Abstract
For acute myeloid leukemia (AML), identification of activating mutations in the FMS-like tyrosine kinase-3 (FLT3) has led to the development of several FLT3-inhibitors. Here we present clinical and next generation sequencing data at the time of progression of a patient on a novel FLT3-inhibitor clinical trial (ASP2215) to show that employing therapeutic interventions with these novel targeted therapies can lead to consequences secondary to selective pressure and clonal evolution of cancer. We describe novel findings alongside data on treatment directed towards actionable aberrations acquired during the process. (Clinical Trial: NCT02014558; registered at: 〈https://clinicaltrials.gov/ct2/show/NCT02014558〉)
Keywords: Acute Leukemia, FMS-like tyrosine kinase-3 (FLT3) inhibitors, ASP2215, Clonal evolution, Philadelphia chromosome (BCR-ABL), Ponatinib
Highlights
-
•
The article reports on a case of AML that underwent clonal evolution.
-
•
We report on novel acquisition of the Philadelphia t(9;22) translocation in AML.
-
•
Next generation sequencing maybe helpful in these refractory/relapse cases.
-
•
Novel FLT3-inhibitor targeted therapies are another option in patients with AML.
-
•
Personalizing cancer treatment based on evolving targets is a viable option.
1. Introduction
The development of kinase inhibitors for the treatment of leukemia has revolutionized the care of these patients. Since the introduction of imatinib for the treatment of chronic myeloid leukemia, multiple other tyrosine kinase inhibitors (TKIs) have become available [1]. Additionally, for acute myeloid leukemia (AML), identification of activating mutations in the FMS-like tyrosine kinase-3 (FLT3) has led to the development of several FLT3-inhibitors [2], [3], [4], [5]. The article herein reports a unique case of AML that underwent clonal evolution while on a novel FLT3-inhibitor clinical trial. Here we show that employing therapeutic interventions with these novel targeted therapies can lead to consequences secondary to selective pressure and clonal evolution of cancer. Personalizing therapy in realtime based on changing targets presents both as challenge and an opportunity. Our work herein presents clinical and next generation sequencing data at the time of progression to illustrate these important concepts stemming from Darwinian evolution [6]. We describe novel findings alongside data on treatment directed towards actionable aberrations acquired during the process.
2. Clinical course and management
Our work focuses on a 23-year-old male who presented with 3 months history of fatigue and easy bruising along with a 2 day history of fever and chest pain. Initial bloodwork revealed a white blood count of 22.0×109/L with 51% circulating blasts, hemoglobin 7.6 g/dL, and a platelet count of 43×109/L. A bone marrow biopsy confirmed a diagnosis of AML. Initial cytogenetic studies identified trisomy 8 in all the twenty metaphases examined. Mutational analysis revealed an internal tandem duplication of the FLT3 gene (FLT3-ITD).
He received standard induction chemotherapy (7+3) with cytarabine (ARA-C; 100 mg/m2 for 7 days) and daunorubicin (DNM; 60 mg/m2 for 3 days). His induction chemotherapy was complicated by severe palatine and uvular necrosis of indeterminate etiology (possible mucormycosis), necessitating aggressive surgical debridement.
Bone marrow biopsy at day 28 demonstrated persistent disease with 10% bone marrow blasts (Fig. 1). Due to his complicated clinical course and the presence of a FLT3-ITD, salvage therapy with 5-azacitidine (5-AZA) and sorafenib (SFN) was instituted.
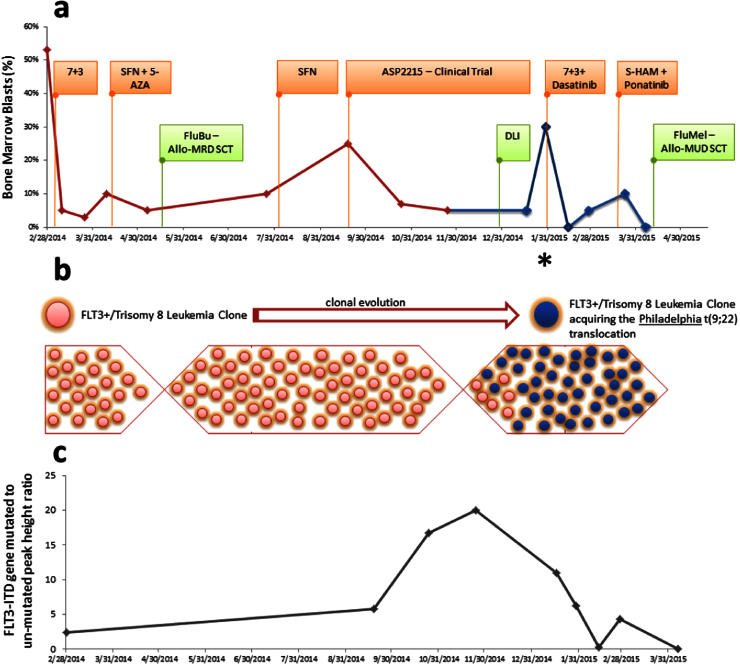
Fig. 1.
Clinical course and various treatments received for our patient with FLT3+/trisomy 8 acute myeloid leukemia (AML). (a) The bone marrow blasts (%) are shown at different time points alongside the treatments received. (b) Schematic representation of selection pressure leading to development of a new clone of FLT3+/Trisomy 8 leukemia with the Philadelphia t(9;22) translocation. (c) The FLT3-ITD gene mutated to un-mutated peak height ratio is shown at different time points. Abbreviations: 7+3 - Induction chemotherapy with an anthracycline and cytarabine (see text for details/doses); SFN – Sorafenib; AZA – 5-Azacitadine; ASP2215 – Novel FLT3 inhibitor (NCT02014558); FluBu – Conditioning chemotherapy with fludarabine and busulfan; FluMel – Conditioning chemotherapy with Fludarabine and melphalan; MRD-SCT – Matched related stem cell transplant; MUD – SCT – Matched unrelated stem cell transplant; S-HAM – sequential high dose Ara-C chemotherapy with mitoxantrone.
With the sorafenib and 5-azacitidine, he achieved morphological remission (<5% blasts). His older brother was found to be a 10/10 HLA match. Subsequently, he underwent a reduced intensity conditioning matched-related donor stem cell transplant (MRD-HCT) with fludarabine and busulfan (FluBu) conditioning chemotherapy.
The patient unfortunately had relapse of his leukemia on day +67 after his allo-HCT (Fig. 1). Sorafenib was briefly reinitiated and then he enrolled in a novel FLT3-inhibitor (ASP2215) clinical trial (NCT02014558) [7]. To potentiate the graft-versus-leukemia (GVL) effect, he received a cryopreserved donor lymphocyte infusion (DLI) during cycle 4 of ASP2215.
Routine surveillance bone marrow biopsy during the fifth cycle (28 day cycle) of ASP2215 therapy demonstrated 5% bone marrow blasts. Cytogenetic studies showed that of the 20 metaphases analyzed all had trisomy 8. However, initially 5 out of 20 and then later 20 out of 20 of these metaphases had also acquired the Philadelphia t(9;22) chromosomal translocation. His bone marrow aspirate/biopsy was also analyzed through a commercial next generation sequencing (NGS) assay (FoundationONE Heme) with results shown in Table 1. Salvage chemotherapy (7+3) was instituted with cytarabine (ARA-C; 100 mg/m2 for 7 days) and Idarubicin (IDA; 12 mg/m2 for 3 days) with the addition of dasatinib 100 mg per oral (PO) daily given the emergence of the BCR-ABL1 clone (the new t(9;22) clone with p210 protein transcript).
Table 1.
Commercial next-generation sequencing (NGS) based assay (FoundationONE Heme) identifying multiple genomic alterations alongside a brief description of potential personalized targeted therapies. The highlighted therapies were employed in this particular case at various time points as shown in Fig. 1.
| Genomic alterations detected* | Potential targeted therapy |
| BCR-ABL1 fusion (p210) | Tyrosine kinase inhibitors (TKIs) such as imatinib, dasatinib, ponatinib, bosutinib, nilotinib and TKIs available through clinical trials |
| FLT3-ITD (E596_Y597ins16) | Tyrosine kinase inhibitors including FLT3 inhibitors such as sorafenib, ponatinib, sunitinib, crenolanib, quizartinib and FLT3 inhibitors available through clinical trials (e.g. ASP2215, E6201) |
| GATA2 (F265fs*15) | – |
| MLL2 (R2635Q) | Targeting DOT1L (a histone methyltransferase) |
| WT1 (R380fs*5, R462Q) | Potential for using DNA methyltransferase (DNMT) inhibitors such as azacitidine and decitabine |
Other variants of unknown significance that were also detected on NGS assay included LRP1B(N4559S), MSH3(A62_P63insAAAPAA) and ROS1(Q291P)
He developed pneumonitis, pericarditis and rapid relapse following reinduction chemotherapy which prompted treatment with sequential-high dose cytarabine (ARA-C 3000 mg/m2 days 1, 2, 8 and 9) and mitoxantrone (10 mg/m2 days 3, 4, 10 and 11) [S-HAM] salvage chemotherapy to achieve aplasia prior to a second allogeneic transplant. Given that the patient's clone had both the FLT3 and the BCR-ABL1 aberrations ponatinib was added since it is a TKI with dual FLT3 and BCR-ABL1 inhibitory activity (Table 1). This was given at the standard dose of 45 mg PO daily for a week prior to chemotherapy and at a reduced dose of 15 mg daily during cytotoxic chemotherapy.
Subsequent bone marrow biopsy prior to the second transplant did not show any morphological or molecular evidence of leukemia. Patient, therefore, underwent conditioning chemotherapy with fludarabine and melphalan (FluMel) prior to a matched unrelated donor allogeneic transplant (MUD-SCT) (Fig. 1). This was complicated by pancreatitis requiring total parenteral nutrition and supportive care for several weeks. Following this, patient now has been transitioned to an outpatient setting with excellent signs of engraftment and is transfusion independent. Day 30, day 60 and now the 6 month chimerism studies show 100% donor and 0% recipient with no peripheral or bone marrow blasts. Given concerns for high risk for relapse, patient was started on ‘maintenance’ ponatinib 15 mg every other day and continues to be on it~8 months post-SCT. The molecular mRNA BCR-ABL1 testing done continue to show no detectable transcripts in the both the patient's blood and bone marrow.
3. Discussion
In this era of novel targeted therapies and personalized cancer medicine, clonal evolution and tumor heterogeneity are important concepts stemming from Darwinian evolution [6]. Our work here illustrates several unique aspects. First we report novel acquisition of the Philadelphia t(9;22) translocation (p210 isotype) while receiving FLT3 inhibitor therapy on a clinical trial with ASP2215. This has not been described previously. There have been cases of AML with emergence of the BCR-ABL1 clone at the time of relapse, but not while receiving FLT3 inhibitor targeted therapy [8], [9]
Second is the challenging aspect of personalizing cancer treatment in the light of evolving therapeutic targets. In this patient's case, there were several potential treatment options to consider as targeted therapy alone or in combination with cytotoxic therapy. For instance, ponatinib is considered both a strong FLT3-inhibitor as well as a pan-BCR-ABL1 inhibitor for Philadelphia chromosome positive leukemia. Its ability to act at both these targets alongside other possible promiscuous interactions, likely helped achieve a deep morphological and molecular response when combined with cytotoxic chemotherapy. Dasatinib has also been shown in cell lines to have FLT3-inhibitor activity but its efficacy in vivo is not known [10]. Choice of the particular targeted therapies was further dictated by what insurance companies would approve of and raises an interesting but challenging issue of coverage of these costly therapies. Furthermore, the significant improvement in performance status while on ASP2215 clinical trial allowed us to consider a second allogeneic transplant.
We interpreted the relapse post-SCT in this patient as clonal evolution. Of note, we did check the initial bone marrow samples with sensitive qualitative and quantitative methods and did not detect the presence of t(9;22) clone. This would make the clonal evolution a more plausible explanation.
The contribution of using a comprehensive method such as next generation sequencing varies from case to case. One may argue that conventional cytogenetics and targeted gene testing may not pick up new or uncommon variants that may be responsible for acquisition of resistance. With these techniques becoming more sensitive and cost-effective, they are likely to be helpful in refractory/relapse cases as the number of targeted therapies also continue to increase. Outcomes of patients in this evolving era of precision-based medicine are likely better as compared to historical cases due to more novel treatment options. Ongoing clinical trials to evaluate the safety and efficacy of novel therapies like ASP2215 in combination with induction and/or consolidation therapies will help quantify their true benefit in first-line and relapse setting. Testing for targetable drivers of oncogenesis and progression can help personalize cancer treatment and increase treatment options for some of these patients.
Conflicts of interest disclosures
The clinical trial component was supported through funding from Astellas Pharma Global Development (APGD). All other support for the research and testing was through division of hematology and center of individualized medicine (CIM), Mayo Clinic, Rochester, MN, USA. No other conflicts of interest to report.
Authors' contributions
Authors PMK and NG wrote the initial draft. Authors MRL, MMP and SKH extensively revised the manuscript and provided additional feedback. All of this was integrated by author PMK to a final draft. All authors approved the final draft for publication. Author MRL is also the site principal investigator for the clinical trial with ASP2215.
Acknowledgments
We are deeply indebted to the patient and his parents for allowing us to share his case. Written informed consent was obtained. Thanks are also due to the multiple care providers and nursing staff who have been diligently taking care of the patient for more than a year now. The clinical trial component was supported through funding from Astellas Pharma Global Development (APGD) Clinical trial number 2215-CL-0101; Clinical trial information: NCT02014558; https://clinicaltrials.gov/show/NCT02014558. All other support for the research and testing was through division of hematology and center of individualized medicine (CIM), Mayo Clinic, Rochester, MN, USA. Thanks are also due to Dr. Abraham, Roshini S., Ph.D., Professor of Laboratory Med/Pathology and Medicine, College of Medicine, Mayo Clinic, Rochester for help and guidance with other testing that was pursued.
References
- 1.Cortes J.E., Kim D.W., Pinilla-Ibarz J. A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. New Engl. J. Med. 2013;369(19):1783–1796. doi: 10.1056/NEJMoa1306494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravandi F., Alattar M.L., Grunwald M.R. Phase 2 study of azacytidine plus sorafenib in patients with acute myeloid leukemia and FLT-3 internal tandem duplication mutation. Blood. 2013;121(23):4655–4662. doi: 10.1182/blood-2013-01-480228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah N.P., Talpaz M., Deininger M.W. Ponatinib in patients with refractory acute myeloid leukaemia: findings from a phase 1 study. Br. J. Haematol. 2013;162(4):548–552. doi: 10.1111/bjh.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarado Y., Kantarjian H.M., Luthra R. Treatment with FLT3 inhibitor in patients with FLT3-mutated acute myeloid leukemia is associated with development of secondary FLT3-tyrosine kinase domain mutations. Cancer. 2014;120(14):2142–2149. doi: 10.1002/cncr.28705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith C.C., Zhang C., Lin K.C. Characterizing and overriding the structural mechanism of the Quizartinib-Resistant FLT3 “Gatekeeper” F691L mutation with PLX3397. Cancer Discov. 2015 doi: 10.1158/2159-8290.CD-15-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greaves M., Maley C.C. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levis M.J., Perl A.E., Altman J.K. Results of a first-in-human, phase I/II trial of ASP2215, a selective, potent inhibitor of FLT3/Axl in patients with relapsed or refractory (R/R) acute myeloid leukemia (AML) J. Clin. Oncol. 2015;33:7003. [Google Scholar]
- 8.Bacher U., Haferlach T., Alpermann T. Subclones with the t(9;22)/BCR-ABL1 rearrangement occur in AML and seem to cooperate with distinct genetic alterations. Br. J. Haematol. 2011;152(6):713–720. doi: 10.1111/j.1365-2141.2010.08472.x. [DOI] [PubMed] [Google Scholar]
- 9.Georgiou G., Efthymiou A., Vardounioti I. Development of acute myeloid leukemia with NPM1 mutation, in Ph-negative clone, during treatment of CML with imatinib. Leukemia. 2012;26(4):824–826. doi: 10.1038/leu.2011.280. [DOI] [PubMed] [Google Scholar]
- 10.Guerrouahen B.S., Futami M., Vaklavas C. Dasatinib inhibits the growth of molecularly heterogeneous myeloid leukemias. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 2010;16(4):1149–1158. doi: 10.1158/1078-0432.CCR-09-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]