Abstract
Purpose
To determine optimal parameters for acquisition and processing of dynamic contrast-enhanced (DCE) MRI to detect small changes in near normal low blood-brain barrier (BBB) permeability.
Methods
Using a contrast-to-noise ratio metric (K-CNR) for Ktrans precision and accuracy, the effects of kinetic model selection, scan duration, temporal resolution, signal drift and length of baseline on the estimation of low permeability values was evaluated with simulations.
Results
The Patlak model was shown to give the highest K-CNR at low Ktrans. The Ktrans transition point, above which other models gave superior results, was highly dependent on scan duration and tissue extravascular extracellular volume fraction (ve). The highest K-CNR for low Ktrans was obtained when Patlak model analysis was combined with long scan times (10-30 minutes), modest temporal resolution (<60 seconds/image), and long baseline scans (1-4 minute). Signal drift as low as 3% was shown to affect the accuracy of Ktrans estimation with Patlak analysis.
Conclusion
DCE acquisition and modeling parameters are interdependent and should be optimized together for the tissue being imaged. Appropriately optimized protocols can detect even the subtlest changes in BBB integrity and may be used to probe the earliest changes in neurodegenerative diseases such as Alzheimer’s disease and Multiple Sclerosis.
Keywords: Ktrans estimation, DCE-MRI, blood brain barrier, parameter optimization, permeability
Introduction
Dynamic contrast-enhanced (DCE) MRI gives quantitative and semi-quantitative information about the integrity of the vascular system (1). It is commonly used to study a number of diseases such as cancer, stroke and multiple sclerosis (MS) (2-6). While originally developed to assess blood-brain barrier (BBB) integrity (7-10), it has also found many applications outside of the brain (3,11-14). The parameter of interest in BBB integrity is the transfer constant Ktrans (15), which describes the transfer rate of molecules from plasma space into interstitial space. Since the BBB permeability to molecules is size and transporter dependent (16-18), interpretation of Ktrans requires consideration of structural and physiological changes at play at the BBB as well as contrast agent (CA) being used to infer Ktrans. Most applications of DCE in the brain have focused on imaging relatively high BBB breakdown (such as observed in malignant tumors or stroke), rather than on subtle changes in the BBB, using small molecule CA. This is in part driven by the fact that CA extravasation in the highly compromised BBB is high, resulting in easier detection of the CA signal. Ktrans values in the intact or subtly damaged BBB are at least one to two orders of magnitude lower than those found when the BBB is highly compromised (8,19,20). Thus, it is much more difficult to measure.
Recent studies indicate a pathophysiological role for the BBB in diabetes (21), cognitive disorders such as mild cognitive impairment (MCI) (20,22), Alzheimer’s disease (AD) (23,24), MS (6,25-28) and chronic traumatic encephalopathy (CTE) (29-31). Although dysfunctional, changes in BBB integrity for these conditions are thought to be subtle and close to intact values (32). Early attempts to measure Ktrans in the normal appearing brain for MCI and AD failed to detect significant changes (33,34). These studies may have been hampered by their relatively long sampling interval and the semi-quantitative nature of their analyses. More recent studies acquired at higher field strengths and using more sophisticated kinetic modeling have determined that the Ktrans of healthy BBB is low but not zero and detected differences in MS patients compared to controls (19,26,28) as well as increases during normal aging and in MCI patients(20). Ktrans from these studies are within a range of previously reported BBB Ktrans values using small inert polar molecules in mammals (32,35,36).
Despite these developments and recent successes (19,20,26,28), optimal methods to collect and analyze DCE data in order to detect subtle changes to BBB integrity remain unclear (1,37,38). This imaging effort poses a distinct set of challenges because the contrast changes are much lower and it can take much longer for detectable leakage to occur. This places considerable demands on the imaging sequence and post-processing used to minimize noise and maximize contrast to detect these subtle changes (25,37).
The few studies that have applied DCE in the low permeability regime have used a wide range of different techniques. Taheri et al introduced a serial T1 mapping technique with low CA concentration (0.025 mmol/kg) injection, finding differences in white matter permeability between controls (Ki ≈ 3 ∗ 10−4 mL·g−1·min−1, Ki = Ktrans/[1-hematocrit]) and MS patients (28). Using a multiple dose injection protocol and a data-driven post-processing, Ingrisch et al reported differences between normal appearing white matter and MS lesions (6). They noted that a two compartment uptake model (8) was appropriate at the low permeability regime. Larsson and colleagues reported that the Patlak model can estimate low vascular permeability well (Ki ≈ 0.1 mL/100mL/min for normal brain tissue) (25,26). They also proposed that the combination of model-free estimation of perfusion (using the Tikhonov’s method, Tik) and the two-compartment exchange model (2CXM) can estimate BBB permeability across a wider range of Ktrans (19). In a subsequent study, Cramer et al compared the ability of Patlak, extended-Tofts and Tik-2CXM to estimate Ktrans, reporting the threshold for accurate Patlak estimation to be Ki ≈ 0.3 mL/100g/min (25). Accuracy of the extended Tofts model was shown to be very sensitive to blood volume and perfusion compared to the other two models. The authors also examined the effects of sampling interval and acquisition duration on Ktrans estimation, finding that the use of short sampling intervals and long acquisition times to be the most accurate. Recent work using the Patlak model with high spatial resolution and short sampling intervals clearly shows that low Ktrans determinations are possible and provide crucial information about BBB dysfunction (20).
While these findings provide some guidelines on acquisition parameters and post-processing methodology, it remains unclear from the published studies under what circumstances these recommendations hold. Most notably, the two parameters that dramatically affect the appropriateness of the Patlak model, imaging time and ve (extravascular extracellular volume fraction), have not been examined. It therefore remains difficult to currently draw specific, quantitative recommendations for ongoing studies. Thus, quantitative analytical methods that simplify the evaluation of how the different experimental factors combine to influence the accuracy, reliability, and ability to resolve changes in the measured Ktrans will greatly facilitate future study design.
Here, we introduce a metric, the contrast-to-noise ratio for Ktrans (subsequently referred to as K-CNR), which enables quantitative comparison of different models to estimate DCE vascular permeability. We demonstrate how K-CNR simplifies model selection analysis at low Ktrans given specific acquisition parameters. Conversely, using the Patlak model as an example, we show how K-CNR can guide the selection of optimized image acquisition parameters to maximize the model’s ability to resolve differences at low Ktrans. The impact of temporal resolution, acquisition time, signal drift and the number of baseline images on K-CNR are examined for a variety of physiological conditions.
Methods
DCE Simulations
A population averaged curve published by Parker et al (39) was modified for use as the simulated arterial input function (AIF). Briefly, the AIF curve is a mixture of two Gaussians plus two exponentials modulated with a sigmoid function:
| (1) |
where Ca is the arterial plasma concentration, An, Tn, and σn are the scaling constants, centers and widths of the nth Gaussian; α and β are the amplitude and decay constant of the exponential; s and τ are the width and center of the sigmoid, respectively. To provide an AIF curve mimicking the behavior in our clinical datasets, different input parameters for Equation 1 were used for our simulations compared to those defined by Parker et al. The amplitude of the first pass bolus was lowered, and the bi-exponential decay constant was altered to accommodate the longer blood half-life of the MultiHance® (Bracco, Milan, Italy) CA used at our site. The parameters were manually adjusted to fit our population-averaged AIF.
Simulated tissue curves were generated by a convolution of the impulse response for a 2CXM with the AIF (40). The 2CXM is described by:
| (2) |
where “⊗” is convolution and F± and K± are defined elsewhere (40). The free parameters used to define the 2CXM are Ktrans, ve, vp and Fp; where Ktrans is the blood-tissue transfer constant, ve is the extravascular extracellular volume fraction, Fp is plasma flow and vp is plasma volume fraction. Unless stated otherwise these parameters were set to ve = 0.05, vp = 0.025, and Fp = 0.130 ml/min/ml assuming 1g = 1ml. These values are within expected physiological ranges (41-45), and similar to average 2CXM values measured in our healthy controls.
The AIF curve and tissue curves were generated at sampling intervals of 100 ms and 1 second respectively. These intervals are significantly faster than the rate of change of the curves and capture all features of the curves; a longer interval was used for the tissue curve to take advantage of its slower rate of change to reduce computation time. The temporal resolution of DCE-MRI studies is known to influence the accuracy of parameter estimation (11,25,46-50). Thus, the generated tissue curves and the AIF were down-sampled to study the effect of temporal resolution on Ktrans analysis. This was done using a Gaussian filter with the standard deviation equal to a third of the sampling window of the desired temporal resolution. This incorporates all the information in the sampling window and simulates the center weighting imposed by k-space sampling.
To evaluate the effect of signal-to-noise ratio (SNR) on DCE analysis, MR-realistic noise was added to achieve the stated SNR levels by adding Gaussian noise to the real and imaginary channels (the MR signal was assumed to be real) and then taking the modulus. The AIF and tissue concentration curves were converted to MRI signal intensity curves using the standard spoiled gradient echo (SPGR) equation at steady state (51), with TR=8.3 ms, FA=15°, blood pre T1=1200 ms, and tissue pre T1=1800 ms. The stated SNR values are the SNR in the tissue prior to CA injection. Unless otherwise stated we defined SNR=30 as this reflects the SNR observed in our clinical datasets. These noise-added signal intensity curves were then converted back into concentration curves and used to calculate the DCE parameters.
Timing alignments between the AIF and sampling window may also affect the accuracy and precision of Ktrans measurement (50), AIF and tissue curve simulation studies were also performed with jitter offsets at 2 second interval increments within each sampling interval. This was only done for datasets with sampling intervals greater than 2 seconds.
Accuracy and precision of Ktrans estimation using different models and acquisition variables for different tissue parameters were assessed using the K-CNR (see below) by adding noise (as described above) and fitting 1000 times per parameter combination. The effect of the number of baseline images was evaluated by generating and fitting curves with baselines ranging from 15 to 240 s prior to CA injection.
To explore the effect of signal drift, a constant ranging from −10% to +10% of the baseline signal was added to the generated AIF and tissue curves prior to noise addition and Ktrans estimation. To simulate drift correction, a noise-added phantom (to serve as an intensity standard) with signal intensity equivalent to half the maximal AIF intensity was used to derive correctional scaling factors across the time series. Robust fitting of the scaling factor versus time curve to a fourth-order polynomial was performed to minimize the noise amplification of using the scaling factor directly. Ktrans was estimated using scale factor-corrected AIF and tissue curves and compared to their uncorrected counterparts.
Clinical studies
Subjects
Participants with no significant history of neurological disease were recruited through the University of Southern California (USC) from May 2013 to May 2014. The study was approved by the USC Institutional Review Board. All participants signed informed consent. We enrolled six participants of both genders with ages 23-47. All procedures were performed at the Keck Medical Center of USC. All participants underwent a medical examination as well as blood draw to ensure appropriate kidney function for CA administration. MRI scans were screened and no subcortical white matter lesions were found on any subjects. MRI scans of the brain were performed on a total of six participants.
Magnetic resonance imaging
All images were obtained on a GE 3T HDXT MR scanner with a standard eight-channel array head coil. Anatomical coronal spin echo T2-weighted scan were first obtained through the hippocampi (TR/TE 1550/97.15 ms, NEX = 1, slice thickness 5 mm with no gap, FOV = 188 × 180 mm, matrix size = 384 × 384). Baseline coronal T1-weighted maps were then acquired using a T1-weighted 3D SPGR pulse sequence and variable flip angle method using flip angles of 2°, 5° and 10°. (TR/TE = 8.29/3.08 ms, NEX = 1, slice thickness 5 mm with no gap, FOV 188 × 180 mm, matrix size 160 × 160). Coronal DCE MRI covering the hippocampi and temporal lobes were acquired using a T1-weighted 3D SPGR pulse sequence (FA = 15°, TR/TE = 8.29/3.09 ms, NEX = 1, FOV 180 × 135 × 60 mm, matrix size 160 × 120 × 12, voxel size was 1.125 × 1.125 × 5 mm3). This sequence was repeated for a total of 16 minutes with an approximate temporal resolution of 15.4 seconds. Multihance (0.05 mmol/kg) was administered intravenously into the antecubital vein using a power injector, at a rate of 3mL/sec followed by a 25mL saline flush, 30 seconds into the DCE scan.
DCE data analysis
Post-processing of the collected MRI data and simulated data was done using in-house DCE processing software implemented in Matlab (The MathWorks Inc., Natick, MA). Plasma concentrations were assumed with Hct=0.45 for all cases. For the acquired data the AIF was obtained from the common carotid artery.
Three different kinetic models were examined: Patlak, extended Tofts, and the 2CXM.
The 2CXM, introduced above, is the most general model considered in this analysis with four free variables. The 2CXM assumes two well-mixed compartments (blood plasma and extracellular extravascular tissue) and allows for general transport back and forth between these compartments. Blood flow is considered finite, so mixing in the capillaries is assumed.
The extended Tofts model is a simplification of the 2CXM, with three free variables it assumes minimal mixing occurs in the capillaries (Fp=∞), and is defined by:
| (3) |
The Patlak model is a simplification of the extended Tofts, with two free variables it assumes minimal back-flux from the tissue back to the arteries, or equivalently that Ca(t)>>C(t). It is defined by:
| (4) |
All these models assume well mixed compartments; a thorough analysis of the mathematics and assumptions has been examined in a recent review (15).
The experimental data were converted to concentration curves using the acquired T1 maps and the standard steady state incoherent equation.
| (5) |
The concentration curves were fitted to equations (2)-(4) using non-linear fitting routines with the trust-region-reflective algorithm in Matlab. To ensure that the fitting converges on a global minimum, tolerances were set to 10−12, 10−, 50, and 50 for the function tolerance, parameter tolerance, maximum iterations, and maximum function evaluations respectively. Multiple starting locations of Ktrans were used during the fit convergence process with the minimal residual used as the final value. Other kinetic parameters were less sensitive to initial starting values; thus, this strategy was not used in those cases.
To assess the impact of different imaging parameters to accurately measure and detect group changes in Ktrans during simulations, we define a K-CNR metric, which estimates the CNR that would be observed for a 10% change in Ktrans:
| (6) |
where Ktrans(true) is the defined Ktrans value used to generate the simulated curve, Ktrans(fitted) is the median of the output Ktrans values obtained by fitting the simulated curve with software. The derivative is the slope of the Ktrans(fitted) versus Ktrans(true), this is multiplied by 0.1·Ktrans(true) to estimate the contrast observed for a 10% change in Ktrans(true). σ is the standard deviation of the 1000 repetitions at that specific Ktrans. The slope was estimated using a Savitzky-Golay filter (52) with a seven point quadratic polynomial kernel to suppress noise in the derivative calculation, and a three point moving average was applied to the K-CNR to reduce noise from the standard deviation term. This metric can be interpreted the same as CNR is normally interpreted, it is a respected measure of precision and contrast, and not directly sensitive to the accuracy of the model.
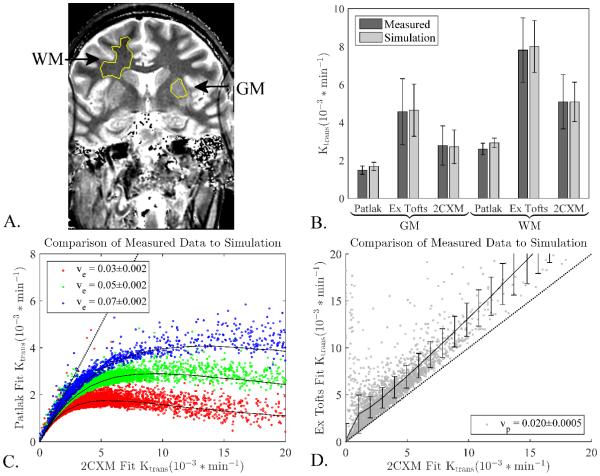
To ensure that the above simulation strategy was capable of generating realistic DCE curves that had the same characteristics of the measured clinical data, a number of steps were taken. First, the simulated curves were visually compared to the measured curves to ensure that no significant features of the curves were different and that the noise levels were similar. Once we had established that the curves were qualitatively the same a quantitative comparison was performed. Regions of interest (ROI) were drawn in normal appearing grey matter (GM) and white matter (WM) regions in control datasets (Figure 1A). The median values for all four parameters in the 2CXM derived from these ROIs were recorded. Simulated DCE curves were then generated using these values and the population AIF as described above. Curve fitting was performed using the three different models on the simulated curves and the fit values from the simulations were compared to the measured values in the subjects for all three different models. This ensured that the population AIF and noise addition used to generate the simulated curves did not significantly alter the characteristics of the curves. It also ensures that the simulated curves reflect the behavior of the measured curves for all three models (2CXM, extended Tofts, and Patlak). The 2CXM was used as it is the most general model; thus provides the best estimate of the true tissue values. Although the plasma flow values derived from this model might not be completely accurate due to the temporal resolution (15.4 seconds) of the DCE MRI data, the calculated median value of 0.13 ml/min/ml (or a blood flow of 0.29 ml/min/ml) is within the expected range (43) and so was considered sufficient for the simulations.
Figure 1.
Simulation results compared to collected data in healthy controls. A) Typical ROIs for a single subject. B) Mean values and standard deviations of measured Ktrans values from GM and WM regions defined in the subjects correspond well to simulated results. Simulated curves were generated and all three models were fit to the simulated data to ensure that the population AIF and generating procedures would give values and noise characteristics similar to the measured data. The similar results for each model show that the simulated curves are representative of the real data. C) Each dot represents a voxel from a healthy control and the black lines are simulation results using the central ve value measured from the clinical data, error bars on simulation results excluded for clarity. D) Same as C, but comparing extended Tofts to 2XCM and using the central vp value measured from the clinical data. Error bars are the standard deviation. The measured voxels follow the simulation results extremely well showing the simulations captures the different behavior of each model. Dotted line indicates slope of unity, i.e. where Patlak or extended Tofts fit is identical to 2CXM fit.
Results
Simulation curve fits with 2CXM and Patlak are consistent with clinical data fits
Comparisons between measured data fits and simulated data fits are shown in Figure 1. For the various models, simulated and measured curves were not significantly different; with Patlak giving values lower than 2CXM, and extended Tofts higher than 2CXM (Figure 1B). The noise characteristics of the models were also accurately reproduced with the simulation matching very close to the measured data, again with extended Tofts showing the highest standard deviation, and Patlak the lowest.
The measured data were also compared against the simulations in a voxel-wise comparison. All brain voxels from the healthy controls were initially considered, then voxels were removed that had poor fits (approximately the top 8% of residuals) or vp > 10% (to eliminate blood vessels). The fitted 2CXM values were compared to the fitted Patlak values for various ve values. Simulation curves comparing 2CXM and Patlak values generated at the corresponding ve were overlaid for comparison (Figure 1C). Simulations corresponded very well to the collected data for the majority of voxels, with the Patlak model showing the expected underestimation at higher Ktrans values for both measured and simulated data. Comparison of 2CXM versus extended Tofts fitted values with their simulated counterparts showed a similar pattern at fixed vp (Figure 1D), further supporting applicability of our simulations to reflect clinically measured data.
Patlak model provides good estimation of true values in low Ktrans regimes
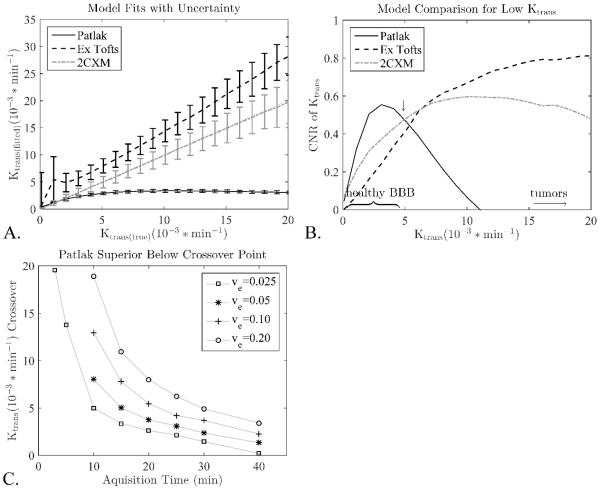
Different models were directly compared by fitting to the same simulated curves and examining their K-CNR (Figure 2A). The Patlak model had the highest K-CNR for low Ktrans values, while extended Tofts and 2CXM had higher K-CNR at higher Ktrans (Figure 2B). The exact crossover point between these two regimes depended on the acquisition time and the ve value (Figure 2C).
Figure 2.
The much lower variance in the Patlak model leads to significantly better K-CNR values compared to the extended Tofts and 2CXM models at low Ktrans. At higher Ktrans significant backflux starts to flatten the Patlak response leading to a loss of contrast and lower CNR values. A) Median fits and standard deviations for the different models, This plot was used to calculate the K-CNR values. B) For a scan time of 15 minutes and a ve=0.05 this crossover occurs around Ktrans=5*10−3/min (arrow). C) Different scans times and ve values lead to different cross over points, below which Patlak outperforms 2CXM model. For clarity only the crossover points for the 2CXM model are shown, the extended Tofts are almost identical.
Effects of acquisition time on Ktrans estimation
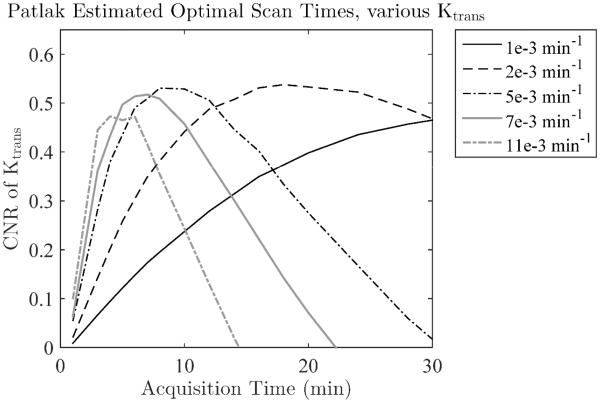
Since the Patlak model had the highest K-CNR in the range of Ktrans values found in intact BBB, the Patlak model was further evaluated to determine the optimal acquisition time. As expected, the optimal acquisition time was found to depend on the Ktrans value: shorter scan times give better K-CNR for higher Ktrans values as this limits the amount of CA backflux that is captured during imaging, which confounds the Patlak Ktrans estimation (Figure 3).
Figure 3.
The optimal scan time depends on the Ktrans value for the Patlak model, as long scan times and high Ktrans will lead to significant backflux and poor results. Scanning longer than 15 minutes is only beneficial for the very lowest Ktrans values, and longer than 5 minutes leads to sharp drops in the K-CNR of Ktrans > 10*10−3/min.
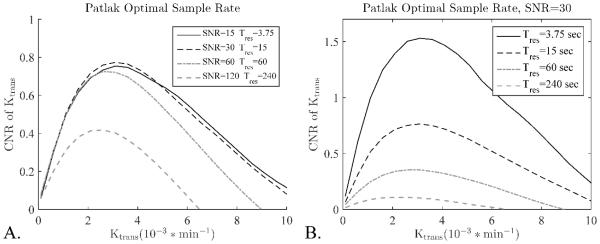
Effects of sampling rate on Ktrans estimation
The effect of sampling rate on K-CNR was evaluated for the Patlak model in two different scenarios. First, the sampling interval was lengthened by a factor and the SNR was increased by the square root of the factor (Figure 4A). This simulates the scenario whereby additional averages were collected or if additional partitions were collected in a 3D scan to increase the field of view. Second, the sampling interval was lengthened with no change to the SNR (Figure 4B). This simulates the scenario whereby more slices were collected in a 2D scan to increase the FOV. When longer sampling intervals lead to increases in SNR, the K-CNR held mostly constant up to a sampling interval of 60 seconds, longer than that it fell significantly (Figure 4A). When changes in sampling interval had no effect on the SNR (SNR held constant), each lengthening in sampling interval caused a steady and significant reduction in the K-CNR (Figure 4B).
Figure 4.
The effect of sampling rate on K-CNR for the Patlak model depends on the effect adjusting the sampling interval has on the SNR of the scan, two different scenarios are considered. A) Lengthened sampling interval increases the SNR, as would happen by adding averages or increasing the matrix size and field of view together. Only small differences are seen between sampling intervals shorter than 60 seconds, while sampling intervals longer than 60 seconds lead to significant K-CNR loss. B) Changing the sampling interval has no effect on SNR (SNR is held constant), as would happen by adding or removing slices in a 2D scan. In this case longer sampling intervals lead to a consistent and significant decrease in the K-CNR.
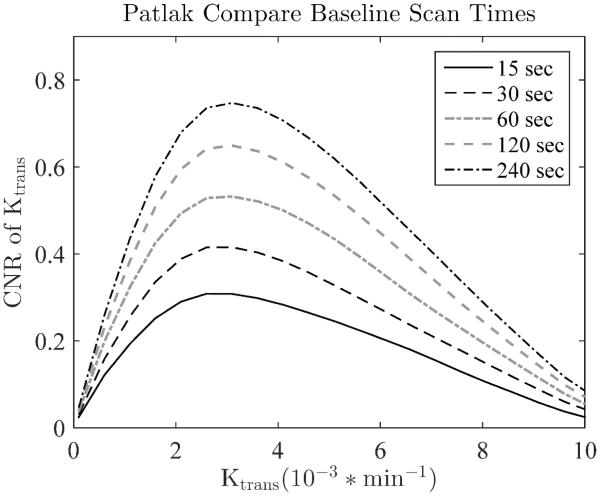
Effects of baseline acquisition duration on Ktrans estimation
The length of the baseline scan was evaluated for its effect on the K-CNR values. From Figure 5, it can be seen that acquiring a longer baseline scan significantly increases the K-CNR of the calculated DCE parameters by reducing the variance of the fit values. Adding as little as 30 seconds to the baseline can increase the K-CNR by approximately 30% (moving from a 30 second to a 60 second baseline).
Figure 5.
The concentration curves for both the AIF and tissue are calculated relative to the baseline images collected before CA injection. Thus increasing the number of baseline images, and therefore baseline SNR, increases the SNR of all calculated values. This is probably the most efficient way to increase the SNR and CNR of the entire DCE study. Temporal resolutions was 15 sec.
Effects of signal drift on Ktrans estimation
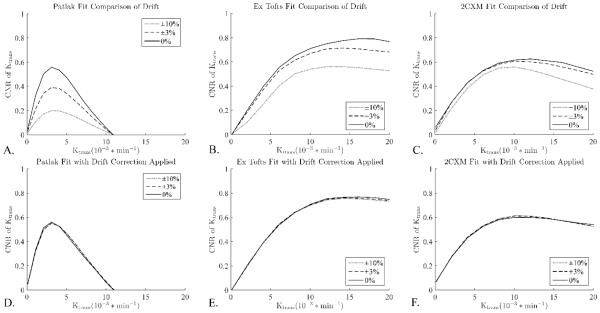
A constant linear drift up or down causes an under or over estimation of Ktrans, respectively. This effect will be consistent in a single study, but across multiple studies variation in the drift will cause variations in the bias it introduces. This will show up as an additional source of variance and uncertainty for inter-study comparisons. To model this, a range of drifts were combined together for analysis. Figures 6 shows the effect of this increased variance on the K-CNR values, the drift causes decreases of K-CNR in all models, especially in the Patlak model (Figure 6A-C). Drift correction using an external intensity standard largely eliminated these discrepancies (Figures 6D-F).
Figure 6.
Drift correction improves precision for inter-study comparison. Patlak is especially sensitive to drift as its lower contrast makes it more sensitive to any increase in variance. Unlike the other variables considered here, signal drift is not a parameter than can be easily controlled or defined a priori. However, it can be largely corrected for using an intensity standard, a small tube of water placed in the FOV whose signal can be used to estimate and remove the drift.
Discussion
The comparison of the measured data to simulations showed excellent correspondence (Figure 1). This shows that the method of simulation curve generation provides realistic DCE curves that are nearly identical to real measured curves in both fitted values and noise characteristics. The fitted Ktrans values from the 2CXM are reflective of more accurate values across the entire Ktrans range; the extended Tofts generally overestimates Ktrans; and Patlak does well at low permeability values (≤5 × 10−3 min−1 ), but underestimates Ktrans at larger values. These findings validate previous studies (6,25).
While this may suggest that one should apply the 2CXM across all Ktrans, it is limited by several factors. The accuracy of the Ktrans values in the extended Tofts and the 2CXM models are strongly affected by the temporal resolution used, likely due to their dependency on accurate perfusion and blood volume estimation (25). Given that 2CXM requires the fitting of four parameters, the potential for curve fitting error is high (25). Past studies examining how other models perform in Ktrans estimation demonstrated that the Patlak model accurately estimates low Ktrans values, consistent with the fact that the model neglects backflux (19,25). To date however, these studies have focused on the accuracy of Ktrans estimations for the respective models. From Figure 2A, we see that Ktrans precision is also important for model selection. Figure 2A shows that, while both 2CXM and Patlak may estimate low Ktrans accurately, the error bounds are much smaller for Patlak. This has practical implications, especially when we wish to differentiate small differences in permeability. In these scenarios we show that Patlak will give a higher K-CNR due to its superior precision, and are able to predict the cross over point where 2CXM or Extended Tofts will start outperforming Patlak. The exact value of the crossover points, and its strong dependence on ve and the acquisition time, has not been shown before. This superior sensitivity has enabled Patlak to detect small regional differences between grey and white matter due to its high precision (20).
The K-CNR metric captures both Ktrans contrast and precision for each model in a single value. This enables facile evaluation of the suitability of different models in estimating Ktrans as a functional of multiple imaging variables. Using the K-CNR metric, we show that the Patlak model clearly outperformed the other two models in the low Ktrans regime due to its high precision (Figure 2). The precision is a direct result of the simplicity of the model, with only two free parameters it is less likely to fit the noise and is therefore very stable. Eventually the high precision is overwhelmed by its Ktrans underestimation at higher Ktrans values, resulting in a sharp drop in K-CNR. While K-CNR offers the ability to easily compare various models, it does not capture all aspects of image quality. In particular it is not the best measure of model accuracy. Despite this shortcoming it is a valuable tool to make clear comparisons across models for a wide variety of parameters.
The sensitivity of the Patlak model to backflux places direct limits on acquisition time, and is dependent on the Ktrans and ve of the tissue being measured. Higher Ktrans values using the Patlak model can be accurately estimated if the acquisition time is kept sufficiently short (Figure 2C). This is generally an acceptable trade off; higher Ktrans does not need long scan times to achieve sufficient K-CNR. However, at higher Ktrans values the other models generally outperform Patlak even if the Patlak acquisition time is sufficiently short. Thus, if the tissue of interest is expected to have high Ktrans then analysis with a different model will generally be more appropriate.
Acquisition times longer than 15 minutes can lead to significant improvement in the K-CNR using the Patlak model, but only for low Ktrans values (Ktrans < 2 * 10−3/min, Figure 3). Long scan times may be beneficial in normal healthy mice where very low Ktrans values have been measured with DCE-MRI using Gd-DTPA (Ki = 0.6±1.9 * 10−3 mL/g/min (53)). However, they should not be necessary in humans, which show higher values in healthy brain ranging from 0.5 – 4.5 * 10−3/min as reported here and elsewhere (19,20,25-28). That scan times of 15 minutes are sufficient is of particular importance in large multicenter trials where extending scan time is extremely difficult.
Generally, the Patlak model shows almost no loss of K-CNR up to a sampling interval of about sixty seconds (Figure 4A). Anything longer leads to a sharp increase in the variance of the fit parameters and a corresponding decrease in K-CNR. However, this K-CNR stability only occurs when the longer sampling interval leads to higher SNR, e.g. if extra partitions are collected to increase the FOV in a 3D scan. If the longer sampling interval does not lead to gains in SNR, e.g. adding more slices to a 2D scan, then K-CNR steadily drops with each decrease in sampling interval (Figure 4B). The use of 3D scans, therefore, gives extra flexibility to increase the FOV and gain more coverage without sacrificing much K-CNR - up to a point. Any increase in FOV in a 2D scan leads to a direct reduction of the K-CNR of the study. It is therefore advisable to use 3D scans whenever possible.
Figure 4A also shows that there is no benefit from running averages during a DCE experiment. The gain in SNR is canceled by the lower sampling interval leading to a net decrease in K-CNR. The extra time points in the short sampling interval scans have a noise reducing effect on fit parameters by adding additional constraints to the fit. Averaging removes these extra constraints and reduces the amount of information extracted from the data curve.
The number of baseline images collected is an acquisition parameter often neglected and rarely optimized in DCE protocols. The number of pre-injection images acquired vary greatly in the literature, from no baseline at all (54), one pre-contrast time point (28), more than 30 seconds (55,56) and a wide time range in between (6,39,57). Too short of a baseline is suboptimal as the concentration curves are calculated partially as the ratio of the pre-injection and post-injection signal intensities. The variance of the ratio will be approximately proportional to the sum of the individual variances, so reducing the variance of the pre-injection scans directly reduces the variance of the concentration curve. This can be a very time efficient way to increase the sensitivity of a study, as adding an extra two minutes to the baseline measurement could potentially double the K-CNR of the study (Figure 5) depending on the initial SNR and acquisition time. Table 1 outlines some typical examples; in some cases it is optimal to spend up to 1/3 of the imaging time collecting the baseline images.
Table 1.
Optimal Baseline Scan Times Differ for High and Low Ktrans
| Total Available Scan Time |
Optimal Baseline Time Ktrans=1e−3/min |
Optimal Baseline Time Ktrans=5e−3/min |
|---|---|---|
| 10 min | 1 min | 2 min |
| 15 min | 3 min | 5 min |
| 30 min | 4 min | >15 min |
| 45 min | 8 min | >30 min |
For a given total scan time, the optimal division of time between pre-contrast (baseline) and post-contrast depends on the tissue Ktrans and the scan duration. Calculated using the Patlak model with ve=0.05. The baseline times are very long for 30-45 minutes and Ktrans=5e−3/min as these scan times extend well beyond the no backflux assumption for the Patlak model, in this situation a different model would give superior results.
Previously, the effects of scanner drift on DCE results were reported to be negligible, due to either ROI averaging of time intensity curves cancelling out those effects, or that the magnitude of the drift is within the uncertainty of Ktrans estimation (25,37). Our results suggest that the effects of signal drift on DCE parameter estimation are significant especially when comparing studies from different scanners or subjects. This is most prominent with Patlak fitting, but is present for all the models examined. Comparing Ktrans derived from curves with different drifts can drastically increase the uncertainty of the Ktrans comparisons (Figure 6), effectively reducing one’s ability to resolve small Ktrans changes. Patlak is especially vulnerable to these types of inter-study errors as its lower contrast is sensitive to increases in variance. Drift correction can essentially eliminate this error. We assume here that the magnitude of the drift across the phantom, tissue and AIF ROIs are the same. In practice, this is likely not the case (58). How this impacts DCE parameter estimation needs to be explored in future studies.
When performing non-linear curve fitting, convergence to a local rather than global minimum is always a concern. This is a significant issue in DCE fitting. In Patlak, which can be linearized, we achieve good results by using the output from a linear fit as the starting location for a non-linear fit. This causes the non-linear fit to converge quickly and reliably giving more accurate values. To avoid local minimum in the other models we use multiple different starting locations, which improves the chances of finding the global minimum. In this regard, the starting value of Ktrans had the largest effect on the final value and it was necessary to try multiple different Ktrans starting values to reliably find the global minimum. It was also quite effective to constrain all the parameters to physiologically realistic values. Specifically, it was helpful to put a lower limit on ve of 1-2%. Without this constraint there was instability at very low Ktrans values; the ve fit tends to fall to zero and the fit Ktrans to rise to relatively high values. This instability at low Ktrans is easily fixed by constraining ve. Other groups have reported difficulty in getting 2CXM to properly converge to accurate values (19). We did not find this to be the case, as our 2CXM and extended Tofts models both converged with both simulated and measured data. Poor convergence has been a criticism of the 2CXM (59), but with proper selection of convergence tolerances, constraints on parameter ranges, and multiple starting locations we have not found this to be an issue.
The values that the 2CXM generates for our measured data in healthy controls largely fell within expected physiological ranges, with the exception of ve. The measured values of ve (3.3% and 5.1% median GM and WM, respectively and 7.4% and 6.4% mean GM and WM, respectively) are significantly lower than the expected value of approximately 15-30% (45) as measured with diffusion techniques. There could be a number of explanations for this difference. First, the measured ve in this study will be lower than the true ve due to the partial albumin binding of the Multihance used in this study. The partial binding to albumin extends the blood half-life of the CA and will cause an underestimation of ve directly proportional to the extent of binding. Unfortunately, it is difficult to estimate the degree that it will be underestimated as the published binding affinities have huge uncertainties so the amount of bound CA could be anywhere from 2%-30% (60,61). Due to the significantly extended blood half-life the binding is probably closer to 30% than 2% and is likely a source of significant underestimation of ve. Second, the extravascular extracellular space sampled by the CA may be different than the diffusion space mentioned above. Restrictions in the diffusion of the CA away from the blood vessels could cause the effective space they inhabit to be smaller than the actual space and give lower ve values. Third, of all four parameters in the 2CXM the curves contain the least amount of information about ve as the low leakage rates do not allow enough time for the extravascular extracellular compartment to fill with CA. Therefore the ve values have a much lower precision than the other parameters, this low precision likely contributes to the low measured values. Finally, deficiencies in measurements techniques, insufficient temporal resolution, or poor curve fitting all are known to bias the DCE values, and it could be that these biases combined with the previously mentioned reasons could cause low ve values to be measured. An accurate measurement of Ktrans should not be taken as an indication that the other fit parameters (ve, vp, and fp) will also be accurate.
The partial albumin binding of Multihance mentioned above could also affect the measured Ktrans values. The magnitude of the change will depend on the binding affinity of Multihance, but as stated above this value is quite ill determined (60,61). Without a reasonable value it is hard to predict if this will be a significant effect. To our knowledge no study has yet examined the differences between different Gd based contrast agents, this could be an important question for future studies.
In conclusion, we have examined the effects of multiple scan parameters and modeling options on the ability to detect small changes in low Ktrans values that can be used to detect subtle changes in BBB permeability. We have defined a K-CNR metric that highlights differences between models. Just as improving image CNR increases our ability to differentiate between regions of interest, improving K-CNR leads to better defined Ktrans values. Using this K-CNR metric, we found that the Patlak model gives the best results in these circumstances when combined with fairly long scan times (10-30 minutes), modest temporal resolution (<60 seconds/image), and long baseline scans (1-4 minutes). These parameters can be used to detect even the subtlest changes in the integrity of the BBB and may be used to probe the earliest changes in neurodegenerative diseases.
Acknowledgement
Funding: This work was funded in part by NIBIB EB000993 (REJ), the Beckman Institute (REJ), R37NS34467 (BVZ), R37AG23084 (BVZ), and R01AG039452 (BVZ).
Bibliography & References Cited
- 1.Chassidim Y, Veksler R, Lublinsky S, Pell GS, Friedman A, Shelef I. Quantitative imaging assessment of blood-brain barrier permeability in humans. Fluids and barriers of the CNS. 2013;10(1):9. doi: 10.1186/2045-8118-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker GJ, Padhani AR. T1-W DCE-MRI: T1-Weighted Dynamic Contrast-Enhanced MRI. Quantitative MRI of the Brain: Measuring Changes Caused by Disease. 2004:341–364. [Google Scholar]
- 3.Hylton N. Dynamic contrast-enhanced magnetic resonance imaging as an imaging biomarker. Journal of clinical oncology. 2006;24(20):3293–3298. doi: 10.1200/JCO.2006.06.8080. [DOI] [PubMed] [Google Scholar]
- 4.Seevinck PR, Deddens LH, Dijkhuizen RM. Magnetic resonance imaging of brain angiogenesis after stroke. Angiogenesis. 2010;13(2):101–111. doi: 10.1007/s10456-010-9174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Connor JP, Jackson A, Parker GJ, Jayson GC. DCE-MRI biomarkers in the clinical evaluation of antiangiogenic and vascular disrupting agents. British journal of cancer. 2007;96(2):189–195. doi: 10.1038/sj.bjc.6603515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingrisch M, Sourbron S, Morhard D, Ertl-Wagner B, Kumpfel T, Hohlfeld R, Reiser M, Glaser C. Quantification of perfusion and permeability in multiple sclerosis: dynamic contrast-enhanced MRI in 3D at 3T. Invest Radiol. 2012;47(4):252–258. doi: 10.1097/RLI.0b013e31823bfc97. [DOI] [PubMed] [Google Scholar]
- 7.Tofts PS, Kermode AG. Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging. 1. Fundamental concepts. Magnetic Resonance in Medicine. 1991;17(2):357–367. doi: 10.1002/mrm.1910170208. [DOI] [PubMed] [Google Scholar]
- 8.Sourbron S, Ingrisch M, Siefert A, Reiser M, Herrmann K. Quantification of cerebral blood flow, cerebral blood volume, and blood–brain-barrier leakage with DCE-MRI. Magnetic Resonance in Medicine. 2009;62(1):205–217. doi: 10.1002/mrm.22005. [DOI] [PubMed] [Google Scholar]
- 9.Kermode A, Tofts P, Thompson A, MacManus D, Rudge P, Kendall B, Kingsley D, Moseley I, Du Boulay E, McDonald W. Heterogeneity of blood-brain barrier changes in multiple sclerosis An MRI study with gadolinium-DTPA enhancement. Neurology. 1990;40(2):229–229. doi: 10.1212/wnl.40.2.229. [DOI] [PubMed] [Google Scholar]
- 10.Larsson HB, Stubgaard M, Frederiksen JL, Jensen M, Henriksen O, Paulson OB. Quantitation of blood-brain barrier defect by magnetic resonance imaging and gadolinium-DTPA in patients with multiple sclerosis and brain tumors. Magn Reson Med. 1990;16(1):117–131. doi: 10.1002/mrm.1910160111. [DOI] [PubMed] [Google Scholar]
- 11.Di Giovanni P, Azlan CA, Ahearn TS, Semple SI, Gilbert FJ, Redpath TW. The accuracy of pharmacokinetic parameter measurement in DCE-MRI of the breast at 3 T. Physics in Medicine and Biology. 2010;55(1):121. doi: 10.1088/0031-9155/55/1/008. [DOI] [PubMed] [Google Scholar]
- 12.Verma S, Turkbey B, Muradyan N, Rajesh A, Cornud F, Haider MA, Choyke PL, Harisinghani M. Overview of dynamic contrast-enhanced MRI in prostate cancer diagnosis and management. AJR American journal of roentgenology. 2012;198(6):1277–1288. doi: 10.2214/AJR.12.8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosen MA, Schnall MD. Dynamic contrast-enhanced magnetic resonance imaging for assessing tumor vascularity and vascular effects of targeted therapies in renal cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13Pt 2(2):770s–776s. doi: 10.1158/1078-0432.CCR-06-1921. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, Folks KD, Guo L, Stockard CR, Fineberg NS, Grizzle WE, George JF, Buchsbaum DJ, Morgan DE, Zinn KR. DCE-MRI detects early vascular response in breast tumor xenografts following anti-DR5 therapy. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging. 2011;13(1):94–103. doi: 10.1007/s11307-010-0320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sourbron SP, Buckley DL. Classic models for dynamic contrast-enhanced MRI. NMR Biomed. 2013;26(8):1004–1027. doi: 10.1002/nbm.2940. [DOI] [PubMed] [Google Scholar]
- 16.Zlokovic B. Cerebrovascular Permeability to Peptides: Manipulations of Transport Systems at the Blood-Brain Barrier. Pharm Res. 1995;12(10):1395–1406. doi: 10.1023/a:1016254514167. [DOI] [PubMed] [Google Scholar]
- 17.Zloković BV, Lipovac MN, Begley DJ, Davson H, Rakić L. Transport of Leucine-Enkephalin Across the Blood-Brain Barrier in the Perfused Guinea Pig Brain. Journal of Neurochemistry. 1987;49(1):310–315. doi: 10.1111/j.1471-4159.1987.tb03431.x. [DOI] [PubMed] [Google Scholar]
- 18.Zlokovic BV, Begley DJ, Chain-Eliash DG. Blood-brain barrier permeability to leucine-enkephalin,d-Alanine2-d-leucine5-enkephalin and their N-terminal amino acid (tyrosine) Brain Research. 1985;336(1):125–132. doi: 10.1016/0006-8993(85)90423-8. [DOI] [PubMed] [Google Scholar]
- 19.Larsson HB, Courivaud F, Rostrup E, Hansen AE. Measurement of brain perfusion, blood volume, and blood-brain barrier permeability, using dynamic contrast-enhanced T(1)-weighted MRI at 3 tesla. Magn Reson Med. 2009;62(5):1270–1281. doi: 10.1002/mrm.22136. [DOI] [PubMed] [Google Scholar]
- 20.Montagne A, Barnes SR, Sweeney MD, Halliday MR, Sagare AP, Zhao Z, Toga AW, Jacobs RE, Liu CY, Amezcua L, Harrington MG, Chui HC, Law M, Zlokovic BV. Blood-Brain Barrier Breakdown in the Aging Human Hippocampus. Neuron. 2015 doi: 10.1016/j.neuron.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starr JM, Wardlaw J, Ferguson K, MacLullich A, Deary IJ, Marshall I. Increased blood-brain barrier permeability in type II diabetes demonstrated by gadolinium magnetic resonance imaging. Journal of neurology, neurosurgery, and psychiatry. 2003;74(1):70–76. doi: 10.1136/jnnp.74.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popescu BO, Toescu EC, Popescu LM, Bajenaru O, Muresanu DF, Schultzberg M, Bogdanovic N. Blood-brain barrier alterations in ageing and dementia. Journal of the neurological sciences. 2009;283(1):99–106. doi: 10.1016/j.jns.2009.02.321. [DOI] [PubMed] [Google Scholar]
- 23.Erickson MA, Banks WA. Blood–brain barrier dysfunction as a cause and consequence of Alzheimer’s disease. Journal of Cerebral Blood Flow & Metabolism. 2013;33(10):1500–1513. doi: 10.1038/jcbfm.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell RD, Zlokovic BV. Neurovascular mechanisms and blood–brain barrier disorder in Alzheimer’s disease. Acta neuropathologica. 2009;118(1):103–113. doi: 10.1007/s00401-009-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cramer SP, Larsson HB. Accurate determination of blood-brain barrier permeability using dynamic contrast-enhanced T1-weighted MRI: a simulation and in vivo study on healthy subjects and multiple sclerosis patients. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2014;34(10):1655–1665. doi: 10.1038/jcbfm.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cramer SP, Simonsen H, Frederiksen JL, Rostrup E, Larsson HB. Abnormal blood-brain barrier permeability in normal appearing white matter in multiple sclerosis investigated by MRI. NeuroImage Clinical. 2014;4:182–189. doi: 10.1016/j.nicl.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taheri S, Gasparovic C, Huisa BN, Adair JC, Edmonds E, Prestopnik J, Grossetete M, Shah NJ, Wills J, Qualls C, Rosenberg GA. Blood-brain barrier permeability abnormalities in vascular cognitive impairment. Stroke. 2011;42(8):2158–2163. doi: 10.1161/STROKEAHA.110.611731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taheri S, Gasparovic C, Shah NJ, Rosenberg GA. Quantitative measurement of blood-brain barrier permeability in human using dynamic contrast-enhanced MRI with fast T1 mapping. Magn Reson Med. 2011;65(4):1036–1042. doi: 10.1002/mrm.22686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchi N, Bazarian JJ, Puvenna V, Janigro M, Ghosh C, Zhong J, Zhu T, Blackman E, Stewart D, Ellis J. Consequences of repeated blood-brain barrier disruption in football players. PloS one. 2013;8(3):e56805. doi: 10.1371/journal.pone.0056805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng TS, Lin AP, Koerte IK, Pasternak O, Liao H, Merugumala S, Bouix S, Shenton ME. Neuroimaging in repetitive brain trauma. Alzheimer's Research & Therapy. 2014;6(1):10. doi: 10.1186/alzrt239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weissberg I, Veksler R, Kamintsky L, Saar-Ashkenazy R, Milikovsky DZ, Shelef I, Friedman A. Imaging Blood-Brain Barrier Dysfunction in Football Players. JAMA neurology. 2014;71(11):1453–1455. doi: 10.1001/jamaneurol.2014.2682. [DOI] [PubMed] [Google Scholar]
- 32.Zlokovic BV. Neurovascular pathways to neurodegeneration in Alzheimer's disease and other disorders. Nat Rev Neurosci. 2011;12(12):723–738. doi: 10.1038/nrn3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starr JM, Farrall AJ, Armitage P, McGurn B, Wardlaw J. Blood-brain barrier permeability in Alzheimer's disease: a case-control MRI study. Psychiatry research. 2009;171(3):232–241. doi: 10.1016/j.pscychresns.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Golob EJ, Su MY. Vascular volume and blood-brain barrier permeability measured by dynamic contrast enhanced MRI in hippocampus and cerebellum of patients with MCI and normal controls. J Magn Reson Imaging. 2006;24(3):695–700. doi: 10.1002/jmri.20669. [DOI] [PubMed] [Google Scholar]
- 35.Bell RD, Winkler EA, Sagare AP, Singh I, LaRue B, Deane R, Zlokovic BV. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron. 2010;68(3):409–427. doi: 10.1016/j.neuron.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deane R, Du Yan S, Submamaryan RK, LaRue B, Jovanovic S, Hogg E, Welch D, Manness L, Lin C, Yu J, Zhu H, Ghiso J, Frangione B, Stern A, Schmidt AM, Armstrong DL, Arnold B, Liliensiek B, Nawroth P, Hofman F, Kindy M, Stern D, Zlokovic B. RAGE mediates amyloid-beta peptide transport across the blood-brain barrier and accumulation in brain. Nature medicine. 2003;9(7):907–913. doi: 10.1038/nm890. [DOI] [PubMed] [Google Scholar]
- 37.Armitage PA, Farrall AJ, Carpenter TK, Doubal FN, Wardlaw JM. Use of dynamic contrast-enhanced MRI to measure subtle blood–brain barrier abnormalities. Magnetic resonance imaging. 2011;29(3):305–314. doi: 10.1016/j.mri.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heye AK, Culling RD, Valdés Hernández MdC, Thrippleton MJ, Wardlaw JM. Assessment of blood–brain barrier disruption using dynamic contrast-enhanced MRI. A systematic review. NeuroImage: Clinical. 2014;6(0):262–274. doi: 10.1016/j.nicl.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker GJM, Roberts C, Macdonald A, Buonaccorsi GA, Cheung S, Buckley DL, Jackson A, Watson Y, Davies K, Jayson GC. Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn Reson Med. 2006;56(5):993–1000. doi: 10.1002/mrm.21066. [DOI] [PubMed] [Google Scholar]
- 40.Sourbron SP, Buckley DL. On the scope and interpretation of the Tofts models for DCE-MRI. Magn Reson Med. 2011;66(3):735–745. doi: 10.1002/mrm.22861. [DOI] [PubMed] [Google Scholar]
- 41.Bender B, Klose U. Cerebrospinal fluid and interstitial fluid volume measurements in the human brain at 3T with EPI. Magn Reson Med. 2009;61(4):834–841. doi: 10.1002/mrm.21915. [DOI] [PubMed] [Google Scholar]
- 42.He X, Yablonskiy DA. Quantitative BOLD: mapping of human cerebral deoxygenated blood volume and oxygen extraction fraction: default state. Magn Reson Med. 2007;57(1):115–126. doi: 10.1002/mrm.21108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leenders KL, Perani D, Lammertsma AA, Heather JD, Buckingham P, Healy MJ, Gibbs JM, Wise RJ, Hatazawa J, Herold S, et al. Cerebral blood flow, blood volume and oxygen utilization. Normal values and effect of age. Brain. 1990;113Pt 1:27–47. doi: 10.1093/brain/113.1.27. [DOI] [PubMed] [Google Scholar]
- 44.Sourbron S, Ingrisch M, Siefert A, Reiser M, Herrmann K. Quantification of cerebral blood flow, cerebral blood volume, and blood-brain-barrier leakage with DCE-MRI. Magn Reson Med. 2009;62(1):205–217. doi: 10.1002/mrm.22005. [DOI] [PubMed] [Google Scholar]
- 45.Sykova E, Nicholson C. Diffusion in brain extracellular space. Physiological reviews. 2008;88(4):1277–1340. doi: 10.1152/physrev.00027.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aerts HJ, Jaspers K, Backes WH. The precision of pharmacokinetic parameters in dynamic contrast-enhanced magnetic resonance imaging: the effect of sampling frequency and duration. Phys Med Biol. 2011;56(17):5665–5678. doi: 10.1088/0031-9155/56/17/013. [DOI] [PubMed] [Google Scholar]
- 47.Kershaw LE, Cheng HL. Temporal resolution and SNR requirements for accurate DCE-MRI data analysis using the AATH model. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2010;64(6):1772–1780. doi: 10.1002/mrm.22573. [DOI] [PubMed] [Google Scholar]
- 48.Lopata RG, Backes WH, van den Bosch PP, van Riel NA. On the identifiability of pharmacokinetic parameters in dynamic contrast-enhanced imaging. Magn Reson Med. 2007;58(2):425–429. doi: 10.1002/mrm.21336. [DOI] [PubMed] [Google Scholar]
- 49.Luypaert R, Sourbron S, Makkat S, de Mey J. Error estimation for perfusion parameters obtained using the two-compartment exchange model in dynamic contrast-enhanced MRI: a simulation study. Phys Med Biol. 2010;55(21):6431–6443. doi: 10.1088/0031-9155/55/21/006. [DOI] [PubMed] [Google Scholar]
- 50.Henderson E, Rutt BK, Lee TY. Temporal sampling requirements for the tracer kinetics modeling of breast disease. Magn Reson Imaging. 1998;16(9):1057–1073. doi: 10.1016/s0730-725x(98)00130-1. [DOI] [PubMed] [Google Scholar]
- 51.Haacke EM, Brown RW, Thompson MR, Venkatesan R. Magnetic resonance imaging: physical principles and sequence design. Wiley; New York: 1999. Fast Imaging in the Steady State; pp. 451–512. [Google Scholar]
- 52.Schafer RW. What is a Savitzky-Golay filter?[lecture notes] Signal Processing Magazine, IEEE. 2011;28(4):111–117. [Google Scholar]
- 53.Ewing JR, Knight RA, Nagaraja TN, Yee JS, Nagesh V, Whitton PA, Li L, Fenstermacher JD. Patlak plots of Gd-DTPA MRI data yield blood-brain transfer constants concordant with those of 14C-sucrose in areas of blood-brain opening. Magn Reson Med. 2003;50(2):283–292. doi: 10.1002/mrm.10524. [DOI] [PubMed] [Google Scholar]
- 54.Bergamino M, Saitta L, Barletta L, Bonzano L, Mancardi GL, Castellan L, Ravetti JL, Roccatagliata L. Measurement of blood-brain barrier permeability with t1-weighted dynamic contrast-enhanced MRI in brain tumors: a comparative study with two different algorithms. ISRN neuroscience. 2013;2013:905279. doi: 10.1155/2013/905279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen HT, Jia G, Shah ZK, Pohar K, Mortazavi A, Zynger DL, Wei L, Yang X, Clark D, Knopp MV. Prediction of chemotherapeutic response in bladder cancer using K-means clustering of dynamic contrast-enhanced (DCE)-MRI pharmacokinetic parameters. Journal of Magnetic Resonance Imaging. 2014;n/a-n/a doi: 10.1002/jmri.24663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dujardin M, Sourbron S, Luypaert R, Verbeelen D, Stadnik T. Quantification of renal perfusion and function on a voxel-by-voxel basis: A feasibility study. Magnetic Resonance in Medicine. 2005;54(4):841–849. doi: 10.1002/mrm.20608. [DOI] [PubMed] [Google Scholar]
- 57.Gill AB, Black RT, Bowden DJ, Priest AN, Graves MJ, Lomas DJ. An investigation into the effects of temporal resolution on hepatic dynamic contrast-enhanced MRI in volunteers and in patients with hepatocellular carcinoma. Phys Med Biol. 2014;59(12):3187–3200. doi: 10.1088/0031-9155/59/12/3187. [DOI] [PubMed] [Google Scholar]
- 58.Smith AM, Lewis BK, Ruttimann UE, Ye FQ, Sinnwell TM, Yang Y, Duyn JH, Frank JA. Investigation of Low Frequency Drift in fMRI Signal. NeuroImage. 1999;9(5):526–533. doi: 10.1006/nimg.1999.0435. [DOI] [PubMed] [Google Scholar]
- 59.Ewing JR, Bagher-Ebadian H. Model selection in measures of vascular parameters using dynamic contrast-enhanced MRI: experimental and clinical applications. NMR Biomed. 2013;26(8):1028–1041. doi: 10.1002/nbm.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Port M, Corot C, Violas X, Robert P, Raynal I, Gagneur G. How to compare the efficiency of albumin-bound and nonalbumin-bound contrast agents in vivo: the concept of dynamic relaxivity. Invest Radiol. 2005;40(9):565–573. doi: 10.1097/01.rli.0000175388.98721.9b. [DOI] [PubMed] [Google Scholar]
- 61.Henrotte V, Vander Elst L, Laurent S, Muller RN. Comprehensive investigation of the non-covalent binding of MRI contrast agents with human serum albumin. Journal of biological inorganic chemistry : JBIC : a publication of the Society of Biological Inorganic Chemistry. 2007;12(6):929–937. doi: 10.1007/s00775-007-0247-5. [DOI] [PubMed] [Google Scholar]