Abstract
The Core Binding Factor (CBF) protein RUNX1 is a master regulator of definitive hematopoiesis, crucial for hematopoietic stem cell (HSC) emergence during ontogeny. RUNX1 also plays vital roles in adult mice, in regulating the correct specification of numerous blood lineages. Akin to the other mammalian Runx genes, Runx1 has two promoters P1 (distal) and P2 (proximal) which generate distinct protein isoforms. The activities and specific relevance of these two promoters in adult hematopoiesis remain to be fully elucidated. Utilizing a dual reporter mouse model we demonstrate that the distal P1 promoter is broadly active in adult hematopoietic stem and progenitor cell (HSPC) populations. By contrast the activity of the proximal P2 promoter is more restricted and its upregulation, in both the immature Lineage- Sca1high cKithigh (LSK) and bipotential Pre-Megakaryocytic/Erythroid Progenitor (PreMegE) populations, coincides with a loss of erythroid (Ery) specification. Accordingly the PreMegE population can be prospectively separated into “pro-erythroid” and “pro-megakaryocyte” populations based on Runx1 P2 activity. Comparative gene expression analyses between Runx1 P2+ and P2- populations indicated that levels of CD34 expression could substitute for P2 activity to distinguish these two cell populations in wild type (WT) bone marrow (BM). Prospective isolation of these two populations will enable the further investigation of molecular mechanisms involved in megakaryocytic/erythroid (Mk/Ery) cell fate decisions. Having characterized the extensive activity of P1, we utilized a P1-GFP homozygous mouse model to analyze the impact of the complete absence of Runx1 P1 expression in adult mice and observed strong defects in the T cell lineage. Finally, we investigated how the leukemic fusion protein AML1-ETO9a might influence Runx1 promoter usage. Short-term AML1-ETO9a induction in BM resulted in preferential P2 upregulation, suggesting its expression may be important to establish a pre-leukemic environment.
Author Summary
The transcription factor RUNX1 is considered a master regulator of adult and embryonic blood cell production. Mutations in RUNX1 cause defects in different blood lineages in human patients and mouse models, including leukemia and blood clotting defects due to a shortage of platelet-producing megakaryocytes. Together with the other RUNX genes present in mammals, RUNX1 is expressed from two promoters, which produce several distinct RNA transcripts and protein isoforms. To investigate the timing and localization of the expression of these two promoters (termed distal and proximal), we created a mouse model with reporter genes expressed under the control of the Runx1 promoters. We previously described the activities of the Runx1 promoters at the initiation of blood production in the developing embryo. We now investigate the output from the two promoters in adult organs, including bone marrow, spleen and thymus. We show here that the distal Runx1 promoter is highly expressed but the proximal promoter is more restricted and in particular marks the point in adult blood production where the red blood cell and megakaryocyte pathways separate. The different proteins produced by these two Runx1 promoters may therefore have different roles in driving the production of these two distinct cell types.
Introduction
Adult hematopoiesis is orchestrated by a series of lineage fate decisions that control the specification of mature erythroid, myeloid and lymphoid blood cells from pluripotent HSCs. RUNX transcription factors play key roles at different stages, activating or repressing transcriptional targets through DNA binding in association with other lineage-specific and ubiquitous transcription factors and cofactors [1,2]. RUNX1 (also known as Acute Myeloid Leukemia 1 or AML1) is a master regulator of definitive hematopoiesis, broadly expressed in HSCs, progenitors and mature populations, with the exception of terminally differentiated erythrocytes [3–5]. RUNX1 activity is vital for the embryonic establishment of normal adult hematopoiesis through the regulation of HSPC emergence in a process termed endothelial-to-hematopoietic transition (EHT) [6–12]. Conditional deletion of Runx1 in adult mice, meanwhile, results in hematological imbalances such as decrease of peripheral blood lymphocytes, expansion of monocytes and granulocytes and impaired T cell maturation [13–15]. RUNX1 is also critical in megakaryocytic maturation and platelet production [16,17]. The requirement for RUNX1 in adult HSC maintenance is more controversial, with assertions of impaired long-term repopulating ability in Runx1-null HSCs due to increased stem cell exhaustion being increasingly challenged [6,18,19].
The importance of normal CBF function extends to malignant hematopoiesis, with RUNX1 or CBFB mutations found in over 20% of acute myeloid and lymphoid leukemia cases [20]. Although impaired RUNX1 activity is frequently important for establishing a pre-leukemic stage, WT RUNX1 protein is nonetheless necessary for maintaining AML1-ETO Acute Myeloid Leukemia (AML) [21,22]. Consequently, the investigation of RUNX1’s expression and function in hematopoiesis is of considerable interest to developmental biologists and clinical researchers alike.
All vertebrate Runx genes contain two alternative promoters, a distal P1 promoter and a proximal P2 promoter thought to represent the initial “primitive” promoter [23–25]. The major protein isoforms produced from the P1 and P2 promoters, RUNX1C and RUNX1B respectively, differ in their N-terminal amino acid sequences; RUNX1C is 14 amino acids longer and begins with the MASDS sequence whereas RUNX1B begins with MRIPV, a feature conserved in mice and humans [26,27]. P2 is the more active promoter at the onset of definitive hematopoiesis in the E7.5 embryo [28,29]. P1 activity is subsequently upregulated, enriched in definitive hematopoietic culture colony-forming unit (CFU-C) populations from E8.5 onwards [29]. Analyses on whole cell populations revealed a remarkable switch to P1-dominant Runx1 expression at the fetal liver stage that is maintained in adult BM populations [28,29]. At this stage P2 activity is only detected in some specific adult hematopoietic subsets. However, the exact cell populations defined by the activities of P1 and P2 remain largely unknown.
To define the activities of the Runx1 promoters in adult HSPCs we utilized a previously described distal-Green Fluorescent Protein (GFP), proximal-truncated human CD4 (hCD4) (P1-GFP::P2-hCD4) dual reporter knock-in mouse line [29]. We observed that all Runx1‐positive adult BM populations expressed P1‐GFP, whereas P2‐hCD4 expression was highly restricted. Phenotypic HSCs expressed solely P1‐GFP, with upregulation of P2‐hCD4 in CD48‐positive multipotent progenitors (MPPs) coinciding with a significant downregulation of erythroid output. We also found that the PreMegE population could be prospectively separated into P2‐hCD4‐ “pro‐erythroid” and P2‐hCD4+ “pro‐megakaryocyte” populations. Global gene expression analyses identified various candidate cell surface markers which were differentially expressed between the two PreMegE subpopulations. Among them, differential expression of the hematopoietic cell antigen CD34 enabled the prospective isolation of CD34- “pro‐erythroid” and CD34+ “pro‐megakaryocyte” PreMegEs from WT BM.
To further investigate the potential functional significance of the dominance of RUNX1C in adult hematopoiesis, we investigated the impact of its absence in adult mice and found it to recapitulate certain phenotypes observed in complete Runx1 knockout mouse models. We observed perturbations in platelet versus erythroid output and altered splenic CD4 SP and CD8 SP specification, suggesting certain lineages were more dependent on specific RUNX1C-associated activity than others. Finally, we probed the potential specific RUNX1 isoform requirements in AML by analyzing the impact of AML1-ETO oncogene expression on Runx1 promoter usage. Interestingly, AML1-ETO expression appeared to promote Runx1 P2 over P1 expression in several HSPC populations, suggesting that the Runx1 isoforms may have specific functions both in normal and malignant hematopoiesis.
Results
Runx1 P1 is the dominant promoter in adult hematopoiesis
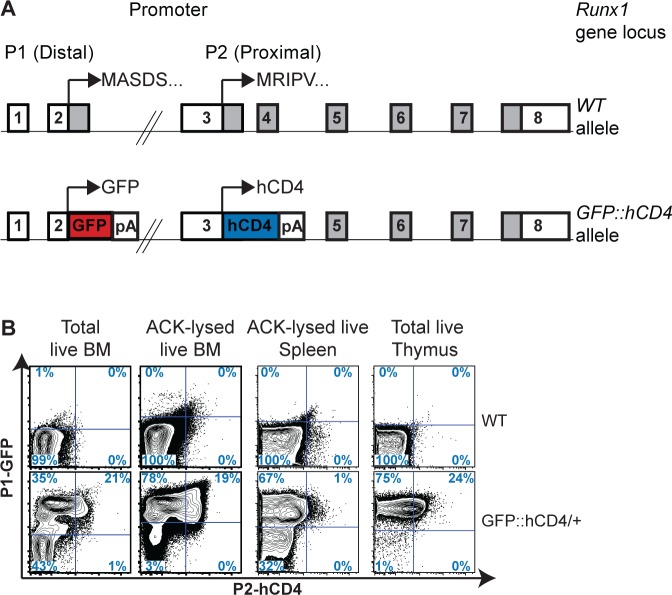
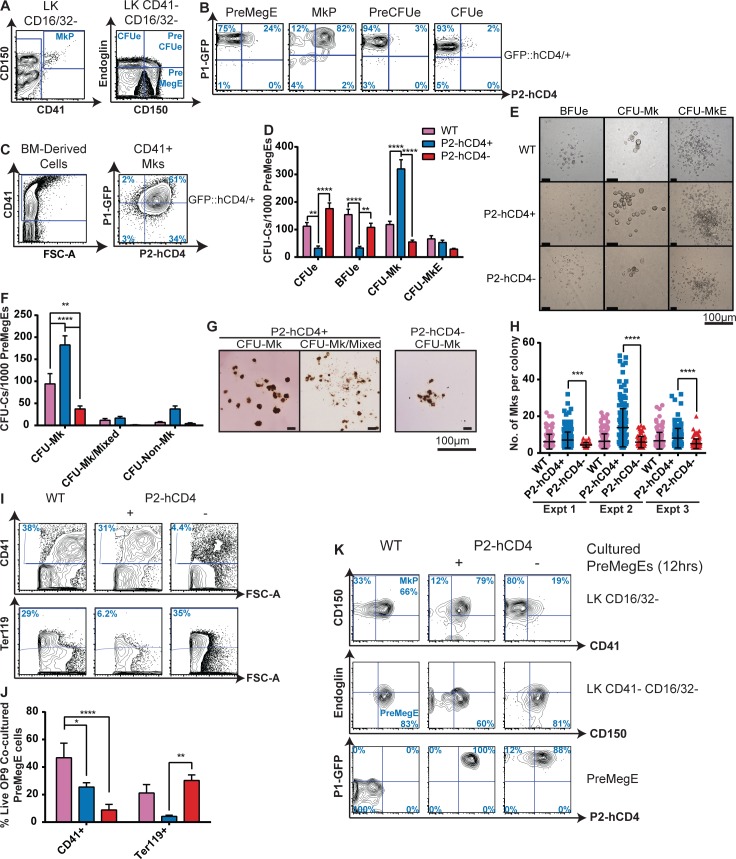
Utilizing the P1-GFP::P2-hCD4 reporter mouse model [29], we traced Runx1 expression for both promoters in vivo at a single cell level in adult mice (with flow cytometry gates based on the WT control tissues) (Fig 1A and 1B). We observed substantial heterogeneity of Runx1 expression within adult BM; approximately 55% of all BM cells were P1-GFP positive, almost 21% co-expressing P2-hCD4 (Fig 1B). Red blood cell lysis (using Ammonium-Chloride-Potassium (ACK) buffer) led to the depletion of P1-GFP- P2-hCD4- cells; 97% of remaining cells expressed P1-GFP with 20% co-expressing P2-hCD4. No P1-GFP- P2-hCD4+ cells were observed. In the spleen, approximately 70% of cells expressed P1-GFP but only 1% co-expressed P2-hCD4, whereas in the thymus almost 100% of cells expressed P1-GFP, a quarter of which also expressed P2-hCD4. Altogether these results establish, in line with other reports, that P1 is the dominant Runx1 promoter in adult hematopoietic populations and that the activity of P2 is much more restricted [28].
Fig 1. Runx1 promoter P1 and P2 expression in adult hematopoietic organs.
(A) Schematic diagrams of the Runx1 WT (top) and P1-GFP::P2-hCD4 dual reporter (GFP::hCD4, bottom) alleles. Expression of GFP is directed by Runx1 promoter P1 and a truncated hCD4 reporter is expressed under the control of Runx1 promoter P2. (B) Contour plots of Runx1 P1-GFP and P2-hCD4 expression in unfractionated total (far left panel) and ACK buffer lysed (middle left panel) adult BM, spleen (middle right panel) and thymus (far right panel) in WT (top) and P1-GFP::P2-hCD4/+ (bottom) mice. Representative data of three independent experiments are shown.
Taking advantage of our reporter model, we pursued a detailed examination of P1-GFP and P2-hCD4 expression in mature lymphoid and erythro-myeloid populations (Figs 2 and S1).
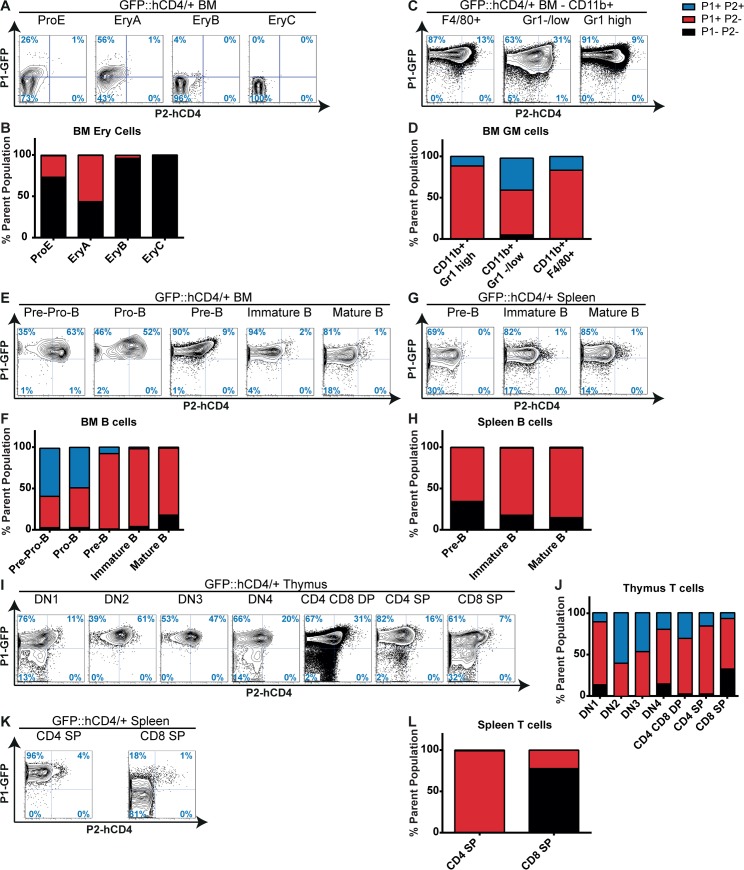
Fig 2. Runx1 P1 and P2 expression in mature hematopoietic lineages.
(A, C, E, G, I, K) Contour plots of Runx1 P1-GFP and P2-hCD4 expression in BM erythroid (A), granulocytic/macrophage (C) and B lymphocyte populations (E), spleen B (G) or T (K) lymphocytes and thymocytes (I), as defined in S1 Fig. (B, D, F, H, J, L) Numbers of Runx1 P1- P2-, P1+ P2- and P1+ P2+ cells as a proportion of defined BM erythroid (B), granulocytic/macrophage (D) and B lymphocyte populations (F), spleen B (H) or T (L) lymphocytes and thymocytes (J). Representative data of three independent experiments are shown.
Runx1 is expressed in definitive erythroid precursors, where it is involved in the regulation of erythroid gene expression as part of a core transcription factor complex, but is subsequently downregulated in mature erythrocytes [3–5,28,30]. Correspondingly, P1-GFP expression was restricted to 26% of the proerythroblast (CD71high Ter119int, ProE), 56% of the basophilic erythroblast (CD71high Ter119high FSChigh, EryA) and 4% of the late basophilic/polychromatic erythroblast (CD71high Ter119high FSClow, EryB) fractions whilst being completely absent in the most mature CD71low Ter119high FSClow (orthochromatic erythroblasts, reticulocytes, red blood cells, EryC) compartment (Figs 2A, 2B, and S1A). P2-hCD4 was expressed in less than 1% of Ter119+ erythroid cells, being apparently entirely dispensable for adult erythropoiesis. The low level of expression from both Runx1 P1 and P2 promoters, particularly the latter, in WT erythroid lineage cells was confirmed at the RNA level by qPCR (S2A Fig).
By contrast to the restricted expression observed in the erythroid lineage, Runx1 P1-GFP was expressed in almost 100% of mature myeloid CD11b+ BM cells (Figs 2C, 2D and S1B). Of these, P2-hCD4 was co-expressed in 12% of Gr1high granulocytes, 39% of Gr1-/low monocytic/immature granulocyte cells and 17% of F4/80+ macrophages. The decreased P2 activity in the more mature granulocytic/macrophage (GM) fractions suggests a diminished role for RUNX1B as myeloid differentiation progresses. This also appears to be the case for terminal lymphoid differentiation, as P2-hCD4 co-expression with P1-GFP was restricted in the B-cell lineage to 58% of the BM Pre-pro-B, almost half (49%) of the Pro-B and just 8% of the Pre-B progenitors (Figs 2E–2H, S1C and S1D). P1-GFP was expressed in over 90% of BM B cell progenitors but was reduced to approximately 80% of mature BM and spleen B cells. Finally, thymic T cells were highly enriched in the P1-GFP+ P2-hCD4- fraction but P2-hCD4 activity appeared to peak in the CD4 CD8 double negative 2 (DN2) fraction at approximately 61% (Figs 2I, 2J and S1E). Interestingly, the more mature spleen CD4 and CD8 single positive (SP) T cell subsets displayed greater heterogeneity than their thymic counterparts; almost 100% of CD4 SP cells express P1-GFP whilst this is the case for only 20% of CD8 SP cells (Figs 2K, 2L and S1F). Relative quantitation of the Runx1 isoforms’ expression revealed comparatively high P1 and P2 activity in the GM, B and T lineages, peaking in the early thymic T cell CD4 CD8 DN population and provides direct evidence that the P1-GFP::P2-hCD4 reporters faithfully represent WT Runx1 expression throughout adult hematopoiesis (S2A–S2C Fig). Overall, P1 clearly dominates, accounting for over 80% of Runx1 expression in all analyzed lineage positive populations. Nonetheless, strong P2 expression was observed in CD11b+ GR1+ GM cells, Pre-pro/pro/pre-B cells and CD4 CD8 DN T cells, decreasing substantially in the more mature IgM+ B and CD4/CD8+ T cells. These results indicate that P1 is the dominant Runx1 promoter in terminally differentiated hematopoietic cells and suggest that downregulation of P2 is required for maturation to occur. We therefore decided to determine whether P2 expression has a greater prominence and significance in immature HSPC subsets.
Upregulation of Runx1 P2 in HSPCs marks a loss of erythroid potential
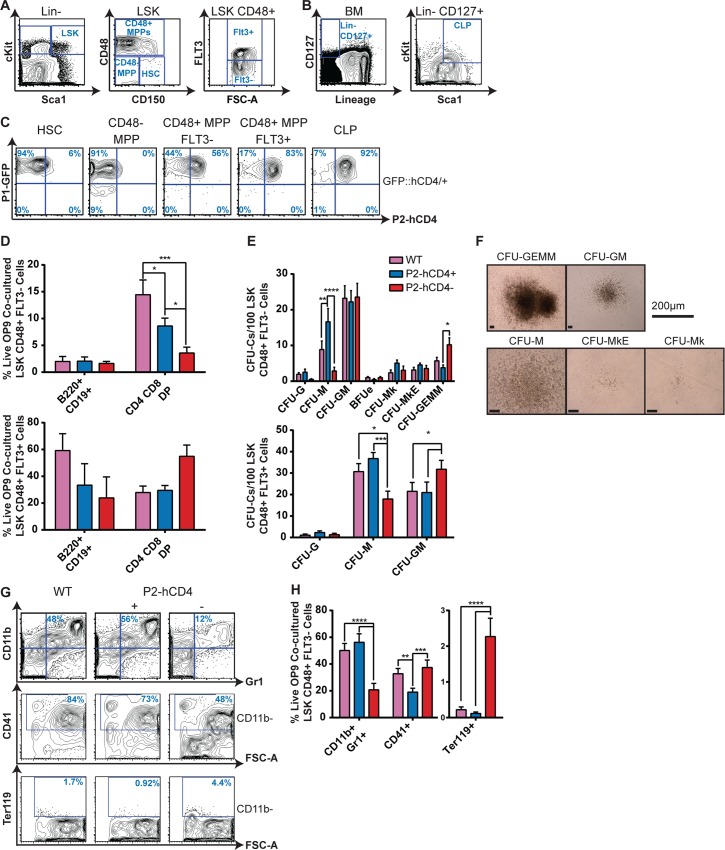
To examine the relative activities of the two Runx1 promoters in the most immature hematopoietic compartments, we separated the LSK fraction into phenotypic HSC and MPP fractions (Fig 3A). We observed that only the P1 promoter was active in the HSCs and CD48- MPPs (Fig 3C). The upregulation of CD48 expression coincides with the loss of long-term repopulating ability, the LSK CD48+ fraction consisting of a mixture of lymphoid and myeloid progenitors with varying multipotentiality. FMS-Like Tyrosine Kinase 3 (FLT3) expression marks a commitment to the GM and lymphoid lineages at the expense of Mk/Ery specification [31,32]. Increased GM/lymphoid lineage commitment appears to coincide with increased P2 activity, as the majority of cells in the lymphoid-primed multipotent progenitor (LMPP)-enriched FLT3+ and the common lymphoid progenitor (CLP, Fig 3B) subsets co-expressed P1-GFP and P2-hCD4 (Fig 3C). Therefore, although P1 is the dominant Runx1 promoter at the onset of adult hematopoiesis, our results suggest that P2 expression imparts or at least reflects distinct lineage commitment decisions in these immature hematopoietic compartments. Consistent with this theory, Runx1 P1 activity, as measured by quantitative RT PCR in WT BM HSPCs, peaked in the WT HSCs and decreased by approximately 50% in the FLT3+ MPPs, coinciding with the substantial increase in Runx1 P2 expression (S3 Fig). The overall result is that total Runx1 expression decreases only modestly in MPPs compared to HSCs but the relative contribution by P1 compared to P2 decreases substantially.
Fig 3. Upregulation of Runx1 P2 in HSPCs marks a loss of erythroid potential.
(A–B) Contour plots showing immature adult BM Lineage (Lin) negative hematopoietic progenitors. (A) The LSK fraction can be separated into distinct HSCs, CD48- MPPs and more mature CD48+ MPPs on the basis of CD150, CD48 and FLT3 expression. (B) The CLP is characterized as Lin- CD127+ cKitlow Sca1low. (C) Representative FACS plots of P1-GFP/P2-hCD4 expression in HSCs, CD48- MPPs, CD48+ FLT3- (LSK48F-) MPPs, CD48+ FLT3+ (LSK48F+) MPPs and CLPs. (D) Numbers of B220+ CD19+ B cells and CD4+ CD8+ DP T cells produced following co-culture of WT, P1+ P2- and P1+ P2+ LSK48F- MPPs (top) and LSK48F+ MPPs (bottom) with OP9 (B cells) or OP9-DL1 (T cells) for 21 days. (n = 4). (E) CFU-C activity of WT, P1+ P2- and P1+ P2+ LSK48F- MPPs (top) and LSK48F+ MPPs (bottom) following culture in pro-myeloid semi-solid methylcellulose-based medium. (LSK48F-, n = 7; LSK48F+, n = 6.) (F) Photographs of representative LSK48F- derived methylcellulose colonies. (G) Representative CD11b/Gr1, CD41 and Ter119 FACS plots of OP9 co-cultured LSK48F- MPPs isolated on day 8. (H) Quantification of CD11b+ Gr1+ granulocyte/macrophage (GM) cells, CD41+ megakaryocytes and Ter119+ erythrocytes in the progenitor/OP9 co-culture assays (n = 5).
When the differences in biological potential were directly assessed in FLT3+ MPPs, P2-hCD4- and P2-hCD4+ subpopulations were capable of relatively similar levels of lymphoid and myeloid differentiation (Fig 3D and 3E bottom), although the increased GM:M CFU-C ratio in the P2-hCD4- subset may suggest it represents a more immature population than its P2-hCD4+ counterpart. However, the difference in lineage output by the FLT3- subsets was more marked; P2-hCD4- LSK CD48+ FLT3- (LSK48F-) MPPs appeared to have reduced T cell output (Fig 3D top) but enhanced multilineage myeloid colony-forming unit potential at the expense of CFU-M output (Fig 3E and 3F). Co-culturing the LSK48F- progenitors with the OP9 murine stromal cell line in myeloid differentiation media revealed that CD11b+/Gr1+ GM output was significantly decreased and CD41+ megakaryocytic (Mk) cell production was slightly increased in the P2-hCD4- fraction as a proportion of total cells (Fig 3G and 3H). As a proportion of non-GM (CD11b-) cells, CD41+ Mk cell output was in fact significantly increased in the P2-hCD4+ LSK48F- fraction. Most strikingly, Ter119+ erythroid cell output was almost entirely restricted to the P2-hCD4- fraction. Our phenotypic characterization of BM HSPCs therefore demonstrate that upregulation of Runx1 P2 not only occurs after loss of HSC activity but also coincides with a substantial decrease in erythroid specification.
To determine whether LSK48F- P2-hCD4- and P2-hCD4+ progenitors arise sequentially or independently in the hematopoietic hierarchy, sorted cells were cultured with pro-myeloid cytokines for up to 18 hours and immunophenotypically characterized (S4A and S4B Fig). Whereas the P2-hCD4+ fraction solely produced P2-hCD4+ LSK cells, P2-hCD4- cultures yielded P2-hCD4- and P2-hCD4+ LSK cells (S4B Fig). In addition, LSK48F- P2-hCD4- cultures produced more phenotypic erythroid (pre-erythroid colony-forming unit, PreCFUe or erythroid colony-forming unit, CFUe) or bi-potential PreMegE progenitors and fewer GM (Pre- Granulocyte-Macrophage progenitor, PreGM or Granulocyte-Macrophage Progenitor, GMP) and megakaryocyte progenitor (MkP) cells compared to LSK48F- P2-hCD4+ cells. Altogether these data demonstrate a hierarchical relationship between an erythroid-biased P2-hCD4- MPP population and increasingly pro-GM/Mk P2-hCD4+ progeny.
Runx1 P2 expression in GM-restricted progenitors enriches for monocyte/macrophage specification
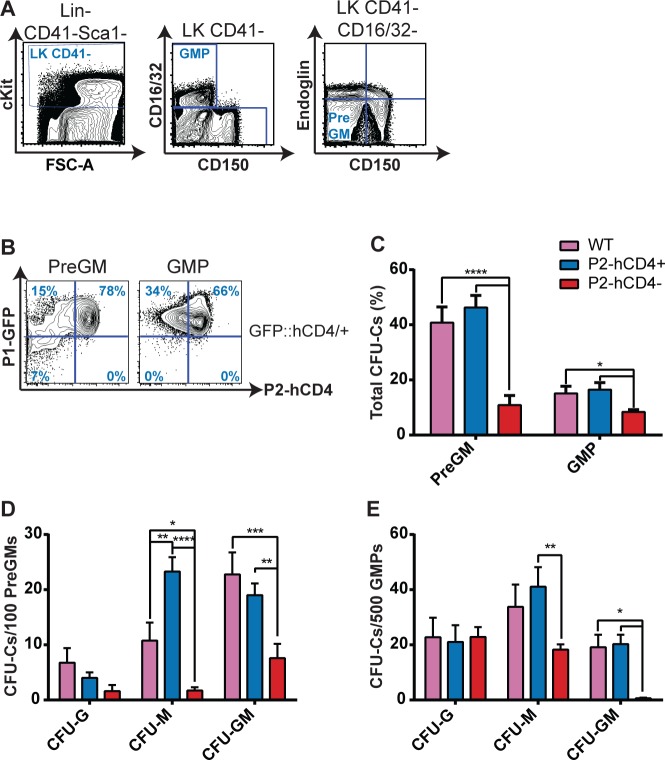
Although Runx1 P2 expression appears to decrease as GM maturation proceeds, its expression in the earliest identified GM-restricted progenitors remained unknown. We found that approximately 80% of PreGMs and 70% of GMPs co-express P1-GFP and P2-hCD4 (Fig 4A and 4B). Moreover, we found P2 expression (as determined by qRT-PCR) to be higher in the PreGM and GMP than other analyzed WT BM HSPC populations but P1 expression was only 50% and 25% of the level observed in HSCs (S3 Fig). High P2 activity therefore appeared to be important for GM lineage commitment and we decided to investigate the functionality of the P2-hCD4+ and minority P2-hCD4- GM progenitor populations. Interestingly, CFU-C activity was significantly higher in the P2-hCD4+ fractions, compared to the P2-hCD4- populations, of both PreGM and GMP populations, reflecting higher CFU-M and CFU-GM frequencies (Fig 4C–4E). In particular, the P2-hCD4- GMP fraction appeared to consist of monopotent granulocytic and monocytic/macrophage progenitors rather than bipotential GM progenitors. Liquid culture of the progenitors confirms an apparent bias against macrophage specification as F4/80+ cell numbers were significantly diminished in P2-hCD4- PreGM and GMP cultures, whereas Gr1high granulocyte output was unaltered (S5A and S5B Fig). Interestingly, production of CD11b- cKit+ FcεR1α+ mast cells was also elevated in P2-hCD4- cultures and more detailed analyses confirmed the absence of P2-hCD4 expression in immunophenotypic mast cell progenitors (MCp, S5A–S5C Fig). The decreased CFU-GM activity of the P2-hCD4- GM progenitors implies that they reside later in the hematopoietic hierarchy than the P2-hCD4+ populations, but in vitro lineage tracing revealed that P2-hCD4- GMPs gave rise to P2-hCD4+ GMPs (S5E Fig). P2-hCD4- PreGMs gave rise to P2-hCD4+ PreGMs and subsequently to P2-hCD4- and P2-hCD4+ GMPs, the latter dominating (S5D Fig). Therefore even within the GM lineage, differential Runx1 promoter activity appears to play a role in or at least correlate with crucial cell fate decisions.
Fig 4. Runx1 P2 expression in GM-restricted progenitors enriches for monocyte/macrophage specification.
(A) Contour plots of adult BM Lin- Sca1- cKit+ (LK) GM progenitors. The GMP and PreGM progenitors can be distinguished on the basis of CD41, CD16/32, SLAMF1 (CD150) and Endoglin expression. (B) Representative FACS plots of P1-GFP/P2-hCD4 expression in PreGM and GMP cells. (C–E) CFU-C activity of WT, P1+ P2- and P1+ P2+ PreGMs and GMPs following culture in pro-myeloid semi-solid methylcellulose-based medium. (C) Total CFU-C numbers (%). (D-E) Granulocyte (CFU-G), macrophage (CFU-M) and granulocyte/macrophage (CFU-GM) colony forming unit numbers per 100 plated PreGMs (D) or 500 plated GMPs (E). n = 4 independent experiments.
Distinct megakaryocytic and erythroid progenitors can be isolated on the basis of Runx1 P2-hCD4 expression
Erythropoiesis and megakaryopoiesis are highly similar developmental pathways, sharing numerous regulatory factors particularly at the point of lineage specification [33,34]. However, there are key differences and the specificity of a megakaryocyte maturation defect in Runx1-null adult BM implicates RUNX1 as a central player in Mk/Ery lineage determination [14]. Moreover, our observation that P2-hCD4- and P2-hCD4+ MPPs have distinct Mk/Ery potential led us to investigate Runx1 promoter activity in Mk/Ery-restricted progenitors further (Fig 5A). We observed that erythroid restricted PreCFUe and CFUe progenitors expressed solely P1-GFP whereas the MkP was chiefly P1-GFP+ P2-hCD4+ (Fig 5B). Because mature megakaryocytes are scarce in adult mice, BM-derived megakaryocytes were obtained by culturing purified MkPs in vitro. CD41-expressing megakaryocytes expressed P2-hCD4 and a large fraction (60%) co-expressed P1-GFP (Fig 5C). Whilst lineage-restricted megakaryocytic and erythroid progenitors were highly homogeneous, the PreMegE fraction, which generates the MkP and PreCFUe populations, was markedly more heterogeneous; approximately 75% express solely P1-GFP whereas the remaining 25% were P1-GFP+ P2-hCD4+ (Fig 5B).
Fig 5. Distinct megakaryocytic and erythroid progenitors can be isolated on the basis of Runx1 P2-hCD4 expression.
(A) Contour plots of adult BM LK Mk/Ery progenitors. The PreMegE, MkP, PreCFUe and CFUe populations can by distinguished on the basis of cKit, CD41, SLAMF1 (CD150), CD16/32 and Endoglin cell surface expression. (B) Representative FACS plots of P1-GFP/P2-hCD4 expression in PreMegE, MkP, PreCFUe and CFUe subsets. (n = 8). (C) P1-GFP/P2-hCD4 expression in BM MkP-derived cultured CD41+ megakaryocytes. (n = 4). (D–H) CFU-C activity of WT, P1+ P2- and P1+ P2+ PreMegEs. Cells were cultured either in pro-myeloid semi-solid methylcellulose-based medium (D) or in pro-megakaryocytic collagen-based MegaCultTM medium (F). Photographs of representative PreMegE-derived methylcellulose (E) and MegaCultTM (G) colonies. (n = 5) (H) Numbers of megakaryocytes per MegaCultTM CFU-Mk colony from 3 independent experiments (mean ± SD, Mann-Whitney U test). (I) Contour plots of OP9 co-cultured PreMegE cells isolated on day 7 and stained with CD41 and Ter119 antibodies. (J) Quantification of CD41+ megakaryocytes and Ter119+ erythrocytes in the PreMegE/OP9 co-culture assays. (n = 5). (K) Representative FACS plots of PreMegE cells following short-term (12 hours) culture in pro-myeloid liquid medium. Top: CD150/CD41 expression of LK CD16/32- progenitor cells. Middle: Endoglin/CD150 expression of LK CD41 negative CD16/32 negative progenitors. Bottom: P1-GFP/P2-hCD4 expression of immunophenotypic PreMegE (LK CD41- CD16/32- Endoglin- CD150+) cells (n = 3).
When compared to the relative homogeneity of the monopotent MkP, PreCFUe and CFUe populations, the heterogeneity of the PreMegE led us to consider the possibility of two functionally distinct and prospectively isolatable PreMegE subsets. We subsequently observed that erythroid CFU-C activity (CFUes and erythroid blast-forming units (BFUes)) was significantly enriched in the P2-hCD4- PreMegE fraction compared to the P2-hCD4+ population (Fig 5D and 5E). By comparison, megakaryocyte CFU-C potential was highly enriched in the P2-hCD4+ fraction (Fig 5D–5H). In the MkP population, the P2-hCD4+ fraction possessed similar megakaryocytic CFU-C activity to its WT counterpart (S6A Fig), suggesting Runx1 haploinsufficiency did not significantly impair megakaryocyte colony formation. In addition to being more numerous, CFU-Mks derived from P2-hCD4+ PreMegEs were also larger than those derived from the P2-hCD4- fraction, the median number of cells per colony being doubled (Fig 5E, 5G and 5H). To determine whether this was a result of increased proliferation in the P2-hCD4+ PreMegE fraction, we analyzed their cell cycle status by measuring 5’ethynyl-2’-deoxyuridine (EdU) incorporation and DNA content (S6B Fig). More P2-hCD4+ PreMegE cells were in the EdU+ DNA Synthesis (S) phase compared to their P2-hCD4- counterparts, suggesting P2-driven RUNX1B expression may confer a proliferative advantage on PreMegE cells.
The distinct megakaryocytic and erythroid potential of the two PreMegE fractions was further confirmed following co-culture with OP9 cells. After 7 days, P2-hCD4+ PreMegE cultures contained significantly more CD41+ megakaryocytes and significantly fewer Ter119+ erythroid cells than the P2-hCD4- PreMegE cultures (Fig 5I and 5J). In addition, we performed clonal analyses by plating single P2-hCD4- and P2-hCD4+ PreMegEs with OP9 (S6C Fig). This demonstrated that although the two fractions had similar clonal output (28% and 24% positive wells respectively) the P2-hCD4+ fraction contained more bi-potent megakaryocytic/erythroid progenitors (42.9% versus 25% “Mk + Ery”) and more monopotent megakaryocyte-producing progenitors (31.4% versus 12.5% “Mk only”) than the P2-hCD4- fraction. The P2-hCD4- fraction was highly enriched for monopotent erythroid-producing progenitors (62.5% “Ery only” compared to 25.7% in the P2-hCD4+ co-cultures). Therefore, as in the immature LSK HSPC compartment, upregulation of P2 expression in PreMegEs appeared to coincide with a loss of erythroid and an enrichment of megakaryocytic specification.
To decipher their relative positions in the hematopoietic hierarchy, P2-hCD4- and P2-hCD4+ PreMegE cells were cultured for up to 12 hours and analyzed (Fig 5K). We observed that P2-hCD4+ PreMegEs made a more rapid transition to an MkP immunophenotype than P2-hCD4- cells. In addition P2-hCD4- PreMegEs gave rise to both P2-hCD4- and P2-hCD4+ fractions in vitro, whereas the P2-hCD4+ fraction did not appear to downregulate P2-hCD4. Taken together, these data suggest the P2-hCD4- PreMegE can be placed earlier in the hematopoietic hierarchy, giving rise to the P2-hCD4+ PreMegE. Interestingly, we also observed that cultured P2-hCD4+ LSK48F- cells produced only P2-hCD4+ immunophenotypic PreMegEs (S4B Fig). The differences in megakaryocytic and erythroid lineage potential in the P2-hCD4 negative and positive LSK48F- fractions may therefore be due to the preferential downstream specification of distinct PreMegE subpopulations.
Global gene expression analysis of Runx1 P2-hCD4 positive and negative PreMegEs enables the identification of WT equivalents
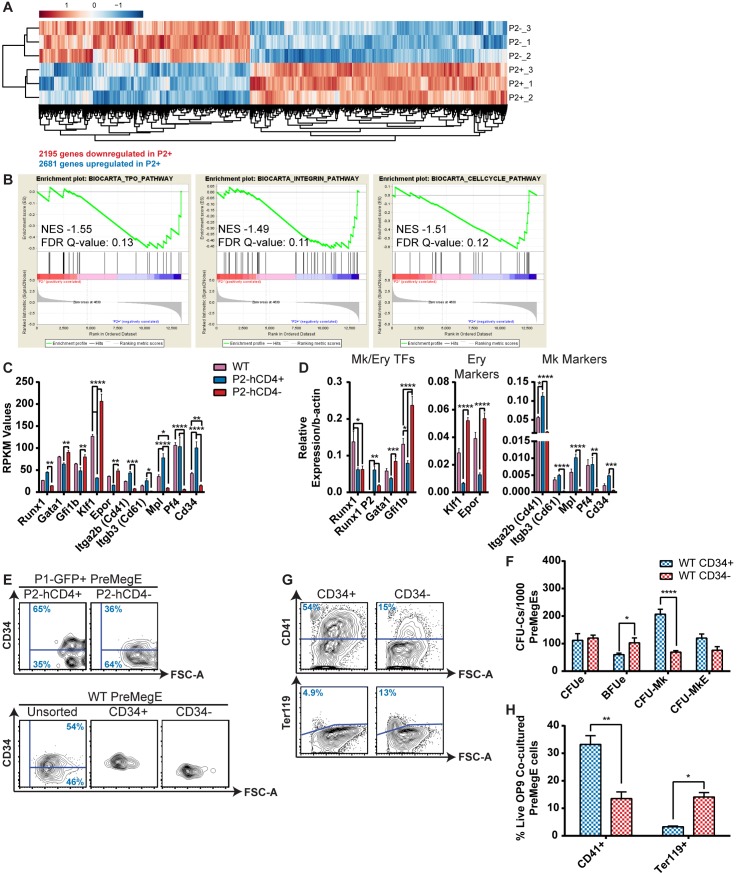
To explore the distinct gene regulatory mechanisms involved in the bifurcation of the Mk/Ery pathway, and to identify candidate genes which may serve as markers to isolate the progenitors in WT BM, we performed global gene expression analysis by RNA-Seq (Fig 6A and S4 Table). The expression patterns of WT, P2-hCD4+ (P2+) and P2-hCD4- (P2-) PreMegE samples were clearly separated based on principal component analysis, with WT cells clustering between the P2+ and P2- samples (S7A Fig). When directly comparing the P2+ and P2- populations, 4876 genes were found to be at least 2-fold differentially expressed (false discovery rate <0.05), 2681 being upregulated in P2+ and 2195 in P2- PreMegEs (Fig 6A and S4 Table). Gene Set Enrichment Analyses (GSEA) revealed a significant correlation between P2-hCD4 expression and activation of the Thrombopoietin (TPO) and Integrin pathways, both of which are crucial for megakaryopoiesis (Figs 6B and S7B–S7D) [35–37]. In line with the observed increased proliferative capacity of P2-hCD4+ PreMegEs, cell cycle regulators were also enriched in this population. Ingenuity pathway analysis (IPA) identified cell migration and blood cell recruitment as highly enriched functions and integrin signaling as the most significant activated pathway in P2-hCD4+ PreMegEs (S7E and S7G Fig). By contrast, functions and pathways associated with cell death and cell cycle inhibition were highly enriched in P2-hCD4- PreMegEs (S7F and S7H Fig).
Fig 6. Global gene expression analysis of Runx1 P2-hCD4+ and P2-hCD4- PreMegEs enables identification of WT equivalents.
(A) Heat map depiction of genes at least 2-fold differentially expressed between P1+ P2- (P2-) and P1+ P2+ (P2+) PreMegE samples, as determined by RNA Seq. Genes in red are upregulated and genes in blue are downregulated. (B) GSEA showing significantly enriched signaling pathways in the gene set upregulated in P2+ PreMegEs relative to P2- PreMegEs. (C) Reads per kilobase per million mapped reads (RPKM) values of selected Mk/Ery- associated genes. (n = 3). (D) Quantitative PCR (qPCR) validation of expression of genes depicted in D. (n = 5). (E) Representative FACS plots of CD34 expression in P2+ and P2- PreMegEs (top) and unsorted, purified CD34+ and purified CD34- WT PreMegEs (bottom). (P1-GFP::P2-hCD4 mice n = 3; WT mice n = 5). (F) CFU-C activity of CD34+ and CD34- PreMegEs following culture in pro-myeloid semi-solid methylcellulose-based medium. (n = 5). (G) Contour plots of OP9 co-cultured CD34+ and CD34- PreMegE cells isolated on day 7 and stained with CD41 and Ter119 antibodies. (H) Quantification of CD41+ megakaryocytes and Ter119+ erythrocytes in the PreMegE/OP9 co-culture assays (n = 4).
To further validate the distinct “pro-megakaryocytic” and “pro-erythroid” phenotypes of each PreMegE population, we screened, and validated by qPCR, the RNA Seq data for the expression of known Mk/Ery regulators and markers (Fig 6C and 6D). Early erythroid-associated factors, including Kruppel-like factor 1 (Klf1) and the Erythropoietin receptor (Epor) were significantly upregulated in P2-hCD4- PreMegEs whereas numerous megakaryocyte-specific markers (Integrin alpha 2b (Itga2b or Cd41), Integrin beta 3 (Itgb3 or Cd61), Myeloproliferative Leukemia Virus Oncogene (Mpl) and Platelet Factor 4 (Pf4)) were enriched in the P2-hCD4+ fraction. Interestingly, the transcription factors GATA binding protein 1 (Gata1) and Growth Factor Independent 1B (Gfi1b) were upregulated in P2-hCD4- PreMegEs. Both factors are crucial for the normal development of both megakaryocytic and erythroid lineages: deletion of either Gata1 or Gfi1b results in an early block in erythropoiesis at the PreCFUe stage whereas megakaryocytic maturation is impaired resulting in the accumulation of undifferentiated megakaryoblasts [38–41]. It would therefore appear that high Gfi1b and/or Gata1 expression promote erythroid specification whereas lower levels would favor megakaryocytic commitment, but ultimately an increase of both would be required for megakaryocytic maturation and thrombopoiesis. It is therefore highly likely that differential expression of Gata1 and Gfi1b at the PreMegE stage plays a role in Mk/Ery lineage determination and their differential expression may be driven by P2-driven RUNX1B.
In order to distinguish “pro-megakaryocytic” and “pro-erythroid” PreMegE subsets in WT mice by alternative means to our reporter line, we screened the list of differentially expressed genes in P2-hCD4+ and P2-hCD4- PreMegEs for cell surface markers with commercially available antibodies validated for use in flow cytometry (S8A Fig). The majority of selected markers had low RPKM values, with the exception of Itgb3 (Cd61) and Cd34 (Figs 6C–6E and S8B). However, CD61 protein expression was not detected on P2-hCD4+ and P2-hCD4- PreMegE cells by flow cytometry (S8C and S8D Fig). By contrast, CD34 expression was approximately 2-fold higher in P2-hCD4+ PreMegEs compared to P2-hCD4- cells, both in terms of numbers of positive cells and median fluorescence intensity (MFI; Figs 6E and S8D). WT CD34+ and CD34- PreMegEs were therefore FACS sorted to >95% purity (Fig 6E) and their lineage output and Runx1 isoform expression elucidated. Importantly, Runx1 P2 expression was substantially higher in the CD34+ PreMegEs compared to the CD34- fraction (S3 Fig). Interestingly, P1 activity was also increased in the CD34+ cells, resulting in a 20% increase in total Runx1 expression. It is therefore unclear how important expression of the RUNX1B isoform is for the promotion of megakaryopoiesis compared to enhanced RUNX1 expression overall. Indeed, P2 transcripts were even more highly expressed in WT MkPs, contributing to the highest levels of total Runx1 in all analyzed HSPCs (S3C Fig). However, P1 expression was in fact decreased in MkPs compared to PreMegEs, offering additional evidence in favor of a specific pro-megakaryopoiesis role for the P2-specified RUNX1B protein.
Akin to P2-hCD4+ PreMegEs, WT CD34+ PreMegEs had enhanced CFU-Mk and diminished BFUe activity compared to CD34- cells (Fig 6F). Myeloid co-culture with OP9 stromal cells confirmed these phenotypes, as CD34+ PreMegEs produced substantially more CD41+ megakaryocytes and fewer Ter119+ erythroid cells (Fig 6G and 6H). Single-cell OP9 co-culture revealed the CD34+ PreMegE compartment was highly enriched for monopotent “Mk only” progenitors (79.2% versus 41.7%) and bipotent “Mk + Ery” progenitors (20.8% versus 8.3%) compared to the CD34- fraction (S8E Fig). By contrast, monopotent “Ery-only” progenitors accounted for 50% of the CD34- PreMegE cultures but were apparently absent from the CD34+ fraction. We have therefore established the existence of prospectively isolatable “pro-megakaryocytic” CD34+ and “pro-erythroid” CD34- PreMegE cells in WT mice.
Deletion of the dominant RUNX1C isoform does not ablate adult hematopoiesis but results in numerous lineage-specific defects
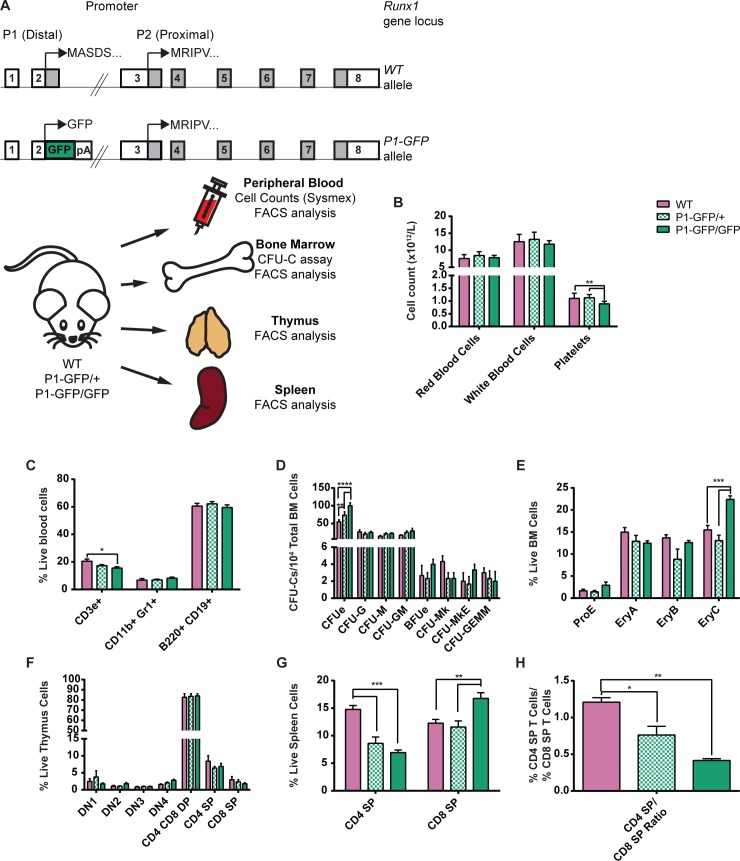
Having established that the P1-directed RUNX1C isoform is expressed throughout adult hematopoiesis, we decided to determine how its absence would impact the overall homeostasis of the adult blood system. Previously, we utilized the P1-GFP homozygous mouse to investigate the requirement for RUNX1C at the onset of hematopoiesis and found it to be dispensable for hematopoietic commitment [29]. However, this may be due to P2 being the dominant promoter at this stage. We therefore analyzed hematopoietic populations in adult WT, RUNX1C heterozygous (P1-GFP/+) and homozygous knockout (P1-GFP/GFP) mice (Fig 7A). Despite the high expression of P1 in erythroid, myeloid and lymphoid progenitors, we observed no significant perturbation of circulating red or white blood cell numbers upon performing automated cell counts (Fig 7B). Erythroid differentiation appeared to be normal, as peripheral blood hematocrit, hemoglobin concentration and the reticulocyte counts of the RUNX1C null mice were unaltered compared to their WT and heterozygous littermates (S9A–S9C Fig). Similarly, myelo-lymphoid cell fate decisions did not appear to be significantly affected, as the proportions of circulating monocytes, neutrophils and lymphocytes were unaffected (S9D-F). However, a modest but significant decrease in platelet numbers was observed in RUNX1C null mice compared to both the WT and heterozygous animals (Fig 7B). Their plateletcrit was also slightly decreased (albeit not to a significant extent) but the mean platelet volume was unaltered (S9G and S9H Fig). This suggests that, unlike in the conditional total Runx1 null adult mouse model, platelet maturation is not impaired but specification may be hampered.
Fig 7. Impact of the absence of P1-directed Runx1 expression on adult hematopoiesis.
(A) Top: Schematic diagrams of the Runx1 WT (top) and P1-GFP (bottom) alleles. Expression of GFP is directed by Runx1 promoter P1 but expression of Runx1 from the P2 promoter remains intact. Bottom: Schematic diagram of the experimental design for the investigation of the impact of Runx1 P1 deletion on adult hematopoiesis. Peripheral blood, BM, thymus and spleen samples were collected from adult WT, P1-GFP heterozygous (P1-GFP/+) and homozygous (P1-GFP/GFP) adult mice. All samples were analyzed for mature blood cell surface marker expression. In addition, blood samples were subjected to automated cell counts (Sysmex) and CFU-C assays were performed on unfractionated BM. (B) Peripheral blood cell counts of WT, P1-GFP/+ and P1-GFP/GFP mice as determined by Sysmex automated cell counting. (C) Numbers of CD3e+ T cells, CD11b+ Gr1+ GM cells and B220+ CD19+ B cells as a proportion of total ACK-lysed blood cells from WT, P1-GFP/+ and P1-GFP/GFP mice. (D) CFU-C activity of WT, P1-GFP/+ and P1-GFP/GFP unfractionated ACK-lysed BM following culture in pro-myeloid semi-solid methylcellulose-based medium. (n = 4.) (E) Numbers of erythroid lineage (ProE, EryA, EryB and EryC) cells as a proportion of live unfractionated BM cells. (F) Numbers of T cell lineage populations as a proportion of live unfractionated thymus cells. (G) Numbers of CD4 SP and CD8 SP T cells as a proportion of live unfractionated spleen cells. (H) Ratio of splenic CD4 SP T cells to splenic CD8 SP T cells (n = 4).
FACS analysis of circulating blood cells and BM confirmed the presence of equal proportions of CD11b+ Gr1+ GM lineage and B220+ CD19+ B lymphoid cells in WT, P1-GFP/+ and P1-GFP/GFP mice (Figs 7C and S10). However, the numbers of CD3ε+ T cells were significantly reduced, suggesting that the absence of RUNX1C partially impairs T cell specification. We therefore analyzed the thymic T cell populations in greater detail and found that CD4/8 DN, DP and SP population numbers were not altered in P1-GFP/GFP mice (Fig 7F). However, the ratio of CD4 SP:CD8 SP T cells in the spleen was severely perturbed, as P1-GFP/GFP mice had considerably fewer CD4 SP and more CD8 SP T cells compared to WT littermates (Fig 7G and 7H). This therefore suggests that RUNX1C is dispensable for the DN to DP transition, observed to be blocked in total Runx1 deficient mice [13]. Nonetheless, the RUNX1C knockout recapitulates the defect in CD4 SP and CD8 SP T cell specification observed in total Runx1+/- mice, clearly demonstrating an important role for P1-driven RUNX1 activity in the T cell lineage [4,42,43].
To determine whether the absence of P1-directed RUNX1C expression impacts adult colony-forming HSPC populations, we performed myeloid CFU-C assays on unfractionated BM from WT, P1-GFP/+ and P1-GFP/GFP mice (Fig 7D). GM, MkE and multilineage GEMM colony numbers were unaffected, but RUNX1C null BM cells produced significantly more erythroid CFUe colonies than either the WT or P1-GFP/+ cultures. FACS analysis of unlysed BM revealed a significant expansion of the EryC population in the RUNX1C null mice, a stage which coincides with almost complete silencing of both the Runx1 P1 and P2 promoters (Fig 7E). In combination with the observed mild thrombocytopenia, it appears that the absence of RUNX1C may favor erythroid specification over megakaryopoiesis, a phenotype observed recently in mouse and human HSPCs depleted for total RUNX1 [44]. Overall, P1-directed RUNX1C activity may be dispensable for normal adult hematopoiesis but its absence nonetheless results in defects reminiscent of total RUNX1 deficiency.
Expression of the AML1-ETO9a oncogene preferentially induces Runx1 P2 expression
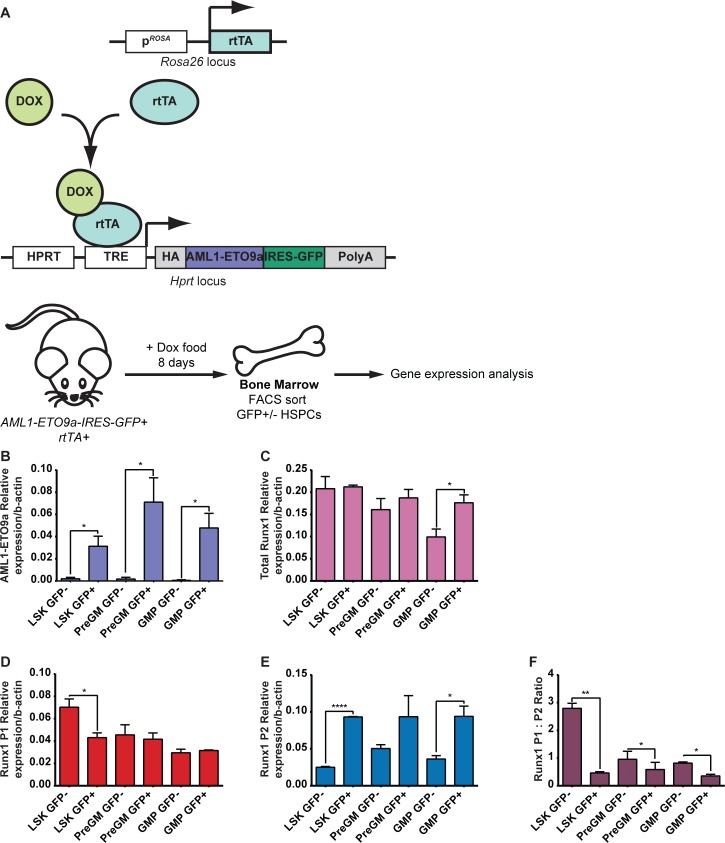
Increasingly it is becoming apparent that, in addition to a more classically defined tumor suppressor role, WT RUNX1 is required for the promotion of leukemogenesis in certain leukemia subtypes. Notably, AML1-ETO-driven CBF AML appears to be dependent on maintaining WT RUNX1 activity [21,22]. However, although AML1-ETO appears to promote RUNX1 expression, it is unclear whether AML1-ETO oncogene expression promotes the expression of one Runx1 promoter over another [45]. To address this question, we utilized a novel mouse model expressing a Doxycycline-inducible AML1-ETO9a transgene under the control of a Tetracycline Responsive Element (TRE, Fig 8A). The AML1-ETO9a oncogenic transcript is expressed in a majority of t(8;21) AML patients studied and encodes a truncated AML1-ETO protein with enhanced leukemogenic potential [46,47]. We therefore took advantage of our ability to induce AML1-ETO expression in adult mice (by administering Doxycycline in the food for 8 days) and studied the impact on Runx1 isoform expression in vivo by isolating AML1-ETO-expressing (AML1-ETO9a-IRES-GFP+) and non-expressing (AML1-ETO9a-IRES-GFP-) BM HSPCs and quantitating Runx1 expression through qRT-PCR (Fig 8A). This allowed us to study the effect of AML1-ETO expression on WT Runx1 expression as one of the earliest events at the initiation of leukemogenesis.
Fig 8. Effect of short-term induction of AML1-ETO9a expression on Runx1 expression in BM HSPCs.
(A) Top: Schematic representation of the Rosa26 and Hprt loci in the Doxycycline-inducible AML1-ETO9a GFP mouse model. The reverse tetracycline-controlled transcriptional activator (rtTA) is constitutively expressed under the control of the Rosa promoter. Upon binding to Doxycycline, the rtTA is capable of binding to and activating a tetracycline responsive element (TRE) located in the ubiquitously expressed Hprt locus, resulting in expression of the hemagglutinin (HA)-tagged AML1-ETO9a::IRES-GFP construct also incorporated into this locus. Bottom: Schematic diagram of the experimental design to induce short-term expression of AML1-ETO9a in adult mice by administering Doxycycline in food for 8 days. Bone marrow cells were the harvested and GFP+ and GFP- HSPC populations were isolated by FACS sorting for RNA extraction and gene expression analysis. (B-E) Gene expression analysis by qPCR of AML1-ETO9a (B), total Runx1 (C), Runx1 P1 (D) and Runx1 P2 (E). (F) Ratio of Runx1 P1:P2 expression (n = 3).
Firstly, we confirmed the presence and absence of AML1-ETO9a expression in BM GFP+ and GFP- HSPCs respectively (Fig 8B). We chose to analyze LSK, PreGM and GMP cells as the immature HSPC and GM-lineage progenitors contain the leukemia propagating cell fraction in numerous AML patient samples and in a previously described AML1-ETO mouse model [48,49]. Whilst in the PreGM, and GMP Runx1 P1 expression was unperturbed by the expression of AML1-ETO, it was in fact decreased by approximately 40% in LSK GFP+ cells compared to GFP- (Fig 8D). In all three HSPC populations, however, the presence of AML1-ETO resulted in an upregulation of Runx1 P2 expression (Fig 8E), albeit not to a significant extent in PreGM cells. This resulted in an increase in total Runx1 expression in the GMP fraction (Fig 8C) but also a significant decrease in the P1:P2 ratio in all three populations, particularly in the LSK compartment, a phenotype associated with enhanced CFU-C activity, particularly in the GM lineage (Fig 8F).
Discussion
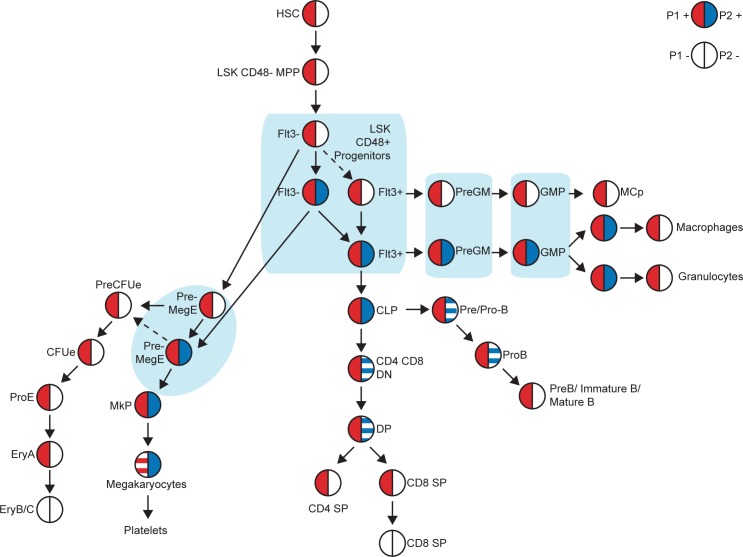
Our understanding of the hematopoietic hierarchy, and of the complexity of cell fate decisions in this system, has been increasingly refined in recent years. For a long time, it was assumed that the most mature shared ancestor for all myeloid populations was the Common Myeloid Progenitor (CMP), until this population was subsequently dissected and shown to be a heterogeneous population containing the PreGM and PreMegE fractions [50,51]. Using the Runx1 P1-GFP::P2-hCD4 dual reporter mouse model, we have now similarly demonstrated further heterogeneity in the PreMegE fraction, prospectively isolating “pro-erythroid” P2- and “pro-megakaryocytic” P2+ fractions (Fig 9). Moreover, we have successfully identified their equivalents in WT BM as being CD34- and CD34+ respectively. CD34, a cell-cell adhesion factor previously characterized as a direct RUNX1 transcriptional target [52] and expressed on vascular-associated tissue and selected HSPCs, was previously used to distinguish the CMP from the Megakaryocyte/Erythroid Progenitor (MEP) [50]. By in vitro cell tracing experiments, we have determined that the P2+ PreMegE lies directly downstream of the P2- PreMegE, apparently contradicting a CMP-based model as this involves downregulation of CD34 expression prior to Mk/Ery lineage commitment. Moreover, we have demonstrated immunophenotypic P2- PreMegEs can be directly derived from P2- LSK48F- MPPs, lending weight to the argument that progenitors lose Mk/Ery potential before separation of the GM and lymphoid pathways [32]. In fact, our model goes further, proposing that erythroid potential is downregulated prior even to megakaryocytic potential, either coinciding with or as a direct result of Runx1 P2 upregulation.
Fig 9. Model of Runx1 P1 and P2 expression in adult megakaryocytic/erythroid lineages.
Runx1 P1 (red) and P2 (blue) expression in adult hematopoietic stem, progenitor and lineage positive cells, as determined using the P1-GFP::P2-hCD4 double knock-in mouse model.
Interestingly, comparative analysis of transcription factor binding motifs by rVISTA [53–56] in the vicinity of the P1 and P2 regions revealed the presence of conserved erythroid transcription factor EKLF (KLF1) motifs in the P1 region but none surrounding P2 (S11A and S11B Fig). By contrast, FLI1-binding motifs are present in both regions. This is interesting as RUNX1 has recently been implicated in regulating the balance of EKLF and FLI1 activity, which promote Ery and Mk output respectively [44]. In addition, EKLF and FLI1 may in fact act upstream of RUNX1, for example with EKLF directly activating Runx1 P1 but not P2 expression, a state which is reinforced by the enhanced EKLF expression in P1+P2- pro-erythroid PreMegEs compared to P1+P2+ pro-megakaryocytic PreMegEs. Analysis of ChIP-Seq data from the mouse ENCODE project [53,54,57,58] also reveal some interesting differences in GATA1 and SCL (TAL1) binding to the P1 and P2 promoter regions in megakaryocytes and erythroblasts (S11C Fig). GATA1 and TAL1 binding appear largely unchanged in the vicinity of the P1 promoter in both cell types. By contrast, GATA1 binding is observed at P2 and GATA1+TAL1 binding approximately 15kb upstream in erythroblasts but not megakaryocytes. It is conceivable, therefore, that GATA1-mediated transcriptional repression of the P2 promoter occurs in the erythroid lineage, whereas the absence of a GATA1-containing complex enables its derepression and recruitment of activating factors instead.
The high number of differentially expressed genes (>4000) in the P2- and P2+ PreMegEs lends credence to the hypothesis that they are derived from distinct progenitor ancestors. Commitment to megakaryocytic or erythroid lineages may even occur earlier, at the HSC level; the P2- and P2+ MPPs may themselves be derived from pro-erythroid and pro-megakaryocyte HSCs respectively as previously described [59–61]. Regardless of this, the increased purification of phenotypically distinct progenitors within the hematopoietic hierarchy afforded by our model will enable the investigation of molecular mechanisms involved in cell fate decisions with significantly greater precision. In fact the role of RUNX1 in lineage commitment was recently expanded to include promotion of megakaryopoiesis over erythropoiesis through repression of KLF1 [44]. Overexpression studies were performed solely using a RUNX1B construct and knockdown was non-isoform specific, so it remained unclear how important the isoform specificity is to the process of megakaryocytic or erythroid lineage commitment. Our investigation of the P1-GFP/GFP model suggests RUNX1C plays a specific role in these lineages, as its absence means circulating platelet numbers are decreased whereas BM CFUes and EryCs are increased. However, we cannot discount the fact that this phenotype may be due to an overall decrease in RUNX1 protein as opposed to the specific loss of RUNX1C and therefore further studies utilizing either targeted mutagenesis of the two Runx1 promoters separately or isoform-specific knockdown whilst not impacting the overall level of RUNX1 would be required to explore this possibility.
As previously mentioned, P1 is the dominant promoter in adult hematopoiesis, being active in all Runx1-expressing populations. P2 expression is far more heterogeneous, confined to immature/progenitor subsets of the GM and lymphoid lineages and megakaryocytes. With the exception of megakaryocytes, it appears that downregulation of P2 is a prerequisite of terminal differentiation of these lineages. We also observed that, at least in myeloid lineages, P2-expression correlates with enhanced CFU-C activity and in the PreMegE specifically with increased proliferation. Numerous cell cycle regulators are upregulated in P2+ PreMegEs, several of which have previously been identified as putative RUNX1-targets. A unique feature of megakaryocytic differentiation is polyploidisation achieved through undergoing numerous abortive cell cycles. Cell cycle activators are therefore highly expressed in these cells, as is Runx1 P2. It is also of interest that P2 expression has previously been observed in newly emerging embryonic HSC-containing hematopoietic clusters but not in the more quiescent BM HSC populations [28]. Despite their distinct roles in hematopoiesis, many parallels have been drawn between the specification of HSCs and megakaryopoiesis [62]. HSCs and MkPs share similar cell surface marker profiles and have numerous regulatory pathways in common [63,64]. These include critical dependence on TPO signaling and hematopoietic transcription factors, including the CBF complex, Ets and HOX-related genes, several of which are upregulated in P2+ PreMegEs (S4 Table) [62,65]. Megakaryocytes appear to have more in common with embryonic than adult HSCs, their production being characterized by CD41 expression and RUNX1-dependency [7,66]. Our observations therefore suggest that expression of P2-driven RUNX1B may actively promote cell cycling, with a role in expanding HSPC numbers and is then downregulated to allow terminal differentiation of the B/T/GM and erythroid lineages. It would be of interest to investigate to what extent RUNX1B, and also RUNX1C, indeed directly regulate different transcriptional targets and the mechanisms through which they may achieve this.
In addition to erythroid progenitors, mast cell progenitor specification did not appear to require P2 expression. Intriguingly, both are lineages which do not appear to be adversely affected by the absence of Runx1; complete ablation of Runx1 in adult mice has no impact on peripheral red blood cell numbers, whereas mast cell development is normal in Runx1 P1-null mice [14,67]. Whether this suggests Runx1 expression is entirely incidental in these lineages will need to be investigated further.
The requirement for WT RUNX1 activity in AML has been extensively studied in recent years. In AML1-ETO CBF AML in particular, a balance of AML1-ETO and RUNX1 expression must be maintained to promote stem cell gene expression and repress differentiation-associated gene expression [68]. Moreover, it appears AML1-ETO may directly regulate Runx1 expression, as depletion of AML1-ETO leads to a decrease in RUNX1 levels in Kasumi1 cells [45]. However, whether expression of P1 or P2 was favored in this context had not been investigated. By utilizing an inducible AML1-ETO mouse model, we were able to establish that AML1-ETO expression resulted in a specific upregulation of Runx1 P2. We have found P2 expression coincides with enhanced CFU activity and proliferation in HSPCs. Ben-Ami et al. previously demonstrated RUNX1 enhances the viability of preleukemic AML1-ETO-expressing cells [21], therefore it may be that RUNX1B activity specifically enhances a preleukemic phenotype in emerging CBF AML leukemia propagating cells. Interestingly, Trombly et al. observed the recruitment of AML1-ETO to P1 and the +23 enhancer but not to P2 in Kasumi1 cells [45]. Therefore, the mechanism of AML1-ETO’s activation of P2 is of considerable interest. AML1-ETO may directly activate P2, potentially via the +23 enhancer or it may instead promote expression of other transcriptional activators which enhance P2 activity. Alternatively, AML1-ETO may directly repress P1, resulting in a compensatory upregulation of P2 by a secondary mechanism. These possibilities will all need to be explored further.
Materials and Methods
Mice
P1-GFP::P2-hCD4 and P1-GFP mice have previously been described [29]. The AML1-ETO9a-IRES-GFP/rtTA mice were generated as follows: HA-tagged AML1-ETO9a cDNAs (provided by the Zhang laboratory [47]) were subcloned into a tet-ON vector in front of an IRES-GFP as described [69]. Ainv18 ES cells [69] (which constitutively express the rtTA under the control of the Rosa26 promoter) were then transfected with this tetracycline-inducible AML1-ETO9a construct by electroporation and stably transfected clones were selected with G418 (0.5mg/ml, Life Technologies) for 10–14 days. Chimeric mice were then generated by injecting AML1-ETO9a-IRES-GFP/rtTA ES cells into C57BL6J blastocysts. To induce AML1-ETO9a-IRES-GFP expression, 12 week-old mice were fed irradiated diet supplemented with 545mg/kg Doxycycline (ssniff Spezialdiäten GmbH) for 8 days prior to humane culling and tissue collection. All animal work was performed under regulations governed by UK Home Office Legislation under the Animals (Scientific Procedures) Act 1986. Details of animal husbandry and tissue collection are listed in S1 File.
Flow cytometry analysis and cell sorting
Dead cells were excluded using either 0.5μg/ml 7-Aminoactinomycin D (7-AAD, eBioscience) or 1μg/ml Hoechst 33258 (Life Technologies). Biotinylated antibody staining was detected by a secondary incubation step with fluorochrome-conjugated Streptavidin. Prior to flow sorting of HSPCs, bone marrow cells stained with biotinylated anti-lineage antibodies were lineage-depleted using anti-biotin-conjugated magnetic beads (Miltenyi) and then stained with additional antibodies, including conjugated streptavidin. Red blood cell depletion was performed by treatment with ACK lysis buffer (154mM ammonium chloride, 9.99mM potassium bicarbonate, 0.110mM EDTA) for 5 minutes at room temperature, followed by quenching with Phosphate-Buffered Saline (PBS).
Details of flow cytometry antibodies and reagents are listed in S1 Table. Details of flow cytometry antibody combinations used for each analysis or sort are listed in S2 Table.
For cell cycle analysis, in vivo EdU incorporation was performed by injecting 1.125mg EdU dissolved in PBS intraperitoneally into adult mice. After two hours, bone marrow was harvested and stained with hematopoietic stem and progenitor cell surface markers as detailed in S2 Table. Cells were then stained using the Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Life Technologies). Total DNA was stained with 1μg/ml FxCycle Violet Stain (Life Technologies).
Cells were analyzed using a LSR-II or LSR-II Fortessa analyzer, a FACSAria-II cell sorter or a FACSAria-III cell sorter (BD).
Automated peripheral blood counts
Tail vein blood (no more than 50μl per mouse) was sampled from 12 week old mice using heparinized end-to-end Micro Pipettes (Vitrex) and analyzed on a Sysmex XT 2000i analyzer, according to the manufacturer’s instructions.
Cell culture
OP9 and OP9-DL1 co-culture
Cells were cultured in 5% CO2 and 5% O2 at 37°C. Mouse OP9 stromal cells were maintained in Alpha-MEM (Lonza) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich), 2mM L-glutamine (Gibco), 1x Penicillin-Streptomycin (Sigma-Aldrich) and were routinely tested for mycoplasma contamination. For myeloid OP9 co-culture PreMegE and LSK48F- MPPs were seeded on mouse OP9 stromal cells (approximately 5000/ml) at a density of approximately 2000 cells/ml in IMDM (Lonza) supplemented with 10% Fetal Bovine Serum (FBS) for Mouse Myeloid Colony-Forming Cells (StemCell Technologies), 0.45mM monothioglycerol (MTG, Sigma-Aldrich), 2mM L-glutamine (Gibco), 1x Penicillin-Streptomycin (100U/ml Penicillin, 100μg/ml Streptomycin, Sigma-Aldrich), 2U/ml erythropoietin (Eprex, Janssen-Cilag Ltd), and medium conditioned by cell lines producing IL3, TPO and Stem Cell Factor (SCF) (1% final concentration). PreMegEs were cultured for 7 days and LSK48F- MPPs for 8 days before being harvested and stained for myeloid cell surface markers (TER119, CD41, GR1 and CD11B as detailed in S2 Table). For single cell co-culture, single PreMegE cells were directly sorted into 96-well plate wells containing 100μl of the above medium and approximately 500 OP9 cells and cultured for 7 days before positive wells were assessed by microscopy and FACS analyzed as above.
For B cell and T cell co-culture LSK48F- MPPs were seeded on mouse OP9 and OP9-DL1 stromal cells respectively at a density of approximately 2000 cells/ml in IMDM supplemented with 20% FBS (Harlan), 2mM L-glutamine, 1x Penicillin-Streptomycin, 5ng/ml FLT3L (PeproTech) and 1ng/ml IL7 (PeproTech). Cultures were passed into fresh media every 4–5 days for 21 days (with IL7 concentrations reduced to 0.25ng/ml in OP9-DL1 co-cultures from day 12 onwards) and harvested and stained for B and T cell surface markers (CD45R (B220), CD19, CD4 and CD8A as detailed in S2 Table).
Short-term culture of HSPCs
Purified PreGM, GMP and PreMegE cells were cultured at 37°C in 5% CO2 and atmospheric O2 for 6–18 hours in pro-myeloid medium (IMDM supplemented with 10% FBS for Mouse Myeloid Colony-Forming Cells, 10% Protein-free hybridoma medium (PFHM-II, Gibco), 180μg/ml Transferrin (Roche Diagnostics), 0.45mM MTG, 50ng/ml ascorbic acid (Sigma-Aldrich), 2mM L-glutamine, 1x Penicillin-Streptomycin, 4U/ml erythropoietin, 5ng/ml IL11 (R&D Systems), 10ng/ml IL6 (R&D Systems), 10ng/ml M-CSF (R&D Systems) and medium conditioned by cell lines producing IL3, GMCSF, TPO and SCF (1% final concentration)). Purified LSK48F- MPPs were cultured for 12–24 hours in Alpha MEM (Lonza) supplemented with 10% FBS, 0.45mM MTG, 2mM L-glutamine, 1x Penicillin-Streptomycin, 1U/ml erythropoietin, and medium conditioned by cell lines producing IL3, TPO and SCF (1% final concentration)) (adapted from [70]). Cells were then harvested and stained with either LSK HSPC markers or myeloid progenitor markers as described in S2 Table.
For longer-term culture (7–11 days), PreGM and GMP cells were cultured in pro-myeloid medium as described above before being harvested and stained with GM and mast lineage markers (CD11B, GR1, F4/80, C-KIT and FCΕR1Α as detailed in S2 Table).
Sorted MkP cells were cultured for 4 days in pro-myeloid medium (see above).
Hematopoietic colony-forming assays
Methylcellulose colony-forming assays were performed as previously described [71], except 10% (FBS) (StemCell Technologies) was added in place of the Fetal Bovine Plasma-derived Serum Platelet Poor (PDS). 200 PreMegEs or 50 LSK48F- MPPs were plated per dish in duplicate. CFUes were scored after 3–4 days and other colonies after 8 days under a microscope (DM IL; Leica). Megakaryocyte-specific colony-forming assays were performed by plating 1000 MkPs or PreMegEs in MegaCultTM medium (StemCell Technologies). Cells were cultured, fixed, stained and analyzed according to the manufacturer’s protocols.
Gene expression
Quantitative PCR
RNA was extracted using the RNeasy Plus Micro Kit (QIAGEN). Complementary DNA was synthesized using the SuperScript III First-Strand Synthesis System (Life Technologies). Quantitative PCR (qPCR) was performed using Universal ProbeLibrary assays (Roche); primers and probes are listed in S3 Table. With the exception of TER119+ erythroid and CD11b+ GR1+ GM lineage cells, expression values were normalized to beta-actin (b-actin). Owing to the significant variation of b-actin in these lineages, total Runx1 expression in these compartments was calculated relative to input and normalized to the level in CD11b+ GR1+ cells. To determine the relative contributions of Runx1 P1 and P2 transcripts to total Runx1 expression, the relative efficiencies of the qPCR primers were calculated in a titration experiment using known quantities of P1 and P2-expressing plasmid template DNA.
RNA sequencing
Tissues from 3 mice were pooled per sample. Total RNA was extracted from purified PreMegEs as described above. Indexed PolyA libraries were prepared using 50ng of total RNA and 16 cycles of amplification in the Agilent SureSelect Strand-Specific RNA Library Prep Kit for Illumina Sequencing (Agilent). Libraries were quantified by qPCR using a KAPA Library Quantification Kit for Illumina platforms (Kapa Biosystems Inc.). Paired-end 75bp sequencing was carried out by clustering 1.7pM of the pooled libraries on a NextSeq 500 sequencer (Illumina Inc.) for 3 biological replicates per population. Details of sequence data analysis are given in S1 File.
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [72] and are accessible through GEO series accession number GSE68958.
Statistical analysis
Flow cytometry plots display the mean values of each indicated population. Unless otherwise indicated, data were evaluated using an Ordinary 2-way ANOVA and expressed as mean ± standard error of the mean (SEM). P<0.05 was considered statistically significant.
*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001
Supporting Information
(Related to Fig 2) (A–F) Contour plots of lineage marker expression in adult BM (A, B, C), spleen (D and F) and thymus (E). (A) CD71/Ter119 expression of live cells (left) and CD71 expression/FSC of Ter119 high cells (right) in BM. (B) CD11b/Gr1 (left) and CD11b/F4/80 (right) expression in live BM cells. (C) B220/CD11b+CD3ε+Ter119 expression in live cells (left), cKit/CD19 expression in B220+ cells (middle) and IgM/IgD expression in B220+ CD19+ cKit- cells (right) in BM. (D) B220/CD19 expression in live cells (left) and IgM/IgD expression in B220+ CD19+ cells (right) in spleen. (E) CD4/CD8a expression in live cells (left) and CD25/CD44 expression in CD4 CD8 double negative (DN) cells (right) in thymus. (F) CD4/CD8a expression in live spleen cells.
(PDF)
(Related to Fig 2) (A-C) Gene expression analysis of Runx1 P1 and Runx1 P2 as a proportion of total Runx1 in erythroid and GM (A), B (B) and T (C) cell lineage populations isolated from WT BM, spleen and thymus. (n = 3.) Owing to the high variability in b-actin expression in mature erythroid and GM blood cells, gene expression values for these populations were normalized to input and expressed relative to CD11b+ Gr1+ cells. Otherwise gene expression is depicted relative to b-actin.
(PDF)
(Related to Figs 3–6) (A-C) Gene expression analysis of total Runx1 (A), Runx1 P1 (B) and Runx1 P2 (C) in Lin- cKit+ HSPC populations isolated from WT BM, normalized to b-actin (n = 3).
(PDF)
(Related to Fig 3) Representative FACS plots of LSK48F- MPPs following 18 hours in vitro myeloid culture. (A) Lin, cKit/Sca1, CD150/CD41, CD16/32 and Endoglin/CD150 expression of WT, P2+ and P2- cultured LSK48F- cells. (B) P1-GFP/P2-hCD4 expression of LSK48F- derived immunophenotypic LSK and PreMegE cells (n = 3).
(PDF)
(Related to Fig 4) (A–B) Top: FACS plots of CD11b/F4/80 and cKit/FcεR1α expression of day 11 cultured PreGM (A) and GMP (B) cells. Bottom: quantification of GM subsets following 7 and 11 days culture. (n = 4). (C) P1-GFP/P2-hCD4 expression in the BM Lin- cKit+ Sca1- FcεR1α- CD27- Ly6c- Integrin Beta7+ MCp. (Representative of 3 independent experiments.) (D-E) Representative FACS plots of PreGMs (D) and GMPs (E) following 12 hours in vitro myeloid culture. (D) CD16/32/CD150, Endoglin/CD150 and P1-GFP/P2-hCD4 expression of PreGM-derived cells. (E) CD16/32/Cd150 and P1-GFP/P2-hCD4 of GMP-derived cells. (n = 3)
(PDF)
(Related to Fig 5) (A) CFU-C activity of WT and P1+ P2+ MkPs following culture in MegaCultTM medium. (n = 5). (B) Cell cycle status of WT, P1+ P2- and P1+ P2+ PreMegEs, as determined by in vivo EdU incorporation and DNA content analysis. (n = 3). (C) Table showing the clonal analysis of P1+ P2- and P1+ P2+ PreMegEs. Shown are the numbers of positive wells at day 7 of OP9 co-culture relative to the numbers of single PreMegE cells plated on day 0 and the proportion of wells that contained CD41+ cells (Mk only), Ter119+ cells (Ery only) or CD41+ and Ter119+ cells (Mk + Ery). Data are compiled from 5 independent experiments.
(PDF)
(Related to Fig 6) (A) Principal component analysis of the RNA Seq expression data from WT, P2- and P2+ PreMegE cells. (B-D) Heat maps depicting expression of gene sets from GSEA plots (displayed in Fig 6B) in P2- and P2+ PreMegE cells. (E–F) Diseases/functions upregulated (E) or downregulated (F) in P2+ PreMegEs compared to P2- PreMegEs as determined by IPA. (G–H) Signaling pathways activated (G) or inhibited (H) in P2+ PreMegEs compared to P2- PreMegEs as determined by IPA.
(PDF)
(Related to Fig 6) (A) Log fold change (counts) of cell surface markers in P2+ PreMegEs compared to P2- PreMegEs as determined by RNA Seq. (B) RPKM values of cell surface markers in WT, P2- and P2+ PreMegEs. (n = 3). (C) Representative FACS plot of CD61 expression in P2+ and P2- PreMegEs. (n = 2). (D) Ratio of median fluorescence intensity (MFI) and percentage of positive cells for CD61 and CD34 protein expression in P2+ PreMegEs compares to P2- PreMegEs, determined by FACS. (n = 2). (E) Table showing the clonal analysis of CD34+ and CD34- WT PreMegEs. Shown are numbers of positive wells at day 7 of OP9 co-culture relative to the numbers of single PreMegE cells plated on day 0 and the proportion of wells that contained CD41+ cells (Mk only), Ter119+ cells (Ery only) or CD41+ and Ter119+ cells (Mk + Ery). Data are compiled from 3 independent experiments.
(PDF)
(Related to Fig 7) (A-H) Quantitation of Hematocrit (A), Total hemoglobin concentration (B), Reticulocyte count (C), Monocyte count (D), Neutrophil count (E), Lymphocyte count (F), Plateletcrit (G) and Mean Platelet Volume (H) in peripheral blood of WT, P1-GFP/+ and P1-GFP/GFP mice. (n = 4)
(PDF)
(Related to Fig 7) (A—B) Numbers of GM (A) and B (B) lineage cells as a proportion of live ACK-lysed BM cells. (C) Numbers of B lineage cells as a proportion of live ACK-lysed spleen cells (n = 4).
(PDF)
(A-B) rVISTA analysis of EKLF, FLI1, GATA1 and TAL1 binding motifs located in regions surrounding Runx1 P1 (A) and Runx1 P2 (B). Top, Blue: All motifs found in the mouse loci. Top, Green: Motifs conserved between mouse and human. Middle: Genetic conservation between mouse and human loci. Bottom: Schematic of mouse Runx1 locus, aligned to conservation map. (C) UCSC Browser tracks of mouse Runx1 locus, featuring GATA1 and TAL1 ChIP-seq data (performed in megakaryocytes and erythroblasts) acquired by the Mouse ENCODE Project, aligned to UCSC Runx1 transcripts and vertebrate genetic conservation data.
(PDF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
Acknowledgments
We thank members of the following facilities of the Cancer Research UK Manchester Institute for technical support: Biological Resources Unit, Advanced Imaging and Flow Cytometry and the Molecular Biology Core Facility.
Data Availability
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO series accession number GSE68958.
Funding Statement
Work in our laboratory is supported by the Leukaemia and Lymphoma Research Foundation (LLR#12037, https://leukaemialymphomaresearch.org.uk/), the Biotechnology and Biological Sciences Research Council (BBSRC# BB/I001794/1, http://www.bbsrc.ac.uk/) and Cancer Research UK (CRUK#A20971, http://www.cancerresearchuk.org/).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Durst KL, Hiebert SW (2004) Role of RUNX family members in transcriptional repression and gene silencing. Oncogene 23: 4220–4224. [DOI] [PubMed] [Google Scholar]
- 2.Yamagata T, Maki K, Mitani K (2005) Runx1/AML1 in normal and abnormal hematopoiesis. Int J Hematol 82: 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Lacaud G, Gore L, Kennedy M, Kouskoff V, Kingsley P, et al. (2002) Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood 100: 458–466. [DOI] [PubMed] [Google Scholar]
- 4.Lorsbach RB, Moore J, Ang SO, Sun W, Lenny N, et al. (2004) Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood 103: 2522–2529. [DOI] [PubMed] [Google Scholar]
- 5.North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MF (2004) Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells 22: 158–168. [DOI] [PubMed] [Google Scholar]
- 6.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA (2009) Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457: 887–891. 10.1038/nature07619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, et al. (2009) The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457: 892–895. 10.1038/nature07679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84: 321–330. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, et al. (1996) Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A 93: 3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lancrin C, Mazan M, Stefanska M, Patel R, Lichtinger M, et al. (2012) GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood 120: 314–322. 10.1182/blood-2011-10-386094 [DOI] [PubMed] [Google Scholar]
- 11.Lie ALM, Marinopoulou E, Li Y, Patel R, Stefanska M, et al. (2014) RUNX1 positively regulates a cell adhesion and migration program in murine hemogenic endothelium prior to blood emergence. Blood 124: e11–20. 10.1182/blood-2014-04-572958 [DOI] [PubMed] [Google Scholar]
- 12.Thambyrajah R, Mazan M, Patel R, Moignard V, Stefanska M, et al. (2015) GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat Cell Biol. [DOI] [PubMed] [Google Scholar]
- 13.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR (2007) The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med 204: 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, et al. (2005) Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichikawa M, Asai T, Chiba S, Kurokawa M, Ogawa S (2004) Runx1/AML-1 ranks as a master regulator of adult hematopoiesis. Cell Cycle 3: 722–724. [PubMed] [Google Scholar]
- 16.Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, et al. (2004) AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med 10: 299–304. [DOI] [PubMed] [Google Scholar]
- 17.Pencovich N, Jaschek R, Dicken J, Amit A, Lotem J, et al. (2013) Cell-autonomous function of Runx1 transcriptionally regulates mouse megakaryocytic maturation. PLoS One 8: e64248 10.1371/journal.pone.0064248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacob B, Osato M, Yamashita N, Wang CQ, Taniuchi I, et al. (2010) Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood 115: 1610–1620. 10.1182/blood-2009-07-232249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai X, Gaudet JJ, Mangan JK, Chen MJ, De Obaldia ME, et al. (2011) Runx1 loss minimally impacts long-term hematopoietic stem cells. PLoS One 6: e28430 10.1371/journal.pone.0028430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naoe T, Kiyoi H (2013) Gene mutations of acute myeloid leukemia in the genome era. Int J Hematol 97: 165–174. 10.1007/s12185-013-1257-4 [DOI] [PubMed] [Google Scholar]
- 21.Ben-Ami O, Friedman D, Leshkowitz D, Goldenberg D, Orlovsky K, et al. (2013) Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep 4: 1131–1143. 10.1016/j.celrep.2013.08.020 [DOI] [PubMed] [Google Scholar]
- 22.Goyama S, Schibler J, Cunningham L, Zhang Y, Rao Y, et al. (2013) Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J Clin Invest 123: 3876–3888. 10.1172/JCI68557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baek D, Davis C, Ewing B, Gordon D, Green P (2007) Characterization and predictive discovery of evolutionarily conserved mammalian alternative promoters. Genome Res 17: 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levanon D, Groner Y (2004) Structure and regulated expression of mammalian RUNX genes. Oncogene 23: 4211–4219. [DOI] [PubMed] [Google Scholar]
- 25.Rennert J, Coffman JA, Mushegian AR, Robertson AJ (2003) The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evol Biol 3: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujita Y, Nishimura M, Taniwaki M, Abe T, Okuda T (2001) Identification of an alternatively spliced form of the mouse AML1/RUNX1 gene transcript AML1c and its expression in early hematopoietic development. Biochem Biophys Res Commun 281: 1248–1255. [DOI] [PubMed] [Google Scholar]
- 27.Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, et al. (1995) Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res 23: 2762–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bee T, Liddiard K, Swiers G, Bickley SR, Vink CS, et al. (2009) Alternative Runx1 promoter usage in mouse developmental hematopoiesis. Blood Cells Mol Dis 43: 35–42. 10.1016/j.bcmd.2009.03.011 [DOI] [PubMed] [Google Scholar]
- 29.Sroczynska P, Lancrin C, Kouskoff V, Lacaud G (2009) The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood 114: 5279–5289. 10.1182/blood-2009-05-222307 [DOI] [PubMed] [Google Scholar]
- 30.van Riel B, Pakozdi T, Brouwer R, Monteiro R, Tuladhar K, et al. (2012) A novel complex, RUNX1-MYEF2, represses hematopoietic genes in erythroid cells. Mol Cell Biol 32: 3814–3822. 10.1128/MCB.05938-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, et al. (2005) SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121: 1109–1121. [DOI] [PubMed] [Google Scholar]
- 32.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, et al. (2005) Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121: 295–306. [DOI] [PubMed] [Google Scholar]
- 33.Dore LC, Crispino JD (2011) Transcription factor networks in erythroid cell and megakaryocyte development. Blood 118: 231–239. 10.1182/blood-2011-04-285981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swiers G, Patient R, Loose M (2006) Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev Biol 294: 525–540. [DOI] [PubMed] [Google Scholar]
- 35.Alexander WS, Roberts AW, Nicola NA, Li R, Metcalf D (1996) Deficiencies in progenitor cells of multiple hematopoietic lineages and defective megakaryocytopoiesis in mice lacking the thrombopoietic receptor c-Mpl. Blood 87: 2162–2170. [PubMed] [Google Scholar]
- 36.Yu M, Cantor AB (2012) Megakaryopoiesis and thrombopoiesis: an update on cytokines and lineage surface markers. Methods Mol Biol 788: 291–303. 10.1007/978-1-61779-307-3_20 [DOI] [PubMed] [Google Scholar]
- 37.Fox NE, Kaushansky K (2005) Engagement of integrin alpha4beta1 enhances thrombopoietin-induced megakaryopoiesis. Exp Hematol 33: 94–99. [DOI] [PubMed] [Google Scholar]
- 38.Foudi A, Kramer DJ, Qin J, Ye D, Behlich AS, et al. (2014) Distinct, strict requirements for Gfi-1b in adult bone marrow red cell and platelet generation. J Exp Med 211: 909–927. 10.1084/jem.20131065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutierrez L, Tsukamoto S, Suzuki M, Yamamoto-Mukai H, Yamamoto M, et al. (2008) Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood 111: 4375–4385. 10.1182/blood-2007-09-115121 [DOI] [PubMed] [Google Scholar]
- 40.Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, et al. (1991) Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349: 257–260. [DOI] [PubMed] [Google Scholar]
- 41.Vyas P, Ault K, Jackson CW, Orkin SH, Shivdasani RA (1999) Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood 93: 2867–2875. [PubMed] [Google Scholar]
- 42.Hayashi K, Natsume W, Watanabe T, Abe N, Iwai N, et al. (2000) Diminution of the AML1 transcription factor function causes differential effects on the fates of CD4 and CD8 single-positive T cells. J Immunol 165: 6816–6824. [DOI] [PubMed] [Google Scholar]
- 43.Sun W, Downing JR (2004) Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood 104: 3565–3572. [DOI] [PubMed] [Google Scholar]
- 44.Kuvardina ON, Herglotz J, Kolodziej S, Kohrs N, Herkt S, et al. (2015) RUNX1 represses the erythroid gene expression program during megakaryocytic differentiation. Blood 125: 3570–3579. 10.1182/blood-2014-11-610519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trombly DJ, Whitfield TW, Padmanabhan S, Gordon JA, Lian JB, et al. (2015) Genome-wide co-occupancy of AML1-ETO and N-CoR defines the t(8;21) AML signature in leukemic cells. BMC Genomics 16: 309 10.1186/s12864-015-1445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao YQ, Chen ZX, He J, Cen JN, Bao XJ, et al. (2007) [Expression of AML1/ETO9a isoform in acute myeloid leukemia-M2 subtype]. Zhonghua Xue Ye Xue Za Zhi 28: 27–29. [PubMed] [Google Scholar]
- 47.Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, et al. (2006) A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med 12: 945–949. [DOI] [PubMed] [Google Scholar]
- 48.Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, et al. (2011) Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell 19: 138–152. 10.1016/j.ccr.2010.12.012 [DOI] [PubMed] [Google Scholar]
- 49.Cabezas-Wallscheid N, Eichwald V, de Graaf J, Lower M, Lehr HA, et al. (2013) Instruction of haematopoietic lineage choices, evolution of transcriptional landscapes and cancer stem cell hierarchies derived from an AML1-ETO mouse model. EMBO Mol Med 5: 1804–1820. 10.1002/emmm.201302661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akashi K, Traver D, Miyamoto T, Weissman IL (2000) A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193–197. [DOI] [PubMed] [Google Scholar]
- 51.Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, et al. (2007) Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1: 428–442. 10.1016/j.stem.2007.07.005 [DOI] [PubMed] [Google Scholar]
- 52.Levantini E, Lee S, Radomska HS, Hetherington CJ, Alberich-Jorda M, et al. (2011) RUNX1 regulates the CD34 gene in haematopoietic stem cells by mediating interactions with a distal regulatory element. EMBO J 30: 4059–4070. 10.1038/emboj.2011.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Consortium EP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mouse EC, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, et al. (2012) An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13: 418 10.1186/gb-2012-13-8-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32: W273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM (2002) rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res 12: 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. (2002) The human genome browser at UCSC. Genome Res 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, et al. (2013) ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res 41: D56–63. 10.1093/nar/gks1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pina C, Fugazza C, Tipping AJ, Brown J, Soneji S, et al. (2012) Inferring rules of lineage commitment in haematopoiesis. Nat Cell Biol 14: 287–294. 10.1038/ncb2442 [DOI] [PubMed] [Google Scholar]
- 60.Sanjuan-Pla A, Macaulay IC, Jensen CT, Woll PS, Luis TC, et al. (2013) Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature 502: 232–236. 10.1038/nature12495 [DOI] [PubMed] [Google Scholar]
- 61.Shin JY, Hu W, Naramura M, Park CY (2014) High c-Kit expression identifies hematopoietic stem cells with impaired self-renewal and megakaryocytic bias. J Exp Med 211: 217–231. 10.1084/jem.20131128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang H, Cantor AB (2009) Common features of megakaryocytes and hematopoietic stem cells: what's the connection? J Cell Biochem 107: 857–864. 10.1002/jcb.22184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakorn TN, Miyamoto T, Weissman IL (2003) Characterization of mouse clonogenic megakaryocyte progenitors. Proc Natl Acad Sci U S A 100: 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spangrude GJ, Heimfeld S, Weissman IL (1988) Purification and characterization of mouse hematopoietic stem cells. Science 241: 58–62. [DOI] [PubMed] [Google Scholar]
- 65.Murone M, Carpenter DA, de Sauvage FJ (1998) Hematopoietic deficiencies in c-mpl and TPO knockout mice. Stem Cells 16: 1–6. [DOI] [PubMed] [Google Scholar]
- 66.Robin C, Ottersbach K, Boisset JC, Oziemlak A, Dzierzak E (2011) CD41 is developmentally regulated and differentially expressed on mouse hematopoietic stem cells. Blood 117: 5088–5091. 10.1182/blood-2011-01-329516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mukai K, BenBarak MJ, Tachibana M, Nishida K, Karasuyama H, et al. (2012) Critical role of P1-Runx1 in mouse basophil development. Blood 120: 76–85. 10.1182/blood-2011-12-399113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ptasinska A, Assi SA, Martinez-Soria N, Imperato MR, Piper J, et al. (2014) Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep 8: 1974–1988. 10.1016/j.celrep.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kyba M, Perlingeiro RC, Daley GQ (2002) HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109: 29–37. [DOI] [PubMed] [Google Scholar]
- 70.Ema H, Morita Y, Yamazaki S, Matsubara A, Seita J, et al. (2006) Adult mouse hematopoietic stem cells: purification and single-cell assays. Nat Protoc 1: 2979–2987. [DOI] [PubMed] [Google Scholar]
- 71.Sroczynska P, Lancrin C, Pearson S, Kouskoff V, Lacaud G (2009) In vitro differentiation of mouse embryonic stem cells as a model of early hematopoietic development. Methods Mol Biol 538: 317–334. 10.1007/978-1-59745-418-6_16 [DOI] [PubMed] [Google Scholar]
- 72.Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Related to Fig 2) (A–F) Contour plots of lineage marker expression in adult BM (A, B, C), spleen (D and F) and thymus (E). (A) CD71/Ter119 expression of live cells (left) and CD71 expression/FSC of Ter119 high cells (right) in BM. (B) CD11b/Gr1 (left) and CD11b/F4/80 (right) expression in live BM cells. (C) B220/CD11b+CD3ε+Ter119 expression in live cells (left), cKit/CD19 expression in B220+ cells (middle) and IgM/IgD expression in B220+ CD19+ cKit- cells (right) in BM. (D) B220/CD19 expression in live cells (left) and IgM/IgD expression in B220+ CD19+ cells (right) in spleen. (E) CD4/CD8a expression in live cells (left) and CD25/CD44 expression in CD4 CD8 double negative (DN) cells (right) in thymus. (F) CD4/CD8a expression in live spleen cells.
(PDF)
(Related to Fig 2) (A-C) Gene expression analysis of Runx1 P1 and Runx1 P2 as a proportion of total Runx1 in erythroid and GM (A), B (B) and T (C) cell lineage populations isolated from WT BM, spleen and thymus. (n = 3.) Owing to the high variability in b-actin expression in mature erythroid and GM blood cells, gene expression values for these populations were normalized to input and expressed relative to CD11b+ Gr1+ cells. Otherwise gene expression is depicted relative to b-actin.
(PDF)
(Related to Figs 3–6) (A-C) Gene expression analysis of total Runx1 (A), Runx1 P1 (B) and Runx1 P2 (C) in Lin- cKit+ HSPC populations isolated from WT BM, normalized to b-actin (n = 3).
(PDF)
(Related to Fig 3) Representative FACS plots of LSK48F- MPPs following 18 hours in vitro myeloid culture. (A) Lin, cKit/Sca1, CD150/CD41, CD16/32 and Endoglin/CD150 expression of WT, P2+ and P2- cultured LSK48F- cells. (B) P1-GFP/P2-hCD4 expression of LSK48F- derived immunophenotypic LSK and PreMegE cells (n = 3).
(PDF)
(Related to Fig 4) (A–B) Top: FACS plots of CD11b/F4/80 and cKit/FcεR1α expression of day 11 cultured PreGM (A) and GMP (B) cells. Bottom: quantification of GM subsets following 7 and 11 days culture. (n = 4). (C) P1-GFP/P2-hCD4 expression in the BM Lin- cKit+ Sca1- FcεR1α- CD27- Ly6c- Integrin Beta7+ MCp. (Representative of 3 independent experiments.) (D-E) Representative FACS plots of PreGMs (D) and GMPs (E) following 12 hours in vitro myeloid culture. (D) CD16/32/CD150, Endoglin/CD150 and P1-GFP/P2-hCD4 expression of PreGM-derived cells. (E) CD16/32/Cd150 and P1-GFP/P2-hCD4 of GMP-derived cells. (n = 3)
(PDF)
(Related to Fig 5) (A) CFU-C activity of WT and P1+ P2+ MkPs following culture in MegaCultTM medium. (n = 5). (B) Cell cycle status of WT, P1+ P2- and P1+ P2+ PreMegEs, as determined by in vivo EdU incorporation and DNA content analysis. (n = 3). (C) Table showing the clonal analysis of P1+ P2- and P1+ P2+ PreMegEs. Shown are the numbers of positive wells at day 7 of OP9 co-culture relative to the numbers of single PreMegE cells plated on day 0 and the proportion of wells that contained CD41+ cells (Mk only), Ter119+ cells (Ery only) or CD41+ and Ter119+ cells (Mk + Ery). Data are compiled from 5 independent experiments.
(PDF)
(Related to Fig 6) (A) Principal component analysis of the RNA Seq expression data from WT, P2- and P2+ PreMegE cells. (B-D) Heat maps depicting expression of gene sets from GSEA plots (displayed in Fig 6B) in P2- and P2+ PreMegE cells. (E–F) Diseases/functions upregulated (E) or downregulated (F) in P2+ PreMegEs compared to P2- PreMegEs as determined by IPA. (G–H) Signaling pathways activated (G) or inhibited (H) in P2+ PreMegEs compared to P2- PreMegEs as determined by IPA.
(PDF)
(Related to Fig 6) (A) Log fold change (counts) of cell surface markers in P2+ PreMegEs compared to P2- PreMegEs as determined by RNA Seq. (B) RPKM values of cell surface markers in WT, P2- and P2+ PreMegEs. (n = 3). (C) Representative FACS plot of CD61 expression in P2+ and P2- PreMegEs. (n = 2). (D) Ratio of median fluorescence intensity (MFI) and percentage of positive cells for CD61 and CD34 protein expression in P2+ PreMegEs compares to P2- PreMegEs, determined by FACS. (n = 2). (E) Table showing the clonal analysis of CD34+ and CD34- WT PreMegEs. Shown are numbers of positive wells at day 7 of OP9 co-culture relative to the numbers of single PreMegE cells plated on day 0 and the proportion of wells that contained CD41+ cells (Mk only), Ter119+ cells (Ery only) or CD41+ and Ter119+ cells (Mk + Ery). Data are compiled from 3 independent experiments.
(PDF)
(Related to Fig 7) (A-H) Quantitation of Hematocrit (A), Total hemoglobin concentration (B), Reticulocyte count (C), Monocyte count (D), Neutrophil count (E), Lymphocyte count (F), Plateletcrit (G) and Mean Platelet Volume (H) in peripheral blood of WT, P1-GFP/+ and P1-GFP/GFP mice. (n = 4)
(PDF)
(Related to Fig 7) (A—B) Numbers of GM (A) and B (B) lineage cells as a proportion of live ACK-lysed BM cells. (C) Numbers of B lineage cells as a proportion of live ACK-lysed spleen cells (n = 4).
(PDF)
(A-B) rVISTA analysis of EKLF, FLI1, GATA1 and TAL1 binding motifs located in regions surrounding Runx1 P1 (A) and Runx1 P2 (B). Top, Blue: All motifs found in the mouse loci. Top, Green: Motifs conserved between mouse and human. Middle: Genetic conservation between mouse and human loci. Bottom: Schematic of mouse Runx1 locus, aligned to conservation map. (C) UCSC Browser tracks of mouse Runx1 locus, featuring GATA1 and TAL1 ChIP-seq data (performed in megakaryocytes and erythroblasts) acquired by the Mouse ENCODE Project, aligned to UCSC Runx1 transcripts and vertebrate genetic conservation data.
(PDF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO series accession number GSE68958.