Abstract
From June 2011 to August 2014, 21 cases of infection by severe fever with thrombocytopenia syndrome bunyavirus (SFTSV) were confirmed in Zhoushan Islands in the Eastern coast of China. To identify the source of SFTSV in Zhoushan Islands, the whole SFTSV genomes were amplified and sequenced from 17 of 21 patients. The L, M, and S genomic segments of these SFTSV strains were phylogenetically analyzed together with those of 188 SFTSV strains available from GenBank. Phylogenetic analysis demonstrated SFTSV could be classified into six genotypes. The genotypes F, A, and D were dominant in mainland China. Additionally, seven types of SFTSV genetic reassortants (abbreviated as AFA, CCD, DDF, DFD, DFF, FAF, and FFA for the L, M and S segments) were identified from 10 strains in mainland China. Genotype B was dominant in Zhoushan Islands, Japan and South Korea, but not found in mainland China. Phylogeographic analysis also revealed South Korea possible be the origin area for genotype B and transmitted into Japan and Zhoushan islands in the later part of 20th century. Therefore, we propose that genotype B isolates were probable transmitted from South Korea to Japan and Zhoushan Islands.
Severe fever with thrombocytopenia syndrome (SFTS) is a new infectious disease that emerged in May 2007 in rural areas of Huaiyangshan Mountain, which links Hubei and Henan Provinces in Central China1,2,3. SFTS is characterized by fever, thrombocytopenia, gastrointestinal symptoms, and leukocytopenia and has a mortality rate of 12.3–20%1,4,5,6,7,8. The causative agent for SFTS was identified as a novel bunyavirus, which was named SFTS bunyavirus (SFTSV) or Huaiyangshan (HYS) virus1,9. SFTSV was also isolated from ticks (Haemaphysalis longicornis) that were collected from domestic animals in the regions where the patients lived3,9. Furthermore, SFTSV fragments were amplified from several samples from domestic animals (e.g., sheep, cows, and dogs)9, suggesting that ticks could serve as a key vector for SFTSV transmission3,9.
SFTSV belongs to the Phlebovirus genus in the Bunyaviridae family1,2,3. Its genome contains three separate RNA segments: S (small), M (middle), and L (large). So far, SFTSV has been isolated only from South Korea, Japan and China. It is prevalent in seven central-eastern provinces of China: Henan, Hubei, Anhui, Jiangsu, Zhejiang, Shandong, and Liaoning1,3,4,9,10. Zhoushan Islands are located in the Eastern coast of China and geographically isolated from other regions of SFTSV prevalence by the sea. In this study, we sequenced SFTSV isolates from Zhoushan Islands and performed detailed phylogeographical analysis of all SFTSV isolates available from this study and GenBank. This led us to propose that the dominant SFTSV genotype B in Zhoushan Islands was transmitted from South Korea rather than from other parts of China.
Results
Demographic and Clinical Characteristics of SFTS patients from the Zhoushan Islands
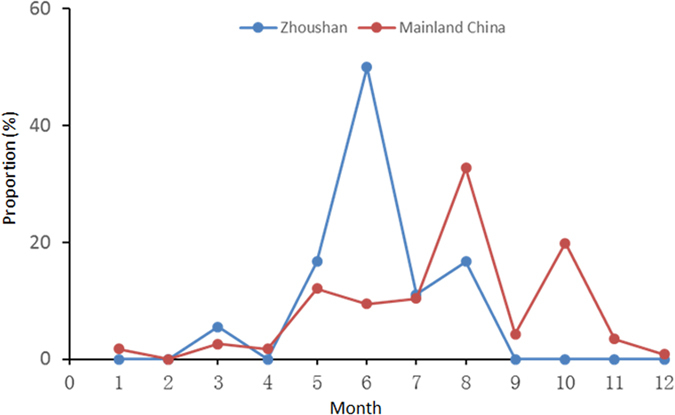
During June 2011–August 2014, 21 patients who displayed major clinical symptoms of acute onset of fever, thrombocytopenia, diarrhea, fatigue, and/or cough for 3 to 11 days were diagnosed as SFTS (Table 1). Infection with SFTSV was confirmed by RT-PCR. Symptoms mainly appeared from May–August, and peaked in June (10/21, 47.6%) (Fig.1). All patients, including 18 farmers, 2 fishermen, and 1 gatekeeper, were local residents of the Zhoushan Islands. Their median age was 67.3 (46–86), with 12 males and 9 females. No patient reported tick bites over the previous six months. Laboratory tests indicated that the most common characteristics were thrombocytopenia (100%) and leukocytopenia (95.2%), with elevated levels of serum alanine aminotransferase (100%), aspartate aminotransferase (100%), creatine kinase (100%), and lactate dehydrogenase (100%) (Table 1). The main clinical phenotypes, including fever, diarrhea, leucopenia, thrombocytopenia, were very consistent between patients in Zhoushan and other regions/country (Supplementary Table S1). Fewer patients in Zhoushan felt weakness and abdominal distension than in other regions/country (Supplementary Table S1).
Table 1. Results on clinical tests in 21 SFTS patients from Zhoushan islands.
| Casenumber | Sex | Age(years) | Occupation | Symptoms | Days withsymptoms | Clinical tests | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WBC(109/L) | PLT(109/L) | HGB(g/L) | AST(U/L) | ALT(U/L) | CK(U/L) | LDH(U/L) | ||||||
| ZS01 | F | 69 | Farmer | Fever with thrombocytopenia | 11 | 4.4 | 35 | 89 | 137 | 57 | 573 | 1336 |
| ZS02 | F | 61 | Farmer | Fever and weakness | 5 | 0.8 | 20 | 106 | 125 | 100 | 245 | 569 |
| ZS03 | M | 62 | Gatekeeper | Fever | 6 | 1 | 35 | 110 | 122 | 77 | 737 | 484 |
| ZS04 | M | 77 | Farmer | Fever, abdominal distension, and feeling restless (10 days) | 3 | 1.7 | 20 | 98 | 556 | 159 | 2635 | 1319 |
| ZS05 | M | 53 | Farmer | Fever and cough | 6 | 1.1 | 22 | 126 | 106 | 128 | 437 | 266 |
| ZS06 | M | 47 | Fishermen | Fever | 5 | 1.9 | 35 | 133 | 715 | 289 | 6161 | 898 |
| ZS07 | M | 70 | Fishermen | Fever and weakness | 5 | 1.4 | 27 | 119 | 91 | 86 | 1129 | 369 |
| ZS08 | F | 80 | Farmer | Fever | 6 | 1.6 | 42 | 127 | 127 | 63 | 515 | 382 |
| ZS09 | M | 54 | Farmer | Pain in back, waist and lower limbs, weakness, and diarrhea | 7 | 1.7 | 46 | 118 | 620 | 239 | 2361 | 660 |
| ZS10 | F | 66 | Farmer | Fever | 7 | 0.6 | 12 | 77 | 437 | 132 | 8495 | 1815 |
| ZS11 | M | 69 | Farmer | Fever and chilly sensations | 7 | 2.6 | 14 | 149 | 379 | 176 | 1722 | 628 |
| ZS12 | M | 46 | Farmer | Fever, cough and diarrhea | 6 | 1.7 | 21 | 134 | 484 | 182 | 2346 | 1478 |
| ZS13 | F | 59 | Farmer | Fever | 4 | 1 | 91 | 108 | 50 | 60 | 122 | 281 |
| ZS14 | F | 85 | Farmer | Fever and diarrhea | 7 | 0.8 | 33 | 76 | 87 | 56 | 734 | 547 |
| ZS15 | F | 70 | Farmer | Fever and diarrhea | 4 | 1.5 | 34 | 78 | 281 | 108 | 319 | 946 |
| ZS16 | M | 81 | Farmer | Fever | 3 | 1.6 | 34 | 96 | 216 | 118 | 653 | 946 |
| ZS17 | F | 86 | Farmer | Fever and diarrhea (5 days) | 7 | 1.67 | 38 | 126 | 592 | 236 | 3252 | 2178 |
| ZS18 | F | 77 | Farmer | Fever | 3 | 0.6 | 35 | 88 | 145 | 61 | 273 | 1187 |
| ZS19 | M | 55 | Farmer | Fever and weakness | 5 | 1.3 | 19 | 89 | 400 | 125 | 3554 | 1594 |
| ZS20 | M | 69 | Farmer | Fever, weakness and diarrhea | 5 | 1.8 | 20 | 135 | 860 | 232 | 3523 | 2415 |
| ZS21 | M | 71 | Farmer | Fever | 9 | 1.1 | 8 | 60 | 1201 | 516 | 1514 | 997 |
Abbreviations: WBC, white blood cell; PLT, platelet; HGB, hemoglobin; AST, aspartate aminotransferase; ALT, alanine aminotransferase; CK, creatine kinase; LDH, lactate dehydrogenase.
Figure 1. Comparison of monthly distributions of reported SFTS cases between Zhoushan and Mainland China.
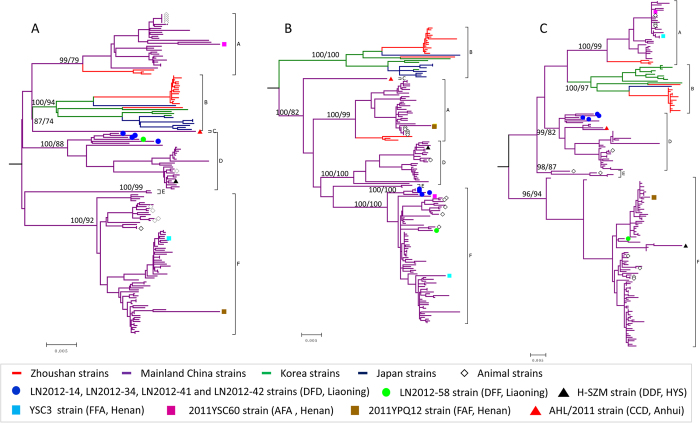
Classification of SFTSV into six genotypes and identification of reassortants
The whole SFTSV genome was successfully amplified and sequenced from 17 of the 21 patients from Zhoushan. SFTSV genomic segments (L, M, and S) obtained in this study were all deposited in GenBank under the following accession numbers: KR017827–KR017845, KR017846–KR017864, and KR017808–KR017826 (Supplementary Table S2).
To investigate the genetic relationship between SFTSV strains from the Zhoushan Islands with those from other parts of China and other countries, ML, NJ and MP trees were constructed based on three SFTSV genomic segments of 17 SFTSV strains reported in this study and 188 strains reported previously. Among these SFTSV strains, 159 of which had all the three genomic segment sequenced (Fig. 2). The topological structures derived from ML, NJ, and MP trees of each genomic segment, as well as the tree-topological structures derived from the three genomic segments (L, M, and S), were similar to each other (Fig. 2 and Supplementary Fig. S1). All SFTSV strains were divided into several major clades with high bootstrap supports of ≥85 in the phylogenetic trees, and most strains clustered within the corresponding clades in separate trees of genomic segments L, M, and S (Fig. 2). Based on the phylogeny (Fig. 2) and mean genetic distances of different clades (See Supplementary Table S3 online), six SFTSV genotypes (A–F) were classified. The mean genetic distances within genotypes were 0.001–0.026, and the distances among different genotypes were 0.035–0.062 (See Supplementary Table S3 online). The S genomic segment appeared to be more divergent than the L and M genomic segments.
Figure 2. Maximum likelihood (ML) phylogenetic trees of SFTSV L(A), M(B), and S(C) genomic segments.
Bootstrap values for ML and NJ tree are shown at corresponding nodes. The red, purple, blue, and green branches represent the strains from Zhoushan, mainland China, Japan, Korea, respectively. The pen diamonds indicate the strains from animals, including ticks, sheep, cows, and dogs. Ten SFTSV reassortants are highlighted by the colored circles, squares, and triangles.
According to the classification, majority (L: 80%, 132/163; M: 81%, 135/166, and S: 84%, 169/201) of SFTSV strains belong to genotype A, D, or F. All strains from Japan and 4 strains from South Korea and 15 strains from Zhoushan (including 14 obtained in this study and 1 retrieved from GenBank) clustered together, and formed the clade of genotype B (L: 17%, 28/163; M: 16%, 28/166, and S: 13%, 28/201) (Fig. 2). Of the 205 strains, 159 had all the three genomic segments sequenced. Whereas majority (93.7%, 149/159) of strains showed consistent genotype classification among the three genomic segments, ten strains had inconsistent genotype results based on the three genomic segments suggesting reassortment (Fig. 2). Among them, four strains (LN2012–14, LN2012–34, LN2012–41, and LN2012–42) had L and S segments of genotype D, but the M segment of genotype F (abbreviated as DFD for the L, M and S segments), The other six cases were DFF (LN2012-58), FFA (YSC3), AFA (2011YSC60), FAF (2011YPQ12), DDF (H-SZM), and CCD (AHL/2011). Therefore, apart from five SFTSV genotypes (A, B, D, E, and F), there were seven reassortment forms.
Comparison of SFTSV genotype distribution in China and neighboring countries
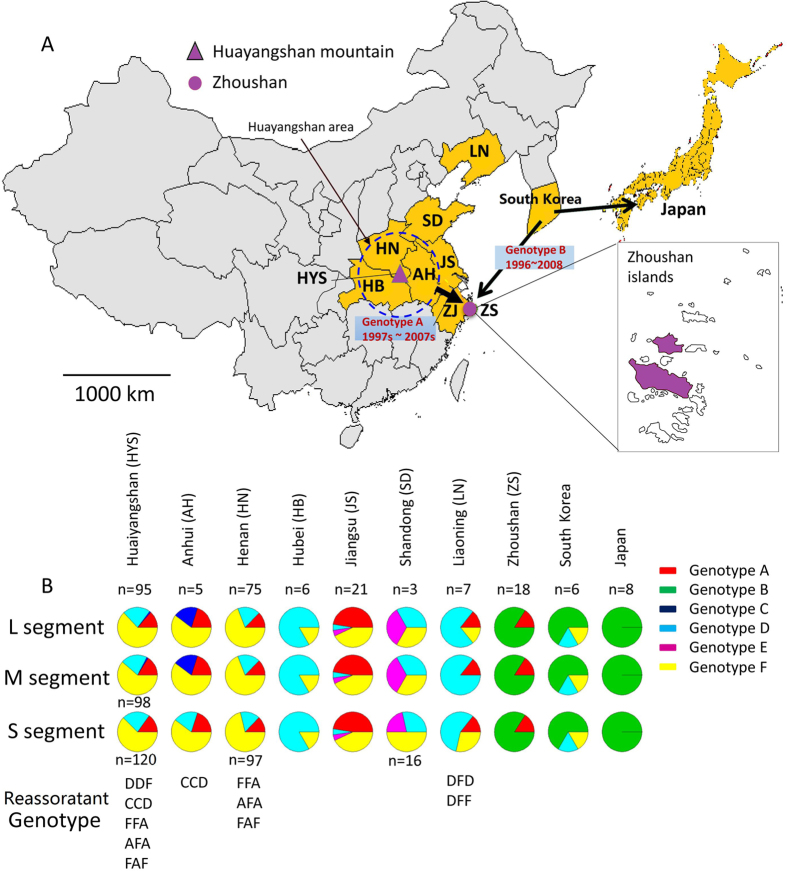
The Huaiyangshan Mountain area is located at the junction of Henan, Hubei, and Anhui Provinces and was considered as a single geographic area in this study (Fig. 3A). Genotypes A, D, and F appeared to be the most common SFTSV in mainland China (Fig. 3B). Genotype F was detected in all six affected areas of China. Genotype D was identified in Huaiyangshan, Henan, Hubei, Jiangsu, and Shangdong. Genotypes D and F were also found in South Korea, but not in the Zhoushan Islands or in Japan. Genotype A was found in all targeted provinces/areas in China except Hubei and Shangdong. In Japan, only genotype B was prevalent, and in South Korea, genotypes B, D, and F co-circulated.
Figure 3. Genotype distribution of SFTSV in China, Korea and Japan.
(A) The location of Zhoushan Islands, (B) Genotype distribution of SFTSV. Arrows indicate the transmission routes of SFTSV genotype B and the transmission routes of SFTSV genotype A from central China to Zhoushan. Two islands where SFTSV was circulating are highlighted in purple. The map was drawn with packages “maps” and “mapdata” in R version 3.1.3(https://cran.r-project.org/web/packages/mapdata/index.html)
Among the pure genotypes, genotype F (43.6%) was the most dominant, followed by genotypes A (20.1%), B (19.5%), and D (15.4%). Genotypes F, A, and D were prevalent in most provinces/areas, and almost all SFTSV strains isolated from animals (e.g., ticks, sheep, cows, and dogs) belonged to these genotypes (A: 45.5%; F: 45.5%; D: 9.0%) (Fig. 3). These findings suggest that the predominant SFTSV genotypes in China had the ability of cross-species transmission between humans and animals (especially ticks). Genotype E was only found in Jiangsu and Shandong, and no pure genotype C strain was found in this study. The SFTSV reassortants were located in Liaoning and Huaiyangshan (including Henan and Anhui).
Genotype B appeared to be only prevalent in islands (Zhoushan and Japan) and/or the Korean peninsula. Apart from genotype B, genotype A was also found in Zhoushan, and genotypes D and F in South Korea.
Tempospatial dynamics of SFTSV and the origin of the Zhoushan strains
Bayesian analysis was performed to estimate the SFTSV evolutionary rates and timescales. The evolutionary rates for the L, M, and S segments of SFTSV were estimated to be 1.87 (95% HPD, 0.84–3.02), 2.84 (95% HPD, 1.53–4.64), and 5.07 (95% HPD, 2.98–7.09) ×10−4 nt substitutions/site/year, respectively (Table 2). Genomic sequences that have exact sample time and geographic area were used to perform the temporal dynamics analyses. Temporal dynamics showed that all SFTSV strains had the time of origin of the most recent common ancestor (tMRCA) at 1868 (95% HPD, 1745–1940), 1867 (95% HPD, 1735–1933), and 1930 (95% HPD, 1880–1963) for the L, M, and S genomic segments, respectively (Table 2, See Supplementary Fig S2 online), suggesting that SFTSV originated during the years 1867–1930. tMRCA of each SFTSV genotype is shown in Table 2. Two SFTSV genotypes (A and B) circulated in Zhoushan. Genotype A of Zhoushan strains cluster together and form a monophyletic lineage with tMRCAs of 1997–2007 (Table 2). Their ancestral geographic states were estimated to be the Huaiyangshan area [posterior probability (PP): 0.99–1, See Supplementary Fig. S2 online], indicating that genotype A was introduced into Zhoushan from central China. For the genotype B strains circulating in Zhoushan, 14 strains (93.3%) formed a monophyletic lineage (Zhoushan linage), and another clustered with a South Korea strain, indicating that genotype B strains were introduced to Zhoushan by two independent events. The inferred ancestral geographic state of the Zhoushan genotype B lineage is South Korea (PP: 0.98–1) and the tMRCA was 1996–2008 (Table 2, See Supplementary Fig. S2 online). Similar to Zhoushan B strains, majority (88.9%) of Japanese genotype B strains formed a monophyletic clade, and another clustered with Zhoushan lineage clade. Their ancestral geographic states were also in South Korea (PP: 0.89–0.94). tMRCA of the Japanese lineage was 1974–1998 (See Supplementary Fig. S2 online).
Table 2. Bayesian estimates of evolutionary parameters based on L, M, and S genomic fragments of SFTSV.
| L segment | M segment | S segment | |
|---|---|---|---|
| Sequence number | 126 | 129 | 166 |
| Sampling time interval | 2007–2014 | 2007–2014 | 2007–2014 |
| Coefficient of variation | 0.26 | 0.36 | 0.80 |
| Evolutionary rate | |||
| (10*-4 nucleotide substitutions/site/year) | 1.87 (0.84–3.02) | 2.84 (1.53–4.64) | 5.07 (2.98–7.09) |
| tMRCA of SFTSV | 1868.5[1745.1–1940] | 1867.6[1735.7–1933.2] | 1930[1880.3–1963.5] |
| tMRCA of SFTSV genotype A | 1933.5[1865.6–1971.8] | 1960.3[1917.5–1984.4] | 1962.2[1934.9–1991.5] |
| tMRCA of SFTSV genotype B | 1901.4[1807.7–1957.5] | 1924[1848.5–1965.6] | 1971.4[1924.6–1982.9] |
| tMRCA of SFTSV genotype D | 1928.4[1857–1966.8] | 1951.5[1895.2–1978.7] | 1982.1[1959.4–1995.5] |
| tMRCA of SFTSV genotype E | 2005.4[1999.6–2008.6] | 2004.3[1996.4–2008.1] | 1987.7[1966.6–2002.7] |
| tMRCA of SFTSV genotype F | 1944.5[1888–1978.2] | 1971.3[1935–1991.7] | 1978.5[1961–1997.3] |
| Zhoushan lineage of genotype A | 1997[1981.4–2005.8] | 1997[1979.3–2006.2] | 2007.3[1999–2011.1] |
| Zhoushan lineage of genotype B | 2004.1[1996.1–2008.6] | 1996.8[1979.9–2005.9] | 2008.2[2004.5–2010.4] |
| Japan lineage of genotype B | 1974[1938.8–1993.3] | 1990.3[1946–1998.2] | 1998.45[1986.9–2005.6] |
The 95% highest posterior density credible regions are given in parentheses.
Discussion
SFTS was first identified in 2007, and its caustic agent was recognized in 2011 as a novel member of bunyavirus (SFTSV)1,2,3. SFTSV circulated in seven central-eastern provinces of China. Recently it was also found in South Korea and Japan11,12,13, which are separated from China by ocean straits. During June 2011, we reported the first SFTSV case in the Zhoushan Islands14. A retrospective review of the local clinic records revealed approximately 15 SFTS-like cases annually in the past decade in the Zhoushan Islands14. Therefore, SFTSV may have been epidemic in the Zhoushan Islands for a long period of time.
In this study, we investigated 100 suspected SFTS cases in the Zhoushan Islands from June 2011 to August 2014, and confirmed 21 as SFTSV infection by RT-PCR (Table 1). Most SFTSV infection in the Zhoushan Islands occurred during May–August and peaked in June. In other areas of China, SFTSV infections often occur during May–October, peaking in August (Fig. 1, See Supplementary Table S2 online). Patients in the Zhoushan Islands displayed similar clinical phenotypes and clinical laboratory-parameters similar to those of patients in the mainland areas (Table 1, Supplementary Table S1 online)1,3,4. However, fewer Zhoushan patients had symptoms of weakness and abdominal distension compared to those in other regions of China and Japan12,15. We were unable to determine whether the clinical symptoms were associated with infection during different SFTSV genotypes since the detail clinical information of each patient reported in previous studies was not available.
Age was an important risk factor for SFTSV infection. The medium age of SFTSV patients in the Zhoushan Islands was 67.3 which similar to that of patients in Japan, and obviously higher than that of patients (52.9–57.2) and fatal cases (62–63) in the mainland areas2,4,12,16. All 21 patients recovered after treatment with symptomatic and supportive therapy under the national guideline for SFTS, which may be attributed to the significant improvement in therapy and patient management.
We amplified and sequenced SFTSV genomic sequences circulating in the Zhoushan Islands, and performed phylogenetic analysis with all available genomic sequences from the mainland areas and surrounding countries (South Korea and Japan). Phylogenies of three genomic segments of SFTSV showed six well-supported clades defined as six SFTSV genotypes A–F (Fig. 2). Among 159 SFTSV strains with all the three genomic segments available, 149 belonged to genotypes A, B, D, E and F. Genotypes F, A, B, and D accounted for majority of SFTSV strains. Genotypes D, F, and A co-circulated in wide geographic regions of mainland China, and also accounted for all strains isolated from animals (e.g. ticks and sheep). Remarkably, genotype B circulated only in Japan, the Korean peninsula and Zhoushan Islands of China (Fig. 3B).
RNA viruses are characterized by a high mutation rate and a high potential of recombination, leading to a high genomic heterogeneity of RNA viruses17. Reassortment of genomic segments is another important mechanism that increases genetic diversity of segmented viruses (e.g., influenza viruses)18. A previous study reported two cases of SFTSV reassorants18,19. Here we identified 10 SFTSV reassortants that cover 7 reassortment forms (AFA, CCD, DDF, DFD, DFF, FAF, and FFA) (Figs 2 and 3), accounting for 6.3% of SFTSV strains (6.3%, 10/159). Except for DFD that was represented by four reassortant strains, each reassortment form had one representative strain. Majority of these reassortants involved genotypes D, F, and A, which may be explained by high prevalence and co-circulation of the three genotypes in SFTS-affected regions.
Three genomic segments of SFTSV appear to have different evolutionary rates. The S segment underwent more rapid evolution than the L and M genomic segments (Table 2). The time of MRCA (tMRCA) of the SFTSV strains was estimated at 1868, 1867, and 1930 based on L, M, and S genomic segments, respectively. Our estimates on origin time of SFTSV are more recent than a previous report19. The most likely reason for the difference was that the restricted molecular clock model was used in the previous study. However, the restricted molecular clock model is not the best fit model to infer the evolution of SFTSV (See Supplementary Table S4 online)20,21,22. The origin time of SFTSV based on L and M segments were very close (1867–1868). Similar to the previous report, the origin time based on the S segment (1930) was obviously more recent than those based on both L and M segments. One possible explanation is that there was no available genotype C sequence in S segment analysis since genotype C may be more ancient than other genotypes, as observed on the temporal dynamic of the L genomic segment (See Supplementary Fig. S2A online).
tMRCA of four predominate genotypes A, B, D, and F were estimated to be 1933–1960, 1901–1924, 1928–1951, and 1944–1971, respectively (Table 2). Genotype B diverged relatively earlier than other genotypes. Geographic origin of all SFTSV genotypes was estimated to most likely be the Huanyangshan area (PP: 0.38–0.82), suggesting that SFTSV spread to other regions of China or surrounding countries from the Huaiyangshan area (Fig. 3A). Two genotypes (A and B) were co-circulating in the Zhoushan Islands. Genotype A strains formed a Zhoushan lineage, having a common geographic origin in the Huaiyangshan area with other genotype A lineages (PP: 0.799–1) (See Supplementary Fig. S2 online). The time for the introduction of genotype A strains into Zhoushan was estimated to be 1997–2007.
Genotype B was only circulating in Zhoushan, South Korea, and Japan. All genotype B strains probably had a geographic origin in South Korea (PP: 0.71–0.82) with tMRCA of 1901–1924 (See Supplementary Fig. S2 online). In the genotype B clade, majority of the Zhoushan and Japanese strains formed their independent lineages and tMRCA of the Zhoushan lineage (1996–2008) was more recent than that of Japanese lineage (1974–1998). Both lineages had a geographic origin of South Korea, implying that the genotype B strain was transmitted from South Korea to the Zhoushan Islands and Japan. In addition, one Japanese strain clustered with the Zhoushan lineage and one Zhoushan strain clustered with one South Korea strain, suggesting at least two independent sea-crossing transmission events of genotype B from South Korea to Zhoushan and from South Korea to Japan. Animals, especially ticks, are the crucial vectors for SFTSV transmission. It is not easy for viruses to spread across geographical barriers, as they do on the continent, because the ocean channels form a natural barrier for most animal reservoirs. International travel increases the possibility of sea-crossing transmission of viruses, and may provide an explanation for the South Korea-to-Japan transmission of SFTSV; however, it is unable to explain how the virus spread to the Zhoushan Islands from South Korea, since almost all SFTSV patients in Zhoushan are local indigenous people and never left the island previously.
Ticks are widely distributed in the world. At least two tick species, Haemaphysa lislongicornis and Rhipicephalus microplucarry1,9, transmit SFTSV and other tick-borne pathogens to other animals, including mammals, land birds, and seabirds23,24,25,26,27. There are many islands in China, South Korea, and Japan, which provide habitats for many migratory seabirds from South Korea to China or other countries, and some ticks with seabirds are dispersed in China, South Korea, and Japan26. The Zhoushan Islands attract many seabirds each year. Some seabirds inhabiting the Zhoushan Islands can serve as an indirect vector to spread SFTSV or other tick-borne viruses. We suspect that the SFTSV genotype B likely transmitted from South Korea to Zhoushan and/or Japan with seabird migration across the oceans. To confirm this hypothesis, the samples of ticks of migratory birds should be collected for the detection and sequence analysis of SFTSV in future. According to this hypotheses, SFTSV genotype B should circulate in some coastal regions in China (e.g., Shandong peninsula and Liaoning), which contain natural habitats for migratory seabirds. Therefore, a molecular epidemiological investigation of SFTSV focusing on Chinese coastal regions will not only shed light on SFTSV evolution and transmission, but also provide valuable information for the prevention and control of SFTS in mainland China.
Methods
Clinical samples and laboratory testing
Among 100 suspected SFTS cases admitted to Zhoushan People’s Hospital during June 2011–August 2014, 21 were diagnosed as SFTS. In these cases the presence of SFTSV genome was detected by reverse transcription-polymerase chain reaction (RT-PCR) (See Supplementary Methods online). Clinical history and physical examination, routine clinical, biochemical, and hematological laboratory results, and acute phase serum samples were collected from all patients.
This study was conducted according to the Helsinki II Declaration and was approved by the ethics committee at the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention. Written informed consent was obtained from the patients.
Phylogenetic analysis
All available SFTSV sequences (including those isolated from animals) were directly downloaded from GenBank or blasted in the GenBank database using known L, M, and S sequences of SFTSV as references. In total, 530 sequences from 205 strains (L: 163; M: 166; S: 201) included 17 strains from Zhoushan Island were obtained (See Supplementary Table S2 online). Sequences generated from this study and retrieved from GenBank were divided into L, M, and S datasets for separate phylogenetic analysis. The sequences in each dataset were aligned using the MUSCLE algorithm implemented in MEGA 6.0 and edited manually28. Maximum likelihood (ML), Maximum Parsimony (MP) and neighbor-joining (NJ) trees were reconstructed by MEGA 6.0. The best-fitting model for ML analysis was determined using JmodelTest29. The ML trees of SFTSV L and M segments were inferred under General Time Reversible model incorporating invariant sites and a gamma distribution (GTR + I + G) and the ML tree of S segment was under Hasegawa-Kishino-Yano (HKY) model. Tree reliability was evaluated by the bootstrap method with 100 replications. For each dataset, mean genetic distances within and between different SFTSV genotypes were calculated using MEGA 6.028.
To estimate SFTSV temporal dynamic, maximum clade credibility (MCC) trees were constructed using a MCMC (Markov Chain Monte Carlo) method implemented in the BEAST v1.8.2 package20,30. The sequences with known sampling time and geographic location were used in the analysis. The evolutionary rates and the times to MRCA (tMRCA) of various nodes on the MCC tree were also estimated using the BEAST package. A relaxed molecular clock with an uncorrelated lognormal distribution and a constant population size model was used in the Bayesian coalescence analysis. The GTR + Γ 4 + I model of nucleotide substitution was used in the analyses of L and M segments, and the HKY + Γ 4 + I model in the analysis of S segment. Statistical uncertainty in parameter estimates was reflected by the 95% highest posterior density (HPD) values. MCMC analysis was run for 200 million generations for L, M, and S segments with sampling every 10,000 generations to achieve parameter convergence and adequate effective sample sizes (ESS > 200). We summarized the trees using Tree Annotator implemented in the BEAST v1.8.2 package. The initial 25% samples were discarded as burn-in, leaving 75% trees per run to produce consistent tree topologies.
Additional Information
How to cite this article: Fu, Y. et al. Phylogeographic analysis of severe fever with thrombocytopenia syndrome virus from Zhoushan Islands, China: implication for transmission across the ocean. Sci. Rep. 6, 19563; doi: 10.1038/srep19563 (2016).
Supplementary Material
Acknowledgments
We especially want to thank Dr. Shuping Tong for critical reading and review of this manuscript. This work was supported by the Special Fund for Health Research in the Public Interest (Grant No. 201202019), the National Key Science & Technology Special Projects on Major Infectious Diseases (Grant No. 2012ZX10004-211 and 2012ZX10004-220), the Public Projects in Zhoushan (Grant No. 2014C31069), and the medical and health system Key Disciplines in Zhejiang province (Grant No. 2012 ZDA044).
Footnotes
Author Contributions Y.F., S.L. and Z.Z. contributed equally to this work, harvesting clinical samples, carrying out the experiments, and bioinformation analysis and prepared figures; S.M. and X.L. carried out the experiments; W.Z. and X.C designed the experiments; C.Z. doing bioinformation analysis and prepared figures; Y.F., Z.Z., C.Z. and X.C. wrote the manuscript. All authors reviewed the manuscript.
References
- Yu X. J. et al. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 364, 1523–1532 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B. et al. Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS Pathog. 7, e1002369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Z. et al. Hemorrhagic fever caused by a novel tick-borne Bunyavirus in Huaiyangshan, China. Zhonghua Liu Xing Bing Xue Za Zhi. 32, 209–220 (2011). [PubMed] [Google Scholar]
- Zhang Y. Z. et al. Hemorrhagic fever caused by a novel Bunyavirus in China: pathogenesis and correlates of fatal outcome. Clin Infect Dis. 54, 527–533 (2012). [DOI] [PubMed] [Google Scholar]
- Liu Y. et al. Person-to-person transmission of severe fever with thrombocytopenia syndrome virus. Vector Borne Zoonotic Dis. 12, 156–160 (2012). [DOI] [PubMed] [Google Scholar]
- Tang X. et al. Human-to-human transmission of severe fever with thrombocytopenia syndrome bunyavirus through contact with infectious blood. J Infect Dis. 207, 736–739 (2013). [DOI] [PubMed] [Google Scholar]
- Gai Z. et al. Person-to-person transmission of severe fever with thrombocytopenia syndrome bunyavirus through blood contact. Clin Infect Dis. 54, 249–252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao C. J. et al. A family cluster of infections by a newly recognized bunyavirus in eastern China, 2007: further evidence of person-to-person transmission. Clin Infect Dis. 53, 1208–1214 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang Y. Z. et al. The ecology, genetic diversity, and phylogeny of Huaiyangshan virus in China. J Virol. 86, 2864–2868 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T. T. et al. Evolutionary and molecular analysis of the emergent severe fever with thrombocytopenia syndrome virus. Epidemics. 5, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S.-M. et al. Severe Fever with Thrombocytopenia Syndrome Virus in Ticks Collected from Humans, South Korea, 2013. Emerg Infect Dis. 20, 1358–1361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T. et al. The First Identification and Retrospective Study of Severe Fever With Thrombocytopenia Syndrome in Japan. J Infect Dis. 209, 816–827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T. et al. Phylogenetic and Geographic Relationships of Severe Fever With Thrombocytopenia Syndrome Virus in China, South Korea, and Japan. J Infect Dis. 212, 889–898 (2015). [DOI] [PubMed] [Google Scholar]
- Li S. et al. Sporadic case infected by severe fever with thrombocytopenia syndrome bunyavirus in a non-epidemic region of China. Biosci Trends. 5, 273–276 (2011). [DOI] [PubMed] [Google Scholar]
- Liu W. et al. Case-fatality ratio and effectiveness of ribavirin therapy among hospitalized patients in china who had severe fever with thrombocytopenia syndrome. Clin Infect Dis. 57, 1292–1299 (2013). [DOI] [PubMed] [Google Scholar]
- Chen H., Hu K., Zou J. & Xiao J. A cluster of cases of human-to-human transmission caused by severe fever with thrombocytopenia syndrome bunyavirus. Int J Infect Dis. 17, e206–208 (2013). [DOI] [PubMed] [Google Scholar]
- Moya A., Holmes E. C. & Gonzalez-Candelas F. The population genetics and evolutionary epidemiology of RNA viruses. Nat Rev Microbiol. 2, 279–288 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E. & Holmes E. C. Why do RNA viruses recombine? Nat Rev Microbiol. 9, 617–626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. et al. The Evolutionary History and Spatiotemporal Dynamics of the Fever, Thrombocytopenia and Leukocytopenia Syndrome Virus (FTLSV) in China. PLoS Negl Trop Dis. 8, e3237 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Suchard M. A., Xie D. & Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry I. M. et al. Unequal Evolutionary Rates in the Human Immunodeficiency Virus Type 1 (HIV-1) Pandemic: the Evolutionary Rate of HIV-1 Slows Down When the Epidemic Rate Increases. J Virol. 81, 10625–10635 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. & Holmes E. C. Avian Influenza Virus Exhibits Rapid Evolutionary Dynamics. Mol Biol Evol. 23, 2336–2341 (2006). [DOI] [PubMed] [Google Scholar]
- Jongejan F. & Uilenberg G. The global importance of ticks. Parasitology. 129 Suppl, S3–14 (2004). [DOI] [PubMed] [Google Scholar]
- Jiang X. L. et al. [Isolation, identification and characterization of SFTS bunyavirus from ticks collected on the surface of domestic animals]. Bing Du Xue Bao. 28, 252–257 (2012). [PubMed] [Google Scholar]
- Estrada-Pena A. & de la Fuente J. The ecology of ticks and epidemiology of tick-borne viral diseases. Antiviral Res. 108, 104–128 (2014). [DOI] [PubMed] [Google Scholar]
- Dietrich M., Gomez-Diaz E. & McCoy K. D. Worldwide distribution and diversity of seabird ticks: implications for the ecology and epidemiology of tick-borne pathogens. Vector Borne Zoonotic Dis. 11, 453–470 (2011). [DOI] [PubMed] [Google Scholar]
- Baneth G. Tick-borne infections of animals and humans: a common ground. Int J Parasitol. 44, 591–596 (2014). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. & Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Ho S. Y. W., Phillips M. J. & Rambaut A. Relaxed Phylogenetics and Dating with Confidence. PLoS Biology. 4, e88 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.