Abstract
Background
The use of antimalarial drugs for prevention and treatment is a major strategy in the prevention of malaria in pregnancy. Although sulphadoxine-pyrimethamine (SP) is currently recommended for intermittent preventive treatment of malaria during pregnancy in Nigeria, previously used drugs for prophylaxis such as chloroquine (CQ) and pyrimethamine are accessible as they are purchased over the counter. This study describes the markers of absence or presence of resistance to quinoline (Pfcrt and Pfmdr 1) and type 1 antifolate antimalarial medicines (Pfdhfr).
Methods
Plasmodium falciparum-positive dried blood spots from pregnant women attending antenatal clinics for the first time during current pregnancy were investigated for the presence of mutations at codons 72–76 of Plasmodium falciparum chloroquine resistance transporter (Pfcrt) gene by real time polymerase chain reaction (PCR) using haplotype-specific probes. PCR followed by sequence analysis was used to identify mutations at codons 86, 184, 1034, 1042 and 1246 of P. falciparum multi-drug resistance-1 (Pfmdr1) gene; and codons 16, 50, 51, 59, 108, 140 and 164 of Pfdhfr gene.
Results
Two haplotypes of Pfcrt (n = 54) were observed: CVMNK 13(24.2%) and CVIET 41 (75.9%) of the samples. The SVMNT haplotype was absent in this population. The Pfmdr1 (n = 28) haplotypes were NYSND 15(53.6%), YYSND 5(17.9%), NFSND 6(21.4%) and YFSND 2(7.1%). The Pfdhfr (n = 15) were ACNCSVI 4(26.7%), and ACICNSVI 1(6.7%) and ACIRNVI 10 (66.7%). The rate of occurrence of Pfcrt 76T, Pfdhfr108N, Pfmdr186Yand184F were 75.9%, 73.3%, 25% and 28.1% respectively. The Pfmdr1 86Y was associated with low parasitaemia (median = 71 parasites/μl, P = 0.024) while Pfcrt 76T was associated with young maternal age (mean 24.1 ± 4.5 years; P = 0.006). The median parasitaemia were similar (P>0.05) in wild and mutant strains of Pfcrt 76, Pfmdr1 184 and Pfdhfr 108. There was no association between gravidity or gestational age of the women and presence of mutations in the Pfcrt, Pfmdr1 or Pfdhfr genes (P>0.05).
Conclusion
Markers of resistance to chloroquine and pyrimethamine were high, whereas cycloguanil-resistance marker was not present in the studied population. The low level of mutations in the Pfmdr1gene indicates likely efficacy of amodiaquine against malaria in pregnancy.
Introduction
The main strategies for the prevention of malaria during pregnancy in Nigeria are by chemoprophylaxis using sulphadoxine-pyrimethamine (SP) and use of long-lasting insecticide treated nets. In spite of the recommendation of two treatment doses of SP for intermittent preventive treatment of malaria during pregnancy[1], other antimalarial medicines such as CQ, pyrimethamine and proguanil are still used in many antenatal clinics. A wide range of un-recommended antimalarial medicines are available and sold over the counter in medicine stores in Nigeria and are accessible to multigravid pregnant women who continued with medications used during their previous pregnancies. Healthcare providers also prescribed un-recommended antimalarials due to poor knowledge on best practices, low confidence on the safety of SP, and challenges in the institution of on-the-job supervision by stakeholders, such as the Ministry of Health, that ensures compliance with recommend malaria in pregnancy guidelines.
Surveillance of antimalarial drug resistance markers in Plasmodium falciparum populations is an important tool in attempts aimed at predicting the level of resistance to antimalarial drugs. Mutations in specific genes of Plasmodium falciparum that confer resistance to antimalarial drugs are selected by sustained drug pressure [2]. The emergence and spread of drug resistance depends, in part, on the number of mutations required to encode resistance and their effects on parasite fitness [3]. Specific multiple point mutations in a gene make up a resistance marker for an antimalarial drug.
The level of susceptibility of P. falciparum strains to quinoline antimalarial drugs such as CQ, amodiaquine, lumefantrine and mefloquine has been attributed to mutations in Pfcrt and Pfmdr1 genes [4,5]. Resistance to type 1 antifolates (pyrimethamine, chlorproguanil, trimethoprim) has been associated with amino acid substitutions in the Pfdhfr gene while resistance to type II antifolates (sulfonamides: sulfadoxine and dapsone) are associated with mutations in P. falciparum dihydropteroate synthase (Pfdhps) gene [6,7,8].
The protective efficacy of SP during pregnancy in Lagos, Nigeria has been reported in a longitudinal study [9], but for ethical reasons, the efficacy of non-recommended antimalarial drugs in pregnancy can only be monitored by the elucidation of markers suggestive of resistance to these medicines. We describe here, single nucleotide polymorphisms (SNP) in Pfcrt, Pfmdr1 and Pfdhfr genes of P. falciparum isolates from asymptomatic pregnant women in Lagos, Nigeria, in an attempt to make some inference on the efficacy of SP and non-recommended antimalarials based on the presence or absence of resistance markers.
Methods
Study samples and area
Mutations in Pfcrt, Pfmdr1and Pfdhfr genes were investigated in 54 dried blood spots from pregnant women positive for falciparum malaria by microscopy but asymptomatic for malaria. All the women were residents of Lagos metropolis who were recruited on their first antenatal visit to 2 hospitals in central and east zones of Lagos state. The study population was part of the base-line study population that was followed up in a parasitological assessment of pregnant women receiving SP in Lagos, Nigeria [9].
The study was conducted in accordance with principles enshrined in the 1964 Declaration of Helsinki as amended as well as provisions encapsulated by the Nigeria Health Research Ethics Code for research involving human participants. Essentially, the aim and procedures for the research was explained to them with provision of willingness to participate or withdraw at any point of the study without affecting the standard care they should receive in the health facilities. Thus, they gave written informed consent to participate using the template approved by the Ethics Committee before the commencement of the study. Enrolment questionnaires and consent documents were reviewed alongside with the protocol submitted to the Ethics Committee. Following National Guideline on the Prevention of Malaria in Pregnancy, pregnant women at booking at the second trimester after first movement of the foetus has been noticed were given standard dose of SP. All consent documents were storedseparately from other study tools in an assess-controlled cabinet. The study protocol was approved by Ethics Committees of the Nigerian Institute of Medical Research, Lagos and College of Medicine of the University of Lagos, Lagos, Nigeria.
Lagos state is situated in the Southwest region of Nigeria bordered by the Atlantic Ocean. Transmission of malaria is moderate and stable in Lagos but peaks during the wet season corresponding to increase in the population of mosquitoes. Lagos has been described as mesoendemic during the dry season [10].
Malaria parasite DNA extraction from dried blood spots
A section of blood spot was cut directly into a 1ml 96 well plate using a sterile Harris Uni-Core hole punch (~3mm diameter). 1mL 0.5% saponin in 1x phosphate buffered saline (PBS) was added into the well and incubated overnight at 37°C to release haemoglobin, leaving parasite DNA on the paper. After a brief centrifugation (4000 rpm for 5 mins) in a plate centrifuge (Eppendorf Centrifuge 5810R) the saponin solution and debris are removed using a vacuum pump. Then 1mL 1x PBS was used to wash the paper twice. 150μL 6% chelex suspension was added to the 96 well plate and plate covered with foil and heat sealed. The plate was incubated in a water bath at 100°C for 25 mins to release parasite DNA from paper. The plate was centrifuged in a plate centrifuge for 2 mins to spin down chelex. The DNA supernatant (~100μL) was transferred to a new plate and stored at -20°C. [11].
Amplification of the Pfcrt gene
A multiplex real-time polymerase chain reaction (RT-PCR) assay using the Rotorgene 3000 platform (Corbett Research, Australia) was used to genotype the Pfcrt gene as described by Sutherland et al. [12]. Briefly, Pfcrt DNA was amplified from each sample using primers Pfcrt F (TGG TAA ATG TGC TCA TGT GTT T) and Pfcrt R (AGT TTC GGA TGT TAC AAA ACT ATA GT) in the presence of each of the three double-labelled probes, TaqMan probes (MWG, Germany), representing the wild-type and the two most common resistance-associated haplotypes at codons 72–76 of Pfcrt. The probes were CVMNK (50FAM-TGT GTA ATG AAT AAA ATT TTT GCT AA-BHQ1), CVIET (50JOE-TGT GTA ATT GAA ACA ATT TTT GCT AA-BHQ1), and SVMNT (50ROX-AGT GTA ATG AAT ACA ATT TTT GCT AA-BHQ2). The reaction conditions were 95°C for 6 minutes; (95°C for 15 seconds, 55°C for 1 minute) x45 cycles. The different fluorescent molecules enabled the detection of the haplotypes present in each sample. Samples were considered positive for a particular genotype if a CT value of 35 cycles or fewer was obtained in at least two independent PCR experiments. The sequence-specific positive control DNA samples, 3D7 (CVMNK), Dd2 (CVIET), and 7G8 (SVMNT) obtained from the Malaria Research and Reference Reagent Resource (MR4, Manassas, Vermont, USA). Nuclease-free water was used as a negative control.
Amplification of Pfmdr1 and Pfdhfr gene fragments
Amplification of the Pfmdr1 gene was performed in three fragments (FR 1, FR 2 and FR 3). The first fragment contained codons 86 and 184; second fragment contained codons 1034 and 1042; and third fragment contained codon 1246. The reaction primers and conditions for FR1, FR2 and FR3 were as described by Humphrey et al. [13]. The following primers and cycling conditions were used. FR 1: Primary reaction primers F: AGGTTGAAAAAGAGTTGAAC and R: ATGACACCACAAACATAAAT; reaction conditions were 94°C for 3 minutes/[94°C for 30 seconds, 45°C for 1 minute, 72°C for 1 minute] x 30 cycles/ 72°C for 5 minutes. The nested reaction primers were F: ACAAAAAGAGTACCGCTGAAT and R: AAACGCAAGTAATACATAAAGTC; reaction condition were the same as primary PCR. FR 2 primers for the primary reaction were F: GCATTTTATAATATGCATACTG and R: GGATTTCATAAAGTCATCAAC; reaction conditions were 94°C for 3 minutes/ [94°C for 30 seconds, 55°C for 1 minute/ 65°C for 40seconds] x 30 cycles/ 65°C for 5minutes/ 15°C for 5 minutes. The nested reaction primers were F: GGTTTAGAAGATTATTTCTGTAA and R: GGATTTCATAAAGTCATCAAC; reaction conditions were the same as the primary reaction. FR3 primary reaction primers were F: CAAACCAATCTGGATCTGCAGAAG and R: CAATGTTGCATCTTCTCTTCC; the reaction conditions were 94°C for 3 minutes/ [94°C for 30 seconds, 56°C for 1 minute, 65°C for 50 seconds] x 30cycles/ 65°C for 5minutes/ 15°C for 5 minutes. The nested reaction primers were F: GATCTGCAGAAGATTATACTG and R: CAATGTTGCATCTTCTCTTCC; the reaction conditions were the same as primary reaction.
The primers used for sequencing of the three fragments were: FR 1 (F: ACAAAAAGAGTACCGCTGAAT and R: AAACGCAAGTAATACATAAAGTC), FR 2 (F: GGTTTAGAAGATTATTTCTGTAA and R: GGATTTCATAAAGTCATCAAC) and FR 3 (F: GATCTGCAGAAGATTATACTG and R: CAATGTTGCATCTTCTCTTCC). The sequencing reaction conditions were 96°C 1 minute [96°C 30 seconds-50°C-60°C 4 minutes] × 26 cycles.
Amplification of Pfdhfr gene involved primers and cycling conditions described by other workers [14,15]. In brief, parasite DNA extracts from blood spots were amplified in a nested polymerase chain reaction. Primary reaction (650 bp) primers were F_Pfdhfr_M1 TTT ATG ATG GAA CAA GTC TGC and R_Pfdhfr_M7 CTA GTA TAT ACA TCG CTA ACA. The reaction conditions were 93°C for 5 5 minutes, [94°C for 30seconds, 54°C for 60seconds, 65°C for 60seconds] x 41cycles, 65°C for 5 minutes, 15°C for 5 minutes. Nested reaction (594 bp) primers were F_Pfdhfr_M3 TGA TGG AAC AAG TCT GCG ACG TT and R_Pfdhfr_M9 CTG GAA AAA ATA CAT CAC ATT CAT ATG. The reaction conditions were 95°C for 5 minutes, [93°C for 30 seconds, 56°C for 30 seconds, 68°C for 75 seconds] x 30 cycles, 75°C for 5 minutes, 4°C hold.
All amplicons of the Pfmdr1 and Pfdhfrgenes were re-amplified in a nested PCR step. The PCR products of nested reactions were separated by gel electrophoresis on a 1.2% agarose gel stained with ethidium bromide to identify amplified bands of DNA under ultra-violet illumination. Amplicons from nested PCR reactions were purified using the QIAquick PCR Purification Kit (QIAGEN, UK) according to manufacturer’s instructions and subjected to di-deoxy fluorescent sequencing (BigDye 3.1, Applied Biosystems, UK) using conditions and sequencing primer pairs described elsewhere (Pearce et al., 2003). The sequence of amplified DNA products was determined using ABI PRISM 3730 Genetic Analyser (Applied Biosystems, UK). ChromasLite® (Technelysium, Australia) was used to analyse the sequence results. The DNA base sequence was converted to peptide sequences using a web-based six frame nucleotide to peptide sequence conversion tool, Transeq® (http://www.ebi.ac.uk/Tools/emboss/transeq/index.html). Detection of single nucleotide polymorphism (SNP) was carried out by aligning the sample protein sequences with reference protein sequence of the Pfmdr1 and Pfdhfr of standard 3D7 (wild) strain of P. falciparum using the online multiple alignment tool, ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Data analysis
The data generated from the study were analyzed using EPIINFOTM Version 3.5.1 statistical software (Center for Disease Control, USA). Frequency tables were generated for resistance markers and haplotypes of genes studied. Comparison of key mutations in the genes versus age, gravidity and gestational age of the women was tested by analysis of variance. Associations between parasitaemia and key mutations were tested for significance by the Kruskal Wallis test.
Results
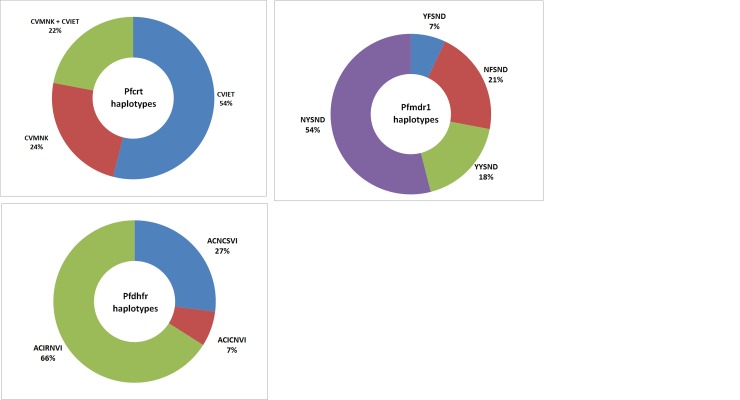
The haplotype of Pfcrt gene was defined by the predicted protein sequence in codons 72–76: CVMNK haplotype is the wild strain while CVIET and SVMNT haplotypes are mutant strains. Thirteen (24.1%) of the 54 evaluable samples had CVMNK strains only, 29(53.7%) had CVIET strains only, while 12(22.2%) were mixed with CVMNK and CVIET. Thus, the mutant strains of P. falciparum were present in 41 (75.9%) of the samples. The SVMNT haplotype of Pfcrt was not found in any of the samples (Fig 1).
Fig 1. Haplotypes of Pfcrt, Pfmdr1 and Pfdhfr genes of P. falciparum isolates from pregnant women.
Pfcrt (n = 54): CVMNK (wild), CVIET (mutant); Pfmdr1 (n = 28): NYSND (wild), YYSND (single mutant), NFSND (single mutant), YFSND (double mutant); Pfdhfr (n = 15): ACNCSVI (wild), ACICNVI (double mutant), ACIRNVI (triple mutant).
The Pfmdr1 haplotype was defined by the protein sequence at codons 86, 184, 1034, 1042 and 1246 for 28 individuals. The NYSND haplotype (wild) had the highest frequency 15(53.6%). Two types of single mutant haplotypes were observed, YYSND 5(17.9%) and NFSND 6(21.4%). The double mutant haplotype YFSND had the least frequency 2(7.1%) (Fig 1).
The Pfdhfr gene haplotype was defined by the protein sequence at codons 16, 50, 51, 59, 108 and 164 for 15 individuals. ACIRNVI (triple mutant) had the highest frequency, 10 (66.7%). The frequency of ACNCSVI (wild) and ACICNSVI (double mutant) were 4(26.7%) and 1(6.7%) respectively Mutations were not observed at codons 16, 50 and 164 in this study. (Fig 1).
The rate of occurrence of key single nucleotide polymorphisms in Pfcrt (76T) and Pfdhfr (108N) were 75.9% and 73.3% respectively. Single nucleotide polymorphisms at codons 86 (86Y) and 184 (184F) of Pfmdr1geneoccurred at the rate of 25% and 28.1% respectively.
There was no statistically significant relationship between median parasitaemia (parasites/μl) and genotype for Pfcrt K76T, Pfmdr1 Y184F, and Pfdhfr S108N, but Pfmdr1 N86Y was associated with a lower parasitaemia than in infections harbouring wild-type N86 (Table 1). The mutant haplotypes of Pfcrt K76T, Pfmdr1 Y184F and Pfmdr1 N86Y occurred more in pregnant women with lower mean gestational age but that was not the case for Pfdhfr S108N. Furthermore, mutations in Pfcrt K76T and Pfmdr1N86Y were more common in women with higher gravidity. However, there was no association between gravidity or gestational age of the women and presence of mutations in the Pfcrt, Pfmdr1 or Pfdhfr genes (P>0.05) (Tables 2 and 3).
Table 1. Relationship between parasitaemia (parasites/μl) and mutations in Pfcrt and Pfmdr1 and Pfdhfr genes.
| Gene | Strain | n | Median Parasitaemia | Interquartile range | Kruskal-Wallis | P |
|---|---|---|---|---|---|---|
| Pfcrt K76T | Wild (76K) | 13 | 127 | 81–527 | 0.904 | 0.342 |
| Mutant (76T) | 41 | 444 | 100–1664 | |||
| Total | 54 | 364 | 88–1680 | |||
| Pfmdr1 N86Y | Wild (86N) | 21 | 476 | 176–2814 | 5.084 | 0.024 |
| Mutant (86Y) | 7 | 71 | 40–475 | |||
| Total | 28 | 400 | 77–2442 | |||
| Pfmdr1 Y184F | Wild (184Y) | 20 | 352.5 | 74–1922 | 0.372 | 0.542 |
| Mutant (184F) | 8 | 947.5 | 128–3197 | |||
| Total | 28 | 400 | 77–2442 | |||
| Pfdhfr S108N | Wild (108S) | 4 | 294.5 | 45–1096 | 0.273 | 0.602 |
| Mutant (108N) | 11 | 149 | 84–1419 | |||
| Total | 15 | 149 | 76–1419 |
Table 2. Relationship between gestational age (months) and mutations in Pfcrt and Pfmdr1 and Pfdhfr genes.
| Gene | Strain | n | Mean gestational age | SD | F | P |
|---|---|---|---|---|---|---|
| Pfcrt K76T | Wild (76K) | 13 | 5.2 | 1.8 | 1.205 | 0.277 |
| Mutant (76T) | 41 | 4.6 | 1.6 | |||
| Total | 54 | 4.7 | 1.6 | |||
| Pfmdr1 N86Y | Wild (86N) | 21 | 5.3 | 1.8 | 1.852 | 0.185 |
| Mutant (86Y) | 7 | 4.3 | 1.1 | |||
| Total | 28 | 5 | 1.7 | |||
| Pfmdr1 Y184F | Wild (184Y) | 20 | 5.2 | 1.6 | 0.637 | 0.432 |
| Mutant (184F) | 8 | 4.6 | 2 | |||
| Total | 28 | 5 | 1.7 | |||
| Pfdhfr S108N | Wild (108S) | 4 | 3.8 | 2.4 | 3.047 | 0.104 |
| Mutant (108N) | 11 | 5.7 | 1.8 | |||
| Total | 15 | 5.2 | 2.1 |
Table 3. Relationship between gravidity and mutations in Pfcrt, Pfmdr1 and Pfdhfr genes.
| Gene | Strain | n | Mean gravidity | SD | F | P |
|---|---|---|---|---|---|---|
| Pfcrt K76T | Wild (76K) | 13 | 1.6 | 1.2 | 1.286 | 0.262 |
| Mutant (76T) | 41 | 2.1 | 1.2 | |||
| Total | 54 | 1.9 | 1.2 | |||
| Pfmdr1 N86Y | Wild (86N) | 21 | 2 | 1.3 | 0.525 | 0.475 |
| Mutant (86Y) | 7 | 2.4 | 1.5 | |||
| Total | 28 | 2.1 | 1.3 | |||
| Pfmdr1 Y184F | Wild (184Y) | 20 | 2.2 | 1.3 | 0.069 | 0.795 |
| Mutant (184F) | 8 | 2 | 1.5 | |||
| Total | 28 | 2.1 | 1.3 | |||
| Pfdhfr S108N | Wild (108S) | 4 | 2.8 | 1.3 | 1.682 | 0.217 |
| Mutant (108N) | 11 | 2 | 0.9 | |||
| Total | 15 | 2.2 | 1 |
Pfcrt K76T mutant occurred more in younger pregnant women compared to the wild-type (P = 0.006). However, the occurrence of the mutant and wild forms of Pfmdr1N86Y, Pfmdr184F and Pfdhfr S108N was similar irrespective of the age of the pregnant women (Table 4).
Table 4. Relationship between age (years) and mutations in Pfcrt and Pfmdr1 and Pfdhfr genes.
| Gene | Strain | n | Mean Age | SD | F | P |
|---|---|---|---|---|---|---|
| Pfcrt K76T | Wild (76K) | 13 | 28.1 | 5 | 8.196 | 0.006 |
| Mutant (76T) | 41 | 24.1 | 4.5 | |||
| Total | 54 | 25.1 | 4.9 | |||
| Pfmdr1 N86Y | Wild (86N) | 21 | 23.7 | 3.8 | 3.077 | 0.091 |
| Mutant (86Y) | 7 | 26.9 | 5.3 | |||
| Total | 28 | 24.5 | 4.3 | |||
| Pfmdr1 Y184F | Wild (184Y) | 20 | 25.4 | 4.4 | 3.592 | 0.069 |
| Mutant (184F) | 8 | 22.1 | 3.2 | |||
| Total | 28 | 24.5 | 4.3 | |||
| Pfdhfr S108N | Wild (108S) | 4 | 24.3 | 4.8 | 0.062 | 0.807 |
| Mutant (108N) | 11 | 24.9 | 4.4 | |||
| Total | 15 | 24.7 | 4.4 |
Discussion
This is the first study to report on antimalarial drug resistance markers in P. falciparum isolates from pregnant women in Nigeria. The high rate of mutant haplotypes of Pfcrt is an indication of sustained CQ pressure in Lagos. This suspicion is supported by the observation in the phase one results of this study that 20% of the pregnant women had taken chloroquine prophylaxis before booking at the various antenatal clinics [16]. High frequency (82.3%) of the mutant form of Pfcrt gene has also been observed in pregnant women taking chloroquine prophylaxis in Senegal [17]. High frequencies of mutant forms of Pfcrt, similar to findings of this study among pregnant women, have been reported by other studies in pre-treatment P. falciparum isolates from children in Southwest Nigeria: 74.0% in Oshogbo, Osun State[18] and 78% in Ibadan, Oyo state [4].
The findings of this study suggests that the use of CQ in this area of Nigeria for either chemoprophylaxis or treatment of malaria during pregnancy may not be effective and should be actively discouraged because it maintains the observed strong selection at the Pfcrt locus [17,19]. In Swaziland, CVIET haplotype increased in frequency from 70% in 1999 to 83% in 2007 in the presence of sustained chloroquine use [20]. The withdrawal of CQ from the open market should be considered to restrict access to CQ. Re-emergence of CQ-sensitive parasites with CVMNK haplotype of Pfcrt gene has been reported following withdrawal of CQ some malaria-endemic countries: Malawi [21,22], Gabon [23], Brazil [24] and China [25]. The absence of SVMNT haplotype observed in this study is consistent with findings of other workers that CVIET is the central determinant of chloroquine resistance in West Africa [26].
Amodiaquine has been reported to select the mutant Pfmdr1 86Y in The Gambia [27], Kenya [28] and Nigeria [4]. Thus, the low level of Pfmdr1 Y86 in this study (25.0%) suggests that the parasites would still be sensitive to amodiaquine for some time. Therapeutic use of drugs such as artesunate-amodiaquine combination in pregnancy is expected to still be effective in Lagos. Moreover, Lekana-Douki et al., [29] reported an association between increase in Pfmdr1 86N and introduction of artemisinin-based combination therapies. In Nigeria, ACTs have been in use since 2005 though not on a large-scale at inception. Molecular surveillance is therefore recommended to monitor the Pfmdr1 haplotypes.
Selection for mefloquine resistance has been associated with a decreased resistance to CQ and an amplification of the Pfmdr1 gene copy numbers. In areas subjected to mefloquine pressure, wild-type Pfmdr1 86N was found more frequently [30]. Although the Pfmdr186N was present in 75% of the isolates in this study, a concrete statement on the resistance cannot be made because the Pfmdr1 gene copy numbers was not determined.
The lack of association between gravidity and mutations in Pfcrt, Pfmdr1 and Pfdhfr codons studied shows that the gravidity of the women did not exert a significant selective pressure. This is consistent with the report that the immune system of semi-immune individuals is able to clear both drug-resistant and sensitive parasites [31].
The Pfmdr1 86Y occurred more at very low parasitaemia. This phenomenon was however not observed with Pfmdr1 and pfdhfr. This report is at variance with the report of Tukwasibwe et al.[32] that in children mutant type malaria parasites were associated with lower virulence, occurring more in asymptomatic children. The immunological differences between children and pregnant women might explain the difference in the findings. However, a major difficulty for our study was the frequent failure to obtain high quality PCR products from DNA sample from our asymptomatic patients, all of whom has low peripheral parasitaemia. This lead to a great reduction in our sample size for some analyses, and future studies should focus on obtaining larger sample sets, to generate more accurate estimates of marker prevalence.
The triple mutant ACIRNVI haplotype of the Pfdhfr gene, which has been reported to be associated with pyrimethamine resistance [33,34], had a high frequency in this population studied. Thus, antimalarial chemoprophylaxis with pyrimethamine in Lagos is not advised. Key mutations such as 16V + 108T and 164L [6,35] associated with high resistance to cycloguanil, the active form of proguanil, were not observed in this study.
Conclusions
High levels of CQ-resistant haplotypes of Pfcrt gene and pyrimethamine-resistant haplotypes of Pfdhfr gene were observed in P. falciparum isolates from pregnant women in Lagos. However, cycloguanil-resistance conferring mutations were not observed in the Pfdhfr gene. The low level of mutations in the Pfmdr1gene suggests the likely efficacy of amodiaquine against malaria in pregnancy, but future, larger studies are required to confirm this.
Acknowledgments
We thank the management and staff of the health facilities where the pregnant women attending Antenatal Clinics were enrolled for this study. Furthermore, we thank the study participants for their consent to participate. We are also grateful for the Laboratory of Colin Sutherland, London School of Tropical Medicine and Hygiene, London, United Kingdom, that provided the platform for the molecular elucidation of the isolates and for hosting Chimere Agomo.
Data Availability
The data for this study (accession number—doi:10.5061/dryad.b3f50), are available in the Dryad Digital Repository: http://datadryad.org/review?doi=doi:10.5061/dryad.b3f50.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Federal Ministry of Health (2005) National guidelines and strategies for malaria prevention and control during pregnancy Federal Ministry of Health, Abuja, Nigeria: pp. 50. [Google Scholar]
- 2.White NJ (2003) Malaria; Cook GC, Zumla A, editors. Edinburgh, United Kingdom: Elsevier Science Limited. 1244 p. [Google Scholar]
- 3.White N (1999) Antimalarial drug resistance and combination chemotherapy. Philosophical transactions of the Royal Society of London Series B, Biological sciences 354: 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Happi CT, Gbotosho GO, Folarin OA, Bolaji OM, Sowunmi A, Kyle DE, et al. (2006) Association between mutations in Plasmodium falciparum chloroquine resistance transporter and P. falciparum multidrug resistance 1 genes and in vivo amodiaquine resistance in P. falciparum malaria-infected children in Nigeria. The American journal of tropical medicine and hygiene 75: 155–161. [PubMed] [Google Scholar]
- 5.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC, et al. (2000) The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Molecular and biochemical parasitology 108: 13–23. [DOI] [PubMed] [Google Scholar]
- 6.Mkulama MA, Chishimba S, Sikalima J, Rouse P, Thuma PE, Mharakurwa S, et al. (2008) Escalating Plasmodium falciparum antifolate drug resistance mutations in Macha, rural Zambia. Malaria journal 7: 87 10.1186/1475-2875-7-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hyde JE (1990) The dihydrofolate reductase-thymidylate synthetase gene in the drug resistance of malaria parasites. Pharmacology & therapeutics 48: 45–59. [DOI] [PubMed] [Google Scholar]
- 8.Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, et al. (2002) Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. The Journal of infectious diseases 185: 380–388. [DOI] [PubMed] [Google Scholar]
- 9.Agomo CO, Oyibo WA, Odukoya-Maije F (2011) Parasitologic Assessment of Two-Dose and Monthly Intermittent Preventive Treatment of Malaria during Pregnancy with Sulphadoxine-Pyrimethamine (IPTP-SP) in Lagos, Nigeria. Malaria research and treatment 2011: 932895 10.4061/2011/932895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aina OO, Agomo CO, Olukosi YA, Okoh HI, Iwalokun BA, Egbuna KN, et al. (2013) Malariometric survey of ibeshe community in ikorodu, lagos state: dry season. Malaria research and treatment 2013: 487250 10.1155/2013/487250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE (1995) Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. The American journal of tropical medicine and hygiene 52: 565–568. [DOI] [PubMed] [Google Scholar]
- 12.Sutherland CJ, Haustein T, Gadalla NB, Armstrong M, Doherty JF, Chiodini PL, et al. (2007) Chloroquine-resistant Plasmodium falciparum infections among UK travellers returning with malaria after chloroquine prophylaxis. Journal of Antimicrobial Chemotherapy 59: 1197–1199. [DOI] [PubMed] [Google Scholar]
- 13.Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJ, Mutabingwa TK, Sutherland CJ, et al. (2007) Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrobial agents and chemotherapy 51: 991–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutherland CJ, Fifer H, Pearce RJ, bin Reza F, Nicholas M, Haustein T, et al. (2009) Novel pfdhps haplotypes among imported cases of Plasmodium falciparum malaria in the United Kingdom. Antimicrobial agents and chemotherapy 53: 3405–3410. 10.1128/AAC.00024-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearce RJ, Drakeley C, Chandramohan D, Mosha F, Roper C (2003) Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of northern Tanzania. Antimicrobial agents and chemotherapy 47: 1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agomo CO, Oyibo WA, Anorlu RI, Agomo PU (2009) Prevalence of malaria in pregnant women in Lagos, South-West Nigeria. The Korean journal of parasitology 47: 179–183. 10.3347/kjp.2009.47.2.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertin G, Ndam NT, Jafari-Guemouri S, Fievet N, Renart E, Sow S, et al. (2005) High prevalence of Plasmodium falciparum pfcrt K76T mutation in pregnant women taking chloroquine prophylaxis in Senegal. The Journal of antimicrobial chemotherapy 55: 788–791. [DOI] [PubMed] [Google Scholar]
- 18.Ojurongbe O, Ogungbamigbe TO, Fagbenro-Beyioku AF, Fendel R, Kremsner PG, Kun JF. (2007) Rapid detection of Pfcrt and Pfmdr1 mutations in Plasmodium falciparum isolates by FRET and in vivo response to chloroquine among children from Osogbo, Nigeria. Malaria journal 6: 41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niang M, Marrama L, Ekala MT, Alioune G, Tall A, Ndiaye JL, et al. (2008) Accumulation of CVIET Pfcrt allele of Plasmodium falciparum in placenta of pregnant women living in an urban area of Dakar, Senegal. The Journal of antimicrobial chemotherapy 62: 921–928. 10.1093/jac/dkn299 [DOI] [PubMed] [Google Scholar]
- 20.Dlamini SV, Beshir K, Sutherland CJ (2010) Markers of anti-malarial drug resistance in Plasmodium falciparum isolates from Swaziland: identification of pfmdr1-86F in natural parasite isolates. Malaria journal 9: 68 10.1186/1475-2875-9-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, et al. (2003) Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. The Journal of infectious diseases 187: 1870–1875. [DOI] [PubMed] [Google Scholar]
- 22.Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, et al. (2006) Return of chloroquine antimalarial efficacy in Malawi. The New England journal of medicine 355: 1959–1966. [DOI] [PubMed] [Google Scholar]
- 23.Schwenke A, Brandts C, Philipps J, Winkler S, Wernsdorfer WH, Kremsner PG. (2001) Declining chloroquine resistance of Plasmodium falciparum in Lambarene, Gabon from 1992 to 1998. Wiener klinische Wochenschrift 113: 63–64. [PubMed] [Google Scholar]
- 24.Gama BE, de Oliveira NK, Zalis MG, de Souza JM, Santos F, Daniel-Ribeiro CT, et al. (2009) Chloroquine and sulphadoxine-pyrimethamine sensitivity of Plasmodium falciparum parasites in a Brazilian endemic area. Malaria journal 8: 156 10.1186/1475-2875-8-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Mu J, Li G, Chen P, Guo X, Fu L, et al. (2005) Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People's Republic of China. The American journal of tropical medicine and hygiene 72: 410–414. [PubMed] [Google Scholar]
- 26.Mita T, Tanabe K, Kita K (2009) Spread and evolution of Plasmodium falciparum drug resistance. Parasitology international 58: 201–209. 10.1016/j.parint.2009.04.004 [DOI] [PubMed] [Google Scholar]
- 27.Duraisingh MT, Drakeley CJ, Muller O, Bailey R, Snounou G, Targett GA, et al. (1997) Evidence for selection for the tyrosine-86 allele of the pfmdr 1 gene of Plasmodium falciparum by chloroquine and amodiaquine. Parasitology 114 (Pt 3): 205–211. [DOI] [PubMed] [Google Scholar]
- 28.Holmgren G, Gil JP, Ferreira PM, Veiga MI, Obonyo CO, Bjorkman A. (2006) Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr1 86Y. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases 6: 309–314. [DOI] [PubMed] [Google Scholar]
- 29.Lekana-Douki JB, Dinzouna Boutamba SD, Zatra R, Zang Edou SE, Ekomy H, Bisvigou U, et al. (2011) Increased prevalence of the Plasmodium falciparum Pfmdr1 86N genotype among field isolates from Franceville, Gabon after replacement of chloroquine by artemether-lumefantrine and artesunate-mefloquine. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases 11: 512–517. 10.1016/j.meegid.2011.01.003 [DOI] [PubMed] [Google Scholar]
- 30.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF (2000) Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403: 906–909. [DOI] [PubMed] [Google Scholar]
- 31.Bloland P (2001) Drug resistance in malaria Geneva, Switzerland: World Health Organization. [Google Scholar]
- 32.Tukwasibwe S, Mugenyi L, Mbogo GW, Nankoberanyi S, Maiteki-Sebuguzi C, Joloba ML, et al. (2014) Differential prevalence of transporter polymorphisms in symptomatic and asymptomatic falciparum malaria infections in Uganda. The Journal of infectious diseases 210: 154–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menegon M, Pearce RJ, Inojosa WO, Pisani V, Abel PM, Matondo A, et al. (2009) Monitoring for multidrug-resistant Plasmodium falciparum isolates and analysis of pyrimethamine resistance evolution in Uige province, Angola. Tropical medicine & international health: TM & IH 14: 1251–1257. [DOI] [PubMed] [Google Scholar]
- 34.Ndiaye D, Daily JP, Sarr O, Ndir O, Gaye O, Mboup S et al. (2005) Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase genes in Senegal. Tropical medicine & international health: TM & IH 10: 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson DS, Milhous WK, Wellems TE (1990) Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proceedings of the National Academy of Sciences of the United States of America 87: 3018–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data for this study (accession number—doi:10.5061/dryad.b3f50), are available in the Dryad Digital Repository: http://datadryad.org/review?doi=doi:10.5061/dryad.b3f50.