Abstract
The maintenance of genome stability is critical for proper cell function, and loss of this stability contributes to many human diseases and developmental disorders. Therefore, cells have evolved partially redundant mechanisms to monitor and protect the genome. One subcellular organelle implicated in the maintenance of genome stability is the centrosome, best known as the primary microtubule organizing center of most animal cells. Centrosomes serve many different roles throughout the cell cycle, and many of those roles, including mitotic spindle assembly, nucleation of the interphase microtubule array, DNA damage response, and efficient cell cycle progression, have been proposed to help maintain genome stability. As a result, the centrosome is itself a highly regulated entity. Here, we review evidence concerning the significance of the centrosome in promoting genome integrity. Recent advances permitting acute and persistent centrosome removal suggest we still have much to learn regarding the specific function and actual importance of centrosomes in different contexts, as well as how cells may compensate for centrosome dysfunction to maintain the integrity of the genome. Although many animal cells survive and proliferate in the absence of centrosomes, they do so aberrantly. Based on these and other studies, we conclude that centrosomes serve as critical, multifunctional organelles that promote genome stability.
Keywords: centrosome, genome stability, chromosomal instability, mitosis, DNA damage, p53, acentrosomal, cell cycle, PCM, aneuploidy, interphase, centrosome separation, asymmetric division
Introduction
During the cell cycle, cells must accurately replicate the entire genome and, in mitosis, segregate that genetic material equally into the two daughter cells. Errors in many different cellular processes can disrupt genome stability leading to a condition known as chromosomal instability (CIN), which has long been believed to be a major contributor to tumor progression in most cancers (Holland and Cleveland, 2009). In this review, we broadly define CIN as defects including aneuploidy, polyploidy, and DNA damage. Understanding the cellular processes and components that ensure the fidelity of events such as DNA replication and mitosis is therefore vital to our understanding of tumorigenesis and normal development. Much of the machinery involved in such processes has been intensely studied for decades, and from this research, centrosomes have emerged as one of the central players in protecting genome stability.
A centrosome is comprised of a central pair of centrioles surrounded by a protein rich matrix called pericentriolar material (PCM). Centrosomes are best known as the primary microtubule (MT) nucleating centers of most cells. For over a century, the highly visible activity and location of the centrosomes during mitosis have supported the widely held belief that MTs nucleated at the centrosomes are essential for formation of the bipolar spindle and the faithful segregation of the chromosomes. Given this vital function, centrosomes are themselves tightly regulated and duplicate only once during the cell cycle in S-phase (Nigg and Stearns, 2011). As cells enter mitosis, the duplicated centrosomes segregate to opposite sides of the nucleus and mature; that is, they augment the complement of PCM proteins that function in nucleating and organizing the bipolar mitotic spindle (Palazzo et al., 2000). These MT arrays span the cell and make direct contact with distinct regions of the chromosomes called kinetochores to facilitate their alignment and faithful segregation (Figure 1). Following mitotic exit, the PCM is shed from the centrosome and each daughter cell inherits a single centrosome associated with a basal level of PCM (Khodjakov and Rieder, 1999). As cells exit mitosis in G1/G0, the centrosome may form a specialized cellular appendage, called the primary cilium, which functions in cell signaling (Nigg and Raff, 2009). Although not discussed in this review, because the primary cilium receives signals that regulate the cell cycle, cilia dysfunction may serve as an additional threat to genome stability.
Figure 1. Centrosomes promote efficient bipolar spindle assembly.
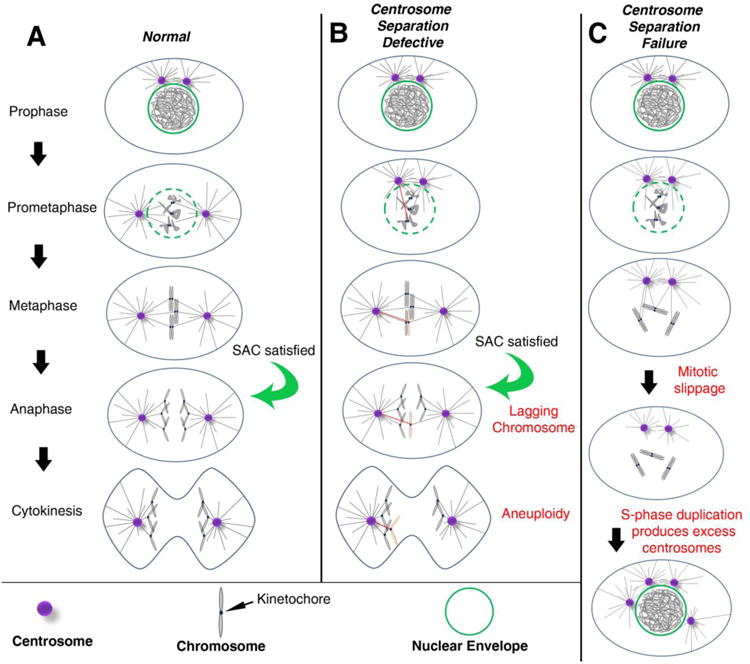
(A) In most cell types, the pair of centrosomes (purple) is closely associated at the entry into mitosis (Prophase). During prophase and prometaphase, centrosomes separate from one another and take up residence on opposite sides of the nucleus. In prometaphase, the nuclear envelope (green) breaks down, allowing MTs (black lines) emanating from the centrosomes to enter the nuclear space, find the kinetochores, and ultimately establish amphitelic attachments (i.e. the kinetochores of sister chromatids are individually attached to MTs from opposing spindle poles). The chromosomes will then congress to the metaphase plate. Once all kinetochores are attached to MTs, the SAC is satisfied, providing the “go anaphase” signal. During anaphase, sister chromatids are segregated towards opposite poles of the spindle. Finally, during telophase, the cytokinetic furrow ingresses, dividing the cell into two daughter cells with identical chromosome complements (abscission and daughter cell formation not depicted). (B) When there are moderate early defects in centrosome separation, the centrosomes may remain closely apposed as the cell enters prometaphase. This is believed to favor incorrect attachments of MTs from both poles to the kinetochore of the same chromatid. The red MT indicates the inappropriate attachment to the affected chromosome (pink). Even in the presence of these merotelic attachments, the SAC can be satisfied, allowing anaphase onset. The opposing forces of MTs from opposite poles on the same kinetochore can result in a lagging chromosome, which, in some cases, will mis-segregate into the wrong daughter cell, resulting in aneuploidy. (C) When centrosomes fail to separate, MTs may not attach to some kinetochores, leaving the SAC unsatisfied. Eventually, mitotic slippage may allow for mitotic exit into G1 without chromosome segregation or cytokinesis, resulting in a tetraploid cell with both centrosomes. In the subsequent S-phase, both centrosomes duplicate, resulting in extra centrosomes.
In this review, we will consider the diversity of mechanisms by which centrosomes have been proposed to promote genome stability. As evidenced by the wide range of topics in this Special Issue, centrosomes have been linked through many different cellular pathways to CIN. In a broad attempt to categorize them, we will group centrosomal functions that occur largely in either mitosis or interphase, as the integrity of the genome is under constant threat throughout the cell cycle. Within those phases of the cell cycle, we will explore the different functions of centrosomes that pertain to protecting genome stability, including bipolar spindle assembly, asymmetric cell division, and cell cycle progression.
Centrosomes are vital for efficient spindle assembly in mitosis
During mitosis, the replicated chromosomes are physically segregated into two newly formed daughter cells via interdependent cytoskeletal and signaling pathways. In spite of this complexity, portioning of the genomic material occurs with remarkably high fidelity. Thus, there is an inextricable link between the machinery that orchestrates mitosis (the mitotic spindle) and genome integrity. In general, studies across divergent species support a role for proper centrosome number, activity, and position in supporting efficient mitotic progression and the maintenance of genomic stability.
Microtubule nucleating centers of spindle assembly
While the centrosome was once considered the de facto MT organizing center (MTOC), numerous studies have since revealed the surprisingly robust ability of animal cells to form bipolar spindles in the complete absence of centrosomes (Heald et al., 1996; Khodjakov et al., 2000; Megraw et al., 2001; Basto et al., 2006). Indeed, most higher plant cells lack centrosomes entirely, instead relying on acentrosomal MT nucleating pathways (Hashimoto, 2013). These findings have led many groups to directly assess the importance of centrosomes in mitosis and also determine the protein complexes that serve as the sources of MTs in the absence of centrosomes (Figure 2A). Thus far, in most cells where centrosomes have been removed, either genetically or physically, mitotic spindle assembly appears to rely heavily on the chromatin-mediated RanGTP pathway and the Augmin complex, which nucleates MTs from preexisting MTs (Goshima et al., 2008; Wainman et al., 2009; Hayward et al., 2014; Poulton et al., 2014). The contribution of Augmin-derived spindle MTs becomes essential when other MT nucleation pathways (i.e. centrosomes or RanGTP) are absent or compromised (Goshima et al., 2008; Wainman et al., 2009; Hayward et al., 2014; Poulton et al., 2014). Another article in this Special Issue discusses the RanGTP pathway in more detail (Lavia, 2016). Importantly, studies also suggest these pathways are active in the presence of centrosomes. In addition to RanGTP and Augmin, recent studies indicate that spindle MTs can arise from acentrosomal MTOCs (aMTOCs), which utilize many of the same PCM components found in normal centrosome-based MTOCs (i.e. Cnn/Cdk5Rap2, Spd-2/Cep192, Asl/Cep152, Pericentrin, γ-Tubulin)(Hornick et al., 2011; Hayward et al., 2014; Baumbach et al., 2015). Given the vital importance of mitotic spindle assembly to genome stability, it is perhaps not so surprising that cells have evolved these semi-redundant and overlapping mechanisms to ensure the fidelity of bipolar mitotic spindle assembly. Yet another means cells possess to ensure accurate spindle formation is the Spindle Assembly Checkpoint (SAC). This highly conserved pathway monitors MT-kinetochore attachment (Foley and Kapoor, 2013). The checkpoint delays anaphase onset until all kinetochores become attached to MTs, thus helping prevent chromosome segregation errors (Musacchio and Salmon, 2007; Foley and Kapoor, 2013). Studies indicate the SAC is instrumental in helping compensate for the absence of centrosomes, where spindle assembly is slow and inefficient (Buffin et al., 2007; Poulton et al., 2014). Nevertheless, despite the identification and ongoing evaluation of multiple acentrosomal MT nucleation pathways and cellular checkpoints, most data continue to support a vital role for centrosomes in building mitotic spindles.
Figure 2. Differential effects of centrosome loss among model systems.
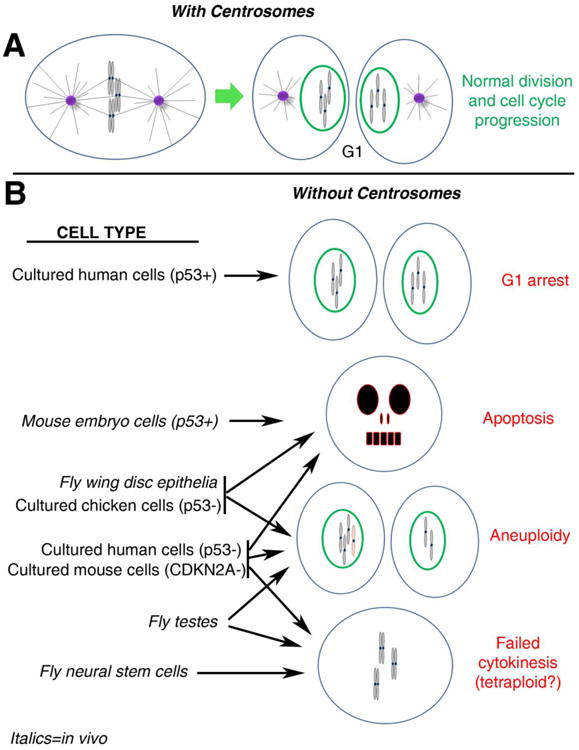
(A) In normal cells with centrosomes, a metaphase cell progresses through cytokinesis (not shown) to produce 2 daughter cells in G1, each with a normal chromosome complement and a single centrosome. To help visualize the consequences of chromosome segregation errors (e.g., aneuploidy), G1 chromosomes are depicted as condensed in this figure, though they would actually decondense upon mitotic exit. (B) The outcomes of mitosis in different cell types depleted of centrosomes are shown. p53 status is indicated for some relevant cell types (if not indicated, p53 is normal). Italics denote cells from in vivo studies. See text for more thorough descriptions of these studies. Not represented is the potential connection between mitotic error and accumulation of DNA damage; as observed in fly wing disc epithelia and cultured chicken cells. Note that lack of a connection to a specific outcome for a particular cell type does not necessarily indicate the outcome does not occur for that cell type. Rather it may simply have not been examined or reported in the relevant studies.
Several recent studies in various model systems have attempted to explicitly characterize the importance of centrosomes in mitotic spindle assembly, accurate chromosome segregation, and maintenance of genome stability. In some respects the findings are quite similar. Acentrosomal cells can typically assemble bipolar spindles and segregate chromosomes, but tend to be very inefficient, resulting in protracted spindle assembly. However, these studies have demonstrated some key differences in the importance of centrosomes among different cell types and species, particularly in their role in maintaining genome stability (Figure 2).
Drosophila mutants for core centrosomal proteins (e.g. cnn, asl, sas-4) survive to adulthood, suggesting centrosomes may be dispensable in most cell types during later development (Megraw et al., 2001; Basto et al., 2006; Blachon et al., 2008). One notable exception in flies is in the early embryo, where the very first divisions of the newly fertilized egg require the presence of functional centrosomes (Stevens et al., 2007; Rodrigues-Martins et al., 2008), probably due to the extremely rapid nature of those divisions (∼10 min), which occur without intervening gap phases. These maternally mutant animals do not develop beyond the first few divisions; however, zygotic mutant animals can be obtained due to maternal provisioning of centrosome-associated proteins necessary to proceed through these early divisions. Early embryos mutant for the key PCM components cnn, asl, or plp can result in aberrant mitotic spindles, centrosome separation defects, and DNA damage (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999; Varmark et al., 2007; Lerit et al., 2015). Similarly, in C. elegans embryos, depletion of the pro-mitotic centrosome maturation factor, Air-1, results in aberrant spindles that drive polyploidy, chromatin bridges, and severe aneuploidy that collectively promote embryonic lethality (Schumacher et al., 1998). Similar results were seen in mutants of another centrosome maturation factor, Spd-5 (Hamill et al., 2002). Centrosomes also play key roles in the mitotic divisions of fly spermatogenesis, where they promote accurate chromosome segregation and cytokinesis, which appear to be vital for male fertility (Bonaccorsi et al., 1998; Li et al., 1998; Rodrigues-Martins et al., 2008). In the developing fly wing disc, while most acentrosomal cells are able to successfully conduct mitosis with no perceivable errors in chromosome segregation, a significant fraction of cells do experience increased rates of aneuploidy and DNA damage, ultimately undergoing JNK-dependent apoptosis. Some of the apoptosis also appears attributable to defects in spindle orientation (Poulton et al., 2014). In contrast, in the developing fly brain, centrosome loss does not lead to a significant increase in aneuploidy or apoptosis (Basto et al., 2006; Buffin et al., 2007; Castellanos et al., 2008). Thus, centrosome dysfunction appears to be particularly detrimental in the early divisions of fly and worm embryos.
Elegant studies using allograft transplants of larval fly brains mutant for core centrosomal proteins into the adult abdomen of normal flies revealed the tumorigenic potential of acentrosomal brain tissue (Castellanos et al., 2008). Importantly, although centrosome loss in the fly brain can result in tumor formation, this appears to result from defects in neural stem cell (NSC) asymmetric division and is not predicated on increased CIN (Castellanos et al., 2008). However, in some of the centrosome mutants examined, most notably sas-4, aneuploidy and polyploidy accumulate over time as the brain tumor-derived cells progress through subsequent generations via allograft assay (Castellanos et al., 2008). It is worth noting that although centrosome loss in the wing disc epithelia does cause CIN, i.e. aneuploidy and DNA damage (Poulton et al., 2014), it does not lead to tumor formation in transplantation assays (Castellanos et al., 2008). It is likely, then, that apoptosis of genomically unstable epithelial cells in the wing disc is one factor limiting their tumor potential (Dekanty et al., 2012). It would be very interesting to determine if blocking apoptosis in acentrosomal wing disc cells can potentiate tumor formation in that tissue.
In cultured chicken cells (DT40 somatic cell line), centrosome loss leads to increased chromosome segregation errors, DNA damage, aneuploidy, and cell death (Sir et al., 2013), which closely parallels findings in fly wing disc epithelia (Figure 2)(Poulton et al., 2014). In both the DT40 cells and fly wing disc cells, it is not clear precisely how centrosome loss leads to DNA damage, though recent studies suggest that chromosome segregation errors can lead to DNA damage (Janssen et al., 2011; Crasta et al., 2012; Zhang et al., 2015).
Similar to the findings in the fly wing imaginal disc and cultured chicken cells, recent studies in mouse embryos and cultured mammalian cells found that centrosome loss in proliferating cells can result in increased rates of apoptosis (Bazzi and Anderson, 2014; Insolera et al., 2014; Wong et al., 2015). However, the mechanisms proposed for the cause of apoptosis in these studies vary significantly. In multiple tissues of the developing mouse embryo, centrosome loss causes a mitotic delay that triggers p53-dependent apoptosis, though no DNA damage or chromosome mis-segregation was detected in those studies (Figure 2)(Bazzi and Anderson, 2014; Insolera et al., 2014). Further work is required to elucidate the mechanism linking mitotic delay to p53 activation and apoptosis in this system. Nevertheless, this work clearly demonstrates that acentrosomal mouse cells can proliferate in vivo. Furthermore, blocking p53-mediated apoptosis restores many aspects of normal development, until defects in cell signaling associated with cilia loss cause embryonic death (Bazzi and Anderson, 2014). In contrast, recent studies in cultured mammalian cells found that mitosis in centrosome depleted cells results in a p53-dependent G1 arrest (see below); however, when p53 is blocked, those cells continue to proliferate, resulting in increased chromosome segregation errors and apoptosis, though no DNA damage was detected (Figure 2)(Lambrus et al., 2015; Wong et al., 2015). Collectively, these studies highlight p53 as a critical determinant of the cellular response to centrosome dysfunction.
Together, these studies indicate that centrosomes are vital components of efficient spindle assembly required for timely progression through mitosis. However, the role for centrosomes in the prevention of DNA damage is more ambiguous, as this phenotype was not observed in mouse embryos or cultured human cells. Nonetheless, most studies suggest that centrosomes serve an important role in accurate chromosome segregation, which promotes genome stability and cell viability, even across widely divergent species (i.e. flies to human).
Proper centrosome separation is essential for genome stability
Centrosomes not only serve as a primary source of MTs that build mitotic spindles, but they also determine the geometry and positioning of the spindle by regulating their own positions within the cell. In most animal cells, the centrosome pair separates in early mitosis and moves to opposite sides of the nucleus to form the bipolar spindle (Figure 1A). This separation of centrosomes is proposed to result from the combined activity of multiple forces acting on MTs emanating from the centrosomes [reviewed in (Tanenbaum and Medema, 2010)]. The proper positioning of centrosomes also serves as a key step in establishing the position and orientation of the cytokinetic furrow. Positioning of the furrow relies on furrow-inducing cues from stable MTs of the spindle midzone that drive constriction of the cortical actomyosin cytoskeleton, as well as the action of cortical inhibitory signals provided by dynamic centrosome-derived astral MTs away from the midzone (Green et al., 2012). Studies from a range of species have revealed that complete loss of centrosomes can disrupt cytokinesis (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001; Piel et al., 2001; Schmutz and Spang, 2005; Basto et al., 2006; Lambrus et al., 2015; Wong et al., 2015), which can lead to tetraploidy.
Numerous studies have demonstrated that moderate disruption of centrosome separation appears to favor the formation of merotelic attachments, where MTs from both spindle poles attach to the same kinetochore, which can increase rates of lagging chromosomes and whole chromosome mis-segregation (Figure 1B)(Cimini et al., 2001; Kaseda et al., 2012; Silkworth et al., 2012; Zhang et al., 2012; Poulton et al., 2013; Nam and van Deursen, 2014; Nam et al., 2015; Prosser et al., 2015). In contrast, when centrosome separation is severely impaired, the duplicated centrosomes remain directly adjacent to one another and produce a monopolar spindle that is insufficient to form complete MT-kinetochore attachments or generate the forces needed to segregate chromosomes (Figure 1C). While monopolar spindles tend to arrest in mitosis through SAC activity (Kapoor et al., 2000; Canman et al., 2003), some cells may exit mitosis (due to mitotic slippage), resulting in tetraploidy (Rieder and Maiato, 2004; Brito and Rieder, 2006; Brito et al., 2008). Tetraploidy not only disrupts the genetic balance, but the resulting tetraploid cell will also exit mitosis with two centrosomes. In the next S-phase, both centrosomes will template formation of a new centrosome, resulting in a cell with excess centrosomes (Figure 1C)(Borel et al., 2002). As discussed in other reviews in this Special Issue, excess centrosomes are problematic for cells and may contribute to CIN and tumor formation (Bolgioni and Ganem, 2016; Cosenza and Kramer, 2016; Nano and Basto, 2016). Remarkably, supernumerary centrosomes and the associated multipolar and error-prone divisions are well-tolerated and intrinsic to the normal development of a few specialized tissues, such as the Drosophila rectal papillae (Fox et al., 2010; Schoenfelder et al., 2014). Investigation of these unusual cells will likely elucidate how cancer cells tolerate centrosome dysfunction and genome instability.
In conclusion, a growing body of experimental data in different model systems and cell types has uncovered a complex and variable relationship between centrosomes and genome stability during mitosis. In most cases, it appears that centrosome dysfunction can threaten genome stability in multiple ways through various mitotic errors—aneuploidy due to chromosome mis-segregation, DNA damage (potentially caused by chromosome mis-segregation), and polyploidy/extra centrosomes caused by mitotic slippage/cytokinesis failure. Furthermore, these types of defects can result from either complete loss of centrosomes, disruption of normal centrosome activity (as in certain PCM mutants), or from defects in centrosome separation.
Centrosomes and interphase cell cycle control
The centrosome cycle is closely entrained with the nuclear cycle. The chromosomes and centrosomes both duplicate in S-phase, and the dramatic oscillations in centrosome composition and activity are tightly linked to the progression of the cell cycle (Figure 3A) (Khodjakov and Rieder, 1999). Thus, the progression of the cell cycle appears to largely dictate centrosome activity. Indeed, it is notable that several cell cycle regulators, including Cdc25, Cyclin E, Cdk2, and p53 to name a few, localize to the centrosome at various stages of the cell cycle (Fukasawa, 2007). It is intriguing to consider the centrosome not merely a passive recipient of instructive cues; rather, the centrosome may itself function to some degree in orchestrating cell cycle progression. In this way, the centrosome may serve to promote local enrichment of substrates, spatially organize complexes, and sequester proteins to prevent unfavorable interactions (Abraham, 2001). For detailed discussion on the role of the centrosome as a cell cycle signaling depot, we refer the reader to several excellent reviews (Doxsey et al., 2005; Fukasawa, 2007; Arquint et al., 2014). Here, we review evidence that implicates centrosomes in permitting cell cycle progression, with an emphasis on the G1 to S phase checkpoint.
Figure 3. Centrosomes as effectors and regulators of cell cycle progression.
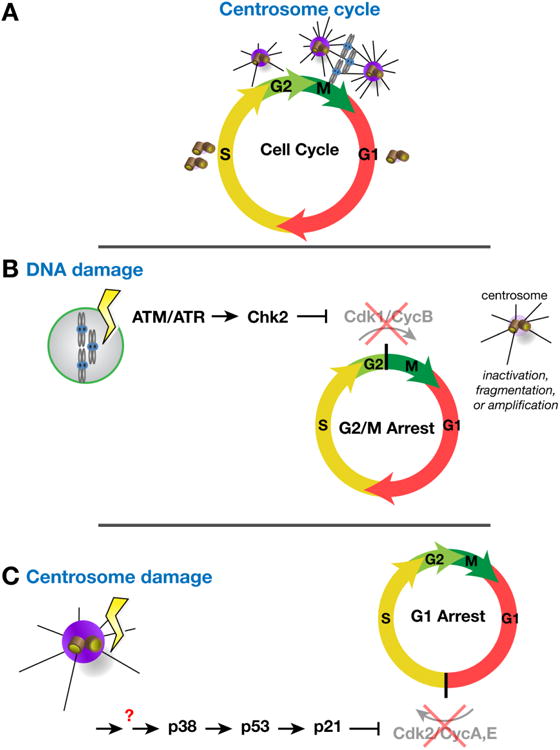
(A) The centrosome cycle is entrained with the cell cycle. Similar to the chromosomes, centrosomes duplicate in S-phase. Centrosome maturation, the recruitment of PCM and concordant increase in MTOC activity, occurs during G2 and into M-phase to support the elaboration of the bipolar mitotic spindle. Centrosome activity attenuates in late mitosis such that each centrosome that segregates to a daughter cell (G1) is associated with a basal level of PCM. (B) Activation of the DNA damage response pathway leads to centrosome dysfunction (e.g., inactivation in Drosophila embryos and fragmentation or amplification in mammalian cells) and triggers a block to mitotic entry. (C) Centrosome dysfunction can itself alter cell cycle progression. In some cells, a centrosome integrity checkpoint impedes interphase progression by triggering a G1 arrest.
The G1 to S checkpoint functions to prepare the cell for replication of the genome. Typically, DNA damage triggers cell cycle arrest to provide time for repair. The G1/S DNA damage checkpoint allows cells to retain DNA replication fidelity, avoid the propagation of deleterious mutations, and prevents the expansion of DNA repeats. Double-stranded DNA breaks (DSBs) are sensed by the kinases ATM (ataxia-telangiectasia mutated) and ATR (ATM and Rad3-related), which activate the effector kinases Checkpoint kinase 1 (Chk1) and Chk2 (Abraham, 2001). Active Chk2 promotes the stabilization and accumulation of p53, which increases p21 activity and suppresses G1 to S progression. Loss of p53, therefore, is sufficient to avoid the G1 checkpoint, often to the detriment of chromosomal stability and cell viability (Kastan et al., 1991).
Genetic analyses of DNA damage checkpoint mutants support a link between DNA damage and centrosome activity (Sibon et al., 2000; Takada et al., 2003; Sakurai et al., 2011). In early syncytial Drosophila embryos, centrosome-nucleated MTs position nuclei at the cortex. DNA damage triggers inactivation of the centrosomes (Figure 3B), effectively diminishing their MTOC activity so as to perturb the fidelity of the subsequent mitosis and displace the damaged nucleus away from the remaining healthy nuclei (Sibon et al., 2000; Takada et al., 2003). This ejection of the damaged nucleus, called “nuclear fallout,” prevents the propagation of damaged nuclei in subsequent cell cycles. Chk2 mediates this DNA damage response and must be activated during interphase prior to its enrichment on centrosomes and spindle poles to drive centrosome inactivation (Takada et al., 2003). Thus, loss of Chk2 permits the accumulation of CIN by allowing damaged nuclei to remain at the embryonic cortex, where they will be propagated in subsequent cell cycles to the detriment of the developing animal. An analogous response occurs in mammalian cells with damaged DNA, where centrosome fragmentation occurs during mitosis (Hut et al., 2003) and Chk1 triggers centrosome amplification in interphase (Inanc et al., 2010; Loffler et al., 2013).
Intriguingly, multiple components of DNA damage pathways localize to centrosomes in a dynamic, cell-cycle dependent manner (Arquint et al., 2014). This subject is addressed elsewhere in this Special Issue (Morrison, 2016); nonetheless, it is worth mentioning that a recent study of centrosome-depleted cells found an intact, functional DNA damage response. Specifically, in both HeLa and 3T3 cells, treatment with the DNA damaging agent doxorubicin results in a G2 arrest, and this DNA damage-induced arrest is unchanged in acentrosomal cells (Wong et al., 2015). Similarly, acentrosomal mouse embryos maintain a normal DNA damage response (Bazzi and Anderson, 2014). Therefore, despite a long history of observations suggesting that centrosomes may help regulate the DNA damage response, functional support for a direct connection remains inconclusive.
The Centrosome Integrity Checkpoint
The importance of centrosomes in cell cycle progression continues to be extensively studied using a variety of centrosome perturbations. Acute centrosome loss via centrosome ablation or microsurgery (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001), as well as transient depletion of centrosome components via RNAi in untransformed human cells (Srsen et al., 2006; Mikule et al., 2007), provided early evidence that disrupting centrosome activity and function results in a G1 arrest (Figure 3C). This G1 arrest is triggered by the suppression of Cdk2 activity caused by p53 accumulation, and removing the Cdk2 inhibitors (p21 or p53) is sufficient to relieve the arrest and permit re-entry into the cell cycle (Mikule et al., 2007). These findings support a model, called the centrosome integrity checkpoint (Heinrichs, 2007), whereby centrosome integrity surveillance is coupled to cell cycle progression. Such a model is attractive because it provides a safety mechanism to abort cellular proliferation in the absence of properly functioning centrosomes. However, the existence of such a checkpoint came under debate after reports that centrosomes could be safely removed without triggering a G1 arrest (Uetake et al., 2007). Importantly, these acentrosomal cells were not completely normal; they were sensitized to additional perturbations that triggered a G1 arrest in the next cell cycle (Uetake et al., 2007).
Several variables may account for the differential response in cell cycle progression observed in these studies, including differences in the degree of centrosome disruption. The RNAi experiments induce a persistent loss of key proteins required for MT nucleation/organization and maintenance of PCM structure (Srsen et al., 2006; Mikule et al., 2007). These factors are still present in surgically manipulated cells, as evident by the de novo centriole assembly apparent in these cells (Uetake et al., 2007). These studies suggest, therefore, that a persistent loss of centrosome integrity, rather than an acute disruption, is required to trigger the checkpoint.
More recent studies have investigated the effects of chronic centrosome disruption/loss in cultured cells. In stable cell lines lacking the centriole components Cep152 or STIL, cell cycle progression and a normal DNA damage response are maintained (Sir et al., 2013). However, these chicken DT40 cells lack the normal p53 response (Ulrich et al., 1992) that is critical for G1 arrest. In lieu of a G1 arrest, these cells are prone to CIN and elevated rates of apoptosis (Sir et al., 2013). Likewise, Drosophila sas-4 mutant NSCs (Basto et al., 2006) and stable cell lines (Lecland et al., 2013) also progress through mitosis without an apparent G1 arrest (Basto et al., 2006), suggesting the effect of loss of centrosome integrity on cell cycle progression is, to some extent, cell type dependent.
The recent development of acute inhibitors of the centriole duplication factor Plk4 provide the strongest evidence in support of the centrosome integrity checkpoint. This approach depletes centrosomes from cultured cells by preventing new centriole assembly. In untransformed human cells that maintain a normal p53 response, the elimination of centrioles through an auxin-inducible degron that targets the destruction of Plk4 (Lambrus et al., 2015) or through the application of a novel Plk4 inhibitor, centrinone (Wong et al., 2015), results in a G1 arrest. This arrest can be subverted by p53 knockdown to restore cell cycle progression (Lambrus et al., 2015; Wong et al., 2015). These elegant studies demonstrate a clear requirement for a p53-dependent centrosome integrity checkpoint in regulating the proliferation of normal human cells.
In conclusion, data from untransformed human cells support a strong link between centrosome integrity and interphase progression from G1 to S phase. While mechanisms that mediate centrosome integrity surveillance remain unknown, centrosome dysfunction typically results in p53-mediated cell cycle arrest and/or sensitizes cells to arrest or death, depending on the cellular context. Future studies should rigorously investigate the link between centrosomes and upstream regulators of the p53 pathway to decipher the signals that instruct the G1 arrest.
Some stem cells maintain a single active interphase centrosome
Most cells attenuate centrosome activity during interphase. For example, many epithelial cells inactivate their centrosomes and reassign MTOC activity to the apical membrane (Figure 4)(Meads and Schroer, 1995; Harris and Peifer, 2007; Feldman and Priess, 2012). The centrosomes within Drosophila interphase NSCs and male germline stem cells (mGSCs) are unique in that one centrosome remains active as an MTOC, whereas the other centrosome is transiently inactivated until mitotic onset (Figure 4)(Rebollo et al., 2007; Rusan and Peifer, 2007; Yamashita et al., 2007). During stem cell asymmetric division, one centrosome is retained within the self-renewing stem cell, while the other centrosome segregates to the daughter cell fated for differentiation. In NSCs, the centrosome that segregates to the differentiating cell sheds its interphase PCM (Rusan and Peifer, 2007), arguing the maintenance of interphase centrosome activity is differentially regulated within stem cells. A number of studies have collectively established a working model of NSC centrosome activity control that requires the activities of Polo kinase/Plk1 (Polo), a highly conserved regulator of centrosome maturation (Sunkel and Glover, 1988), the Polo substrate Centrobin (Cnb)(Januschke et al., 2011; Januschke et al., 2013), and negative regulators, like Pericentrin-like protein (PLP)(Lerit and Rusan, 2013) and Bld 10/Cep135 (Singh et al., 2014). Future research is needed to determine if the same molecular players function in asymmetric centrosome maturation in mGSCs and other stem cell models.
Figure 4. Strategies for regulating centrosome activity.
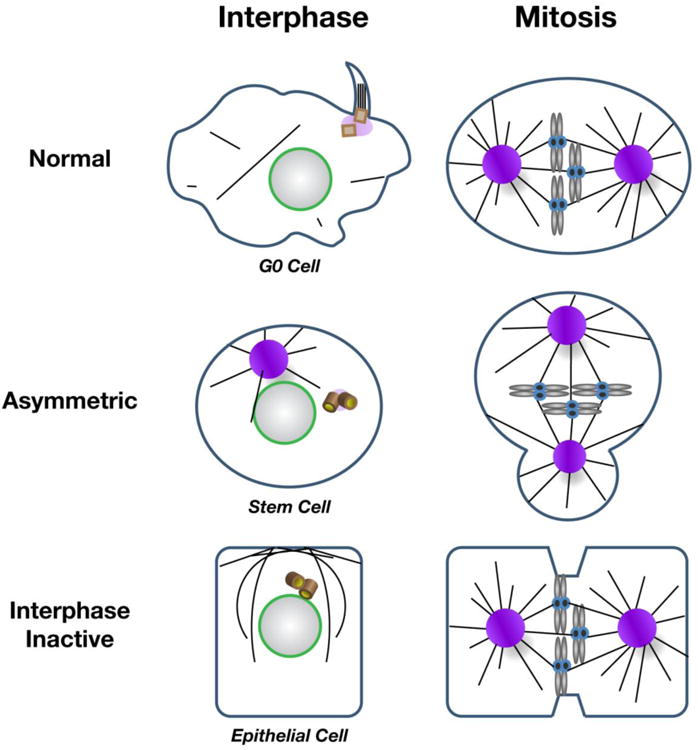
(A) MT nucleation is differentially regulated in various cell types. In many normal mammalian cells, quiescent (G0) cells form primary cilia during interphase when the centriole docks at the plasma membrane. Cilia are reabsorbed by the cell prior to mitotic onset, when centrosomes function as MTOCs and contribute to spindle formation. Variations on interphase centrosome activity control are observed in Drosophila asymmetrically dividing stem cells, which display asymmetric interphase centrosome activity (one off, one on). During mitosis, both centrosomes are active in stem cells. In contrast, Drosophila and C. elegans epithelial cells deregulate interphase centrosome activity and MTs are nucleated from the apical cortex. Centrosomes mature, or become active, upon mitotic onset. Centrioles (brown barrels) denote inactive centrosomes.
Recent work has highlighted the importance of interphase centrosome asymmetry in ensuring proper centrosome separation required for genome stability. In NSCs, the precocious activation of the second centrosome, either through ectopic localization of Cnb (Januschke et al., 2013) or through mutation of either PLP (Lerit and Rusan, 2013) or Bld10 (Singh et al., 2014) results in two active centrosomes that remain anchored to the apical cortex until just after nuclear envelope breakdown (NEB). Studies from cultured mammalian PtK cells indicate failure to separate centrosomes prior to NEB promotes merotelic attachments of MTs to kinetochores and drives elevated rates of chromosome mis-segregation (Silkworth et al., 2012). Therefore, additional research is needed to determine whether interphase centrosome asymmetry may be generally important for normal centrosome separation and genome stability in models of asymmetric cell division.
Collectively, these studies indicate centrosome activity is differentially regulated in different cellular contexts with a single active interphase centrosome maintained in some stem cell models. With respect to genome stability, the selective activation and inactivation of one or both centrosomes must be tightly regulated in space and time to avoid mis-attachments of MTs to the chromosomes. Thus, centrosome activity control must be tightly entrained to centrosome position to permit proper mitotic spindle assembly for normal cell cycle progression. Additional studies are required to parse apart the molecular coordination of centrosome position and activity control.
Concluding Remarks
The centrosome has been the topic of intense study since its initial discovery over a century ago. Long appreciated for its role in orchestrating the precise events of mitotic division, subsequent work has suggested intriguing roles in cell cycle progression and the DNA damage response. Researchers have long recognized the redundant capabilities of cells to nucleate and organize MTs independent of centrosomes. We now have a far greater molecular understanding of several of these acentrosomal MTOCs and a growing sense of the interconnectedness of these systems, which will continue to come into focus in the coming years. Certainly, the centrosome is not essential in all contexts. Entire organisms function and thrive in the absence of centrosomes (e.g. higher plants). Yet, for a number of cellular systems, including asymmetrically dividing stem cells, the early embryos of many (but not all) animals, and most normal human cells, the centrosome is critical for normal cell division and genome stability. Even those cells that continue to proliferate following centrosome disruption are not completely normal. Often, their cell cycles are prolonged and they are more sensitive to secondary stresses, including external stressors, like photodamage, and internal stressors, like second site mutations. These observations indicate that although the cell compensates for centrosome loss (e.g. through alternative MT nucleating pathways), its ability to do so is often limited. Going forward, it will be interesting to decipher the crosstalk between the various MT nucleating pathways beyond their coordinated use of the same effector molecules, such as γ-Tubulin. Some of these studies are already well underway.
Studies from a broad range of species and cell types highlight significant differences in the cellular response to centrosome loss. The relatively consistent differences between studies conducted in vivo and those performed in cultured cells suggest that acentrosomal cultured mammalian cells are more prone to checkpoint induced cell cycle arrest, whereas acentrosomal cells in whole animals typically continue to divide. The fact that acentrosomal fly cells, both in vivo and in culture, continue to divide without any reported cell cycle arrest suggests those cells may generally have much weaker checkpoints. Nevertheless, what these most recent studies convincingly indicate is that centrosomes are not specifically and directly required to progress through the cell cycle. Rather, it appears that centrosome loss can trigger dramatic cellular responses (e.g. a G1 arrest checkpoint in cultured cells), but when those checkpoints are removed, the cells can complete and re-enter the cell cycle repeatedly. Importantly, most studies also indicate that these proliferating acentrosomal cells are error-prone, particularly in mitosis, and many of those cells ultimately converge on the same phenotype as mouse and fly cells in vivo—apoptosis. Thus, while centrosomes may not be strictly required for mitosis or cell cycle progression, they clearly have important roles in efficient spindle assembly, which, in most cell types examined thus far, helps promote accurate chromosome segregation, cytokinesis, and maintenance of genome stability.
Acknowledgments
We thank Nasser M. Rusan, Mark Peifer, and Erich Kushner for critical comments. DAL is supported by a Lenfant Biomedical Postdoctoral Fellowship and a NHLBI Career Transition Award (1K22HL126922). JSP is supported by the Peifer lab grant NIH R01GM067236.
Abbreviations
- CIN
chromosomal instability
- MT
microtubule
- MTOC
microtubule organizing center
- aMTOC
acentriolar microtubule organizing center
- PCM
pericentriolar material
- RNAi
RNA interference
- NEB
nuclear envelope breakdown
- SAC
spindle assembly checkpoint
- NSC
neural stem cell
- mGSC
male germline stem cell
References
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Arquint C, Gabryjonczyk AM, Nigg EA. Centrosomes as signalling centres. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Baumbach J, Novak ZA, Raff JW, Wainman A. Dissecting the function and assembly of acentriolar microtubule organizing centers in Drosophila cells in vivo. PLoS Genet. 2015;11:e1005261. doi: 10.1371/journal.pgen.1005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H, Anderson KV. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci U S A. 2014;111:E1491–1500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S, Giansanti MG, Gatti M. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J Cell Biol. 1998;142:751–761. doi: 10.1083/jcb.142.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel F, Lohez OD, Lacroix FB, Margolis RL. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc Natl Acad Sci U S A. 2002;99:9819–9824. doi: 10.1073/pnas.152205299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Yang Z, Rieder CL. Microtubules do not promote mitotic slippage when the spindle assembly checkpoint cannot be satisfied. J Cell Biol. 2008;182:623–629. doi: 10.1083/jcb.200805072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E, Emre D, Karess RE. Flies without a spindle checkpoint. Nat Cell Biol. 2007;9:565–572. doi: 10.1038/ncb1570. [DOI] [PubMed] [Google Scholar]
- Canman JC, Cameron LA, Maddox PS, Straight A, Tirnauer JS, Mitchison TJ, Fang G, Kapoor TM, Salmon ED. Determining the position of the cell division plane. Nature. 2003;424:1074–1078. doi: 10.1038/nature01860. [DOI] [PubMed] [Google Scholar]
- Castellanos E, Dominguez P, Gonzalez C. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr Biol. 2008;18:1209–1214. doi: 10.1016/j.cub.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekanty A, Barrio L, Muzzopappa M, Auer H, Milan M. Aneuploidy-induced delaminating cells drive tumorigenesis in Drosophila epithelia. Proc Natl Acad Sci U S A. 2012;109:20549–20554. doi: 10.1073/pnas.1206675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Feldman JL, Priess JR. A role for the centrosome and PAR-3 in the hand-off of MTOC function during epithelial polarization. Curr Biol. 2012;22:575–582. doi: 10.1016/j.cub.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Gall JG, Spradling AC. Error-prone polyploid mitosis during normal Drosophila development. Genes Dev. 2010;24:2294–2302. doi: 10.1101/gad.1952710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. aPKC controls microtubule organization to balance adherens junction symmetry and planar polarity during development. Dev Cell. 2007;12:727–738. doi: 10.1016/j.devcel.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T. A ring for all: gamma-tubulin-containing nucleation complexes in acentrosomal plant microtubule arrays. Curr Opin Plant Biol. 2013;16:698–703. doi: 10.1016/j.pbi.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Hayward D, Metz J, Pellacani C, Wakefield JG. Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev Cell. 2014;28:81–93. doi: 10.1016/j.devcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heinrichs A. A centrosome-integrity checkpoint. Nat Rev Mol Cell Bio. 2007;8:98–98. [Google Scholar]
- Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornick JE, Mader CC, Tribble EK, Bagne CC, Vaughan KT, Shaw SL, Hinchcliffe EH. Amphiastral mitotic spindle assembly in vertebrate cells lacking centrosomes. Curr Biol. 2011;21:598–605. doi: 10.1016/j.cub.2011.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hut HM, Lemstra W, Blaauw EH, Van Cappellen GW, Kampinga HH, Sibon OC. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol Biol Cell. 2003;14:1993–2004. doi: 10.1091/mbc.E02-08-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanc B, Dodson H, Morrison CG. A centrosome-autonomous signal that involves centriole disengagement permits centrosome duplication in G2 phase after DNA damage. Mol Biol Cell. 2010;21:3866–3877. doi: 10.1091/mbc.E10-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insolera R, Bazzi H, Shao W, Anderson KV, Shi SH. Cortical neurogenesis in the absence of centrioles. Nat Neurosci. 2014;17:1528–1535. doi: 10.1038/nn.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- Januschke J, Llamazares S, Reina J, Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Reina J, Llamazares S, Bertran T, Rossi F, Roig J, Gonzalez C. Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat Cell Biol. 2013;15:241–248. doi: 10.1038/ncb2671. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaseda K, McAinsh AD, Cross RA. Dual pathway spindle assembly increases both the speed and the fidelity of mitosis. Biol Open. 2012;1:12–18. doi: 10.1242/bio.2011012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus BG, Uetake Y, Clutario KM, Daggubati V, Snyder M, Sluder G, Holland AJ. p53 protects against genome instability following centriole duplication failure. J Cell Biol. 2015;210:63–77. doi: 10.1083/jcb.201502089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecland N, Debec A, Delmas A, Moutinho-Pereira S, Malmanche N, Bouissou A, Dupre C, Jourdan A, Raynaud-Messina B, Maiato H, et al. Establishment and mitotic characterization of new Drosophila acentriolar cell lines from DSas-4 mutant. Biol Open. 2013;2:314–323. doi: 10.1242/bio.20133327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit DA, Jordan HA, Poulton JS, Fagerstrom CJ, Galletta BJ, Peifer M, Rusan NM. Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function. J Cell Biol. 2015;210:79–97. doi: 10.1083/jcb.201503117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit DA, Rusan NM. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J Cell Biol. 2013;202:1013–1022. doi: 10.1083/jcb.201303141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Xu EY, Cecil JK, Turner FR, Megraw TL, Kaufman TC. Drosophila centrosomin protein is required for male meiosis and assembly of the flagellar axoneme. J Cell Biol. 1998;141:455–467. doi: 10.1083/jcb.141.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler H, Fechter A, Liu FY, Poppelreuther S, Kramer A. DNA damage-induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene. 2013;32:2963–2972. doi: 10.1038/onc.2012.310. [DOI] [PubMed] [Google Scholar]
- Meads T, Schroer TA. Polarity and nucleation of microtubules in polarized epithelial cells. Cell Motil Cytoskeleton. 1995;32:273–288. doi: 10.1002/cm.970320404. [DOI] [PubMed] [Google Scholar]
- Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Megraw TL, Li K, Kao LR, Kaufman TC. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nam HJ, Naylor RM, van Deursen JM. Centrosome dynamics as a source of chromosomal instability. Trends Cell Biol. 2015;25:65–73. doi: 10.1016/j.tcb.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HJ, van Deursen JM. Cyclin B2 and p53 control proper timing of centrosome separation. Nat Cell Biol. 2014;16:538–549. doi: 10.1038/ncb2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo RE, Vogel JM, Schnackenberg BJ, Hull DR, Wu X. Centrosome maturation. Curr Top Dev Biol. 2000;49:449–470. doi: 10.1016/s0070-2153(99)49021-0. [DOI] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- Poulton JS, Cuningham JC, Peifer M. Acentrosomal Drosophila epithelial cells exhibit abnormal cell division, leading to cell death and compensatory proliferation. Dev Cell. 2014;30:731–745. doi: 10.1016/j.devcel.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton JS, Mu FW, Roberts DM, Peifer M. APC2 and Axin promote mitotic fidelity by facilitating centrosome separation and cytoskeletal regulation. Development. 2013;140:4226–4236. doi: 10.1242/dev.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser SL, Sahota NK, Pelletier L, Morrison CG, Fry AM. Nek5 promotes centrosome integrity in interphase and loss of centrosome cohesion in mitosis. J Cell Biol. 2015;209:339–348. doi: 10.1083/jcb.201412099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. From centriole biogenesis to cellular function: centrioles are essential for cell division at critical developmental stages. Cell Cycle. 2008;7:11–16. doi: 10.4161/cc.7.1.5226. [DOI] [PubMed] [Google Scholar]
- Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H, Okado M, Ito F, Kawasaki K. Anaphase DNA bridges induced by lack of RecQ5 in Drosophila syncytial embryos. FEBS Lett. 2011;585:1923–1928. doi: 10.1016/j.febslet.2011.04.074. [DOI] [PubMed] [Google Scholar]
- Schmutz C, Spang A. Knockdown of the centrosomal component SAS-5 results in defects in nuclear morphology in Caenorhabditis elegans. Eur J Cell Biol. 2005;84:75–82. doi: 10.1016/j.ejcb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Schoenfelder KP, Montague RA, Paramore SV, Lennox AL, Mahowald AP, Fox DT. Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development. 2014;141:3551–3560. doi: 10.1242/dev.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher JM, Ashcroft N, Donovan PJ, Golden A. A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos. Development. 1998;125:4391–4402. doi: 10.1242/dev.125.22.4391. [DOI] [PubMed] [Google Scholar]
- Sibon OC, Kelkar A, Lemstra W, Theurkauf WE. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell. 2012;23:401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Ramdas Nair A, Cabernard C. The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr Biol. 2014;24:1548–1555. doi: 10.1016/j.cub.2014.05.050. [DOI] [PubMed] [Google Scholar]
- Sir JH, Putz M, Daly O, Morrison CG, Dunning M, Kilmartin JV, Gergely F. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J Cell Biol. 2013;203:747–756. doi: 10.1083/jcb.201309038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srsen V, Gnadt N, Dammermann A, Merdes A. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J Cell Biol. 2006;174:625–630. doi: 10.1083/jcb.200606051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Raposo AA, Basto R, St Johnston D, Raff JW. From stem cell to embryo without centrioles. Curr Biol. 2007;17:1498–1503. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel CE, Glover DM. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci. 1988;89(Pt 1):25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- Takada S, Kelkar A, Theurkauf WE. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell. 2003;113:87–99. doi: 10.1016/s0092-8674(03)00202-2. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich E, Boehmelt G, Bird A, Beug H. Immortalization of conditionally transformed chicken cells: loss of normal p53 expression is an early step that is independent of cell transformation. Genes Dev. 1992;6:876–887. doi: 10.1101/gad.6.5.876. [DOI] [PubMed] [Google Scholar]
- Vaizel-Ohayon D, Schejter ED. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr Biol. 1999;9:889–898. doi: 10.1016/s0960-9822(99)80393-5. [DOI] [PubMed] [Google Scholar]
- Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr Biol. 2007;17:1735–1745. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Wainman A, Buster DW, Duncan T, Metz J, Ma A, Sharp D, Wakefield JG. A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev. 2009;23:1876–1881. doi: 10.1101/gad.532209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW, et al. Cell biology. Reversible centriole depletion with an inhibitor of Pololike kinase 4. Science. 2015;348:1155–1160. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Foreman O, Wigle DA, Kosari F, Vasmatzis G, Salisbury JL, van Deursen J, Galardy PJ. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J Clin Invest. 2012;122:4362–4374. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]


