Abstract
Despite identification of several sex-determining genes in non-mammalian vertebrates, their detailed molecular cascades of sex determination/differentiation are not known. Here, we used a novel RNAi to characterise the molecular mechanism of Dmy (the sex-determining gene of medaka)-mediated masculinity in XY fish. Dmy knockdown (Dmy-KD) suppressed male pathway (Gsdf, Sox9a2, etc.) and favoured female cascade (Rspo1, etc.) in embryonic XY gonads, resulting in a fertile male-to-female sex-reversal. Gsdf, Sox9a2, and Rspo1 directly interacted with Dmy, and co-injection of Gsdf and Sox9a2 re-established masculinity in XY-Dmy-KD transgenics, insinuating that Dmy initiates masculinity by stimulating and suppressing Gsdf/Sox9a2 and Rspo1 expression, respectively. Gonadal expression of Wt1a starts prior to Dmy and didn’t change upon Dmy-KD. Furthermore, Wt1a stimulated the promoter activity of Dmy, suggesting Wt1a as a regulator of Dmy. These findings provide new insights into the role of vertebrate sex-determining genes associated with the molecular interplay between the male and female pathways.
Sex determination is the process that determines the sex of an organism. The signals that control this decision are usually categorized as genetic or environmental. In the genetic sex determination, four master sex-determining genes have been previously identified: SRY/Sry, most mammals including humans1; Dmy/Dmrt1Y, medaka2,3; Dmrt1, chicken4; Dm-W, Xenopus laevis5. More recently, novel autosomal sex-determining genes were reported in five teleost species: Amhy, Odontesthes hatcheri6; Gsdf, Oryzias luzonensis7; Amhr2, Takifugu rubripes8; SdY, Oncorhynchus mykiss9 and Sox3, Oryzias dancena10. In the past decade, it has become clear that although the upstream sex-determining signals are diverse, they often act through more ancient downstream regulatory hierarchies11.
Japanese medaka (Oryzias latipes) is one of the best-studied species among non-mammalian vertebrates with respect to sex determination and differentiation. Sex determination in medaka is strictly genetic, with a male heterogametic (XX/XY) system2. The testicular morphogenesis in medaka is clearly distinctive from the ovarian morphogenesis during the early stages of gonadal differentiation12. It is generally accepted that the number of germ cells, more specifically the proliferative mitosis and subsequent occurrence of meiosis in females and mitotic arrest in males by the day of hatching, are the first sign of morphological sex differentiation in medaka12. Thus, the day of hatching is an important time point on which the morphological sex differentiation becomes apparent in the medaka XY and XX gonads not only in the form of germ cell proliferation, but also meiosis. Furthermore, in the QurtE strain of Japanese medaka, genetic sex of the embryos can be identified by the presence or absence of leucophores on the head region from 2 days after fertilization (daf)13. Leucophores are present in the head region of XY embryos but not in XX embryos13.
In medaka, the Y chromosome harbours the master sex-determining gene (primary testis determinant), Dmy/Dmrt1bY2,3. A mutation due to a single nucleotide insertion in the exon 3 of Dmy ORF leads to a truncated Dmy, which gives rise to spontaneous sex reversed XY female medaka2,14. The overexpression of Dmy resulted in the induction of testis differentiation and subsequent male development in XX (genetically female) medaka. Moreover, gripNA induced knockdown of Dmy by our group15 showed signs of increased germ cell number and meiotic progression, as early as hatching day (indicators of feminization) in XY medaka. However, no studies have yet revealed the course of action of Dmy on somatic and germ cells that induces the male sexual differentiation. Although several male-associated genes like Gsdf (gonadal soma derived factor, a unique member of the TGF-β family)16, Sox9a2 (also known as Sox9b, the orthologue of tetrapod Sox9)17,18, and Dmrt119 have been suggested to be downstream of Dmy, no specific targets have been identified for Dmy. On the contrary, even though the key ovary determinant in medaka has not been defined, R-spondins, estrogen receptor β2 (ERβ2) and Foxl2 have been shown to express specifically in XX gonads during sex determination and differentiation20,21,22,23.
Although several fish sex-determining genes have been identified by now, no elaborate knockdown study of these sex-determining genes is available. Therefore, it is of critical importance to study the effect of reduced Dmy expression on sex differentiation and maturation in medaka. Although ZFN/TALEN/CRISPER-based knockouts are gaining popularity, each one has their own advantages and disadvantages24. Antisense (AS) RNA has long been considered a promising technique for treating disease25. There are several mechanisms by which AS RNA may exert its effect, such as de-stabilization of endogenous mRNA26, production of small RNAs27 and activation of the siRNA/miRNA pathway27,28. Medaka and zebrafish (Danio rario) are excellent models for studying developmental processes, because of several advantages over higher vertebrates. For example, these egg-laying teleosts are small, have a rapid generation time and produce large number of offsprings. Also, their external fertilization and well-documented developmental processes makes gene manipulation studies much easier to perform. In the present study using medaka, we have developed an AS DNA vector-based approach to overcome the shortcomings of the effect of Dmy knockdown. Upon knockdown of Dmy, we were successful in generating a functional sex-reversed XY medaka with full fertilization potential. The gonadal sex reversal mechanism in medaka was associated with simultaneous suppression and induction of downstream male-associated and female-associated genes, respectively.
Results
Trans-generational antisense DNA-dependent knockdown in medaka
The present study used a 220 bp antisense DMY construct (neighbouring the DM domain) expressing short AS-RNA (Supplementary Fig. 1), which was designed and evaluated using E-RNAi (http://www.dkfz.de/signaling/e-rnai3//), to silence the Dmy gene in medaka. Gene specificity and possible off-target effects of the pmDMY-AS-construct on the downstream genes Dmrt1, Sox9a2, Gsdf and Sf1 were studied in cell cultures co-transfected with pCMV-DMY, pCMV-DMRT1, pCMV-SOX9a2, pCMV-GSDF, and pCMV-SF1. Dose-dependent suppression of the Dmy (92% at 100 ng group) and Dmrt1 (43.75% at 100 ng group) transcript were documented in this experiment. However, the expression of the other three genes (Sox9a2, Gsdf and Sf1) remained unaffected (Fig. 1A).
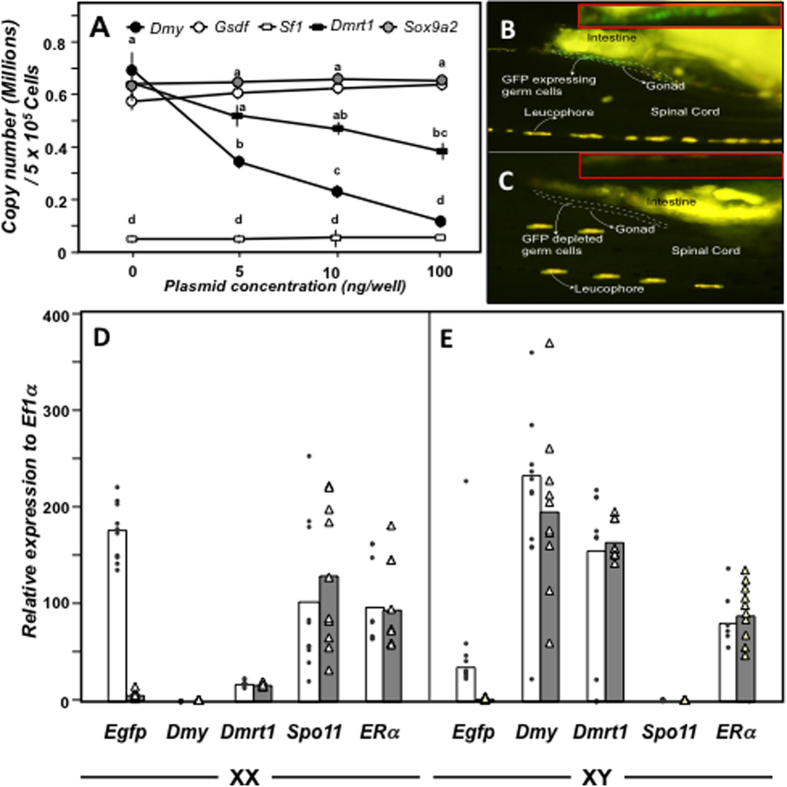
Figure 1. In vitro and in vivo validation of knockdown strategy.
(A) In vitro validation. The specificity and off target effects of pmDMY-AS construct was validated in vitro using COS7 cells via co-transfection of pmDMY-AS construct and the ORF plasmids of either Dmy (specific, solid circles), Dmrt1 (partially specific, solid squared), Sox9a2 (non-specific, grey circles) Gsdf (non specific, open circles) or Sf1 (non specific, open squared). The mean absolute copy numbers of respective gene per 5 ng of RNA are plotted in the graph along with SEM. The significance is indicated by a, b, c, in which different letters indicate the significant difference from other group at p < 0.05. (B,C) In vivo validation. Olvas-eGFP medaka were electroporated with pEGFP-AS plasmid (carries an antisense eGFP sequence) and continuous microscopic visualization (from 3days after fertilization (daf) to 5 days after hatching (dah)) of GFP expression in individual embryos was performed to ascertain the changes of GFP production in control (B, 6 daf) and pEGFP-KD (pEGFP-AS electroporated) groups (C, 6 daf). (D,E) Sharp fall in Gfp mRNA production was assessed through real-time PCR at 20 dah (days after hatching). Non-specific effects of pEGFP-AS construct were analysed by measuring the mRNA amount of Dmy, Dmrt1, Spo11, and ERα. Data are presented as both individual values (with dots and triangles) and mean of 10 individual (white and grey columns for control and knockdown groups, respectively) fish of each sex.
To confirm the knockdown effect of the AS technique in vivo, Gfp expression was knocked down in olvas-eGFP transgenic medaka (expresses GFP in the germ cells) one-two cell stage embryos29 by electroporation, using a pEGFP-AS plasmid (contains 320 bp antisense GFP sequence and expected to knockdown the GFP expression)30. Microscopic observations from 3 daf indicated a reduction of Gfp expression in gonads of electroporated embryos compared to negative controls (Fig. 1B,C). Real-time PCR using 20 dah (days after hatching) XX and XY fish demonstrated unchanged expression of other genes including Dmy, Gsdf, Dmrt1, Spo11, and estrogen receptor (ER) α (Fig. 1D,E). A group of pEGFP- knockdown (KD) fish was raised to adulthood to check whether the present method had some effect on gonad maintenance and secondary sexual character development in the long run. No side effect was observed with reference to growth and development of secondary sexual characteristics (Supplementary Table 1).
Our knockdown strategy is expected to transcribe AS RNA, which might be cleaved by endogenous RNA processing machineries (e.g., dicer, RNaseH, etc.) and produce a pronounced knockdown effect. To elucidate the molecular mechanism of knockdown action, small-RNAs were isolated from sex-reversed F1 generation fish (mentioned later in detail) of Dmy knockdown (Dmy-KD) group. Northern blotting (using same 220 bp Dmy fragment as a probe) detected a strong signal at 60 base (b) and a weak signal in the 18–28 b region, only in Dmy-KD lane samples (Supplementary Fig. 2). Cloning and sequencing analysis (details in Supplementary Materials and Methods) of these bands revealed several fragments of both 60–70 b and 18–28 b were derived from the long Dmy AS sequence (Supplementary Fig. 2) and rest were randomly distributed throughout the Dmy mRNA sequence. We also sequenced the larger fragments (>100 bp size fragments) to find out any evidence for alternative splicing associated transcriptional rate inhibition. Both control-XY and Dmy-KD-XY fish had similar pattern of large RNA (>600 bp) fragmentation except the fact that, only Dmy-KD samples contained a trail for different size samples (most of them had portion of ORF/5UTR/3UTR but none showed any evidence for alternative splicing, especially for intronic sequences, joined or duplicated exons), and relative occurrence of full ORF was also significantly less (1/123.4 clones) than their control counterpart (1/4.7 clones). These observations suggest that the AS RNA-expressing construct mainly destabilizes the mRNA probably via small interfering RNA (siRNA)/micro RNA (miRNA) pathway to block the expression of Dmy (Supplementary Fig. 2D).
To test the above hypothesis about mode of knockdown, three representative shDNA (small hairpin DNA, each of 38 bp) were artificially synthesized using a Block-it™ pol II miR RNAi expression vector kit (Invitrogen, USA) following the manufacturer’s instructions. Precautions were taken while designing the shDNA constructs in order to make sure that all three shDNA fragments did not overlap each other and also belonged from the same targeted 220 bp Dmy region (used to prepare the pmDMY-AS construct). All these shDNA plasmids (pmCMV-DMYsh1, pmCMV-DMYsh2 and pmCMV-DMYsh3) were electroporated in one or two-cell medaka embryos in all possible combinations. pmDMY-AS and pEGFP-AS were used as positive and negative controls, respectively. Despite Dmy knockdown, the singularly injected pmCMV-DMYsh1, 2 or 3 specimens failed to show any significant phenotypic effect in terms of increased meiosis (Supplementary Table 2). The occurrence of mitosis and meiosis in male gonads increased approximately 2 fold upon double knockdown of pmCMV-DMYsh1 and 2. Triple knockdown using three short hairpin constructs, pmCMV-DMYsh1, 2 and 3, gave maximum knockdown comparable with pmDMY-KD (Supplementary Table 2). This suggests that, our pmDMY-AS construct reduces the mRNA stability and produces several small RNA, which in turn amplifies the knockdown effect.
Functional sex reversal by Dmy knockdown
After checking the knockdown efficiency by real-time PCR analysis of several sex related genes including Dmy (discussed later), gonadal histology of Dmy-KD XY embryos was performed to assess the changes at the phenotypic level caused by Dmy suppression. Cells in early stages of meiosis were observed in XY gonads even at 0 dah and gradually increased at 5 and 10 dah (Fig. 2A–D). To observe the duration of the silencing effect, the electroporated XY fish were stained with haematoxylin and eosin (HE) at 50 dah (Fig. 2E–G). 50% of the fish were found to have fully grown ovaries containing variable numbers of oocytes, while 20% had testicular tissue in the gonad (Supplementary Table 3).
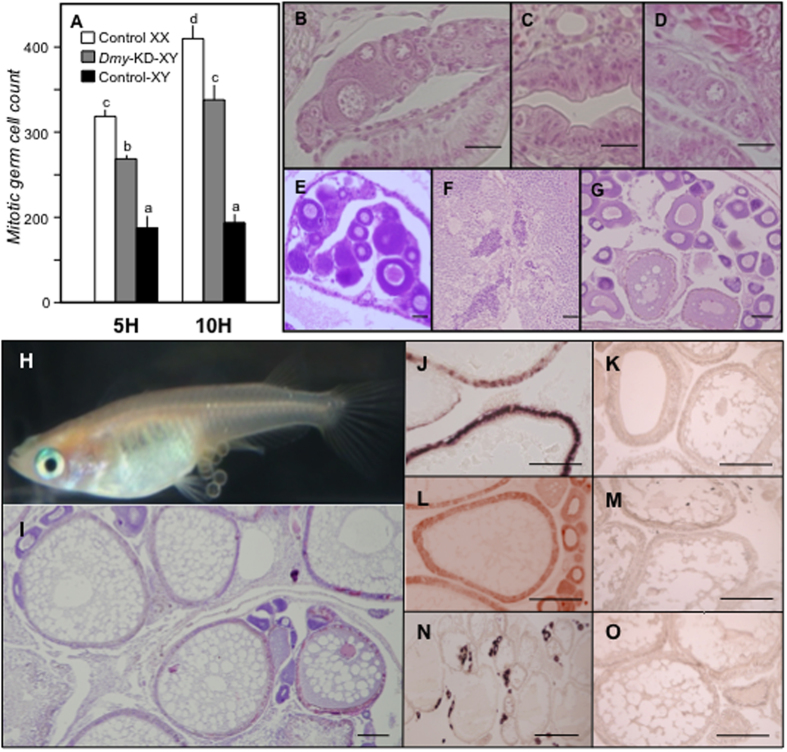
Figure 2. Effect of Dmy knockdown on gonad development.
(A) The total number of mitotic germ cells counted (control XX, white; Dmy-KD XY, grey; control XY, black) at 5 dah (5H) and 10 dah (10H), was used as a marker for gonadal sex. Data are presented as mean of 10 individuals from both control and Dmy-KD groups. Error bars indicate SEM. Letters (a-c) above the bars indicate that these groups differ significantly (p < 0.05) from each other. (B–G) The candidate HE-stained sections of control XX females (B,E), control XY males (C,F), and Dmy-KD XY fish (D,G). Female-like gonad development is seen in Dmy-KD XY fish, which later forms a fully developed ovary. Note: B, C, and D represent gonads at 10 dah, while E, F, and G represent gonadal structure at 50 dah. (H,I) Dmy knockdown directed femininity was persistent during adulthood, which was confirmed by female-like external appearances (H) and gonad structure (I). The femininity was further confirmed by ISH with female (Cyp19a1, J; Foxl2, L; Rspo1, N), and male (Dmy, K; Cyp11a, M); Cyp11b, O) associated genes. (Scale bars, 20 μm.)
Further, genotyping was carried out and the sex-reversed Dmy-KD XY F0 females (Fig. 2H,I) were raised to maturity and five individuals were mated with normal males to test their mating and breeding ability. Although all the five F0 fish gave rise to viable progeny, complete AS construct was integrated into the genome of two specimens (here after considered as founder fish), as evidenced by the amplification of the transgene sequence (using a vector specific forward primer and a Dmy fragment specific reverse primer) from genomic DNA isolated from caudal fin clips (data not shown). We did not find any significant differences between progenies from these two founder fish (Supplementary Table 4). Hence, the progenies generated from these two F0 phenotypic females were used for further analysis.
In two specimens of F1 generation fish (out of ten checked by HE staining), meiotic cells were detected at 10 dah stage, which indicated that the antisense Dmy transgene (CMV-DmyAS) had passed to the next generation. However, the low level of gene transmission might be associated with the crossing method, where control XY fish were used as male partner. The remaining F1 progeny were raised to adulthood till the development of secondary sexual characters. Three sex reversed XY females (confirmed by the presence of male-specific leucophore (Y linked character) and Dmy-transgene amplification) from the F1 group were similarly analysed for mating and breeding behaviour. All tested fish were fully fertile and further mated to produce viable progeny (Supplementary Table 3). Similar analyses were performed in each generation before mating the animals for progeny production. The fecundity (egg number) of the Dmy-KD XY females was also quite similar to that of control XX females in different generations (from F2-F5, Supplementary table 5). We used the F3 fish to generate pmDMY-KD line in which the Dmy mRNA expression was very low (measured by real-time PCR in embryonic gonads) from early development period.
Down-regulation of male-associated genes by Dmy knockdown
To determine the effect of Dmy knockdown, real-time PCR was performed using samples from 5 daf (days after fertilization) to adulthood. As expected, at early stages, transient Dmy knockdown reduced the Dmy expression by 66–92%, even from 5 daf (p < 0.05). At 5 daf, there was slight reduction in the expression of other male-associated genes like Gsdf and Sox9a2. At 0 dah, the reduction rate of Sox9a2 expression was much larger than Gsdf. At 5 and 10 dah, both Gsdf and Sox9a2, resulted in approximately 80–90% reduction compared to controls (p < 0.05) (Fig. 3A). Similar trend was also observed in the adults. Until 10 dah, there was no Dmrt1 expression in both the knockdown and control groups. However, in the knockdown group, Dmrt1 expression was drastically reduced in adulthood (p < 0.05) (Supplementary Figs 3, 4). All these results were further substantiated using in situ hybridisation (ISH). Among the fully-grown Dmy-KD XY fish, only the partially sex reversed fish showed low expression (p < 0.05) of Dmy, Gsdf and the spermatogonial markers (Cyp11a and Cyp11b) (Fig. 2K,M,O & S3). On the other hand, the fully developed pmDMY-KD XY ovary did not show any sign of spermatogonial marker gene expression but had female-like expression of Gsdf and other male dominated genes (p < 0.05) (Supplementary Fig. 3).
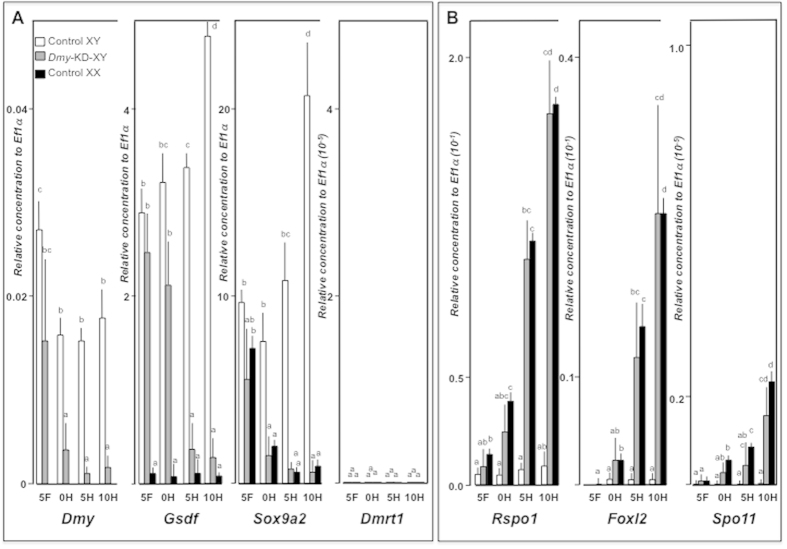
Figure 3. Dmy-knockdown induced transcriptional alterations of male and female associated genes.
(A,B) Ontogenic changes in the expression of male (Dmy, Gsdf, Sox9a2, and Dmrt1) (A) and female (Rspo1, Foxl2, and Spo11) (B) associated genes from 5 daf (5F) to 10 dah (10H) were analysed by real-time PCR. Data were normalised with Ef1a expression. Each column in the graph represents mean ± SEM of 10 individuals of control XY (white columns) and Dmy-knockdown XY (grey columns) fish. Letters above the bars indicate that these groups differ significantly (p < 0.05) from each other.
Up-regulation of female-associated genes by Dmy knockdown
To determine the effect of Dmy knockdown on the expression of female-associated genes, real-time PCR was carried out using Dmy-KD fish. Rspo1 showed increased expression at 5 daf in knockdown fish, which became more evident at later stages in comparison to control-XY groups (p < 0.05). However, Foxl2 could only be differentiated from 0 dah onwards. Interestingly, when compared to controls, all these female-specific genes were markedly upregulated from 5 dah (p < 0.05) (Fig. 3B). In adults, except for four fish, all showed a substantial increase in the expression of female-associated genes compared to their respective controls (Supplementary Fig. 3). Dmy-KD-induced feminization was further confirmed by ISH (Fig. 2J,L,N). All the sex-reversed Dmy-KD XY fish (both partial and complete) showed elevated Spo11, Foxl2, Rspo1, and Cyp19a1 (ovarian aromatase) expression (Supplementary Fig. 3), while unchanged fish had male-alike or no expression of these genes.
Role of Wt1a in the regulation of Dmy expression
In mammals, Wt1 is known to be an important regulator of Sry. Duplicated copies of Wt1 (Wt1a and Wt1b) have been reported in teleosts, including medaka31. In order to understand the isoform-specific effects of Wt1 on Dmy regulation, we characterised the expression patterns of Dmy, Wt1a and Wt1b during gonadal differentiation in both XY and XX fish, including the F3 Dmy KD transgenics (pmDMY-KD XY). Wt1a starts its zygotic expression from stage 12 (10 hours after fertilization, haf) (Fig. 4A), while Wt1b sets in from stage 25 (2 daf) (data not shown). Interestingly, Dmy initiated its zygotic expression from stage 18 (1 daf) (Fig. 4A), which was much later than Wt1a expression but earlier than Wt1b expression. This gave rise to a possibility of Wt1a as a candidate regulator of Dmy in medaka. Thereafter, we examined the effect of Dmy knockdown on Wt1a expression. Our real-time PCR and whole-mount ISH data showed identical expression between pmDMY-KD XY and control XY embryos at different stages (Fig. 4B–D).
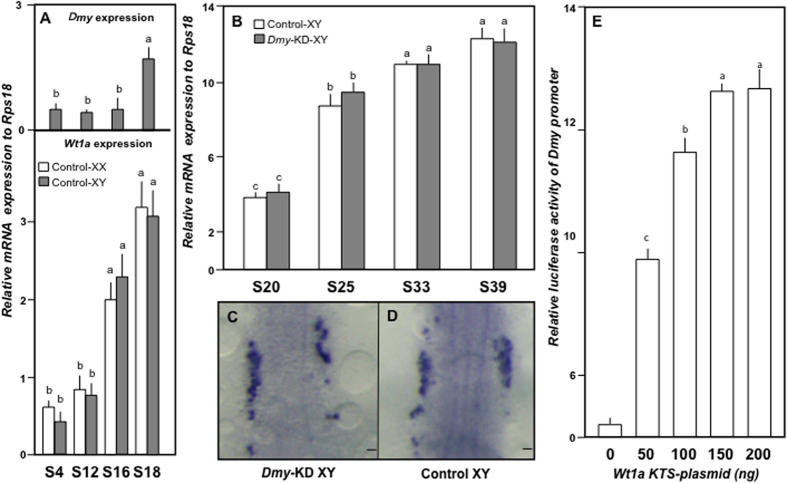
Figure 4. Wt1, an upstream candidate for Dmy regulation.
(A) Dmy and Wt1a expression were analysed using several early stage samples (S4-S18) to assess the precise start point of zygotic expression (XX, white columns and XY, grey columns). (B) Differences in Wt1a expression between control (white columns) and Dmy-KD (grey columns) XY embryos during gonadal differentiation (S20-S39) were also analysed by real-time PCR. (C,D) Wt1a specific whole mount ISH of Dmy-KD XY (C) and control XY (D) embryos (stage 22) showed no virtual difference. (E) Luciferase assays using HEK293 cells showed a dose dependent activation of Dmy promoter by Wt1a KTS (−). Graphical data are presented as an average of three independent experiments, each containing triplicates of individual sampling group. Error bars represent SEM. Letters (a–c) above the bars indicate that these groups differ significantly from each other at p < 0.05.
In the subsequent experiment, we found a highly conserved Wt1-binding site (GAGGGGGGAG) in the Dmy promoter region32. Reportedly, in mammals, Wt1 KTS (+) alternatively spliced isoform regulates the SRY transcription in a dose responsive manner32. To determine the role of Wt1(s) in the regulation of Dmy expression, a luciferase assay was performed using four Wt1-ORF plasmids (Wt1a KTS (+/−) and Wt1b KTS (+/−)) and Dmy promoter construct. To our surprise, only the Wt1a KTS (−) isoform showed dose-dependent activation of Dmy promoter-driven luciferase production (Fig. 4E).
Rescue of Dmy knockdown effects by co-overexpression of Gsdf and Sox9a2
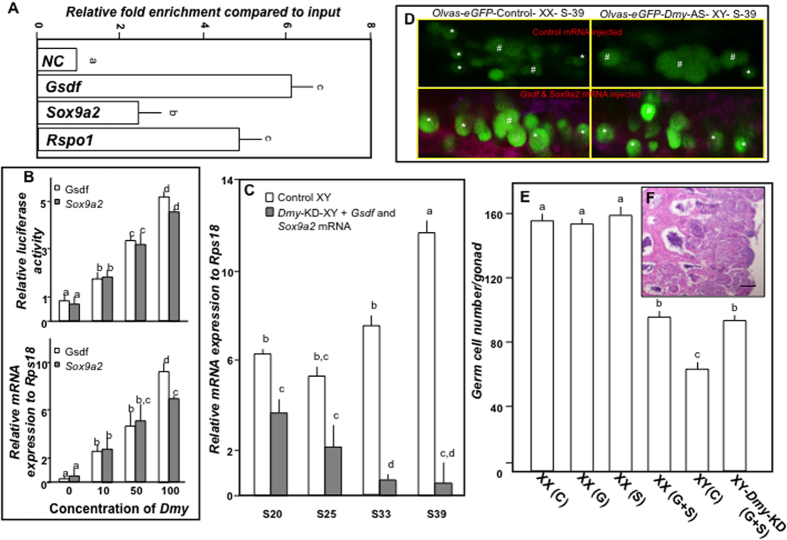
To find out the direct downstream target of Dmy, we performed chromatin immuno-precipitation (ChIP) using Dmy (tagged with eGFP) mRNA overexpressed 6 daf medaka embryos. Since we observed differential but significant Dmy knockdown dependent down-regulation of Gsdf and Sox9a2 and up-regulation of Rspo1, the potential downstream targets for Dmy, these genes were chosen as candidates for ChIP analysis. The primers were selected based on the availability of Dmy binding site (wacawtgtwk)33 in both upstream (−20 kb) and downstream (+10 kb) region of candidate genes. We found strong Gsdf and Rspo1 (4–6 and 3.5–5 fold, respectively, depending on the area tested), but weak Sox9a2 (1.59–2.87 fold) enrichment from 6 daf Dmy-GFP mRNA-injected embryonic samples (details in Supplementary Materials and Methods), determined by ChIP, using GFP antibody (Fig. 5A, Supplementary Fig. 5). Recently we published that, Rspo1 promoter activity was inversely regulated by Dmy34. To get a better idea about Dmy association in Gsdf and Sox9a2 regulation, we performed in vitro promoter assay of both Gsdf and Sox9a2 promoters using OL32 cell line. Although the Ol32 cells possess slight Dmy, Gsdf and Sox9a2 transcription of its own, the luciferase activity of both promoters and transcription of Gsdf and Sox9a2 showed significant increment after Dmy overexpression, in a dose dependent manner (Fig. 5B). Owing to the transcriptional history of medaka and our data, it is highly likely that Dmy directly promotes the transcriptions of both Gsdf and Sox9a2, and simultaneously suppresses Rspo1 to maintain the masculinity. If these assumptions are correct, then Gsdf and Sox9a2 will alone or together be sufficient to rescue the Dmy knockdown effect. To prove that, Gsdf (tagged with mCherry) and Sox9a2 (tagged with cyan) mRNA were singularly injected or co-injected into both olvas-eGFP XX and olvas-eGFP Dmy-KD XY embryos (F3 generation). Although singular injections failed to completely suppress meiosis and proliferative mitosis, co-injection re-established the male phenotype in the XY gonad, leading to the complete formation of testis (Fig. 5C–F, Supplementary Fig. 6). However, the amount of Dmy remained substantially low (Fig. 5C). Interestingly, the Gsdf and Sox9a2 co-injected control olvas-eGFP XX embryos were also redirected towards male type gonadal development, i.e. restriction in germ cell proliferation (Fig. 5D,E).
Figure 5. Gsdf and Sox9a2 are potential downstream targets of Dmy.
(A) In vivo chromatin immunoprecipitation analysis of candidate Dmy targets. Relative expression to control input was calculated using real-time PCR and plotted on the graph. Data are presented as means ± SEM of 4 separate experiments and significances (p < 0.05) are denoted by different letters. (B) Effects of Dmy overexpression on Gsdf and Sox9a2 promoter activity (upper panel) and mRNA transcription (lower panel) were ascertained. (C) Differences in Dmy expression between control (white columns) and olvas-eGFP-Dmy-KD co-injected with Gsdf-cherry and Sox9a2-cyan (grey columns) XY embryos during gonadal differentiation (S20-S39) were assessed using real-time PCR. (D) Representative images showing the germ cell distribution in Olvas-eGFP-control XX, Olvas-eGFP-Dmy-KD-XY embryos at S39 after control (upper panel) and Gsdf/Sox9a2 (lower panel) mRNA injection. Isolated and cluster germ cells are marked with ‘*’ and ‘#’, respectively. Note: Clustered germ cells are representative of female type gonadal development (N = 9/ group). (E) The total number of germ cells was plotted (Y-axis) against different injected groups (X-axis). Data are presented as means of 10 individuals (error bars represent SEM). Different letters (a–d) indicate significant differences from other groups at p < 0.05. Note: C, G, S, and G + S represents PBS, Gsdf-cherry, Sox9a2-cyan, and Gsdf-cherry/Sox9a2-cyan mRNA injected embryos, respectively. (F) The gonadal sexuality of olvas-eGFP-Dmy-KD embryos co-overexpressed with Gsdf-Sox9a2 (N = 7) was assessed at maturity. (Scale bar, 20 μm).
Discussion
In the present study, we developed a unique, relatively stable, cost-effective and gene-specific RNAi delivery system, capable of producing multiple small RNA against a specific gene, Dmy, in medaka. A DNA vector-based approach was used, owing to its many advantages over other approaches, like better stability, prolonged retention, ease of transmission and cost effectiveness35,36. The long AS molecule produced by this vector was useful in inhibiting the gene expression by promoting degradation of double-stranded RNA, perhaps by Dicer and miRNA/siRNA mechanism37. Availability of several kinds of unique small RNA helped us to hypothesize that AS-RNA were processed by the cell machinery to generate several small RNA (60–70 nucleotides) and siRNA (18–28 nucleotides) molecules27,28 and increase the silencing efficiency. Moreover, due to continuous transcription, long AS-RNAs were expected to be constitutively transcribed in vivo, thus blocking the translation of messenger RNA before being degraded38. The hypothesis was further confirmed by specific knockdown of Gfp expression in olvas-eGFP transgenic medaka. The specificity of Gfp knockdown was undoubtedly confirmed by the microscopic reduction of Gfp expression in the gonads. However, unlike shrimp39, in the present study, we did not observe any suppression in growth and reproduction of knockdown fish.
This newly developed knockdown technology was used to investigate the function of the zygotic gene product, Dmy, in medaka throughout its life span. Upon knockdown of Dmy, we were successful in generating a functional sex-reversed XY medaka with full fertilization potential. The marked suppression of Dmy, even at 5 daf, suggests the efficacy of knockdown. Being the master sex determination regulator in medaka, it is very important to know the detailed role of Dmy during gonadal determination, development and maturation. In medaka, early appearance of progressive mitosis and meiosis is the first sign of morphological sex determination in females. However, in males, the occurrence of Dmy arrests mitosis and drives the male sex determination. In the present study, we could also observe meiosis in Dmy-KD XY gonads even at 0 dah, confirming the previous findings which showed a surge in Scp3 production and mitotic proliferation in XY DMY-gripNA and morpholino knockdown medaka15,40.
Knockdown of Dmy also induced marked reduction of two major male-associated autosomal genes, Gsdf and Sox9a2, during early gonadal sex differentiation in XY fish. We have previously shown that a rise in Gsdf expression is seen specifically in genetic males around 6 daf, almost the same time at which Dmy expression takes the peak16. Importantly, Dmy and Gsdf were found to be co-localized in the same somatic cells in the XY gonads16. Although there was very little reduction in Gsdf expression at 5 daf after knockdown of Dmy, a marked reduction was evident at 5 and 10 dah. Similarly, ChIP and promoter analysis also depicted direct regulation between Gsdf and Dmy. This suggests that male oriented gonadal fate determination was initiated by Dmy expression and further progressed by Dmy and Gsdf in a co-operative manner16. This also supports our previous hypothesis that Gsdf is a potential target of Dmy in male gonad development in XY Japanese medaka (O. latipes)16. Intriguingly, recent findings indicate that Gsdf is the master sex-determining gene in the Hainan medaka, Oryzias curvinotus, a closely related species of medaka7. Although Dmy is not present in O. luzonensis, the spatial and temporal expression pattern of Gsdf is closely correlated to Dmy in medaka16 and the expression patterns of more downstream genes in testicular differentiation, such as Sox9a2 and Dmrt1, are similar in both medaka species7,23,41. Therefore, O. latipes and O. luzonensis share a common sex differentiation pathway downstream of Gsdf and, if high Gsdf expression is achieved during sex differentiation, then the XX embryo develops as a male without Dmy7.
In a number of higher vertebrates, Sox9 has been shown to play a major conserved role in testis determination. In mice, from 10.5 dpc, the Y-linked sex-determining gene Sry is expressed in XY genital ridges and initiates Sox9 expression and testis differentiation42. Knocking-down of Sry expression resulted in significant decrease in the Sox9 expression and gonadal feminization of mouse embryos43. Similarly, in chicken, Sox9 expression was significantly reduced in Dmrt1 knockdown genetically male (ZZ) gonads relative to controls4. In medaka, Sox9a2 (a duplicated homologue of mammalian Sox9) is initially expressed in somatic cells of both sexes, but upregulated in testicular somatic cells and down regulated in the XX gonads, at 10 to 30 dah17. Contrastingly, Nakamura et al.18, using both transgenic and chimeric Sox9a2 (Sox9b) medaka mutants, reported that Sox9a2 is required for germ cell proliferation and survival, but not for testis determination17. However, we previously depicted a significant elevation of Sox9a2 at 10 dah XY fish, suggesting its role in testis development17,18. In the present study, upon Dmy knockdown, there was a marked reduction of Sox9a2 expression even from 0 dah onward. These results suggest that Sox9a2 is downstream of Dmy and possesses some role in the early stages of testis differentiation. Although significantly lower, the precipitation of Sox9a2 DNA in Dmy-ChIP experiment suggests a direct link between Sox9a2 and Dmy, which is substantiated by our promoter analysis data. Further in-depth analysis is required to clarify the role of Dmy-Sox9a2 interaction in testis differentiation and development in medaka.
As stated above, the expression of two major male-associated genes, Gsdf and Sox9a2, were markedly reduced in Dmy-KD XY fish. In the present study, the availability of F3 Dmy-KD transgenics gave us the option to study the downstream targets of Dmy by overexpressing candidate down-stream genes such as Gsdf and Sox9a2. Our results showed that although individual overexpression of either Gsdf or Sox9a2 induced partial recovery of male phenotype, co-overexpression of these two genes could fully re-establish the male pathway, producing fertile sperm in Dmy-KD fish. These results, along with the ChIP data, suggest some parallel and essential role of these two genes in testicular differentiation and development. Since Gsdf and Sox9a2 are expressed in the same somatic cells in medaka XY gonads, it is quite possible that, like mammalian Sry/Sox9-Fgf9 regulation11, Dmy could simultaneously induce both Gsdf and Sox9a2, which in turn influences each other to maintain masculinity. However, no such evidence has been reported till date in medaka.
Unlike Gsdf and Sox9a2, in XY medaka gonad, Dmrt1 expression is initiated only after the formation of testis (20–30 dah). Recently, we found that Dmrt1 mutant XY medaka develops gonad that initially appear to be males but later becomes fertile XY females44. These results indicate that Dmrt1 is essential to maintain testicular identity after Dmy-triggered male differentiation pathway. In the present study, we found a drastic reduction of Dmrt1 expression in sex-reversed XY adults, further confirming the significance of Dmrt1 in maintaining testicular differentiation and development.
In this study, Dmy knockdown caused complete suppression of male-associated genes in XY gonads, and generated sex-reversed XY fertile females, which thereby highlights the essentiality of female/ovarian-associated genes in Dmy-reduced condition. Although the key ovarian determinant has not yet been identified, we now know that a number of female-associated factors, such as canonical Wnt signaling through Wnt4, Rspo1 and β-catenin, and Foxl2, are critical for ovarian differentiation in vertebrates45,46. Recently, much attention has been paid to the role of Rspo-activated signaling pathway in early sex determination and differentiation. Rspo1 displays a conserved, female-specific increase in expression in several vertebrate species33,47. In medaka, Rspo1 is predominantly expressed in XX gonads during sex differentiation, immediately preceding the first signs of ovarian differentiation, suggesting that Rspo1 is a key player in medaka ovarian differentiation20. In the present study, we observed significant elevation in Rspo1 expression at 0 dah in Dmy-KD XY fish. It is important to note that at this stage Dmy expression is already markedly down regulated, but Gsdf expression is still elevated. These findings suggest that Dmy expression, but not Gsdf expression, in males can block the Rspo1 signalling. It has been proposed that the fate of the developing mammalian gonad involves a tug of war between the Sry/Sox9-induced Fgf9 signalling (male promoting) versus the Wnt signalling (female promoting)46. In this connection, it is important to note that, Rspo1 directly interacts with Dmy (as evidenced by ChIP) and Rspo1 expression is localized to somatic cells surrounding germ cells as well as germ cells during early developmental stages20. Further studies are required to determine whether a similar antagonistic system (the Dmy/Gsdf signalling in males versus the Rspo1 signalling in females) also applies in medaka.
Foxl2 is also known to be involved in ovarian differentiation in a number of vertebrate species45. More recently, FOXL2 was identified as a bona fide female sex-determining gene in goat48. In medaka, Foxl2 is expressed only in the female gonads during gonadal differentiation, which starts around the day of hatching. In Dmy-KD XY fish, Foxl2 expression was markedly upregulated at 5 dah, a time that coincides well with the marked fall in Gsdf expression in the Dmy-KD fish. These findings may suggest that there is a counter interaction between Gsdf and Foxl2, but this remains to be elucidated.
The important role of the Rspo1 and Foxl2 appears to be conserved from fish to mammals. In medaka, Rspo1 expression precedes Foxl2 by 2–3 days. It is important to determine how these two genes interact to coordinate ovarian determination and differentiation. We recently showed that in medaka, the expression profiles of Rspo1–3 in gonads were markedly upregulated during a short period of oestrogenic exposure in 0 dah XY fish20, and the expression of ERβ2, but neither ERα nor ERβ1, exhibited marked increase in XX embryos between 6 and 8 daf with a peak at 7 daf21. It was also shown that Foxl2 upregulated Cyp19a1 gene transcription in a female-specific manner in other fish22. Thus, it is possible that oestrogen may be an intervening factor between Rspo1 and Foxl2 during early ovarian differentiation in medaka.
Although knockdown of Dmy gave rise to functional XY sex reversal, the exact molecular mechanisms involved in the regulation of Dmy expression in fish sex determination, is still vaguely understood. In mammals, a number of genes have been implicated as potential regulators of Sry expression, such as Gata2, Gata4, M33, and Wt142. The present study focuses on the role of one of these genes, Wt1, in the regulation of Dmy during early testicular differentiation. In medaka, Wt1a is one of the primary genes expressed in somatic cells (precursor cells for Dmy expression) of bipotential gonads and first appears at Stage 12, preceding the zygotic expression of Dmy31. This highlights the importance of Wt1a in the regulation of Dmy. Unlike male and female-associated genes, Wt1a expression showed no change between control and Dmy-KD XY fish, suggesting that Wt1a is upstream of Dmy. Confirming this hypothesis, our results showed that Wt1a enhances the promoter activity of Dmy in a dose-dependent manner. Similarly, in mammals, the Wt1a KTS (−) and KTS (+) isoforms acts on transcription and RNA stability of Sry, respectively49,50. Taken together, these data suggests a conserved role of Wt1a in regulating the two sex-determining genes, Sry and Dmy. It is of interest to determine whether Wt1 can also regulate the expression of chicken Dmrt14, Xenopus laevis Dm-W5 and other novel fish specific sex determining genes, i.e. Gsdf, Amh, Amhr2, SdY, and Sox36,7,8,9.
In conclusion, the new gene-specific transgenic RNAi technology developed for medaka has made it possible for the first time to investigate the function of Dmy in XY fish throughout its life span. A distinct shift in gene expression patterns from Gsdf/Sox9a2 to Rspo1/Foxl2 occurs in the germ cells surrounding somatic cells of Dmy-KD XY embryos, leading to a functional male to female sex reversal. Further studies, using this Dmy knockdown line, along with other related transgenics/mutants, are required to understand the 1) exact roles of Gsdf and Sox9a2 in testicular differentiation and development and 2) life long tug of war between male/female associated genes in determining or maintaining the gonadal sex. The probable limitations of this method concerning the sterility of homozygous and heterozygous animals and associated hurdles of transgenic line maintenance need to be critically investigated, using fertility-associated genes.
Materials and Methods
Ethics statement
The studies were carried out in accordance with the Institutional Ethics Committee of National Institute for Basic Biology, Okazaki, Japan, strictly adhering to the guidelines set for the usage of animals by this committee. All in vivo experiments and fish maintenance were conducted following protocols and procedures approved by the Institutional Animal Care and Use committee at National Institute for Basic Biology, Japan. All surgery was performed under Tricaine-S anesthesia, and all efforts were made to minimize suffering.
Experimental procedures
Detailed description of plasmid constructs, gene knockdown in Cos7 cells, experimental animals, knockdown of Dmy expression in medaka embryos, sampling, histology and in situ hybridisation, quantification of changes in gene expression by real-time PCR, assessment of the trans-generational knockdown effect, characterisation of small RNAs, promoter analysis of Dmy, Immunoprecipitation analysis and rescue of Dmy knockdown effect can be found in Supplementary Materials and Methods. Primer sequences for quantitative PCR and ISH probes are provided in Supplementary table 6.
Additional Information
How to cite this article: Chakraborty, T. et al. Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci. Rep. 6, 19480; doi: 10.1038/srep19480 (2016).
Supplementary Material
Acknowledgments
We thank Dr. M. Matsuda (Utsunomiya University, Japan), Dr. Y. Ogino (National Institute for Basic Biology, Japan) and Dr. P. Krisnan (ICAR, India) for their help. We extend our gratitude to Dr. Hiroshi Mitani for providing different medaka cell lines. QurtE and olvas-eGFP transgenic strain of medaka was provided by National Bio Resource Project Medaka (NBRP Medaka) of Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. This work was in part supported by Grants from Science and Technology Agency (SORST Program) and the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Footnotes
The authors declare no competing financial interests.
Author Contributions T.C. & L.Z. contributed equally to this work. T.C., A.C. & Y.N. conceived the idea. T.C. & L.Z. performed all the experiments and statistical analysis. T.C., T.I. & Y.N. provided the necessary equipments, chemicals etc. for the experiment. T.C., T.I. & Y.N. wrote the manuscript.
References
- Sinclair A. H. et al. A gene from the human sex determining region encodes a protein with homology to a conserver DNA-binding motif. Nature 346(6281), 240–4 (1990). [DOI] [PubMed] [Google Scholar]
- Matsuda M. et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417(6888), 559–63 (2002). [DOI] [PubMed] [Google Scholar]
- Nanda I. et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99(18), 11778–83 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A. et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461(7261), 267–71 (2009). [DOI] [PubMed] [Google Scholar]
- Yoshimoto S. et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus leavis. Proc. Natl. Acad. Sci. USA 105(7), 2469–74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori R. S. et al. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. USA, 109(8), 2955–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myosho T. et al. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191(1), 163–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T. et al. A Trans-species missense SNP in Amhr2 is associated with sex determination in the Tiger Pufferfish, Takufugu rubripes (Fugu). PLoS Genet. 8(7), e1002798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A. et al. An immune-related gene evolved into the master sex determining gene in rainbow trout Oncorhynchus mykiss. Curr. Biol. 22(15), 1423–8 (2012). [DOI] [PubMed] [Google Scholar]
- Takehana Y. et al. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Comm. 5, 4157 (2014). [DOI] [PubMed] [Google Scholar]
- Clinton K. M. & Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Gen. 13(3), 163–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. et al. Two DM domain genes, DMY and DMRT1, involved in testicular differentiation and development in the medaka, Oryzias latipes. Dev. Dyn. 231(3), 1518–1526 (2004). [DOI] [PubMed] [Google Scholar]
- Wada H. et al. Sex- Linked inheritance of the lf locus in the medaka fish (Oryzias latipes). Zool. Sci. 15(1), 123–126 (1998). [DOI] [PubMed] [Google Scholar]
- Okate H. et al. The medaka sex-determining gene DMY acquire a novel temporal expression pattern after duplication of DMRT1. Genesis 46(12), 719–23 (2008). [DOI] [PubMed] [Google Scholar]
- Paul-Prasanth B. et al. Knock-down of DMY initiates female pathway in the genetic male medaka, Oryzias latipes. Biochem. Biophys. Res. Commun. 351(4), 815–9.(2006). [DOI] [PubMed] [Google Scholar]
- Shibata Y. et al. Expression of gonadal soma derived factor (GSDF) is spatially and temporally correlated with early testicular differentiation in medaka. Gene Expr. Patterns 10(6), 283–9 (2010). [DOI] [PubMed] [Google Scholar]
- Nakamoto M. et al. Testicular type Sox9 is not involved in sex determination but might be in the development of testicular structure in medaka, Oryzias latipes. Biochem. Biophys. Res. Commun. 333(3), 729–36 (2005). [DOI] [PubMed] [Google Scholar]
- Nakamura S. et al. Analysis of medaka sox9 orthologue reveals a conserved role in germ cell maintenance. PLoS One 7(1), e29982 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. et al. Two DM domain genes, DMY and DMRT1, involved in testicular differentiation and development in the medaka, Oryzias latipes. Dev. Dyn. 231(3), 1518–26 (2004). [DOI] [PubMed] [Google Scholar]
- Zhou L. Y. et al. R-spondins are involved in the ovarian differentiation in a teleost, medaka (Oryzias latipes). BMC Dev. Biol. 12, 36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T. et al. Differential expression of three estrogen receptor subtype mRNAs in gonads and liver from embryos to adults of the medaka, Oryzias latipes. Mol. Cell. Endocrinol. 333(1), 47–54 (2011). [DOI] [PubMed] [Google Scholar]
- Wang D. S. et al. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with Ad4 binding protein/steroidogenic factor 1. Mol. Endocrinol. 21(3), 712–25 (2007). [DOI] [PubMed] [Google Scholar]
- Nakamoto M. et al. Gonadal sex differentiation and expression of Sox9a2, Dmrt1, and Foxl2 in Oryzias luzonensis. Genesis 47(5), 289–299 (2009). [DOI] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A. & Barbas C. F. III ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31(7), 397–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Davidkova G. & Zhou L. W. Antisense RNA gene therapy for studying and modulating biological processes. Cell Mol. Life Sci. 55(3), 334–58 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T. Baas M. & Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 21(11), 1944–54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg M. S. et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 12(2), 256–62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J., Jung S., Keller S., Gregory R. & Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11(3), 228–34 (2009). [DOI] [PubMed] [Google Scholar]
- Tanaka M., Kinoshita M., Kobayashi D. & Nagahama Y. Establishment of medaka (Oryzias latipes) transgenic lines with the expression of green fluorescent protein fluorescence exclusively in germ cells: a useful model to monitor germ cells in a live vertebrate. Proc. Natl. Acad. Sci. USA 98(5), 2544–9 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetler H. A. Peck S. L. & Muir W. M. High efficiency production of germ line transgenic Japanese medaka (Oryzias latipes) by electroporation with direct current-shifted radio frequency pulses. Transgenic Res. 12(4), 413–24 (2003). [DOI] [PubMed] [Google Scholar]
- Kluver N. et al. Regulatory back-up circuit of medaka Wt1 co-orthologs ensures PGC maintenance. Dev. Biol. 325(1), 179–88 (2009). [DOI] [PubMed] [Google Scholar]
- Hossain A. & Saunders G. F. The human sex-determining gene SRY is a direct target of WT1. J. Biol. Chem. 276(20), 16817–23 (2001). [DOI] [PubMed] [Google Scholar]
- Herpin A. et al. Divergent expression regulation of gonad development genes in medaka shows incomplete conservation of the downstream regulatory network of vertebrate sex determination. Mol. Biol. Evol. 30(10), 2328–2346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. Y. et al. Rspo1-activated signalling molecules are sufficient to induce ovarian differentiation in XY medaka (Oryzias latipes). Sci. Rep. (Accepted; SREP-15-07077B). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R. G., Ghodgaonkar M. M., Affar E. L. B. & Shah G. M. DNA vector-based RNAi approach for stable depletion of poly (ADP-ribose) polymerase-1. Biochem. Biophys. Res. Commun. 331(1), 167–74 (2005). [DOI] [PubMed] [Google Scholar]
- Chen Y. et al. Nanoparticles modified with tumor targeting scFv deliver siRNA and miRNA for cancer therapy. Mol. Ther. 18(9), 1650–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilenchik M. et al. Antisense RNA down-regulation of bcl-xL expression in prostate cancer cells leads to diminished rates of cellular proliferation and resistance to cytotoxic chemotherapeutic agents. Cancer Res. 62(7), 2175–83 (2002). [PubMed] [Google Scholar]
- Clayton J. RNA interference: the silent treatment. Nature 431(7008), 599–605 (2004). [DOI] [PubMed] [Google Scholar]
- Krisnan P. et al. DNA constructs expressing long-hairpin RNA (lhRNA) protect Penaeus monodon against White Spot Syndrome Virus. Vaccine 27(29), 3849–55 (2009). [DOI] [PubMed] [Google Scholar]
- Herpin A. et al. Inhibition of primordial germ cell proliferation by the medaka male determining gene Dmrt1bY. BMC Dev. Biol. 7, 99 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S. et al. Hyperproliferation of mitotically active germ cells due to defective anti-Müllerian hormone signaling mediates sex reversal in medaka. Development 139(13), 2283–7 (2012). [DOI] [PubMed] [Google Scholar]
- Svingen T. & Koopman P. Building the mammalian testis: origins, differentiation and assembly of the component cell populations. Genes Dev. 27(22), 2409–26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Yu A. B., Zhu H. B. & Lin X. K. Effective Silencing of Sry Gene with RNA Interference in Developing Mouse Embryos Resulted in Feminization of XY Gonad. J. Biomed. Biotechnol. 2012(343891), 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama H. et al. Dmrt1 mutation causes a male to female sex reversal after the sex determination by Dmy in the medaka. Chromosome Res. 20(1), 163–76 (2012). [DOI] [PubMed] [Google Scholar]
- Baron D. et al. An evolutionary and functional analysis of Foxl2 in rainbow trout gonad differentiation. J. Mol. Endocrinol. 33(3), 705–15 (2004). [DOI] [PubMed] [Google Scholar]
- Cutting A., Chu J. & Smith C. A. Just how conserved is vertebrate sex determination? Dev Dyn. 242(4), 380–7 (2013). [DOI] [PubMed] [Google Scholar]
- Smith C. A. et al. Cloning and expression of R-Spondin1 in different vertebrates suggests a conserved role in ovarian development. BMC Dev. Biol. 8, 72 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger. L. et al. Foxl2 is a female sex-determining gene in the goat. Curr. Biol. 24(4), 404–8 (2014). [DOI] [PubMed] [Google Scholar]
- Caricasole A. et al. RNA binding by the Wilms tumor suppressor zinc finger proteins. Proc. Natl. Acad. Sci. USA. 93(15), 7562–6 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford S. T. et al. A cell autonomous role for WT1 in regulating Sry in vivo. Human Mol. Gen. 18(18), 3429–38 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.