Abstract
Species identification of Nocardia is not straightforward due to rapidly evolving taxonomy, insufficient discriminatory power of conventional phenotypic methods and also of single gene locus analysis including 16S rRNA gene sequencing. Here we evaluated the ability of a 5-locus (16S rRNA, gyrB, secA1, hsp65 and rpoB) multilocus sequence analysis (MLSA) approach as well as that of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) in comparison with sequencing of the 5’-end 606 bp partial 16S rRNA gene to provide identification of 25 clinical isolates of Nocardia. The 5’-end 606 bp 16S rRNA gene sequencing successfully assigned 24 of 25 (96%) clinical isolates to species level, namely Nocardia cyriacigeorgica (n = 12, 48%), N. farcinica (n = 9, 36%), N. abscessus (n = 2, 8%) and N. otitidiscaviarum (n = 1, 4%). MLSA showed concordance with 16S rRNA gene sequencing results for the same 24 isolates. However, MLSA was able to identify the remaining isolate as N. wallacei, and clustered N. cyriacigeorgica into three subgroups. None of the clinical isolates were correctly identified to the species level by MALDI-TOF MS analysis using the manufacturer-provided database. A small “in-house” spectral database was established incorporating spectra of five clinical isolates representing the five species identified in this study. After complementation with the “in-house” database, of the remaining 20 isolates, 19 (95%) were correctly identified to species level (score ≥ 2.00) and one (an N. abscessus strain) to genus level (score ≥ 1.70 and < 2.00). In summary, MLSA showed superior discriminatory power compared with the 5’-end 606 bp partial 16S rRNA gene sequencing for species identification of Nocardia. MALDI-TOF MS can provide rapid and accurate identification but is reliant on a robust mass spectra database.
Introduction
Nocardia species are ubiquitous environmental bacteria that cause suppurative infections in humans, including in the lung, central nervous system and skin [1]. Nocardiosis typically occurs in immunosuppressed patients such as organ and stem cell transplantation, and malignancy, but also affects immunocompetent hosts [2–4]. Since there are species-specific differences with regard to geography and disease patterns, identification of Nocardia to species level is important to determine both epidemiology and clinical associations.
Conventional culture identification of Nocardia based on phenotypic methods and distinct (but limited) antimicrobial susceptibility profiles lacks sufficient discriminatory power, is slow and time-consuming and requires staff expertise [5, 6]. Thus identification of Nocardia isolates by molecular methods is increasingly used, of which PCR-based methods combined with DNA sequencing are the most popular. Of these, 16S rRNA gene sequencing is considered the “gold-standard” [6, 7]. However, 16S rRNA gene sequencing is unable to distinguish certain closely related species due to insufficient interspecies gene polymorphisms [6, 8], whilst also unable to resolve certain species, e.g. Nocardia nova due to the presence of multiple yet different copies of this gene [9]. To overcome this limitation, other gene polymorphisms have been evaluated, such as those within the β-subunit of the type II DNA topoisomerase gene (gyrB) [10, 11], the subunit A of the SecA preprotein translocase gene (secA1) [12, 13], the 65-kDa heat shock protein gene (hsp65) [14, 15], and the β subunit of DNA-dependent RNA polymerase gene (rpoB) [11]. A multilocus sequence analysis (MLSA) scheme analyzing sequence polymorphisms of multiple Nocardia genes was reported to be accurate not only for the identification of known Nocardia species but the unveiling of novel species [16].
Other than genomic approaches, proteomic methods, most notably matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) method, have also been evaluated to identify Nocardia species in clinical laboratories. However, the presence of aliphatic acids in the cell wall of Nocardia has posed an obstacle in achieving satisfactory protein profiles. Furthermore, although improvements are continuing, many databases provided by commercial MALDI-TOF MS systems contain only a limited number of archived Nocardia spectral profiles [17, 18]. Previous studies have reported identification to species level in only 14.9–80.4% of isolates [18–23]. Complementation with profiles provided by “in-house” databases, which in turn relies on knowledge of local epidemiology, hence may assist with identification to the species, or even, genus level.
In the present study, we firstly evaluated a published MLSA scheme [16] employing polymorphisms in the 16S rRNA, gyrB, secA1, hsp65 and rpoB loci for the identification of clinical Nocardia isolates in our laboratory; and secondly, determined the ability of MALDI-TOF MS for species assignment. Results were compared to those obtained by the 5’-end 606 bp 16S rRNA gene sequencing. An “in-house” database of Nocardia protein profiles was established and evaluated for its ability to complement a commercial database for identification of Nocardia species.
Materials and Methods
Ethics
The study was approved by the Human Research Ethics Committee of Peking Union Medical College Hospital (PUMCHBC-C-2-Q01-1). Written informed consent was obtained from patients for the use of the samples in research.
Nocardia strains and reference sequences
Twenty-five clinical strains were studied in the evaluation of the ability of an MLSA scheme [16] and MALDI-TOF MS to provide species identification. Isolates were cultured from patients admitted to the Peking Union Medical College Hospital from January 2009 to January 2015 (Table 1). All isolates were identified by standard phenotypic methods [24]. Isolates were stored at -80°C and subcultured on Columbia blood agar for 72 h to 96 h at 37°C to ensure adequate growth before study.
Table 1. Nocardia isolates examined (n = 25) in the present study.
| Strains no. | Age (years) | Gender | Medical department | Specimen type | Immuno-Compromised (Yes/No) |
|---|---|---|---|---|---|
| PUNC001 | 53 | Male | Outpatient | PICC Drainage | No |
| PUNC002 | 70 | Female | Gastroenterology | Sputum | No |
| PUNC003 | 53 | Male | Immunology | Lung tissue | Yes |
| PUNC004 | 64 | Male | Respiratory | Subcutaneous nodule | Yes |
| PUNC005 | 72 | Male | Nephrology | Sputum | Yes |
| PUNC006 | 31 | Female | Emergency | Sputum | Yes |
| PUNC007 | 43 | Female | Thoracic Surgery | Sputum | No |
| PUNC008 | 50 | Male | Emergency | Hydrothorax fluid | Yes |
| PUNC009 | 67 | Male | Emergency | Sputum | No |
| PUNC010 | 70 | Male | Respiratory | Sputum | Yes |
| PUNC011 | 70 | Male | Respiratory | Sputum | No |
| PUNC012 | 51 | Male | Respiratory | Sputum | No |
| PUNC013 | 76 | Male | Respiratory | Sputum | No |
| PUNC014 | 45 | Female | Respiratory | Lung tissue | No |
| PUNC015 | 61 | Female | Immunology | Hydrothorax fluid | Yes |
| PUNC016 | 81 | Male | Outpatient | Sputum | No |
| PUNC017 | 71 | Male | Respiratory | Sputum | Yes |
| PUNC018 | 56 | Female | Respiratory | BALF | Yes |
| PUNC019 | 53 | Male | Orthopedics | Pus | No |
| PUNC020 | 46 | Female | Outpatient | Pus | No |
| PUNC021 | 57 | Male | Immunology | Sputum | Yes |
| PUNC022 | 32 | Female | Respiratory | BALF | Yes |
| PUNC023 | 45 | Male | Emergency | Sputum | No |
| PUNC024 | 37 | Male | Emergency | Pus | No |
| PUNC025 | 65 | Male | Emergency | Sputum | No |
Abbreviations: PICC, peripherally inserted central catheter; BALF, bronchoalveolar lavage fluid; SLE, systemic lupus erythematosus; COPD, chronic obstructive pulmonary disease.
In addition, the GenBank sequences of 16S rRNA, gyrB, secA1, hsp65 and rpoB loci corresponding to 20 Nocardia type strains were studied as the “validation cohort” to determine the ability of the MLSA typing scheme [7, 16] to identify Nocardia clinical isolates collected (see S1 Table).
DNA extraction, PCR amplification and sequencing
DNA extraction of all isolates was performed as previously described [8]. The Nocardia 16S rRNA gene was amplified using the primer pair 27F and 1522R [25]. The Nocardia gyrB, secA1, hsp65 and rpoB genes were amplified as previously described [16]. In all cases, amplified PCR products were sequenced in both directions using the amplification primers on the ABI 3730XL platform (Applied Biosystems, Foster City, CA).
Species identification by the 5’-end 606 bp partial 16S rRNA gene sequencing and MLSA
Molecular-based species identification was initially carried out by analysis of the 5’-end 606 bp partial 16S rRNA gene sequences as described using a percentage similarity (or identity) score of ≥ 99% as the criterion to strictly classify an isolate to species level [7]. MLSA was then carried out examining nucleotide polymorphisms in the partial 16S rRNA gene at 5’-end (462 bp), as well as the secA1 (445 bp), gyrB (482 bp), rpoB (400 bp) and hsp65 (401 bp) genes [16]. The concatenated gyrB-16S-secA1-hsp65-rpoB sequences (2190 bp) were then used for phylogenetic analysis by the maximum-likelihood algorithm based on the Tamura-Nei model [26] with 1000 bootstrap replication to ensure robustness using MEGA software (version 6.0, MEGA Inc., Englewood, NJ). DnaSP software (version 5.1, University of Barcelona, Spain) was used to assess the haplotypes of each gene [27]. Determining of potential subtypes of N. cyriacigeorgica isolates was further carried out by analysis of the Nocardia hsp65 gene as previously described [28, 29].
Establishment and validation of an “in-house” Nocardia mass spectrum database for MALDI-TOF MS
All Nocardia isolates was pretreated using a mechanical disruption-ethanol extraction method as described by Dunne et al. [30]. MALDI-TOF MS analysis of 25 clinical Nocardia strains was first carried by the Bruker Autoflex Speed TOF/TOF MS system using Biotyper software version 3.1 (db 4613, Bruker Daltonics, Billerica, USA) according to the manufacturer’s instructions. A spectral score of < 1.70 was considered not to provide reliable identification. A score of ≥ 1.70 but < 2.00 indicated identification at the genus level, and a score of ≥ 2.00 was considered identification at the species level [18].
Because the identification results using the manufacturer-provided Biotyper database was unsatisfactory (see “Results” below), five of the 25 clinical isolates representing five different Nocardia species (as identified by DNA sequencing in the present study—strain ID no. PUNC001 (Nocardia farcinica), PUNC002 (Nocardia abscessus), PUNC006 (Nocardia cyriacigeorgica), PUNC020 (Nocardia wallacei), and PUNC024 (Nocardia otitidiscaviarum); Table 2) were used to generate an “in-house” mass spectrum fingerprint database following the methodology of Segawa et al. [18]. Each isolate was analyzed in replicates of eight (ie. inoculated in eight spots on the target plate), and each spot was interrogated three times. This procedure was repeated at least once on a different occasion under identical test conditions. The resulting data were averaged using the Biotyper software v3.1 (Bruker Daltonics), a single mass spectrum representing each isolate was generated, and the resultant profiles established as a small “in-house” database.
Table 2. Identification results of 25 Nocardia clinical isolates by DNA sequencing and MALDI-TOF MS after interrogation against the Biotyper database (version 3.1) and the “in-house” database.
| Strain ID no. | Identification by MLSA | Identification by conventional methods | Identification by MALDI-TOF MSa | |
|---|---|---|---|---|
| First reported species | Score | |||
| Isolates used for establishment of “in-house” mass spectra database (no. = 5) | ||||
| PUNC001a | N. farcinica | Nocardia sp. | N/A | N/A |
| PUNC002a | N. abscessus | Nocardia sp. | N/A | N/A |
| PUNC006a | N. cyriacigeorgica | Nocardia sp. | N/A | N/A |
| PUNC020a | N. wallacei | Nocardia sp. | N/A | N/A |
| PUNC024a | N. otitidiscaviarum | N. otitidiscaviarum | N/A | N/A |
| Isolates used for validation of the MALDI-TOF MS systema (no. = 20) | ||||
| PUNC003 | N. cyriacigeorgica | N. cyriacigeorgica | N. cyriacigeorgica | 2.160 |
| PUNC004 | N. farcinica | Nocardia sp. | N. farcinica | 2.332 |
| PUNC005 | N. cyriacigeorgica | Nocardia sp. | N. cyriacigeorgica | 2.129 |
| PUNC007 | N. cyriacigeorgica | N. asteroides | N. cyriacigeorgica | 2.393 |
| PUNC008 | N. farcinica | Nocardia sp. | N. farcinica | 2.411 |
| PUNC009 | N. farcinica | N. farcinica | N. farcinica | 2.284 |
| PUNC010 | N. cyriacigeorgica | N. cyriacigeorgica | N. cyriacigeorgica | 2.011 |
| PUNC011 | N. cyriacigeorgica | N. cyriacigeorgica | N. cyriacigeorgica | 2.232 |
| PUNC012 | N. farcinica | Nocardia sp. | N. farcinica | 2.196 |
| PUNC013 | N. cyriacigeorgica | N. cyriacigeorgica | N. cyriacigeorgica | 2.126 |
| PUNC014 | N. farcinica | N. farcinica | N. farcinica | 2.124 |
| PUNC015 | N. farcinica | N. farcinica | N. farcinica | 2.055 |
| PUNC016 | N. cyriacigeorgica | N. cyriacigeorgica | N. cyriacigeorgica | 2.422 |
| PUNC017 | N. abscessus | N. abscessus | N. abscessus | 1.894 |
| PUNC018 | N. cyriacigeorgica | Nocardia sp. | N. cyriacigeorgica | 2.307 |
| PUNC019 | N. cyriacigeorgica | N. cyriacigeorgica | N. cyriacigeorgica | 2.145 |
| PUNC021 | N. cyriacigeorgica | Nocardia sp. | N. cyriacigeorgica | 2.079 |
| PUNC022 | N. farcinica | Nocardia sp. | N. farcinica | 2.498 |
| PUNC023 | N. farcinica | N. farcinica | N. farcinica | 2.431 |
| PUNC025 | N. cyriacigeorgica | Nocardia sp. | N. cyriacigeorgica | 2.052 |
Abbreviation: N/A, not applicable.
a Results by using the Bruker Biotyper version 3.1 (Bruker Daltonics) complementation with the “in-house” database (see Materials and Methods for detail).
The 20 remaining clinical isolates were then analyzed against the Biotyper version 3.1 database (Bruker Daltonics) complemented with the “in-house” database. The technicians performing the analysis were blinded to the DNA sequencing results to avoid potential bias.
A main spectrum profile (MSP) dendrogram was further constructed using spectra of 25 Nocardia clinical strains in this study along with 37 reference spectra of 32 Nocardia species already contained in the Biotyper version 3.1 database for analysis of genetic relatedness.
Review for publications on validation of MALDI-TOF MS in identification of Nocardia species
Key words “Nocardia” and “matrix-assisted laser desorption ionization-time of flight mass spectrometry” were used to search related publications in English archived in NCBI PubMed database (http://www.ncbi.nlm.nih.gov/pubmed, till Dec 31st, 2015). All complete articles of the search (16 publications in all) were screened. After excluding case reports, six publications were reviewed in detail.
GenBank accession numbers
The DNA sequences of the full-length 16S rRNA, secA1, gyrB, rpoB and hsp65 genes of 25 Nocardia isolates studied have been submitted to GenBank database (accession numbers KT985911 to KT985935, KU052160 to KU052184, KU052085 to KU052109, KU052135 to KU052159 and KU052110 to KU052134 for the above five genes, respectively, S2 Table).
Results
Identification by the 5’-end 606 bp partial 16S rRNA gene sequencing
Of 25 Nocardia clinical strains, 24 isolates representing four Nocardia species were identified unambiguously by the 5’-end 606 bp partial 16S rRNA gene sequencing. N. cyriacigeorgica (n = 12, 48%) and N. farcinica (n = 9, 36%) were the most common species, followed by N. abscessus (n = 2, 8%) and N. otitidiscaviarum (n = 1, 4%). However, the 5’-end 606 bp partial 16S rRNA gene sequence of strain PUNC020 showed 99% sequence similarity to both N. wallacei type strain DSM 45136T (606/606, 100%) and Nocardia transvalensis type strain NRRL B-16037T (600/606, 99.0%), though was identical to the former.
MLSA
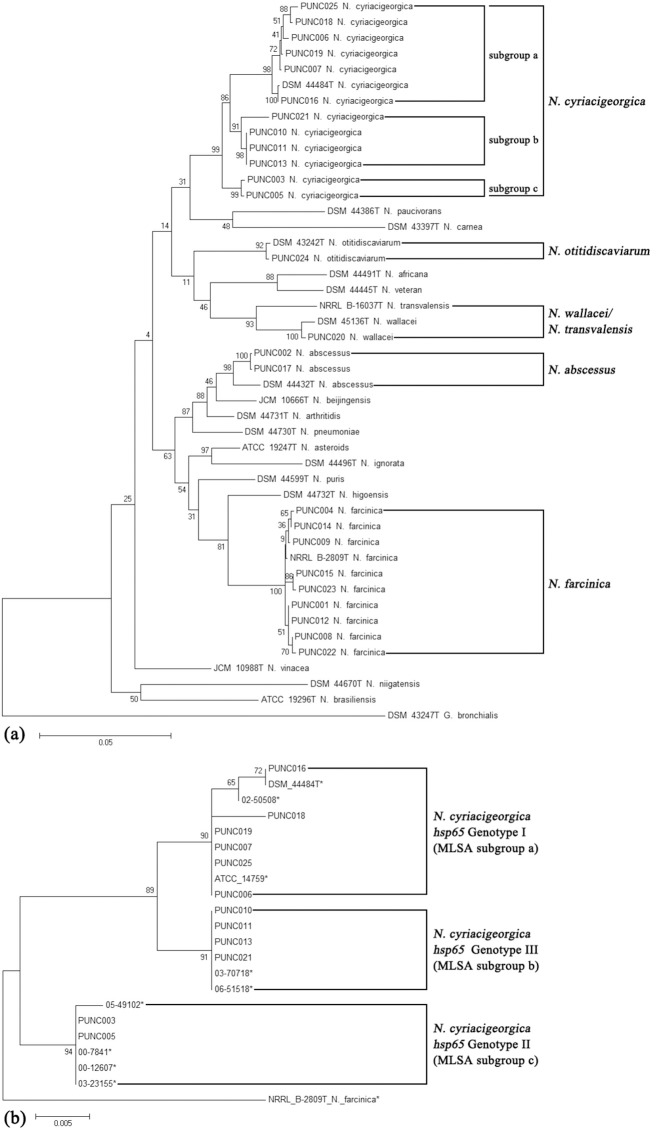
Phylogenetic analysis of concatenated gyrB-16S-secA1-hsp65-rpoB sequences obtained from the 25 clinical isolates and 20 Nocardia type strain sequences clearly revealed five major clusters with bootstrap threshold values of ≥ 90% (Fig 1). Each cluster corresponded to a unique Nocardia species, ie. N. cyriacigeorgica, N. otitidiscaviarum, N. farcinica, and N. abscessus), except for clinical strain PUNC020 that was clustered together with type strains of N. wallacei and N. transvalensis. As strain PUNC020 was more phylogenetically closely related to N. wallacei strain DSM 45136T compared with N. transvalensis strain NRRL B-16037T (Fig 1), the isolate was assigned as N. wallacei.
Fig 1. Phylogenetic trees are shown and were conducted using the maximum-likelihood method.
Fig 1a. Phylogenetic tree based on the concatenated gyrB-16S-secA1-hsp65-rpoB sequences of 25 Nocardia clinical isolates and 20 Nocardia type strains, using Gordonia bronchialis strain ATCC 25592T as an outgroup (S1 Table). Fig 1b. Phylogenetic tree based on the hsp65 gene sequences of 12 N. cyriacigeorgica clinical isolates and nine Nocardia cyriacigeorgica isolates whose genotypes have previously been determined by Schlaberg et al. [29] using the sequence of a Nocardia farcinica strain as an outgroup.
Twelve N. cyriacigeorgica clinical isolates could further be classified into three subgroups: subgroup a (n = 6), b (n = 4) and c (n = 2), (Fig 1A). Comparing with results obtained by N. cyriacigeorgica genotyping using sequence analysis of the hsp65 gene [28, 29], isolates belonging to N. cyriacigeorgica MLSA subgroups a, b and c corresponded to N. cyriacigeorgica hsp65 genotype I, III and II, respectively (Fig 1B).
Overall, it was observed that the rpoB, gyrB and secA1 genes were the most polymorphic (14 to 16 haplotypes identified amongst 25 clinical strains, haplotype diversity ≥ 0.950), followed by hsp65 (10 haplotypes, haplotype diversity = 0.837), whilst the partial 16S rRNA gene was the least polymorphic (five haplotypes, haplotype diversity = 0.657) (Table 3 and S2 Table). Different degrees of intra-species micro-heterogeneity of different genes were also observed (Table 3). N. cyriacigeorgica isolates generally exhibited higher intra-species diversity than other species e.g. N. farcinica (Table 3).
Table 3. Genetic polymorphisms contained within the 16S rRNA, gyrB, secA1, hsp65 and rpoB genes for 25 clinical Nocardia isolates studied.
| Characters | gyrB | 16S rRNA gene | secA1 | hsp65 | rpoB |
|---|---|---|---|---|---|
| All clinical isolates (n = 25) | |||||
| No. of haplotypes | 16 | 5 | 14 | 10 | 16 |
| Haplotype diversity | 0.957 | 0.657 | 0.950 | 0.837 | 0.963 |
| N. cyriacigeorgica (n = 12) | |||||
| No. of haplotypes | 8 | 1 | 6 | 5 | 8 |
| Haplotype diversity | 0. 924 | N/A | 0. 848 | 0. 803 | 0. 924 |
| N. farcinica (n = 9) | |||||
| No. of haplotypes | 4 | 1 | 5 | 1 | 5 |
| Haplotype diversity | 0. 778 | N/A | 0. 889 | N/A | 0. 861 |
Abbreviation: N/A, not applicable.
Improvement of MALDI-TOF MS identification capacity by the “in-house” database
Of the five species identified by the 5’-end 606 bp partial 16S rRNA gene sequencing and MLSA from the present study which we selected to establish a mini “in-house” database, representation of four species (N. cyriacigeorgica, N. farcinica, N. abscessus and N. otitidiscaviarum) was already included in the Biotyper version 3.1 database (Bruker Daltonics); however, N. wallacei was not represented. When all 25 clinical isolates were interrogated using only the Biotyper database, only eight of 25 (32%) isolates was identified to genus level, none (0%) to species level, and 17 isolates were not identified (68%).
Using the “in-house” database to complement the Biotyper database, 19 of 20 (95%) Nocardia clinical isolates were correctly identified to species level with scores of ≥ 2.0 (Table 2). N. abscessus strain PUNC017, which yielded a MALDI-TOF MS score of 1.894, was identified to genus level (Table 2).
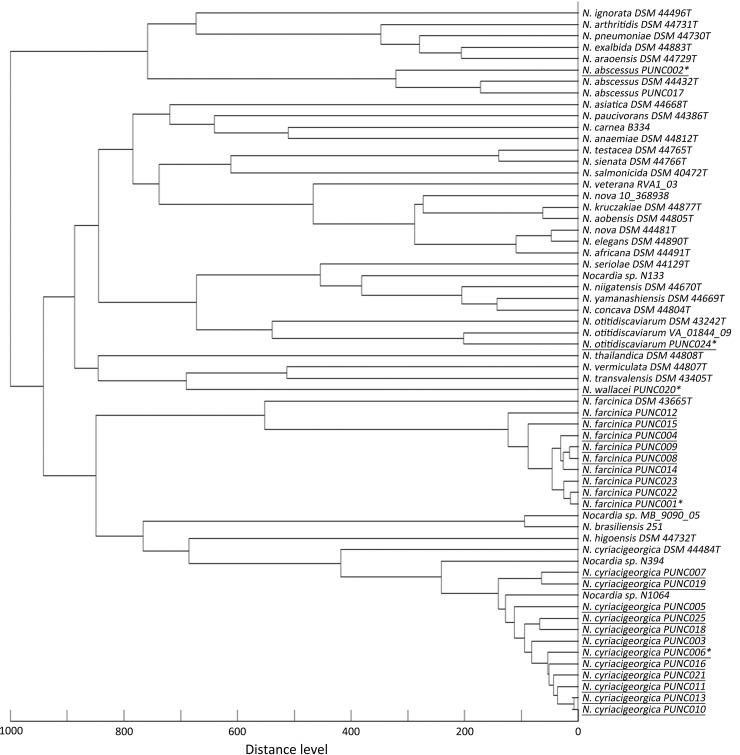
The MSP dendrogram indicated that the spectral profile of clinical isolates were distinct from the corresponding reference spectra for its species in the Biotyper database (Fig 2). The mass spectra differences between N. abscessus strain PUNC002 (for establishment of the “in-house” database) and N. abscessus strain PUNC017 (for test of the “in-house” database) also resulted in failure in identifying strain PUNC017 to specie level using the “in-house” database (Fig 2). Therefore, the mass spectra of strain PUNC017 was incorporated into the “in-house” database for future study. Similar divergence of mass spectra were also noted in N. otitidiscaviarum for clinical strain PUNC024 versus strain VA_01844_09 and type strain DSM 43242T that included in the commercial database (Fig 2), which led to failure of identifying strain PUNC024 to species level using the commercial database.
Fig 2. The main spectrum profile (MSP) dendrogram constructed using spectra of 25 Nocardia clinical strains along with 37 reference spectra of 32 Nocardia species contained in the original Biotyper database (version 3.1; Bruker).
Strain identification numbers of clinical isolates collected in the present study are shown underlined, and isolates used for establishment of the “in-house” database was labeled with asterisks.
Literature review for Nocardia identification by MALDI-TOF MS
Of six studies reviewed, five had evaluated the Bruker Biotyper commercial database (version 3.1 or 3.0.2) and one, an Andromas (Paris, France) system (Table 4). The Biotyper database only correctly identified 14.9–52.8% of isolates to species level and an additional 6.9–52.7% of isolates to genus level. The performance of Andromas system (Paris, France) was better (80.4% to species level and 17.6% to genus level). Three studies further developed local “in-house” databases, and the correct identification rate was significantly improved to 79.1–90.6% to species level and an additional 4.7–11.5% to genus level (Table 4).
Table 4. Review of publications for evaluation of MALDI-TOF MS for the identification of Nocardia species.
| Commercial DB | "In-house" DB | |||||||
|---|---|---|---|---|---|---|---|---|
| System | Reference method | ID criteria to species/genus | Ni/Ns for DB validation | DB version | Correct ID to species/genus | Ni/Ns for DB development | Correct ID to species/genus | Reference |
| Bruker Autoflex Speed | MLSA | ≥ 2.0/< 2.0 and ≥ 1.7 | See notea | BioTyper ver. 3.1 | 0%/32.0%a | 5/5 | 95.0%/5.0%a | This study |
| Bruker microflex LT | 16S rRNA gene and conventional | ≥ 2.0/< 2.0 and ≥ 1.7 | 148/15 | BioTyper ver. 3.1 | 41.9%/15.5% | 232/53 | 89.9%/4.7% | [23] |
| Bruker (Model not specified) | 16S rRNA, hsp65 and secA1 genes | ≥ 1.9/< 1.9 and ≥ 1.7 | 87/25 | BioTyper ver. 3.1 | 52.8%/6.9% | 13/13 | 82.8%/11.5% | [22] |
| Bruker microflex LT | 16S rRNA gene and conventional | ≥ 2.0/< 2.0 and ≥ 1.7 | 64/22 | BioTyper ver. 3.1 | 15.6%/25.0% | 192/73 | 90.6%/9.4% | [18] |
| Bruker microflex LT | 16S rRNA and secA1 genes | ≥ 2.0/< 2.0 and ≥ 1.7 | 74/14 | BioTyper ver. 3.1 | 14.9%/52.7% | ND | ND | [19] |
| Andromas | 16S rRNA, hsp65 genes, conventional | See noteb | 51/12 | See noteb | 80.4%/17.6% | ND | ND | [21] |
| Bruker microflex LT | 16S rRNA gene and conventional | ≥ 2.0/< 2.0 and ≥ 1.7 | 43/9 | BioTyper ver. 3.0.2 | 23.3%/20.9% | 110/17 | 79.1%/9.3% | [20] |
Abbreviations: ID, identification; Ni/Ns, number of isolates/number of species; DB, database; MLSA, multilocus sequence analysis.
a In the present study, all 25 Nocardia clinical isolates collected were involved for validation of the commercial Bruker Biotyper database. While for the "in-house" database, five of the 25 Nocardia isolates were used for databased development, and the rest 20 isolates were used for "in-house" database validation. See details in Methods section.
b An Andromas database (database version not specified) was employed in the study by Farfour et al. Criteria for correct identification to species level: percentage of common peaks is ≥ 68% and more than 10% difference between the first two best-match species. Criteria for correct identification to genus level: percentage of common peaks is ≥ 68% yet less than 10% difference between the first two best-match species.
A proposed algorithm for species identification of Nocardia in clinical laboratories
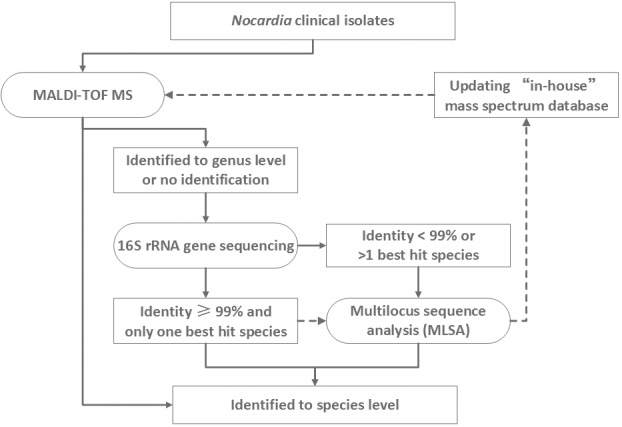
Although the MLSA scheme was definitively able to assign species, identify novel species, and to differentiate within species, it has the disadvantage of relatively high costs, and requiring staff time for analysis of results. We propose an algorithm for species identification of Nocardia in the clinical laboratory (Fig 3), recommending that identification by MALDI-TOF MS using a database complemented by “in-house” protein profiles be attempted in the initial instance. For those isolates that cannot be identified to species level by MALDI-TOF MS, the 5’-end 606 bp partial 16S rRNA sequencing may be attempted. For any isolates with < 99% gene sequence similarity to a known archived strain sequence or those with two or more “best hit” species identities, MLSA could be undertaken for a definitive identification.
Fig 3. Proposed algorithm for laboratory identification of Nocardia isolates (solid lines) and establishment and expansion of ongoing “in-house” mass spectrum database (dashed lines).
Discussion
To date, more than 50 species of pathogenic Nocardia have been identified [31]. Since there are differences in prevalence of species between geographic regions, knowledge of species distribution is important. In the present study, N. cyriacigeorgica and N. farcinica were the most commonly encountered species, similar to that in Canada [16], but in contrast to a report from Taiwan where Nocardia brasiliensis was the most common [32]. In comparison, Nocardia asteroides was reported to be predominant in Switzerland [31], while N. nova was predominant in Australia [7].
Most studies examining identification of Nocardia have indicated that molecular techniques are required to achieve species assignment [6, 16, 31, 32]. Although polymorphisms in the 5’-end partial 16S rRNA gene is widely used to discriminate between Nocardia species, misidentification may occur due to high sequence similarity between certain species [6, 7, 16, 33], as exemplified by inability to distinguish N. wallacei from N. transvalensis in the present study, or due to multiple different copies of this gene, such as that in N. nova [9]. N. wallacei is very closely related to N. transvalensis and indeed was previously classified within the N. transvalensis complex. Using a more discriminatory method, in this case, a 5-locus MLSA, N. wallacei was assigned as an individual species, which supported findings by Conville et al. [12].
In this context then, MLSA has been carried out as an alternative technique for the identification of Nocardia. The most commonly used genes have been the 16S rRNA, secA1, gyrB, rpoB and hsp65 genes. Different studies may employ different combination of the genes [11, 16, 34], yet it is notable that no single gene locus sequencing is sufficient to resolve a substantial number of Nocardia species [16]. In addition, the MLSA scheme can be used for examining intra-species genetic diversity [10, 11, 13]. Using this approach, Carrasco et al. has reported that species N. nova had highest intra-species heterogeneity, followed by N. cyriacigeorgica and N. abscessus, while N. farcinica was more conserved [11]. Although the diversity of N. abscessus and N. nova were not compared in this study due to only a limited number of isolates studied, we similarly found genetic heterogeneity within N. cyriacigeorgica where three subgroups were identified. Of note, these subgroups correspond to three hsp65 genotypes proposed in previous studies [28, 29]. In the diagnostic algorithm proposed (Fig 3) for laboratory identification of Nocardia, MLSA has good clinical utility in definitive confirmative of species.
Application of sequencing-based methods in the routine work of clinical laboratories is restricted by high costs and the need for on-site sequencing facilities which potentially affect turn-around times. The recent introduction of MALDI-TOF MS platforms in clinical laboratories has revolutionized diagnostic microbiology [35]. However, its application in identification of Nocardia species required extra protein-extraction pretreatment of isolates [17, 30], and more importantly, may be limited by insufficient archiving of reference spectra within commercial MALDI-TOF MS databases [17–20]. In this study, the MSP analysis indicated significant differences between spectra of the clinical isolates studied and reference spectra of corresponding species contained in the Biotyper version 3.1 database (Bruker Daltonics), which yielded suboptimal results in species identification when used as the only database. However, the establishment of even a small “in-house” database significantly improved the identification capacity of the MALDI-TOF MS system, as also found previously [18, 20, 22, 23]. To improve the identification capacity of MALDI-TOF MS, it is important for MS databases to contain more reference mass spectra from type strains of different bacteria species, and also spectra representing different strains of the same species [35], as strains of the same species may have divergent mass spectra e.g. the case of N. abscessus clinical strain PUNC002 and PUNC017 found in the present study. With an ongoing expanded “in-house” database representing species that are the more frequently encountered, the utility and practicality of MALDI-TOF MS for Nocardia identification will be improved. However, before expanding any “in-house” mass spectrum database, we recommend that Nocardia isolates be analyzed by MLSA to ensure the high-quality of the “in-house” database (Fig 3). Moreover, comparing to MLSA, the MALDI-TOF MS system was unable to cluster N. cyriacigeorgica into subgroups. In this regard, MLSA is superior to MALDI-TOF MS for subgrouping N. cyriacigeorgica. Further studies addressing differences in MS profiles between N. cyriacigeorgica strains are warranted.
The major limitation of the current study is that relatively few isolates representing a small number of species was examined, and all isolates were from a single center and hence may not be generalizable across centers in China. Continuous expansion of the MALDI-TOF MS databases to include more species is necessary.
Conclusions
In conclusion, we have evaluated MLSA scheme and MALDI-TOF MS for identification of Nocardia clinical isolates. MLSA showed superior discriminatory power compared with the 5’-end 606bp partial 16S rRNA gene sequencing for species identification of Nocardia. MALDI-TOF MS has good utility in rapidly and accurately providing species identification but is contingent on building up a robust library of reference spectra.
Supporting Information
(DOCX)
(DOCX)
Data Availability
The DNA sequences of the 16S rRNA, secA1, gyrB, rpoB and hsp65 genes of 25 Nocardia isolates studied have been submitted to GenBank database (accession numbers KT985911 to KT985935, KU052160 to KU052184, KU052085 to KU052109, KU052135 to KU052159 and KU052110 to KU052134 for the above five genes, respectively.
Funding Statement
The study was supported by a National Research Special Fund for Public Welfare Industry of Health of China (grant number 201402001, http://www.nhfpc.gov.cn) and a National High Technology Research and Development Program of China (the “863” Program, grant number 2013AA020203, http://program.most.gov.cn). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Minero MV, Marin M, Cercenado E, Rabadan PM, Bouza E, Munoz P. Nocardiosis at the turn of the century. Medicine (Baltimore). 2009;88(4):250–61. 10.1097/MD.0b013e3181afa1c8 . [DOI] [PubMed] [Google Scholar]
- 2.Clark NM, Practice ASTIDCo. Nocardia in solid organ transplant recipients. Am J Transplant. 2009;9 Suppl 4:S70–7. [DOI] [PubMed] [Google Scholar]
- 3.Wang HL, Seo YH, LaSala PR, Tarrand JJ, Han XY. Nocardiosis in 132 patients with cancer: microbiological and clinical analyses. Am J Clin Pathol. 2014;142(4):513–23. 10.1309/AJCPW84AFTUWMHYU . [DOI] [PubMed] [Google Scholar]
- 4.Zhou L, Liu H, Yuan F, Yuan S. Disseminated nocardiosis in a patient with nephrotic syndrome following HIV infection. Exp Ther Med. 2014;8(4):1142–4. 10.3892/etm.2014.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth A, Andrees S, Kroppenstedt RM, Harmsen D, Mauch H. Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals underspeciation and division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. J Clin Microbiol. 2003;41(2):851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ Jr. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev. 2006;19(2):259–82. 10.1128/CMR.19.2.259–282.2006. 16614249; PubMed Central PMCID: PMC1471991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong F, Chen SC, Chen X, Sintchenko V, Halliday C, Cai L, et al. Assignment of reference 5'-end 16S rDNA sequences and species-specific sequence polymorphisms improves species identification of Nocardia. Open Microbiol J. 2009;3:97–105. 10.2174/1874285800903010097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao M, Kong F, Sorrell TC, Cao Y, Lee OC, Liu Y, et al. Identification of pathogenic Nocardia species by reverse line blot hybridization targeting the 16S rRNA and 16S-23S rRNA gene spacer regions. J Clin Microbiol. 2010;48(2):503–11. 10.1128/JCM.01761-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conville PS, Witebsky FG. Multiple copies of the 16S rRNA gene in Nocardia nova isolates and implications for sequence-based identification procedures. J Clin Microbiol. 2005;43(6):2881–5. 10.1128/JCM.43.6.2881-2885.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrasco G, Valdezate S, Garrido N, Medina-Pascual MJ, Villalon P, Saez-Nieto JA. gyrB analysis as a tool for identifying Nocardia species and exploring their phylogeny. J Clin Microbiol. 2015;53(3):997–1001. 10.1128/JCM.03072-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrasco G, Valdezate S, Garrido N, Villalon P, Medina-Pascual MJ, Saez-Nieto JA. Identification, typing, and phylogenetic relationships of the main clinical Nocardia species in Spain according to their gyrB and rpoB genes. J Clin Microbiol. 2013;51(11):3602–8. 10.1128/JCM.00515-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conville PS, Zelazny AM, Witebsky FG. Analysis of secA1 gene sequences for identification of Nocardia species. J Clin Microbiol. 2006;44(8):2760–6. 10.1128/JCM.00155-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong F, Wang H, Zhang E, Sintchenko V, Xiao M, Sorrell TC, et al. secA1 gene sequence polymorphisms for species identification of Nocardia species and recognition of intraspecies genetic diversity. J Clin Microbiol. 2010;48(11):3928–34. 10.1128/JCM.01113-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Nava V, Couble A, Devulder G, Flandrois JP, Boiron P, Laurent F. Use of PCR-restriction enzyme pattern analysis and sequencing database for hsp65 gene-based identification of Nocardia species. J Clin Microbiol. 2006;44(2):536–46. 10.1128/JCM.44.2.536-546.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin X, Liang S, Sun X, Luo S, Wang Z, Li R. Ocular nocardiosis: HSP65 gene sequencing for species identification of Nocardia spp. Am J Ophthalmol. 2007;144(4):570–3. 10.1016/j.ajo.2007.06.031 . [DOI] [PubMed] [Google Scholar]
- 16.McTaggart LR, Richardson SE, Witkowska M, Zhang SX. Phylogeny and identification of Nocardia species on the basis of multilocus sequence analysis. J Clin Microbiol. 2010;48(12):4525–33. 10.1128/JCM.00883-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toyokawa M, Kimura K, Nishi I, Sunada A, Ueda A, Sakata T, et al. Reliable and reproducible method for rapid identification of Nocardia species by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi. 2013;24(1):1–8. . [PubMed] [Google Scholar]
- 18.Segawa S, Nishimura M, Sogawa K, Tsuchida S, Murata S, Watanabe M, et al. Identification of Nocardia species using matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Clin Proteomics. 2015;12(1):6 10.1186/s12014-015-9078-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsueh PR, Lee TF, Du SH, Teng SH, Liao CH, Sheng WH, et al. Bruker biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Nocardia, Rhodococcus, Kocuria, Gordonia, Tsukamurella, and Listeria species. J Clin Microbiol. 2014;52(7):2371–9. 10.1128/JCM.00456-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verroken A, Janssens M, Berhin C, Bogaerts P, Huang TD, Wauters G, et al. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Nocardia species. J Clin Microbiol. 2010;48(11):4015–21. 10.1128/JCM.01234-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farfour E, Leto J, Barritault M, Barberis C, Meyer J, Dauphin B, et al. Evaluation of the Andromas matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of aerobically growing Gram-positive bacilli. J Clin Microbiol. 2012;50(8):2702–7. 10.1128/JCM.00368-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khot PD, Bird BA, Durrant RJ, Fisher MA. Identification of Nocardia Species by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J Clin Microbiol. 2015;53(10):3366–9. 10.1128/JCM.00780-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckwalter SP, Olson SL, Connelly BJ, Lucas BC, Rodning AA, Walchak RC, et al. Evaluation of MALDI-TOF Mass Spectrometry for the Identification of Mycobacterium species, Nocardia species and Other Aerobic Actinomycetes. J Clin Microbiol. 2015. 10.1128/JCM.02128-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Versalovic J, American Society for Microbiology. Manual of clinical microbiology. 10th ed. Washington, DC: ASM Press; 2011. [Google Scholar]
- 25.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62(2):625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–26. . [DOI] [PubMed] [Google Scholar]
- 27.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–2. 10.1093/bioinformatics/btp187 . [DOI] [PubMed] [Google Scholar]
- 28.Rudramurthy SM, Honnavar P, Kaur H, Samanta P, Ray P, Ghosh A, et al. Molecular identification of clinical Nocardia isolates from India. J Med Microbiol. 2015;64(10):1216–25. 10.1099/jmm.0.000143 . [DOI] [PubMed] [Google Scholar]
- 29.Schlaberg R, Huard RC, Della-Latta P. Nocardia cyriacigeorgica, an emerging pathogen in the United States. J Clin Microbiol. 2008;46(1):265–73. 10.1128/JCM.00937-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunne WM Jr, Doing K, Miller E, Miller E, Moreno E, Baghli M, et al. Rapid inactivation of Mycobacterium and Nocardia species before identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2014;52(10):3654–9. 10.1128/JCM.01728-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambrosioni J, Lew D, Garbino J . Nocardiosis: updated clinical review and experience at a tertiary center. Infection. 2010;38(2):89–97. 10.1007/s15010-009-9193-9 . [DOI] [PubMed] [Google Scholar]
- 32.Liu WL, Lai CC, Ko WC, Chen YH, Tang HJ, Huang YL, et al. Clinical and microbiological characteristics of infections caused by various Nocardia species in Taiwan: a multicenter study from 1998 to 2010. Eur J Clin Microbiol Infect Dis. 2011;30(11):1341–7. 10.1007/s10096-011-1227-9 . [DOI] [PubMed] [Google Scholar]
- 33.Tamura T, Matsuzawa T, Oji S, Ichikawa N, Hosoyama A, Katsumata H, et al. A genome sequence-based approach to taxonomy of the genus Nocardia. Antonie Van Leeuwenhoek. 2012;102(3):481–91. 10.1007/s10482-012-9780-5 . [DOI] [PubMed] [Google Scholar]
- 34.Baio PV, Ramos JN, dos Santos LS, Soriano MF, Ladeira EM, Souza MC, et al. Molecular identification of Nocardia isolates from clinical samples and an overview of human nocardiosis in Brazil. PLoS Negl Trop Dis. 2013;7(12):e2573 10.1371/journal.pntd.0002573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015;6:791 10.3389/fmicb.2015.00791 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
The DNA sequences of the 16S rRNA, secA1, gyrB, rpoB and hsp65 genes of 25 Nocardia isolates studied have been submitted to GenBank database (accession numbers KT985911 to KT985935, KU052160 to KU052184, KU052085 to KU052109, KU052135 to KU052159 and KU052110 to KU052134 for the above five genes, respectively.