < 0.00001). Finally, pTK-MSR1-3x had 1.9 to 2.2-fold higher activity than pTK-MSR1-4x; this difference was significant in both cell lines tested (HeLa t = 4.75, p < 0.00008; RPE-1 t = 9.54, p < 0.00001). Subsequently, the MSR1 element was cloned into a pGL3-basic vector in 2-, 3- or 4-copies (Δ1-MSR1-2x, Δ1-MSR1-3x, Δ1-MSR1-4x, respectively). This analysis demonstrated that the isolated MSR1 cluster did not have intrinsic promoter activity, regardless of copy number (Fig. 3E, Table 1).
Figure 3.
(A–D) Analysis of CNV of MSR1 in pTK promoter vector; the repeat cluster was cloned in forward strand orientation (A,C) and reverse strand orientation (B,D). This demonstrated a stepwise decrease in reporter activity with increasing MSR1 copy number. (E) Analysis of CNV of MSR1 in a pGL3-empty vector, demonstrating that the MSR1 elements have no intrinsic promoter activity. Data is presented as the mean ratio of the vector construct to pTK, a basic promoter vector, together with one standard deviation. The positive (pTK) and negative (pGL3) controls are also shown; x-axis – fold-induction over pTK; y-axis – vector.
These experiments demonstrate that modulation of gene expression by MSR1 is not gene-specific, but it can control expression of any promoter element. In other words, the MSR1 element is a generic molecular switch that can control the expression of any gene in which it is contained. It is important to note that this effect is the reverse of the effect observed with the natural PRPF31 promoter where increasing copy number increased reporter activity. This possibly suggests that the effect of the MSR1 cluster is dependent on the spatial relation of the repeats and the promoter, but not their orientation. It will be necessary to perform further reporter assays, with the MSR1 clusters at various genetic distances from the core promoter to fully explore this interesting hypothesis. In addition, we conclusively demonstrate that the MSR1 repeat is not a promoter element but a modulator of gene transcription.
Genomic features of MSR1 elements
Since MSR1 elements are widespread in the human genome (RepeatMasker browser; www.repeatmasker.org), we sought to examine how they might affect gene expression broadly. We examined population-level gene expression of four groups of chromosome 19 genes (i) genes containing an MSR1 cluster in the promoter, (ii) genes containing an MSR1 cluster in intron 1, (iii) genes containing an MSR1 cluster in other introns, and (iv) genes that did not contain an MSR1 cluster. Analysis of gene expression of the cognate group of genes in 270 control individuals, with published data from HapMap Phase II19, demonstrated that genes containing an MSR1 cluster in the promoter showed greater population-level variability, estimated as the mean standard deviation, than did genes without a MSR1 cluster (p < 0.04) (Fig. 4). Second, genes containing an MSR1 cluster in intron 1 also showed a significantly greater variability than non-MSR1 containing genes (p < 0.04) (Fig. 4). There was no observable difference between non-MSR1 containing genes and those with MSR1 elements in introns other than intron 1 (p = 0.23). These findings are consistent with the hypothesis that MSR1 elements regulate gene expression and that CNVs of MSR1 leads to the increased variation in gene expression. We believe that these variations represent an under-estimate of the true effect of MSR1 because it is unlikely that CNVs occurs in all MSR1 clusters in groups (i) and (ii).
Figure 4. Analysis of population gene expression data demonstrated that genes containing an MSR1 cluster in the promoter or intron 1 had greater average variability than genes that did not contain an MSR1 cluster.
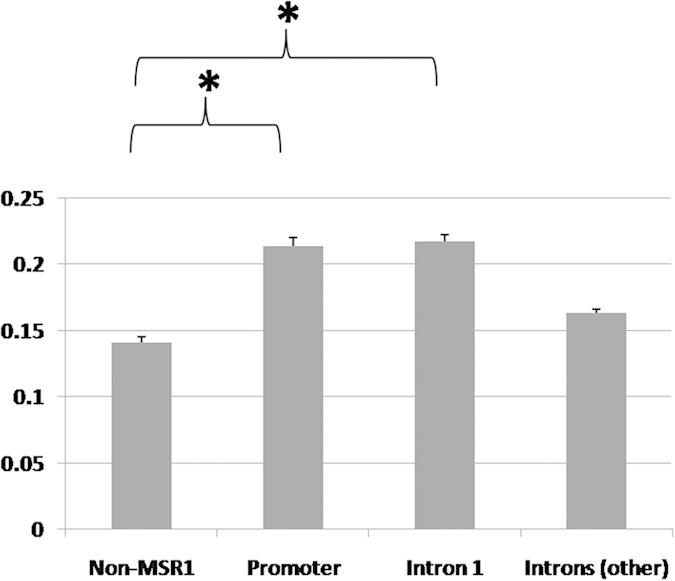
Data is presented as the mean variability of each gene group (mean standard deviation) ± SEM; an asterisk indicates statistical significance.
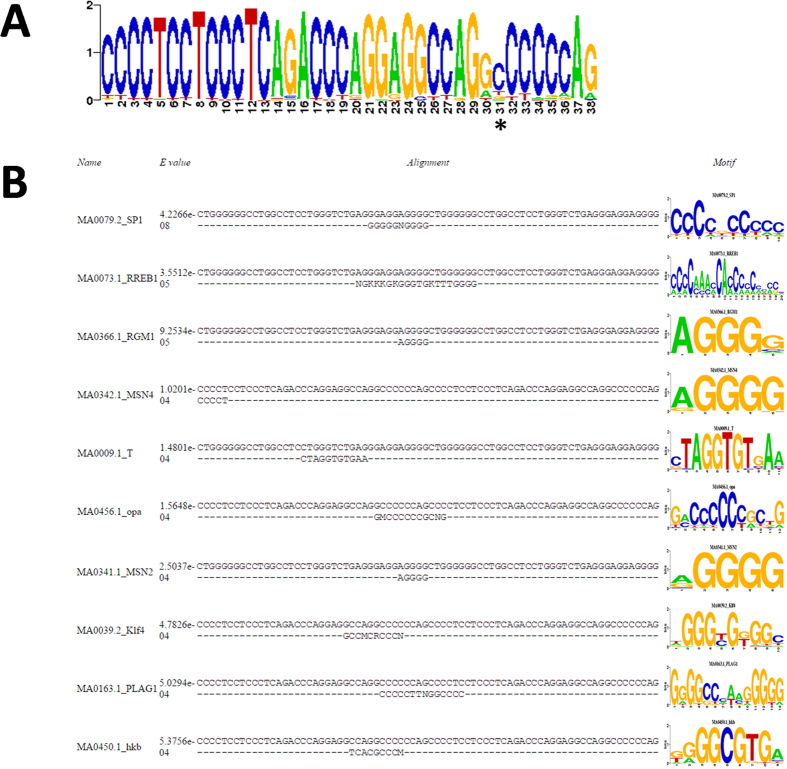
To understand how the MSR1 effect might be mediated, possibly through binding of a specific RNA or protein, we generated a consensus sequence positional weight matrix (PWM) using 100 randomly selected MSR1 elements from chromosome 19 (Fig. 5A). We used these sequences to identify potential transcription factor binding sites (TFBS), including at junctional binding sites when two or more MSR1 elements are present (Fig. 5B, Table 2). This demonstrated that there are a small number of putative TFBS within the MSR1 sequence, although only three potential sites for human transcription factors (SP1, RREB1, PLAG1). Further, the MSR1 sequence did not support the contention that it was a CpG island, so that epigenetic regulation by CpG methylation is unlikely to contribute to its gene expression effect.
Figure 5.
(A) Positional weight matrix of consensus sequence of MSR1 element, derived from 100 randomly selected MSR1 elements on chromosome 19. The asterisk indicates a C nucleotide that is frequently absent. (B) Prediction of TFBS using PWM of two consecutive MSR1 elements, allowing demonstration of junctional binding sites.
Table 2. Bioinformatic prediction of TFBSs based on consensus PWM of two consecutive MSR1 elements.
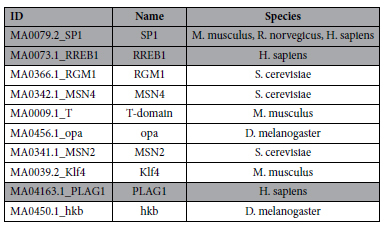
The three factors highlighted in grey represent human transcription factors.
Discussion
The major finding from our study is that CNV in an MSR1 repeat element upstream of PRPF31 modulates its transcriptional activity and thereby controls the phenotypic incomplete penetrance in PRPF31-associated adRP. This modulation, as a decrease in gene expression, occurs in all individuals that have at least one copy of the 3-repeat MSR1 allele. Although analysis of PRPF31 gene expression in the retina has not been performed – and would be admittedly challenging – it is a specific prediction of our findings. In PRPF31 mutation carriers there is one wildtype and one mutant allele; these mutant alleles are either loss-of-function or functional hypomorphs. Consequently, if such mutation bearers are homozygous for the 3-repeat MSR1 allele, PRPF31 gene expression is reduced for both the mutant and wildtype allele, and these individuals can be symptomatic. In 3-repeat heterozygotes, gene expression will be reduced for the PRPF31 allele in cis with the 3-repeat allele and so will have an impact on PRPF31 gene expression either through the wildtype allele or further decrease for the PRPF31 mutant. Our study confirms the theory that linked regulatory elements can modulate human disease penetrance1.
The primary benefit of this study is that our data provides an avenue for more accurate genetic counseling of families affected by PRPF31-associated adRP. The negative predictive value of a test assessing only the MSR1 copy number was estimated at up to 100%, meaning that if a PRPF31 mutation carrier also carries a 4-copy allele in cis to the wildtype allele, we can say with very high certainty that they will not develop clinically relevant disease. Clearly, we need a replicate study before such quantitative statements can be clinically used. Nevertheless, the predictive accuracy of the test could be improved by integrating data relating to other SNPs that have been linked to the incomplete penetrance phenotype and/or expression QTLs (quantitative trait loci) for PRPF31.
Our investigations, however, have revealed a disparity between the dramatic effect observed in vitro (with up to 115-fold change in expression levels) and the modest effect seen in vivo14,15. Furthermore, a reasonable proportion of asymptomatic individuals carry two copies of the 3-copy allele and, in 3-copy allele homozygotes, in vivo expression cannot be completely abrogated, as homozygous mutation of PRPF31 is embryonic lethal. Thus, additional genes and other factors must also play a role in the incomplete penetrance phenotype of PRPF31-associated adRP. These observations are in agreement with previous work suggesting polygenic control of gene expression and multiple modifying factors15,16. Other modifying factors might include negative regulation by CNOT3 and distant regulators on chromosome 1416,20. Finally, prior to nonsense-mediated decay of mutant PRPF31 there is some increased expression of both PRPF31 alleles in asymptomatic mutation carrying individuals21, suggesting that trans-acting factors might also control PRPF31 expression.
What is the mechanism by which MSR1 elements exert their effect? Our work provides some initial clues. First, we demonstrate that the MSR1 element cluster is a generic modulator of basal promoter activity without possessing any intrinsic promoter activity. Second, like all classical enhancers, the effect of copy number is independent of orientation but dependent on the spatial relationship of the MSR1 cluster to the core promoter. Third, MSR1 elements throughout the genome that are close to the promoter or reside in intron 1 also affect gene expression. Consequently, by the classical definition, the MSR1 element acts as a modulator (enhancer/suppressor) rather than a repeat sequence that has been co-opted as a modulatory element.
Bioinformatics analyses show that the repeat element contains sequences that can putatively bind specific transcription factors including two sites generated when multiple MSR1 sequences are consecutively arranged in a cluster (junctional sites), so that CNVs of the elements might allow for differential binding and differential gene expression through TF-mediated enhancer activity. On the other hand, given that when cloned upstream to pTK basic promoter, variable copy number of the MSR1 repeat had the inverse effect, it seems more likely that mechanisms other than transcription factor binding might be involved. Since the element is not a site for CpG methylation, other epigenetic mechanisms might be considered. Given that MSR1 acts as an enhancer, we feel that it is very likely that DNA looping is critical to its effect on gene expression. This is supported by the dramatic effect of the spatial relation of the MSR1 cluster to the promoter. Further experiments to test these various hypotheses are necessary in order to fully understand how MSR1 modulates gene expression.
It is clear that MSR1 elements are likely to be an important genome-wide regulator of gene expression and it will be critical to establish which of the MSR1 clusters exhibit CNV in different human populations. The repeat element is a 36–38 bp minisatellite repeat that exists in highest density on chromosome 19q. The element appears to have arisen relatively recently in evolution, after the divergence of avian and mammalian lineages, and locus-specific conservation of elements has been reported22,23. It is crucial to examine whether CNV of elements underlies the population-level gene expression variability we observed at the large numbers of MSR1 containing loci across chromosome 19 and elsewhere. Examples of MSR1 CNV do exist: the kallikrein locus has two MSR1 clusters that have shown CNV, one within KLK14 and one within KLK424. Intriguingly, correlations between MSR1 copy number and breast and prostate adenocarcinoma were shown24. Given that aberrant kallikrein gene expression is key to the pathogenesis in many tumours, we predict that MSR1 CNVs might underlie increased susceptibility to cancer in these patients. In addition, hypermutation of MSR1 elements within cancer cells might further promote gene dysregulation. Finally, a cluster of MSR1 elements within the TNNI3 promoter, conserved in both human and mouse has been reported23,25. Some authors have suggested selection pressure to maintain the elements as part of the functional TNNI3 promoter since deletion of this degenerate cluster altered core TNNI3 promoter activity in the mouse25,26.
These results allows us to hypothesize that variability of gene expression due to MSR1 CNV will be a modifier in many monogenic diseases and an important risk factor for the development of a multitude of polygenic disorders.
Materials and Methods
Amplification and cloning of BiP-MSR
The BiP-MSR fragment was amplified using template DNA from two individuals harbouring a 112 kb deletion at chromosome 19q13 and who were, therefore, hemizygous for the entire region of interest18. One individual (RP15011) was symptomatic, therefore representing a lower-expressing allele; this allele carried 3-copies of MSR1. The other individual (III.7) was asymptomatic, therefore representing a higher-expressing allele and this person carried 4-copies of MSR1. Blunt-ended PCR was performed using KOD polymerase (Novagen) and the following primer pair: GAAGAATCGTTTGGAACCCG and GATGTGGCCACCAAATAG (1398 bp, hg 19 co-ordinates chr19:54617996-54619393). pGL3-basic vector (Promega) was digested using SmaI restriction enzyme. The BiP-MSR insert was ligated into the digested vector and vectors of interest transformed into DH5-α cells; plasmids were isolated using standard Miniprep methods.
Dual-luciferase reporter assay was performed in HeLa and RPE-1 cell lines, as described in full previously17. Briefly, HeLa and RPE-1 cells were grown to 80–90% confluence and co-transfected with pGL3-MSR and pRenilla; each transfection was performed in quadruplicate. The cells were grown for 48 h, then lysed and dual-luciferase reporter assay was performed as instructed by the kit manufacturer (Promega). For each fragment, at least three separate transfections were performed. On each plate, a modified pGL3 plasmid containing basic TK promoter (pTK) was used as a positive control and pGL3-basic vector as a negative control. Data were disregarded if the pRenilla values for repeats varied more than ± 10%.The data were firstly analyzed by calculating the ratio between pGL3-MSR and pRenilla; this calculation standardized for cell number and transfection efficiency. This ratio was then compared with the average value of the positive control (pTK), to generate the pGL3-MSR1:pTK ratio. Statistical significance was assessed using two-tailed Mann-Whitney U-test. The full dataset of luciferase assays are shown in Supplementary Data 2.
Assessment of CNV in asymptomatic and symptomatic mutation carrying individuals
Genomic DNA was extracted from peripheral blood samples of PRPF31 mutation-carrying individuals under standard conditions (72 individuals: 42 symptomatic and 29 asymptomatic). Informed consent had been obtained from all patients and asymptomatic relatives used in this study, and research was performed according to the tenets of the Declaration of Helsinki. The use of human DNA samples had been approved by the Moorfields Eye Hospital and UCL Institute of Ophthalmology Ethics Committee. PCR was performed using one labelled primer (5′FAM-GTTAGGGGTTTGGACTGC) and an unlabelled reverse primer (GATGTGGCCACCAAATAG). Labelled PCR product was sized using GeneScan 500 LIZ size standard (Invitrogen) on ABI3700 and data was analyzed on GeneMarker v1.7 (SoftGenetics LLC).
Studies in different ethnic populations
All family trees affected by PRPF31-associated adRP with stated ethnicity that were published prior to January 2014 were assimilated, along with unpublished families from our laboratory. The asymptomatic and symptomatic mutation carriers were identified. Statistical significance was ascertained by comparison of two independent proportions (z-test).
CNV of MSR1 element was assayed as described above in genomic DNA of individuals of different ethnicity (Caucasian n = 283, Oriental n = 71). The DNA were from ECACC control panels of DNA, as well as samples collected from unrelated, healthy controls at Moorfields Eye Hospital, London, UK.
Analysis in pTK and pGL3-basic vectors
To generate pTK-MSR1, pTK vector (Promega) was digested with SmaI restriction enzyme under standard conditions. PCR products (2- 3- or 4-copies of MSR1) were amplified from the BiP-MSR1 construct of individual III.7; these products were purified and the insert was ligated in both orientations into the digested vector and transformed as previously described. To generate Δ1-MSR1 constructs, pGL3-basic vector (Promega) underwent the same process as that of pTK-MSR1. Luciferase assay was performed in RPE-1 and HeLa cell lines, as previously described for BiP-MSR.
Analysis of population-level gene expression variability
The expression of chromosome 19 genes containing MSR1 elements in (i) promoter (ii) intron 1 or (iii) other introns was compared to a random sample of chromosome 19 genes that did not contain MSR1 elements (chosen from an alphabetical list using a random number generator). Gene expression data from the 270 individuals of various ethnicities from the HapMap phase II project was downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo)19. The mean and standard deviation of each gene of interest was calculated. The mean standard deviation (e.g. variability) and standard error of the mean (SEM) were then calculated for the four study groups (promoter, intron 1, other introns, non-MSR1). Statistical significance was assessed using Student’s T-test for two independent means.
Bioinformatic analysis of TFBSs and epigenetic regulation
The sequence of 100 randomly selected MSR1 elements was obtained and the frequency of nucleotides at each position was recorded to generate a consensus positional weight matrix. TFBSs were identified using a consecutive series of two consensus MSR1 PWMs in the STAMP database on routine settings (available at http://www.benoslab.pitt.edu/stamp)27. CpG island analysis was performed using the EMBOSS Newcpgreport tool (available at http://www.ebi.ac.uk/Tools/seqstats/emboss_newcpgreport/)28.
Additional Information
How to cite this article: Rose, A. M. et al. Transcriptional regulation of PRPF31 gene expression by MSR1 repeat elements causes incomplete penetrance in retinitis pigmentosa. Sci. Rep. 6, 19450; doi: 10.1038/srep19450 (2016).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge grant funding from Rosetrees Trust (M68), The Astor Foundation, The Butterfield Trust, RP Fighting Blindness, Fight for Sight, Foundation Fighting Blindness USA, National Institute of Health Research BRC for Ophthalmology and The Special Trustees of Moorfields Eye Hospital London.
Footnotes
AMR holds UK patent number PCT/GB2014/050107.
Author Contributions Study design – A.M.R.; experimental work – A.M.R., A.Z.S., G.V., C.R. and A.K.; data analysis and interpretation – A.M.R., A.K., C.R., A.C. and S.S.B.; preparation and review of manuscript – A.M.R., A.Z.S., C.R., A.C. and S.S.B.
References
- Chakravarti A. & Kapoor A. Genetics- Mendelian puzzles. Science. 335, 930–931 (2012). [DOI] [PubMed] [Google Scholar]
- Al-Maghtheh M. et al. Evidence for a major retinitis pigmentosa locus on 19q13.4 (RP11) and association with a unique bimodal expressivity phenotype. Am J Hum Genet. 59, 864–871 (1996). [PMC free article] [PubMed] [Google Scholar]
- Vithana E. N. et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell. 8, 375–381 (2001). [DOI] [PubMed] [Google Scholar]
- Weidenhammer E. M., Ruiz-Noriega M. & Woolford J. L. Jr. Prp31p promotes the association of the U4/U6 × U5 tri-snRNP with prespliceosomes to form spliceosomes in Saccharomyces cerevisiae. Mol Cell Biol. 17, 3580–3588 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O. V., Makarov E. M., Liu S., Vornlocher H. P. & Lührmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21, 1148–1157 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Rauhut R., Vornlocher H. P. & Lührmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA. 12, 1418–1430 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. et al. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 316, 115–120 (2007). [DOI] [PubMed] [Google Scholar]
- Waseem N. H., al. Mutations in the gene coding for the pre-mRNA splicing factor, PRPF31, in patients with autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 48, 1330–1334 (2007). [DOI] [PubMed] [Google Scholar]
- Vithana E., Al-Maghtheh M., Bhattacharya S. S. & Inglehearn C. F. RP11 is the second most common locus for dominant retinitis pigmentosa. J. Med. Genet. 35, 174–175 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audo I. et al. Prevalence and novelty of PRPF31 mutations in French autosomal dominant rod-cone dystrophy patients and a review of published reports. BMC Med. Genet. 11, 145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F. et al. Novel PRPF31 mutations associated with Chinese autosomal dominant retinitis pigmentosa patients. Mol. Vis. 18, 3021 (2012). [PMC free article] [PubMed] [Google Scholar]
- McGee T. L., Devoto M., Ott J., Berson E. L. & Dryja T. P. Evidence that the penetrance of mutations at the RP11 locus causing dominant retinitis pigmentosa is influenced by a gene linked to the homologous RP11 allele. Am. J. Hum. Genet. 61, 1059–1066 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A. M. et al. Dominant PRPF31 mutations are hypostatic to a recessive CNOT3 polymorphism in retinitis pigmentosa: a novel phenomenon of “linked trans-acting epistasis”. Ann. Hum. Genet. 78, 62–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithana E. N. et al. Expression of PRPF31 mRNA in patients with autosomal dominant retinitis pigmentosa: a molecular clue for incomplete penetrance? Invest. Ophthalmol. Vis. Sci. 44, 4204–4209 (2003). [DOI] [PubMed] [Google Scholar]
- Rivolta C. et al. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum. Mutat. 27, 644–653 (2006). [DOI] [PubMed] [Google Scholar]
- Rio Frio T., Civic N., Ransijn A., Beckmann J. S. & Rivolta C. Two trans-acting eQTLs modulate the penetrance of PRPF31 mutations. Hum. Mol. Genet. 17, 3154–3165 (2008). [DOI] [PubMed] [Google Scholar]
- Rose A. M. et al. Expression of PRPF31 and TFPT: regulation in health and retinal disease. Hum. Mol. Genet. 21, 4126–4137 (2012). [DOI] [PubMed] [Google Scholar]
- Rose A. M., Mukhopadhyay R., Webster A. R., Bhattacharya S. S.& Waseem N. H. A 112 kb deletion in chromosome 19q13.42 leads to retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 52, 6597–6603 (2011). [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 449, 851–61. (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini G., Rose A. M., Shah A. Z., Bhattacharya S. S. & Rivolta C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 8, e1003040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio Frio T. et al. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J Clin Invest. 118, 1519–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das H. K., Jackson C. L., Miller D. A., Leff T. & Breslow J. L. The human apolipoprotein C-II gene sequence contains a novel chromosome 19-specific minisatellite in its third intron. J Biol Chem. 262, 4787–93 (1987). [PubMed] [Google Scholar]
- Bhavsar P. K., Brand N. J., Yacoub M. H. & Barton P. J. Isolation and characterization of the human cardiac troponin I gene (TNNI3). Genomics. 35, 11–23 (1996). [DOI] [PubMed] [Google Scholar]
- Yousef G. M., Bharaj B. S., Yu H., Poulopoulos J. & Diamandis E. P. Sequence analysis of the human kallikrein gene locus identifies a unique polymorphic minisatellite element. Biochem. Biophys. Res. Commun. 285, 1321–1329 (2001). [DOI] [PubMed] [Google Scholar]
- Cullen M. E., Dellow K. A. & Barton P. J. Structure and regulation of human troponin genes. Mol. Cell. Biochem. 263, 81–90 (2004). [DOI] [PubMed] [Google Scholar]
- Di Lisi R. et al. Combinatorial cis-acting elements control tissue-specific activation of the cardiac troponin I gene in vitro and in vivo. J. Biol. Chem. 273, 25371–25380 (1998). [DOI] [PubMed] [Google Scholar]
- Mahony S.& Benos P. V. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 35, W253–8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H. et al. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 41, W597–600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.